Abstract
The continued development of in vitro systems that accurately emulate human response to drugs or chemical agents will impact drug development, our understanding of chemical toxicity, and enhance our ability to respond to threats from chemical or biological agents. A promising technology is to build microscale replicas of humans that capture essential elements of physiology, pharmacology and/or toxicology (microphysiological systems). Here, we review progress on systems for microscale models of mammalian systems that include two or more integrated cellular components. These systems are described as a “Body-on-a-Chip.”, and utilize the concept of physiologically-based pharmacokinetic (PBPK) modeling in the design. These microscale systems can also be used as model systems to predict whole-body responses to drugs as well as study the mechanism of action of drugs using PBPK analysis. In this review, we provide examples of various approaches to construct such systems with a focus on their physiological usefulness and various approaches to measure responses (e.g. chemical, electrical, or mechanical force and cellular viability and morphology). While the goal is to predict human response, other mammalian cell types can be utilized with the same principle to predict animal response. These systems will be evaluated on their potential to be physiologically accurate, to provide effective and efficient platform for analytics with accessibility to a wide range of users, for ease of incorporation of analytics, functional for weeks to months, and the ability to replicate previously observed human responses.
1. Introduction
Early stage drug discovery relies on evaluating thousands of compounds with regard to their efficacy as well as toxicity. Before drugs are tested in small animals, tests utilizing in vitro cell culture models are conducted. These cell culture models often consist of primary cells or cells derived from immortalized cell lines. However, this approach has limitations since it cannot match the complexity of the human body, which consists of multiple tissues, each of them having multiple cell types typically in a complex architecture. The interactions between multiple tissues results in a complex, time-dependent concentration profile of a drug. A mathematical modeling technique called pharmacokinetics (PK) modeling is applied to analyze and predict the dynamics of these multi-organ interactions and resulting time dependent concentration profile of a drug and its metabolites.
While conventional in vitro cell culture systems typically utilize only a single cell type, co-culture of cell types from different tissues can predict aspects of inter-organ communication via soluble metabolites. Simple co-culture systems that mimic the first pass metabolism of drugs consist of transwells where Caco-2 cells are cultured on the membrane, and liver cells are cultured in the well beneath (1). A more complex system that confined different cell types to wells within a cell culture dish was capable of showing efficacy and toxicity with six cell types that were in communication with each other via a common medium (2). These systems are capable of mimicking an inter-organ relationship at a level that does not take into account the tissue sizes in comparison to each other. A further disadvantage is that the liquid to cell ratio is much larger than in vivo, and these limitations produce inaccuracies when evaluating drugs because they change the in vivo PK of a drug and its metabolites. For example, based on in vitro studies, flavonoids-rich foods have long been thought to play health-promoting effect through antioxidant capacity, but a recent study indicates that due to low bioavailability and fast metabolism, the plasma concentration of flavonoids is very low, which casts doubt on the previous notions of benefit (3).
Micro- and nanofabrication techniques have enabled the development of on-chip tissue cultures. Several systems with multiple organs-on-chip have been developed, taking advantage of the opportunity to tailor the chemical and physical cell culture environments within microfluidic cell culture chambers. Furthermore, based on the development of individual organ-on-a-chip systems, microscale systems containing multiple organs, therefore capable of reproducing the interaction between the organs, have appeared (4–7). Known as body-on-a-chip or human-on-a-chip systems, these platforms allow for the accurate control of cell numbers and dimensions of culture chambers. When organ volume ratios become more physiologic metabolite concentrations (in relation to one another) are more likely to be accurate. If microfluidic technology is employed, fluidic channels can connect the cell culture chambers the same way blood vessels connect different organs. Another advantage of microfluidic cell culture systems is that they provide a liquid to cell ratio that is closer to that found in vivo, and also exhibit shear stress that is closer to physiologic, constituting a more tissue-like cellular environment. These advantages offer a better possibility for body-on-a-chip systems to reproduce the PK profile of a drug similar to what would be observed in the human body.
In this article, the concept of using the PK modeling approach to design and operate body-on-a-chip systems to reproduce the multi-organ interactions is described. A subgroup of PK modeling techniques known as physiologically-based pharmacokinetic (PBPK) modeling is particularly useful for designing such systems, as this modeling approach is based on the physiological consideration of the human anatomy. We will describe recent reports of these preliminary body-on-a-chip systems and what type of medical and biological questions they can address. In addition, state-of-the-art technologies for on-chip measurement of various physiological responses of cells are discussed as well as engineering challenges that need to be overcome to answer difficult questions in medicine and biology.
2. PBPK and PD models
2.1 PBPK modeling
Pharmacokinetics is the science of drug absorption, distribution, metabolism and elimination, or more specifically the quantification of those processes, leading to an understanding, interpretation, and prediction of blood concentration-time profiles (8). The purpose of PK modeling is to mathematically quantify the amount of drugs in different parts of the body, and gain insight into the kinetics of drug distribution. In addition, it is often useful to describe the complex process of absorption, distribution, metabolism and elimination, collectively known as ADME, of drugs after administration with simple terms such as area-under-the-curve, peak concentration, and half-life. Hence it is a very useful tool for analyzing the complex interplay of convection, diffusion and reaction. However, the difficulty of building a physiologically accurate model puts a limitation on the wider usage of this method.
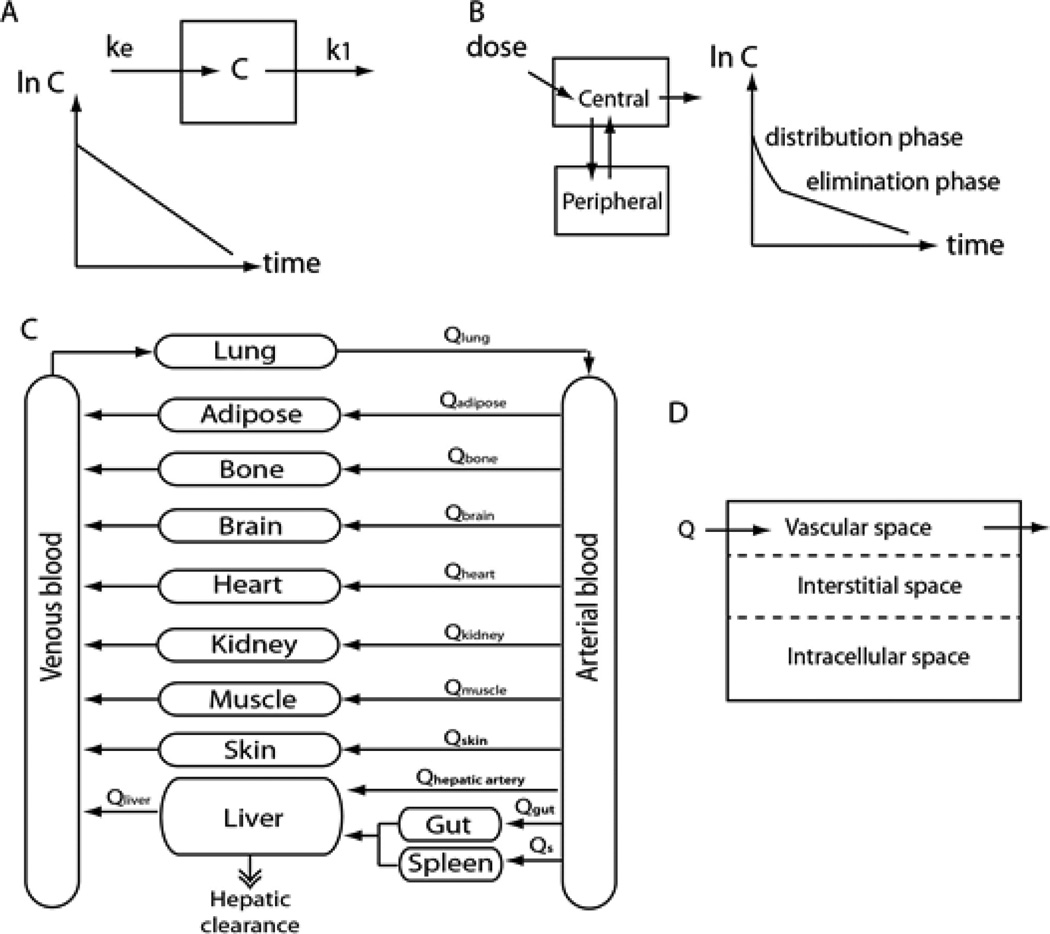
PK models can be classified as noncompartmental and compartmental models with varying degrees of complexity. The simplest form of PK model among compartmental PK models is what is known as a one-compartment model, where the whole body is represented by a single compartment with an input and an output. It is a very simple and empirical model, but often is sufficient to describe the pharmacokinetics of a drug (9). The fundamental assumption underlying the one-compartment model is that rapid equilibrium is achieved within the body after administration. Establishing a mass balance equation on the compartment and solving the differential equations results in the concentration profile expressed as an exponential function of time (Figure 1(a), equation 1). The semilog plot of the concentration versus time gives a straight line with negative slope, where the value of the slope is − k.
| Eqn 1 |
Figure 1.
(A) one-compartment model (B) two-compartment model (C) PBPK model (D) diffusion-limited model.
Many drugs do not reach equilibrium rapidly, and require distribution time. In this case, the one-compartment model does not provide an accurate representation of the drug’s PK. A two-compartment model can be used to describe the distribution time of such drugs (10). It consists of a central compartment and peripheral compartment with flow between the two. The central compartment can be viewed as a lumped sum of rapidly-perfused tissues, such as the liver, kidney, heart and lung, whereas the peripheral compartment can be viewed as a lumped sum of slowly-perfused tissues, such as the muscle, fat and skin. Using a similar approach as the one-compartment model, one can set up a mass balance equation for the two compartments and solve the coupled differential equations, obtaining a sum of two exponential terms. The two exponential terms can be interpreted as the distribution phase and the elimination phase with different slopes (Figure 1(B)).
These simple compartment models are useful for extracting PK parameters such as clearance and half-life. However, these models are experimental and do not reflect the physiology of the body. As a result, these models are less suitable for extrapolation or providing mechanistic insight. A more comprehensive model with a physiological basis can be developed by representing actual organs as separate compartments. This type of PK model is known as a physiologically-based pharmacokinetic (PBPK) model (11). In PBPK modeling, organs in the body are represented as compartments connected with hypothetical blood flow (Figure 1(C)). Setting up mass balance equations for each compartment gives a set of ordinary differential equations that can be solved numerically. In these mass balance equations, physiological parameters such as blood flow rates and organ volumes are used. This approach has a more mechanistic basis as it follows the actual anatomy of the body, and can be useful for extrapolation. Also, since it’s based on the actual physiology, it can be a useful tool for studying the mechanism underlying the ADME properties of a drug. Sometimes, additional definition of drug transport in the tissue space is needed. For example, if the transport of a drug is limited mainly by blood perfusion, the compartment can be assumed to be well-mixed. This is usually the case with lipophilic drugs, while on the other hand, if the diffusion of the drug in the tissue space is limiting, the drug is permeability-limited. In such a case, the organ compartment may be divided into extracellular, interstitial, and intracellular space where the model needs to account for diffusion between the spaces (Figure 1(D)). Also, in many situations, not all of the organs in the body are included in the model. Often several organs are lumped into a single compartment, based on their similarity. For example, organs can be grouped into rapidly-perfused tissues and slowly-perfused tissues. This lumping of organs into a single compartment will introduce an empirical aspect into the PBPK model. Simpler one or two-compartment models and more complex PBPK models both have advantages and disadvantages. Which type of models to use for certain drugs depends on many factors such as complexity of the drug’s mechanism of action, availability of PK data, and the purpose of the PK modeling.
The development of a PBPK model is often limited by the difficulty of finding accurate parameters for the model. Physiological parameters such as organ volumes and blood flow rates can be easily found in the literature (12), although individual variations have to be considered in some cases. Finding the enzyme kinetic parameters or clearance rates is usually more difficult. In some cases, in vitro enzyme kinetic parameters are measured and extrapolated to in vivo values while in other cases, kinetic parameters may have to be fitted to experimental data. Various model systems exist for providing the information on the metabolic profile and rate for organs. These models vary in their complexity and in vivo relevance (13). For example, as the liver is the main organ responsible for metabolic reactions, the liver tissue slice provides the most realistic representation of the liver because it maintains physiological tissue architecture as well as contains nonparenchymal cells. However, this model suffers from the lack of blood supply and has a short viability time. Primary hepatocytes also offer good prediction value for in vivo metabolic profiles, but suffer from the absence of nonparenchymal cells and loss of activity upon cell culture (13). At the other end of the spectrum are the cell-free systems such as isolated cytochrome P450 enzymes or liver microsomes, which are easy to use but only provide partial metabolic activity of the liver.
Another important set of parameters are tissue-plasma partition coefficients, which refers to the relative distribution of a drug between the tissue and the blood plasma. It can be either experimentally measured, or estimated based on its hydrophobicity data (14). Although all these parameters can sometimes be estimated with a reasonable accuracy, often it is necessary to fit the parameters to experimental data. Therefore, having a sufficient amount of experimental data is very important for building an accurate PBPK model. Unfortunately, researchers often have to deal with a scarcity of data for many important parameters. Plasma concentration data are available in many cases, but drug concentration from an individual organ is rarely available. Due to this lack of human data, PBPK modeling often relies on animal models where extrapolation between the animal data to the human model is necessary (15). Another issue with building an accurate PBPK model is that the PK of a drug can be affected by the disease conditions of an organism, and population variations can also exist (16). A more complete review on the construction of PBPK models and their application can be found in other review papers (11, 17).
2.2 PBPK-PD models
Pharmacodynamics (PD) is the study of the pharmacological effect of drugs in the body. In PD modeling, the pharmacological effect is viewed as a function of drug concentration. Diverse variations of PD models exist, depending on the mechanism of a drug’s action. For example, the effect of the β1 receptor blocker metroprolol increases with increasing concentration (18). In such a case, the effect can be a linear function of concentration. However, linear effect function should be used with care, since it does not define the maximum effect, and implies that the effect will increase indefinitely with increasing concentration. Alternatively, the effect can be defined with a more complex function that can have a maximum effect value. In this model, often called the Emax model, the effect will increase in a nonlinear manner with increasing concentration. Some drugs exert their effect through a complex pathway, resulting in a time-delayed, irreversible effect. Chemotherapeutic agents can be a prominent example of such drugs. In this case, the growth of cells can be modeled with a cell proliferation model, where the rate of cell growth is a linear function of cell number. The effect of drugs on the cell growth rate can be incorporated into the cell proliferation model (19). A more comprehensive review on the diversity of PD models can be found in a review paper by Mager et al. (20).
PD models can be coupled with PK models, and the combined PK-PD model can be used to predict or analyze the physiological effect of a drug at a given dose. In the combined model, the PK profile of a drug gives the drug concentration at a specific dose, and the drug concentration can be used to obtain the amount of physiological effect. Therefore, an integrated PK-PD model gives the time-dependent changes of the physiological outcome of a drug at a specific dose. For example, a PBPK-PD model was used to simulate the tumor killing efficacy of an anticancer agent in a body-on-a-chip model system (21). Using this model, the effect of the patient’s liver enzyme activity on the efficacy of the anticancer agent was studied, and the optimal combination ratio of the drug with another drug was found. In another study, a PBPK-PD model was developed to study the effect of a carbaryl insecticide. In this study, a Bayesian approach was used for the estimation of parameters for use in the model. Timchalk et al. studied the metabolic interaction of chlorpyrifos and diazinon insecticides with a PBPK-PD model (22, 23). Several review papers are available for more complete review of integrated PK-PD modeling approach (24–26).
2.3 PBPK-guided design of body-on-a-chip systems
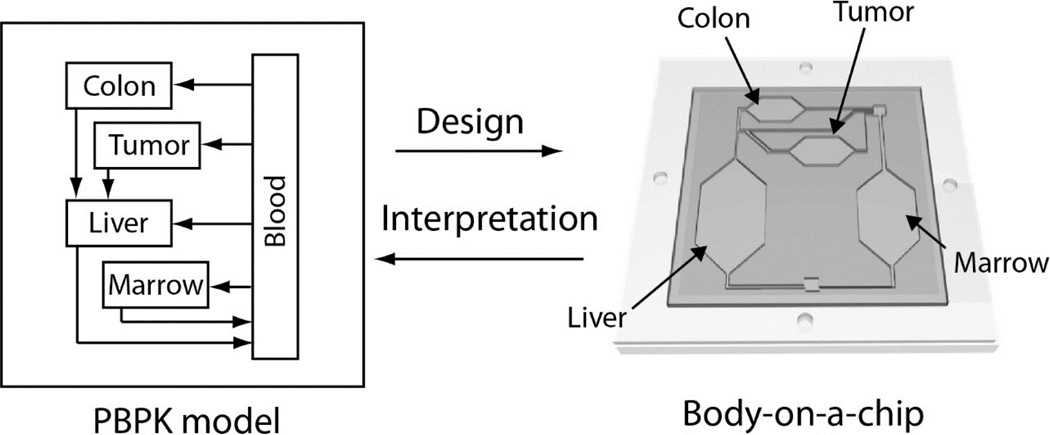
Body-on-a-chip, also known as human-on-a-chip, systems aim to emulate the whole-body response of a human to drugs. Several proof-of-concept studies have demonstrated interactions for up to four organs, as will be introduced later. Most proposed body-on-a-chip systems do not truly represent a complete human body, and it is reasonable and desirable to design the system so that a specific aspect of drug’s action will be observed. For example, a body-on-a-chip system can be designed to study the mechanism of toxicity in specific organs, or the effect of specific genotypes on the response to drugs. In this perspective, knowledge of human physiology and the pharmacokinetics is crucial in designing the system correctly so that the intended phenomena is observed, most importantly in a manner similar to the human body. PBPK modeling can be an ideal tool for designing the relevant physical systems (Figure 2). The main advantage of PBPK model guided design of ‘body-on-a-chip’ systems is that one can predict the PK profile of xenobiotics in the chip before the fabrication step, allowing adjustments in the relative chamber sizes, flow rates, cell numbers, etc.
Figure 2.
Design of body-on-a-chip devices based on PBPK modeling principles.
The first step in designing these systems is to choose target organs that are important in emulating the human response to a particular compound or class of compounds to be evaluated. This involves as much knowledge of a drug’s mechanism of action as possible. To start, the prominent site(s) of organ toxicity is important and needs to be included. The primary routes of administration and elimination also need to be included in any model system. For orally administered drugs, absorption in the gastrointestinal tract is essential. If a drug is excreted mainly through the kidney, the filtration function of the kidney should be considered. In general, the liver is frequently involved in drug metabolism and should be included if this is indicated. In a similar manner to PBPK modeling, other less important organs can be grouped in a single compartment, but care should be taken to ensure that this lumping of organs does not change the PK of drugs or affect the action of drugs significantly by overlooking a key contribution to toxicity or clearance.
Ideally, the size of the compartments should be proportional to the size of the corresponding organs, but it can be difficult to match the relative sizes of organs for certain models, although it does not necessarily mean that the size of microscale structures such as cells and barriers must be matched. For example, if a tumor compartment is to be added, the size of a tumor is generally much smaller than other major organs such as the liver (21). It is more important to correctly scale different organs, according to their function so that phenomena of interest occur in a manner similar to the human body. The scaling issue arises from the fact that all organ functions do not scale linearly with organ sizes. In fact, different organ functions will scale differently depending on their mechanism, for example, gas exchange in the lung is closely related to the surface area, whereas liver enzyme activity is related more closely to the mass rather than the surface area. The issue of scaling can have a profound effect on the performance of body-on-a-chip systems, especially with smaller sized chips, and this increases with increasing numbers of organs that need to be emulated.
The issue of scaling in body-on-a-chip systems has been discussed in recent studies (7, 27). One method to address this issue is to use a conventional scaling approach that is frequently used to scale animals with humans. Allometry studies the relationship between the body size and organ physiology between different species (28). In allometric scaling, an exponential power relationship is used to scale organ functions to body mass:
| Eqn 2 |
, where Y is the physiological function, M is the mass, α is the scaling exponent, and Y0 is a constant. One example is the scaling of basal metabolic rate (BMR), which is known to follow a 3/4 power law (29). Ahluwalia published several articles describing the design of a microfluidic multi-compartment model using allometric principles (30, 31). This method has proven useful for systems with four or less compartments. However, it could cause unacceptably large deviations when more organs are involved, since the allometric coefficients are different for each organ. Another complication is that cells cultured in vitro might not follow the same principles as observed in vivo. In cases of scaling between different animals, allometric scaling has shown limitations in predicting the clearance rate, and empirically determined allometric scaling laws might not always hold for human-on-a-chip systems, especially at higher organ numbers.
Recently, another scaling approach has been introduced, where the organ compartments on a chip are scaled to match the basal metabolic rate of the cells in vivo. In this approach, the authors assumed that the metabolic rate of cells could be controlled by controlling the oxygen delivery rate, rather than supplying the system with excess oxygen. In this approach, also termed ‘functional scaling’, organ sizes are chosen to match the relative organ functional activity (7). The authors classified the organs into two groups, ‘functionally two dimensional’ and ‘functionally three dimensional’ organs. These classifications are based on whether an organ’s primary function is related to the mass or the surface area. For example, storage function of fat cells is considered a 3D based function, whereas filtration function of the kidney would be a 2D based function. However, as the authors noted, such an approach could result in abnormally large volumes for 2D scaled organs, and further adjustment may be needed. Wikswo provides a more comprehensive demonstration of this functional scaling approach for several organs (7). Another simpler, yet useful scaling approach is to use organ volumes and residence times (32). In this approach, organ sizes are scaled relative to their volumes, and flow rates into each organ are designed to yield fluid residence times that correspond to organs in the human body. Such an approach ensures that cells in the chamber are exposed to chemicals or growth factors for the same amount of time as they would be exposed to in vivo. If the in vitro cells mimic in vivo cell metabolism, the degree of conversion should be similar. This method was largely initiated by Shuler’s group at Cornell University, and has proven useful (32–35). While all these methods have pros and cons, perhaps a more important prerequisite is that the cells representing specific organs have physiological activity at least comparable to their in vivo counterparts.
Here, we will briefly describe the design process of body-on-a-chip systems using the organ volume and residence time principles. The first step in designing a body-on-a-chip is to determine the size of the organ chambers. Ideally, in this step, the actual sizes of different organs are reflected so that the relative sizes are kept constant. The next step is to calculate the flow rates for each chamber, which is necessary to achieve approximate residence times closer to the physiological values in the human body. Obtaining flow rates allows the calculation of the fraction of fluid that each chamber will receive, which can be used to determine the length, width, and height of the channels connecting the chambers. When determining the geometry of the channels, some compromises may be necessary to achieve realistic operation. For example, the channel depth needs to be sufficient to allow mammalian cells to be cultured with unimpeded flow, and preferably less than 200 µm to ensure the robust fabrication in standard microfabricated devices.
The calculation of the flow pattern in a microfluidic network used to emulate flow in the human body relies on simple fluid dynamics principles. A microfluidic network can be viewed as analogous to an electrical circuit, where the hydrodynamic pressure is equivalent to the voltage, the volumetric flow rate to the current, and the hydrodynamic resistance to the electrical resistance (36). The hydrodynamic resistance is largely dependent on the geometry of the fluidic channel, such as width, height and length, and the property of the fluid, particularly viscosity.
3. Emulating multi-organ interactions on a chip
3.1 Organ-on-a-chip for individual organs
During the past 1–5 years, many systems have been developed with the aim of reproducing specific organ functions, such as the blood vessels (37, 38), liver (39, 40), gastrointestinal tract (41, 42), blood-brain-barrier (43–45), lung (46, 47), kidney (48–50) and heart (51, 52). Specific systems to emulate biological processes, such as metastasis (53), vasculogenesis (54), angiogenesis (55), and chemotaxis (56) have also been developed. In many of these systems, researchers reported observations of better tissue-like function or a more functional behavior from the cells cultured in the devices than cells in 2D culture, demonstrating another facet of the usefulness of these systems. One example is the lung-on-a-chip device developed by the Ingber group at Harvard University (47). In this study, human alveolar epithelial cells and pulmonary microvascular endothelial cells were cultured on both sides of a porous membrane and placed under constant strain to mimick the stretching action of the lung. This device was further used to reproduce drug-induced pulmonary edema (57). Natarajan et al. reported a patterned cardiac system integrated with a microelectrical array which allowed conduction velocity to be measured without dispersion losses and was shown to reproduce drug effects seen for in vivo studies (52). Bhatia et al. reported a liver-on-a-chip system, where a co-culture of hepatocytes and fibroblasts in a precisely defined area resulted in enhanced liverspecific functions as measured by mRNA expression and metabolizing enzyme activities (40). In a subsequent study, the same system was used for reproducing the life cycle of hepatitis C virus (58). More details of these organ-on-a-chip systems for individual organs are covered in several review articles (4, 6, 59).
3.2 Connecting multiple organs
While these organ-on-a-chip systems provide excellent platforms for studying biological phenomenon within individual organs, most biological phenomenon involves interactions between multiple organs. For instance, glucose metabolism is based on a highly complex interplay between the liver, pancreas, muscle, fat and the brain, connected by the blood flow (60). Flavonoid quercetin, which is one of the most widely consumed flavonoids in the human diet, found in fruits and vegetable (61), is known to exert its antidiabetic effect through the modulation of inflammatory response in the adipose tissue (62), by inhibiting glucose uptake in the gut (63), and by stimulating insulin secretion in the pancreas (64). It is obvious that studying this complex, time-dependent phenomenon with a system representing a single organ is impossible.
In fact, attempts to observe the interaction between multiple cell types have existed for many years, such as the co-culture of gut epithelial Caco-2 cells and hepatocytes in both sides of a transwell (1), or culturing multiple cell types in a confined space within a single well (2). However, these examples are not accurate models of the human body because of unrealistic cell-to-liquid ratios, inaccurate relative tissue sizes, and lack of transport phenomena. In addition, Caco-2 cells lack appropriate levels of several important metabolizing enzymes present in gut epithelia (65). Microfluidic techniques allows for culturing cells in compartmentalized chambers with controlled interactions between each chamber in a physiologic relationship. The fluidic channels connecting the chamber can be viewed at a basic level as blood vessels connecting the organs. One of the main interests in pharmaceutical science is the efficacy and the toxicity of drugs after undergoing liver metabolism. Several microscale systems for reproducing this process have been reported. Lee et al. encapsulated P450 enzymes in a hydrogel to allow reaction with a drug, and the metabolites formed were used for a cellular toxicity assay (66), where different cell lines were cultured in separate compartments in a single well. By sharing the culture medium, a drug metabolized in one compartment can diffuse into another compartment and affect other cells (2). In addition, a microfluidic system for incubating precision-cut tissue slices was also developed (67). In another variation of the system, intestinal and liver slices were treated with a drug, resulting in secretion of growth factors from the intestinal slice, which down-regulated the metabolizing enzyme in the liver slice (68).
These proof-of-concept studies successfully demonstrated the possibility of using microtechnology to study simple interactions between organs. More complex systems have also been developed (32, 69). These systems were aimed at reproducing the pharmacokinetics of a substance observed in the body, and were designed based on pharmacokinetic modeling principles. These early body-on-a-chip systems were used to study the mechanism of naphthalene toxicity, which involves the interplay between the liver, lung and the fat. In subsequent studies, body-on-a-chip systems with similar design principles were used to evaluate the drug mixtures for multidrug resistance (35), the transport of acetaminophen in the gastrointestinal tract and subsequent liver toxicity (42), and the effect of an anti-cancer agent on the tumor and the bone marrow compartment after metabolism (33). In the study by Tatosian et al., a PK model that corresponds to the body-on-a-chip system was built to predict the pharmacokinetic profile of the chip, which compared favorably with experimental result (35). This study demonstrated that PK models describing body-on-a-chip systems can be formulated and that the mathematical model also enables the quantitative study of the mechanism of a drug’s action (35, 70). For example in a study by Sung et al., a PK-PD model describing a drug’s action in a chip was developed, which was used to extract parameters describing the kinetics of cell death after treatment with chemotherapeutic agents.
Several research groups have reported the development of body-on-a-chip systems with a similar concept, but with different design principles. Zhang et al. reported a multi-channel 3D microfluidic system, where aggregates of four different cell types (representing liver, lung, adipose and kidney) were cultured (71). The notable feature in this device was that tissue-specific environments were maintained for each organ compartment by encapsulating the cells in a gelatin sphere. Ahaluwalia et al. published a series of papers describing a “multicompartmental modular bioreactor”, a microfluidic system with separate microchambers for multiple cell types interconnected with microfluidic channels (30, 31). In a recent study, this group reported the development of a three-chamber system, representing the liver, fat and blood vessels, and observed changes in glucose and fatty acid metabolism in a hyperglycaemia condition (72). Wagner et al reported the development of a multi-organ-on-a-chip for long term cultivation of 3D human liver cells and skin tissue (73). Interestingly, this research group developed various versions of the multi-organ-on-a-chip systems with a different number of chambers, up to seven compartments. Although, the two-chamber model was mainly used for toxicity studies, it indicated the possibility of developing body-on-a-chip systems with more than four chambers.
4. Measuring responses
The functional responses of the cultured tissue when treated with drugs or subjected to engineered stimuli need to subsequently be identified and measured. Most of the body-on-a-chip systems have integrated measurement technologies and this section describes the development and results for some of these functional responses.
4.1 Cell viability
The benefits of body-on-a-chip systems will be fully realized as more analytical tools are incorporated onto the chip to assess cellular conditions. Microscale systems contain limited fluid volume and low numbers of sometimes inaccessible cells, complicating detection and analysis with conventional methods. Endpoint measurements of cell function may allow access to a cell’s physiological state, but may also require disintegration of the system for cell removal, essentially terminating the experiment. Some optical measures, such as live/dead stains (33, 35) are useful in determining minimal requirements for cell survival, but may not provide sufficient insight into cellular mechanisms involved in drug response. Advances in body-on-a-chip system design are moving towards non-invasive, on-chip microscale measurements, allowing for assessment of real-time dynamic interactions of multiple organ systems. For example, an optical detection and analysis system has been developed for real-time analysis of 3D cultures in a microfluidic environment (74). A measurement of electrically active cells has also been analyzed by incorporation of electrode arrays into the chip design (75). One of the challenges for body-on-a-chip systems development will be incorporation of multiple measurement approaches for simultaneous and real-time analysis of all organs in the system.
4.2 Mechanical forces
Mechanical forces drive numerous cardiovascular processes such as morphogenesis during embryonic development and tissue remodeling in adulthood (76). In the myocardium, tissue elasticity shows significant regional variation during heart disease (77). Therefore, a study of the contractile function of cardiac tissue is important. Similarly, an evaluation of the active tension generated by cultured skeletal muscle cells or tissues (78, 79) can be used to study skeletal muscle physiology and test drugs for skeletal muscle-related diseases. A wide range of methods have been used to measure the force generated by these cells. These methods have been applied both at a single cell level and also on sheets or films of cells. The force and stress developed in single cardiac myocytes have been indirectly measured in terms of cell shortening and velocity (80), while other studies have directly measured force using microdevices such as cantilever beams made of thin steel foils (81), measurement of contractile force with the voltage-clamp method (82), or force regulation by Ca2+ (83). Traction force microscopy is another technique that has been used to investigate forces produced by migrating myofibroblasts (84). A mechanical coupling between polymer micropillars and cardiomyocytes has been used to estimate force generated by cardiomyocytes by observing the displacement of the oscillating micropillars (85). A fully submersible piezoelectric force transducer has been implemented for high fidelity force measurements from cardiac myocytes (86). The study of single cells provide useful insights, but the importance of the community size of the cells for obtaining stable and reliable results for drug discovery has also been proven (87). The contraction stress in micropatterned cardiovascular tissue on thin flexible polydimethylsiloxane (PDMS) sheets has been measured (76) and the contractile force of high-density cardiomyocytes on 3D grooved surfaces of a PDMS cantilever has also been evaluated. The deflection of thin film elastomers caused by the contraction of cardiac microtissue on the surface has been used to calculate diastolic and systolic stresses. Cardiac myocytes cultured on a stretchable substrate and stretched in longitudinal and transverse directions have been used to measure contractile stress generation (88).
Tissue culture protocols developed over the years can be applied to expand proliferating myoblasts to large numbers and to differentiate them into contractile post-mitotic muscle fibers (89, 90). Numerous protocols are available to engineer 3D skeletal muscle systems capable of generating contractile forces in a directed manner (91). Most of the 3D skeletal muscle systems are based on synthetic (92) or biopolymer (93) exogenous matrices that function as scaffolds in which the cell population is seeded. The mechanical forces generated by these cells are measured by electronic force transducers (94), or other methods based on optical imaging of cell contraction within these systems (95). The complexity of the 3D muscles systems can make integration with other complementary organ systems difficult. In 2D muscle systems, the desired orientation of muscle cells can be achieved by surface chemistry modification (90). The use of Si-MEMS cantilever combined with an atomic force microscope (AFM) detection apparatus (79) to measure cantilever deflection provides a non-invasive evaluation of forces generated by a 2D muscle system. The active tension of single myotubes has been measured by the horizontal displacement of a Si-MEMS cantilever and then evaluated by image analysis (96). The simplicity of the two dimensional muscle systems enables integration with other complementary cell types such as Schwann cells, motoneurons and sensory neurons.
4.3 Electrical signals
On-chip electrical characterization of the physiological properties of single cells or cell layers is of relevant importance for toxicology studies and drug development. In particular, the ability to electrically characterize the transport of biological or pharmacological molecules through a layer of cells and study cellcell interactions within networks of cells is critical to the development of in vitro systems for the study and treatment of pathologic conditions. A relevant example is the assessment of the integrity and functionality of barrier tissues. Barrier tissues serve as functional barriers to regulate the transport of ions and macromolecules through the interface of fluid compartments to maintain distinct homeostatic environments (97). Disruption of the barrier tissue integrity leads to increases in permeability and loss of barrier functionality, and it is associated with the development and progression of several diseases, such as gastrointestinal autoimmune diseases or neuroinflammatory diseases (98, 99). Trans endothelial electrical resistance (TEER) measurement is a well-established method to assess the integrity of barrier tissues. TEER measurements are traditionally performed using commercially available systems, normally consisting of a pair of Ag/AgCl electrodes, to record the low frequency resistance of confluent cell monolayers grown on porous membranes (100, 101). These conventional handheld systems, which require manual insertion of the electrodes into the culture well, have the disadvantage of being discontinuous and low throughput. To overcome these limitations and achieve reliable and continuous measurements, several groups have explored ways of including electrodes within a microfluidic system, either by integration with surface embedded microelectrodes or by simply utilizing micron-sized wire electrodes, to enable TEER measurements by measuring the resistance and capacitance (impedance) of the cell monolayer (102, 103). For example, a microfluidic model of the blood-brain barrier (BBB) with integrated electrodes for TEER measurements has been developed (43). In this system, with a porous polycarbonate membrane sandwiched between two PDMS channels, the authors were able to culture the cells under flow stimulation and measure the response of the barrier to histamine treatment.
A similar concept was recently reported to modulate the barrier function by shear stress or biochemical stimulation and assess the barrier integrity for up to 7 days (44). An alternative approach to the use of integrated electrodes was the use of an organic electrochemical transistor for high temporal resolution monitoring of the barrier tissue integrity of a Caco-2 cell layer after introduction of destructive species (104). A bio-impedance chip to monitor the integrity of the bronchial epithelium has been reported (103), that utilizes an array of 8 pairs of dot-ring planar electrodes to measure the impedance changes of the cell monolayer in response to addition of permeabilizing solutions. The results indicated good correlation with data from conventional TEER measurements. As an alternative to TEER measurements, the permeability of a membrane can be evaluated using Lucifer yellow as an indicator of the membrane integrity. In comparing the suitability of using Lucifer yellow dye method vs TEER measurement it should be noted that TEER is a nondestructive approach that can monitor the membrane integrity over a period of time and is more suitable for automation.
The electrophysiological proprieties of electrically excitable cells, such as neurons and cardiomyocytes, and the analysis of toxicology effects of different compounds on the spontaneous activity of these cells, have been extensively studied using 2D arrays of surface embedded microelectrodes, usually referred as multi electrode arrays or microelectrode arrays (MEAs) (105, 106). These systems are capable of measuring extracellular field potential changes from multiple cells simultaneously and independently, and provide information of cell network activity at high spatial and temporal resolution (107, 108). Additionally, they are compatible with chemical and physical surface modification techniques to guide the growth of cultured cells and record electrical signals along specific patterns on top of the array (109). Recordings of neuronal electrical activity obtained with a dual compartment device for physical segregation and fluidic isolation of two distinct neuronal populations connected through neuritis propagation have been reported (110). The non-invasive nature of the measurements with MEAs makes these systems ideal for studying long term in vitro drug testing and stimulation (111, 112) and to reproduce disease models (113). For example, the effects of 1-Heptanol, a gap junction blocker, and Sparfloxacin, a fluoroquinone antibiotic, on conduction velocity and refractory period after action potential of surface patterned cardiomyocytes were recently investigated using MEAs (52).
4.4 Chemical analysis
In traditional tissue engineering, the presence of a target protein or biomarker is determined by classical methods such as Western blot and antibody-based enzyme-linked immunosorbent assays, which require sample volumes in the range of micro-litres to milliliters. However, the small volume requirements as determined by functional scaling for body-on-a-chip systems places constraints on the analytical methods to characterize organ functions and drug responses. Some of these limitations can be overcome by microfluidic assays of analytical methods such as Western blotting (114) and polymerase chain reaction (115). Most of these microfluidic implementations have not yet been fully developed and standardized.
Mass spectrometry (MS) together with high-performance liquid chromatography (LC) or gas chromatography (GC) based metabolomics analysis is increasingly becoming the method of choice for new drug discovery and development. The use of gas chromatography is limited (116) to a small set of biological molecules that are volatile or can be derivatized to make them volatile. A HPLC separation can be time consuming with multiple separation steps required in some cases. In the case of body-on-a-chip systems, where there are multiple experimental parameters being varied at a time, there is a need for a more dynamic measurement of physiological variables. These requirements could be met by the ion-mobility mass spectrometry (IM-MS) approach where a low-vacuum gas-phase electrophoresis can measure molecular cross-sections in microseconds to milliseconds, followed by MS analysis in microseconds. When compared to HPLC\MS systems, IM-MS approach can handle sample volumes as low as 100 nL (117) and provides near-real-time analysis (118).
5. Issues and challenges
5.1 Scaling issues
The issue of scaling in designing body-on-a-chip devices has been discussed earlier, with details of scaling methods that have been applied to fabricate a variety of systems. In the early stages of development of body-on-a-chip systems, cell culture models with multiple cell types in microwells connected by a static layer of fluid were utilized for toxicity analysis from five interacting organs (27). With the development in microfabrication technology, body-on-a-chip systems have made significant progress in recreating human organ mimics in a microfabricated format. One of the critical systems engineering challenges for the success of body-on-a-chip technology is to design the scale of the interacting organs to match the human physiological conditions. Any drug screening tests performed on a body-on-a-chip with physiologically unrealistic inter-organ scaling cannot reliably be extended to human responses. If the relative size of each organ on the chip is not correctly matched, the drug metabolites from one organ could be transmitted to other organs in extremely high or low concentrations that are not physiologically realistic. For example, if a liver and a lung were coupled without physiologically realistic inter-organ scaling, the liver might not respond significantly to drugs or toxins metabolized by the lung, e.g., the conversion of angiotensin I into angiotensin II (117).
5.2 High-throughput analysis
The acceptance of microfluidic body-on-a-chip systems by the biological research community depends on the ease of integration of these systems with the existing laboratory protocols as well as ease of use. In some cases, the tissue content of these devices would have to be removed to perform biochemical assays outside the system. Most of the microfluidic body-on-a-chip systems require special skills to incorporate the cells into the compartments and operate the system. For example, when compared to using a multiwell plate, the simple step of replacing the culture medium in these systems would require careful handling of the microfluidic tubing connections and attention to removal of any air bubbles generated while introducing the culture medium. The advantages of a microfluidics system can be negated by a lack of user friendliness, but this challenge can be addressed by improving collaborations between the biologists and engineers designing these devices. A microfluidic system design approach that allows body-on-a-chip systems to function with traditional biological tools used in analysis such as pipettors and plate readers can make their use more prevalent in the life sciences. A microfluidic cell culture platform designed to be compatible with existing plate readers common to both academic and industrial labs allows for high throughput assay when compared to microscopy analysis and manual cell counting (119). Microfluidic systems can be designed to define the cell-to-media volume ratio (32), but in devices that use monolayer cell cultures it may be impractical since a minimal flow path (20 µm deep) is needed for robust operation (120). A tube-less microfluidic system to which cell culture media can be delivered using a 96-tip automated liquid handling robot has demonstrated one avenue in which high throughput lab automation can be achieved (121). The compatibility of body-on-a-chip systems with fully automated detection systems is essential for high throughput screening where numerous specific chemical compounds would need to be tested in a short time duration. However, these systems have to be simple, flexible and user-friendly to have an increased impact and to enable widespread acceptance in the field of toxicology and drug development.
5.3 Detection and analysis
The development of body-on-a-chip systems with integrated assay techniques can facilitate the dynamic measurement and collection of statistically significant data utilizing the functional output parameters of the cultured tissues. Many current biological assays that require large sample volumes are not compatible with the small dimensions and cell numbers that are typically observed in these systems. The small number of cells leads to smaller quantities of biomarkers that need to be detected and increases the sensitivity requirement of the analytical detection methods applied in these devices. The transient nature of some of these biomarkers in vivo can make it challenging to capture and detect them within a body-on-a-chip system. Also, removal of recirculating fluid required for any external assays is complicated by the low fluid volumes used in body-on-a-chip systems. To maintain constant fluid volume and operation in a device with total fluid recirculation requires replacement of the fluid taken for sampling. Normally in a physiological system about 25% of the fluid can be replaced per day based on fluid loss in urine, sweat or often fluid excretions, so this needs to be considered in determining device operating procedures.
Considering these limitations to external measurement of biomarkers the development of microfluidics based assay techniques integrated with body-on-a-chip systems would be required. Microfluidic implementations of many of these biological assays exist, but are not standardized nor as user friendly as bench top analyzers. Both optical and electrical detection techniques have been widely used in these devices due to their ease of integration and compatibility with microfluidic implementation. The optical transparency of many substrate materials used for body-on-a-chip systems is suitable for optical measurements. For example, a fluorescent measurement based assay for individual cells was performed for cystolic calcium transients induced in multiple single T cells in parallel (122). It is also important to consider the biocompatibility of the substrate materials used for body-on-a-chip systems. Polymers such as PDMS are optically transparent but any uncured oligomers can leech into solution and the PDMS bulk can absorb small molecules from culture media (123). Most electrical measurement methods can be integrated within these systems by using embedded microelectrodes (52, 113). A cytometric fluorescence detection system has been used to measure the real-time dynamics of cell death (124), enzymatic activity of liver cells (125), and migration of cells in 3D extracellular matrix (74).
The application of embedded microelectrodes within body-on-a-chip system also enables a wider range of electrical measurements for determining the concentrations of specific biomolecules and of viable cells. A microfluidic glucose biosensor based on cyclic voltammetry and amperometry techniques was used to study the electrochemical properties of immobilized glucose oxidase on Au/Ni/copper electrodes (126). Many recent important advances in label free assays of proteins using a number of electrical methods have been reported (127), but have not been widely incorporated within body-on-a-chip systems yet.
5.4 Disease models and Cell Sources
At present, the majority of the disease models are based on standard cell culture protocols or various animal models. These traditional models often do not adequately represent human diseases and is a major cause of drug failure during the drug development process, particularly in the later stages. The traditional animal testing approach is expensive and also faces an ethical dilemma as to why animals should be sacrificed when the drug testing results on animals most often do not translate to humans. Thus, there is a need for more representative models for diseases that can be evaluated with body-on-a-chip systems. The need for disease models is particularly critical when considering genetic background, neurological diseases, diseases with an auto-immune component and cancer. The choice of cell source for these models, whether it is of human or animal origin, depends on the research problem that is to be solved. The progress in current stem cell research provides unique and numerous opportunities for the development of disease models for body-on-a-chip systems. There are numerous sources of stem cells, such as embryonic stem cells (eSC), induced pluripotent stem cells (iPSC), cell lines, or primary cells and each of these cell types have their pros and cons when applied in developing body-on-a-chip systems. The development of certain diseases depends on the genetic background of the patient and in such cases a human cell source that has the inherent genetic mutations and variations would be more critical for the study. Human cell lines are widely available, but they are not suitable for patient specific drug testing. Primary cells are patient-specific, but are difficult to acquire and may not survive in culture for longer periods of time to enable selection of an optimal strategy for drug treatment. iPSC can comparatively be easy to generate and are especially useful for generating cell lines with genomes that are predisposed to disease, in particular when genetically inherited disease affects tissues such as the heart and brain that cannot be easily accessed (128). One of the hurdles in using iPSC is that the differentiated cells can be immature and the disease phenotypes may not be evident (128). Maturation protocols are under development for many cell types to overcome this hurdle. The potential for disease modeling and therapeutic interventions, especially using human iPSC for neural and cardiac disorders, may enable a new type of patient for the 21st century (128).
5.5 Integration for true multi-organ dynamics
The integration of multiple organs within microfluidic systems for drug and toxicity testing was discussed earlier. It was pointed out how simply integrating different cell types without taking into account physiologic and metabolic aspects of the interaction between the different organs is not truly representative of complex dynamics of living organisms. To develop in vitro models that accurately mimic human physiology, several aspects have to be considered. The system has to be designed so that each integrated organ tissue shows the same functionality as shown by its in vivo counterpart. To correctly predict the final effects of the drug, the pharmacokinetic process has to be represented in vitro. This implies the necessity to integrate within the platform system the specific organs involved in these functions. While any organ may be important, the liver, kidney, heart, lung, and brain are key organs and the GI tract is particularly important to control entry of drugs into the circulation. Co-culture systems to study the interaction of liver and kidney under the effect of an anticancerous drug and a perfusion system for the sequential perfusion of intestinal and liver tissue have been recently reported (68, 129), but the development of an in vitro system to accurately predict the complete PK of drugs still remains a challenge, particularly when a large number of organs may be involved.
Another critical aspect in the development of an authentic representation of the human body is that the model should take missing organs into account (7). This means that when an organ is not considered to be of primary importance for the representation of the mechanism under study, it is possible to not directly represent it in the model, and a simplified physiological representation of the missing organs can be included instead (130). This approach is similar to that in a PBPK model where other tissues are placed in compartments such as rapidly perfused or slowly perfused tissues. This simplified physiological representation still would need to be tested and validated by mathematical and physical models. This approach can be considered a suitable compromise between accuracy of the model and simplicity of its implementation, as a physiologically relevant representation of the complete human body can be obtained without overcomplicating the system.
5.6 Cell Culture Medium, Scaffolds and Surface Modifications
To develop a body-on-a-chip system for accurate prediction of human responses to drugs and chemical agents, in vitro culture conditions must mimic the in vivo physiological environment of the cells and microtissues. Typically, in vitro media formulations consist of a basal medium, supplemented with nutrients and growth factors that are essential and/or useful to the cell type. Animal sera, such as fetal bovine serum (FBS), have been commonly used as media supplements in cell culture research. However, due to the undefined and variable composition of serum, as well as the ethical concerns with the production and use of animal-derived components in cell culture, there is an increasing demand for serum-free and chemically-defined media formulations as a good cell culture practice (131). Each cell type has its own unique media supplement requirements to promote cell adhesion, differentiation, proliferation and growth. One of the challenges in the development of a body-on-a-chip system is the establishment of a suitable universal medium that enables retention of cellular phenotype and function, and provides effective humoral communication between the multiple cell and tissue types. A serum-free culture medium in a defined system was first established for the study of hippocampal neuronal differentiation (132) and in vitro culture of motoneurons (133). Patterned cardiomyocytes on microelectrode arrays (52) in a serum-free medium have been developed to demonstrate electrophysiological function and response to cardioactive substances. An in vitro cell culture model consisting of a serum-free medium and defined culture systems have also been used to promote differentiation of skeletal muscle (90). A common media has been developed for co-culture of motoneurons with skeletal muscle to form robust neuromuscular junctions (134–136). An in vitro model with define medium (137) has been developed for oligodendrocyte differentiation and maturation in co-culture with embryonic motoneurons. An optimized basal culture medium has been developed for successful culture of four human cell types (71) optimized in a 3D microfluidic cell culture system. In a scenario where drug-plasma protein binding is being studied, binding proteins such as albumin could be artificially added to a serum free universal media. The complexity of body-on-chip systems will continue to increase with the addition of more organ types and will increase the demand for a universal cell culture medium.
Similar to the need for a serum free universal medium, there is also a need to replace poorly defined animal based extracellular surfaces such as matrigel with synthetic materials (138) that can be used to control the profileration and differentiation of cells into viable tissues. Surface modifications can be achieved with silane coatings as well as utilizing peptides and other materials to allow proper cell growth, proliferation and longevity. In order for new scaffold materials to produce a 3D tissue that is morphologically and functionally similar to in vivo tissue, it should be highly porous with interconnecting pores that allow cell growth and flow of waste and nutrient materials, be biodegradable at a controlled rate that matches growth of cells, and have a surface that promotes attachment, proliferation and differentiation (139). An ideal scaffold system should also provide all the biochemical and biomechanical cues that in vivo tissues receive (139). The design of cost-effective scaffold materials and surface modifications that closely mimic the extracellular matrix and provides suitable growth factors and environmental stimuli to direct tissue growth can be challenging, considering the many subtleties involved.
6. Conclusion
Body-on-a-chip systems present an attractive experimental platform for studying medical and biological questions that cannot be answered with conventional macroscale in vitro systems. There are two major advantages of body-on-a-chip systems over existing cell-based methods. 1) By utilizing micro- and nano-technology, in vivo tissue-like cellular environments can be provided, which leads to more authentic and realistic behavior of the cells. 2) When combined with a PBPK modeling approach, body-on-a-chip systems can provide information on quantitative, time-dependent phenomena arising from the interaction of multiple organs in the body. For a long time, animal models have been an important tool for experiments that cannot be readily performed on humans. However, animal models always carry potential ethical issues, and the extrapolation of results from animal models to human remains a problem. Considering that body-on-achip technologies still face many challenges and issues, and currently developed systems are still in proof-ofconcept studies, animal models are still the closest system to model a human body. However, body-on-a-chip systems have a great potential of working as ‘improved’ in vitro systems, which are closer to the human or animals than conventional in vitro systems. Another value of body-on-a-chip systems is that they can function as a tool for de-convolution of various environmental factors present in vivo. For example, one might be concerned about the exact effect of 3D tissue architecture, mechanical stress, or chemical signals from other tissues, or combinations of those factors on cells, but it is a question that is difficult to answer. Body-on-a-chip systems can be an ideal tool for answering such questions.
Acknowledgments
JHS acknowledges research reported in this publication was supported by the National Research Foundation of Korea (NRF, Grant no. 2011-0013862), KFRI (Korea Food Research Institute, grant no: E0121705), 2012 Hongik University Research Fund. MLS acknowledges research reported in this publication was supported by the NSF (CBET-1106153) and the National Center For Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH) Microphysiological Systems (UH2TR000516-02SI). JJH research reported in this publication was supported by the NCATS NIH Microphysiological Systems (UH2TR000516), NINDS R01NS050452 and NIBIB R01EB009429. “The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
Statement of Author Contributions and Acknowledgments: All authors participated in writing and reviewing of the manuscript. MLS and JJH provided the title and the general outline of the manuscript, and wrote the abstract and the conclusion of the manuscript, MBE wrote the introduction, JHS wrote the ‘PBPK models’ and ‘multi-organ-on-a-chip sections’, WC, CB, and BS wrote the ‘measuring responses’ and ‘issues and challenges’ sections.
References
- 1.Lau YY, Chen YH, Liu TT, Li C, Cui X, White RE, Cheng KC. Evaluation of a novel in vitro Caco-hepatocyte hybrid system for predicting in vivo oral bioavailability. Drug Metab Dispos. 2004;32:937–942. [PubMed] [Google Scholar]
- 2.Li AP, Bode C, Sakai Y. A novel in vitro system, the integrated discrete multiple organ cell culture (IdMOC) system, for the evaluation of human drug toxicity: comparative cytotoxicity of tamoxifen towards normal human cells from five major organs and MCF-7 adenocarcinoma breast cancer cells. Chem Biol Interact. 2004;150:129–136. doi: 10.1016/j.cbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Lotito SB, Frei B. Consumption of flavonoid-rich foods and increased plasma antioxidant capacity in humans: cause, consequence, or epiphenomenon? Free Radic Biol Med. 2006;41:1727–1746. doi: 10.1016/j.freeradbiomed.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 4.Sung JH, Esch MB, Prot JM, Long CJ, Smith A, Hickman JJ, Shuler ML. Microfabricated mammalian organ systems and their integration into models of whole animals and humans. Lab Chip. 2013;13:1201–1212. doi: 10.1039/c3lc41017j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sung JH, Esch MB, Shuler ML. Integration of in silico and in vitro platforms for pharmacokinetic-pharmacodynamic modeling. Expert Opin Drug Metab Toxicol. 2010;6:1063–1081. doi: 10.1517/17425255.2010.496251. [DOI] [PubMed] [Google Scholar]
- 6.Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE. Microengineered physiological biomimicry: organs-on-chips. Lab Chip. 2012;12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 7.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip. 2013;13:3496–3511. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aarons L. Physiologically based pharmacokinetic modelling: a sound mechanistic basis is needed. Br J Clin Pharmacol. 2005;60:581–583. doi: 10.1111/j.1365-2125.2005.02560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarley KD, Bunge AL. Physiologically relevant one-compartment pharmacokinetic models for skin. 1. Development Of models. J Pharm Sci. 1998;87:1264. [PubMed] [Google Scholar]
- 10.Czock D, Keller F, Rasche FM, Haussler U. Pharmacokinetics and pharmacodynamics of systemically administered glucocorticoids. Clin Pharmacokinet. 2005;44:61–98. doi: 10.2165/00003088-200544010-00003. [DOI] [PubMed] [Google Scholar]
- 11.Himmelstein KJ, Lutz RJ. A review of the applications of physiologically based pharmacokinetic modeling. J Pharmacokinet Pharmacodyn. 1979;7:127–145. doi: 10.1007/BF01059734. [DOI] [PubMed] [Google Scholar]
- 12.Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharm Res. 1993;10:1093–1095. doi: 10.1023/a:1018943613122. [DOI] [PubMed] [Google Scholar]
- 13.van Midwoud PM, Verpoorte E, Groothuis GM. Microfluidic devices for in vitro studies on liver drug metabolism and toxicity. Integr Biol (Camb) 2011;3:509–521. doi: 10.1039/c0ib00119h. [DOI] [PubMed] [Google Scholar]
- 14.Poulin P, Theil FP. Generic physiologically based pharmacokinetic models of drug disposition. J Pharm Sci. 2002;91:1358–1370. doi: 10.1002/jps.10128. [DOI] [PubMed] [Google Scholar]
- 15.Benowitz N, Forsyth RP, Melmon KL, Rowland M. Effects of hemorrhage and sympathomimetic drug administration. Clin Pharmacol Ther. 1974;16:99–109. doi: 10.1002/cpt1974161part199. [DOI] [PubMed] [Google Scholar]
- 16.Aarons L. Population pharmacokinetics: theory and practice. Br J Clin Pharmacol. 1991;32:669–670. [PMC free article] [PubMed] [Google Scholar]
- 17.Nestorov I. Whole body pharmacokinetic models. Clin Pharmacokinet. 2003;42:883–908. doi: 10.2165/00003088-200342100-00002. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie RH, Morgan DJ, Horowitz JD. Myocardial effect compartment modeling of metoprolol and sotalol: importance of myocardial subsite drug concentration. J Pharm Sci. 1998;87:177–182. doi: 10.1021/js9702776. [DOI] [PubMed] [Google Scholar]
- 19.Lobo ED, Balthasar JP. Pharmacokinetic-pharmacodynamic modeling of methotrexate-induced toxicity in mice. J Pharm Sci. 2003;92:1654–1664. doi: 10.1002/jps.10431. [DOI] [PubMed] [Google Scholar]
- 20.Mager DE, Wyska E, Jusko WJ. Diversity of mechanism-based pharmacodynamic models. Drug Metab Dispos. 2003;31:510–518. doi: 10.1124/dmd.31.5.510. [DOI] [PubMed] [Google Scholar]
- 21.Sung JH, Dhiman A, Shuler ML. A combined pharmacokinetic-pharmacodynamic (PK-PD) model for tumor growth in the rat with UFT administration. J Pharm Sci. 2009;98:1885–1904. doi: 10.1002/jps.21536. [DOI] [PubMed] [Google Scholar]
- 22.Timchalk C, Nolan RJ, Mendrala AL, Dittenber DA, Brzak KA, Mattsson JL. A Physiologically based pharmacokinetic and pharmacodynamic (PBPK/PD) model for the organophosphate insecticide chlorpyrifos in rats and humans. Toxicol Sci. 2002;66:34–53. doi: 10.1093/toxsci/66.1.34. [DOI] [PubMed] [Google Scholar]
- 23.Timchalk C, Poet TS. Development of a physiologically based pharmacokinetic and pharmacodynamic model to determine dosimetry and cholinesterase inhibition for a binary mixture of chlorpyrifos and diazinon in the rat. Neurotoxicology. 2008;29:428–443. doi: 10.1016/j.neuro.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Derendorf H, Lesko LJ, Chaikin P, Colburn WA, Lee P, Miller R, Powell R, Rhodes G, Stanski D, Venitz J. Pharmacokinetic/pharmacodynamic modeling in drug research and development. J Clin Pharmacol. 2000;40:1399–1418. [PubMed] [Google Scholar]
- 25.Csajka C, Verotta D. Pharmacokinetic-pharmacodynamic modelling: history and perspectives. J Pharmacokinet Pharmacodyn. 2006;33:227–279. doi: 10.1007/s10928-005-9002-0. [DOI] [PubMed] [Google Scholar]
- 26.Agoram BM, Martin SW, van der Graaf PH. The role of mechanism-based pharmacokinetic-pharmacodynamic (PK-PD) modelling in translational research of biologics. Drug Discov Today. 2007;12:1018–1024. doi: 10.1016/j.drudis.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Moraes C, Labuz JM, Leung BM, Inoue M, Chun TH, Takayama S. On being the right size: scaling effects in designing a human-on-a-chip. Integr Biol (Camb) 2013;5:1149–1161. doi: 10.1039/c3ib40040a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich PB, Tjoelker MG, Machado JL, Oleksyn J. Universal scaling of respiratory metabolism, size and nitrogen in plants. Nature. 2006;439:457–461. doi: 10.1038/nature04282. [DOI] [PubMed] [Google Scholar]
- 29.West GB, Woodruff WH, Brown JH. Allometric scaling of metabolic rate from molecules and mitochondria to cells and mammals. Proc Natl Acad Sci U S A. 2002;99:2473–2478. doi: 10.1073/pnas.012579799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazzei D, Guzzardi MA, Giusti S, Ahluwalia A. A low shear stress modular bioreactor for connected cell culture under high flow rates. Biotechnol Bioeng. 2010;106:127–137. doi: 10.1002/bit.22671. [DOI] [PubMed] [Google Scholar]
- 31.Vozzi F, Heinrich JM, Bader A, Ahluwalia AD. Connected Culture of Murine Hepatocytes and HUVEC in a Multicompartmental Bioreactor. Tissue Eng Part A. 2009;15:1291–1299. doi: 10.1089/ten.tea.2008.0066. [DOI] [PubMed] [Google Scholar]
- 32.Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, Shuler ML. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol Prog. 2004;20:338–345. doi: 10.1021/bp034077d. [DOI] [PubMed] [Google Scholar]
- 33.Sung JH, Shuler ML. A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip. 2009;9:1385–1394. doi: 10.1039/b901377f. [DOI] [PubMed] [Google Scholar]
- 34.Sung JH, Shuler ML. Pharmacokinetic-pharmacodynamic models on a chip. In: Nahmias Y, S B, editors. Methods in Bioengineering: Microdevices in Biology and Medicine: Artech House. 2009. [Google Scholar]
- 35.Tatosian DA, Shuler ML. A novel system for evaluation of drug mixtures for potential efficacy in treating multidrug resistant cancers. Biotechnol Bioeng. 2009;103:187–198. doi: 10.1002/bit.22219. [DOI] [PubMed] [Google Scholar]
- 36.Oh KW, Lee K, Ahn B, Furlani EP. Design of pressure-driven microfluidic networks using electric circuit analogy. Lab Chip. 2012;12:515–545. doi: 10.1039/c2lc20799k. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Lee H, Chung M, Jeon NL. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip. 2013;13:1489–1500. doi: 10.1039/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- 38.Moya ML, Hsu YH, Lee AP, Hughes CC, George SC. In vitro perfused human capillary networks. Tissue Eng Part C Methods. 2013;19:730–737. doi: 10.1089/ten.tec.2012.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen JW, Khetani SR, Bhatia SN. In vitro zonation and toxicity in a hepatocyte bioreactor. Toxicol Sci. 2005;84:110–119. doi: 10.1093/toxsci/kfi052. [DOI] [PubMed] [Google Scholar]
- 40.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 41.Kim J, Hegde M, Jayaraman A. Co-culture of epithelial cells and bacteria for investigating host-pathogen interactions. Lab Chip. 2010;10:43–50. doi: 10.1039/b911367c. [DOI] [PubMed] [Google Scholar]
- 42.Mahler GJ, Esch MB, Glahn RP, Shuler ML. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol Bioeng. 2009;104:193–205. doi: 10.1002/bit.22366. [DOI] [PubMed] [Google Scholar]
- 43.Booth R, Kim H. Characterization of a microfluidic in vitro model of the blood-brain barrier (muBBB) Lab Chip. 2012;12:1784–1792. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- 44.Griep LM, Wolbers F, de Wagenaar B, ter Braak PM, Weksler BB, Romero IA, Couraud PO, Vermes I, van der Meer AD, van den Berg A. BBB on chip: microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed Microdevices. 2013;15:145–150. doi: 10.1007/s10544-012-9699-7. [DOI] [PubMed] [Google Scholar]
- 45.Prabhakarpandian B, Shen MC, Nichols JB, Mills IR, Sidoryk-Wegrzynowicz M, Aschner M, Pant K. SyM-BBB: a microfluidic Blood Brain Barrier model. Lab Chip. 2013;13:1093–1101. doi: 10.1039/c2lc41208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller C, George S, Niklason L. Developing a tissue-engineered model of the human bronchiole. J Tissue Eng Regen Med. 2010;4:619–627. doi: 10.1002/term.277. [DOI] [PubMed] [Google Scholar]
- 47.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang KJ, Cho HS, Kang do H, Bae WG, Kwon TH, Suh KY. Fluid-shear-stress-induced translocation of aquaporin-2 and reorganization of actin cytoskeleton in renal tubular epithelial cells. Integr Biol (Camb) 2011;3:134–141. doi: 10.1039/c0ib00018c. [DOI] [PubMed] [Google Scholar]
- 49.Jang KJ, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh KY, Ingber DE. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol (Camb) 2013;5:1119–1129. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 50.Jang KJ, Suh KY. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip. 2010;10:36–42. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- 51.Chen MB, Srigunapalan S, Wheeler AR, Simmons CA. A 3D microfluidic platform incorporating methacrylated gelatin hydrogels to study physiological cardiovascular cell-cell interactions. Lab Chip. 2013;13:2591–2598. doi: 10.1039/c3lc00051f. [DOI] [PubMed] [Google Scholar]
- 52.Natarajan A, Stancescu M, Dhir V, Armstrong C, Sommerhage F, Hickman JJ, Molnar P. Patterned cardiomyocytes on microelectrode arrays as a functional, high information content drug screening platform. Biomaterials. 2011;32:4267–4274. doi: 10.1016/j.biomaterials.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song JW, Cavnar SP, Walker AC, Luker KE, Gupta M, Tung YC, Luker GD, Takayama S. Microfluidic endothelium for studying the intravascular adhesion of metastatic breast cancer cells. PloS One. 2009;4:e5756. doi: 10.1371/journal.pone.0005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu YH, Moya ML, Hughes CC, George SC, Lee AP. A microfluidic platform for generating large-scale nearly identical human microphysiological vascularized tissue arrays. Lab Chip. 2013;13:2990–2998. doi: 10.1039/c3lc50424g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, Lopez JA, Stroock AD. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci U S A. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang H, Kim BJ, Kim YS, Suarez SS, Wu M. Different migration patterns of sea urchin and mouse sperm revealed by a microfluidic chemotaxis device. PloS One. 2013;8:e60587. doi: 10.1371/journal.pone.0060587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, Ingber DE. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med. 2012;4:159ra47. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ploss A, Khetani SR, Jones CT, Syder AJ, Trehan K, Gaysinskaya VA, Mu K, Ritola K, Rice CM, Bhatia SN. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci U S A. 2010;107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moraes C, Mehta G, Lesher-Perez SC, Takayama S. Organs-on-a-chip: a focus on compartmentalized microdevices. Ann Biomed Eng. 2012;40:1211–1227. doi: 10.1007/s10439-011-0455-6. [DOI] [PubMed] [Google Scholar]
- 60.Landersdorfer CB, Jusko WJ. Pharmacokinetic/pharmacodynamic modelling in diabetes mellitus. Clin Pharmacokinet. 2008;47:417–448. doi: 10.2165/00003088-200847070-00001. [DOI] [PubMed] [Google Scholar]
- 61.Stefek M, Karasu C. Eye lens in aging and diabetes: effect of quercetin. Rejuvenation Res. 2011;14:525–534. doi: 10.1089/rej.2011.1170. [DOI] [PubMed] [Google Scholar]
- 62.Siriwardhana N, Kalupahana NS, Cekanova M, LeMieux M, Greer B, Moustaid-Moussa N. Modulation of adipose tissue inflammation by bioactive food compounds. J Nutr Biochem. 2013;24:613–623. doi: 10.1016/j.jnutbio.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 63.Manzano S, Williamson G. Polyphenols and phenolic acids from strawberry and apple decrease glucose uptake and transport by human intestinal Caco-2 cells. Mol Nutr Food Res. 2010;54:1773–1780. doi: 10.1002/mnfr.201000019. [DOI] [PubMed] [Google Scholar]
- 64.Bardy G, Virsolvy A, Quignard JF, Ravier MA, Bertrand G, Dalle S, Cros G, Magous R, Richard S, Oiry C. Quercetin induces insulin secretion by direct activation of L-type calcium channels in pancreatic beta cells. Br J Pharmacol. 2013;169:1102–1113. doi: 10.1111/bph.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Artursson P, Palm K, Luthman K. Caco-2 monolayers in experimental and theoretical predictions of drug transport. Adv Drug Deliv Rev. 2001;46:27–43. doi: 10.1016/s0169-409x(00)00128-9. [DOI] [PubMed] [Google Scholar]
- 66.Lee MY, Kumar RA, Sukumaran SM, Hogg MG, Clark DS, Dordick JS. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc Natl Acad Sci U S A. 2008;105:59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Midwoud PM, Groothuis GM, Merema MT, Verpoorte E. Microfluidic biochip for the perifusion of precision-cut rat liver slices for metabolism and toxicology studies. Biotechnol Bioeng. 2010;105:184–194. doi: 10.1002/bit.22516. [DOI] [PubMed] [Google Scholar]
- 68.van Midwoud PM, Merema MT, Verpoorte E, Groothuis GM. A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip. 2010;10:2778–2786. doi: 10.1039/c0lc00043d. [DOI] [PubMed] [Google Scholar]
- 69.Viravaidya K, Sin A, Shuler ML. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol Prog. 2004;20:316–323. doi: 10.1021/bp0341996. [DOI] [PubMed] [Google Scholar]
- 70.Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic-pharmacodynamic (PKPD) model on a chip. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 71.Zhang C, Zhao Z, Abdul Rahim NA, van Noort D, Yu H. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip. 2009;9:3185–3192. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- 72.Iori E, Vinci B, Murphy E, Marescotti MC, Avogaro A, Ahluwalia A. Glucose and fatty acid metabolism in a 3 tissue in-vitro model challenged with normo- and hyperglycaemia. PloS One. 2012;7:e34704. doi: 10.1371/journal.pone.0034704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wagner I, Materne EM, Brincker S, Sussbier U, Fradrich C, Busek M, Sonntag F, Sakharov DA, Trushkin EV, Tonevitsky AG, Lauster R, Marx U. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip. 2013;13:3538–3547. doi: 10.1039/c3lc50234a. [DOI] [PubMed] [Google Scholar]
- 74.Choi JR, Sung JH, Shuler ML, Kim D. Investigation of portable in situ fluorescence optical detection for microfluidic 3D cell culture assays. Opt Lett. 2010;35:1374–1376. doi: 10.1364/OL.35.001374. [DOI] [PubMed] [Google Scholar]
- 75.Natarajan A, Molnar P, Sieverdes K, Jamshidi A, Hickman JJ. Microelectrode array recordings of cardiac action potentials as a high throughput method to evaluate pesticide toxicity. Toxicol In Vitro. 2006;20:375–381. doi: 10.1016/j.tiv.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 76.Alford PW, Feinberg AW, Sheehy SP, Parker KK. Biohybrid thin films for measuring contractility in engineered cardiovascular muscle. Biomaterials. 2010;31:3613–3621. doi: 10.1016/j.biomaterials.2010.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herrmann KL, McCulloch AD, Omens JH. Glycated collagen cross-linking alters cardiac mechanics in volume-overload hypertrophy. Am J Physiol Heart Circ Physiol. 2003;284:H1277–H1284. doi: 10.1152/ajpheart.00168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shimizu K, Fujita H, Nagamori E. Evaluation systems of generated forces of skeletal muscle cell-based bio-actuators. J Biosci Bioeng. 2013;115:115–121. doi: 10.1016/j.jbiosc.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 79.Wilson K, Das M, Wahl KJ, Colton RJ, Hickman J. Measurement of contractile stress generated by cultured rat muscle on silicon cantilevers for toxin detection and muscle performance enhancement. PloS One. 2010;5:e11042. doi: 10.1371/journal.pone.0011042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fraticelli A, Josephson R, Danziger R, Lakatta E, Spurgeon H. Morphological and contractile characteristics of rat cardiac myocytes from maturation to senescence. Am J Physiol. 1989;257:H259–H265. doi: 10.1152/ajpheart.1989.257.1.H259. [DOI] [PubMed] [Google Scholar]
- 81.Tasche C, Meyhofer E, Brenner B. A force transducer for measuring mechanical properties of single cardiac myocytes. Am J Physiol. 1999;277:H240–H2408. doi: 10.1152/ajpheart.1999.277.6.H2400. [DOI] [PubMed] [Google Scholar]
- 82.Shepherd N, Vornanen M, Isenberg G. Force measurements from voltage-clamped guinea pig ventricular myocytes. Am J Physiol. 1990;258:H452–H459. doi: 10.1152/ajpheart.1990.258.2.H452. [DOI] [PubMed] [Google Scholar]
- 83.Brandt PW, Colomo F, Piroddi N, Poggesi C, Tesi C. Force regulation by Ca2+ in skinned single cardiac myocytes of frog. Biophys J. 1998;74:1994–2004. doi: 10.1016/S0006-3495(98)77906-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marganski WA, De Biase VM, Burgess ML, Dembo M. Demonstration of altered fibroblast contractile activity in hypertensive heart disease. Cardiovasc Res. 2003;60:547–556. doi: 10.1016/j.cardiores.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 85.Tanaka Y, Morishima K, Shimizu T, Kikuchi A, Yamato M, Okano T, Kitamori T. Demonstration of a PDMS-based bio-microactuator using cultured cardiomyocytes to drive polymer micropillars. Lab Chip. 2006;6:230–235. doi: 10.1039/b512099c. [DOI] [PubMed] [Google Scholar]
- 86.Lin G, Palmer RE, Pister KS, Roos KP. Miniature heart cell force transducer system implemented in MEMS technology. IEEE Trans Biomed Eng. 2001;48:996–1006. doi: 10.1109/10.942589. [DOI] [PubMed] [Google Scholar]
- 87.Kaneko T, Kojima K, Yasuda K. An on-chip cardiomyocyte cell network assay for stable drug screening regarding community effect of cell network size. Analyst. 2007;132:892–898. doi: 10.1039/b704961g. [DOI] [PubMed] [Google Scholar]
- 88.McCain ML, Sheehy SP, Grosberg A, Goss JA, Parker KK. Recapitulating maladaptive, multiscale remodeling of failing myocardium on a chip. Proc Natl Acad Sci U S A. 2013;110:9770–9775. doi: 10.1073/pnas.1304913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vandenburgh H. High-content drug screening with engineered musculoskeletal tissues. Tissue Eng Part B Rev. 2010;16:55–64. doi: 10.1089/ten.teb.2009.0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Das M, Gregory CA, Molnar P, Riedel LM, Wilson K, Hickman JJ. A defined system to allow skeletal muscle differentiation and subsequent integration with silicon microstructures. Biomaterials. 2006;27:4374–4380. doi: 10.1016/j.biomaterials.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 91.Liao H, Zhou GQ. Development and progress of engineering of skeletal muscle tissue. Tissue Eng Part B Rev. 2009;15:319–331. doi: 10.1089/ten.teb.2009.0092. [DOI] [PubMed] [Google Scholar]
- 92.Levenberg S, Rouwkema J, Macdonald M, Garfein ES, Kohane DS, Darland DC, Marini R, van Blitterswijk CA, Mulligan RC, D'Amore PA, Langer R. Engineering vascularized skeletal muscle tissue. Nat Biotechnol. 2005;23:879–874. doi: 10.1038/nbt1109. [DOI] [PubMed] [Google Scholar]
- 93.Hinds S, Bian W, Dennis RG, Bursac N. The role of extracellular matrix composition in structure and function of bioengineered skeletal muscle. Biomaterials. 2011;32:3575–3583. doi: 10.1016/j.biomaterials.2011.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Powell CA, Smiley BL, Mills J, Vandenburgh HH. Mechanical stimulation improves tissue-engineered human skeletal muscle. Am J Physiol Cell Physiol. 2002;283:C1557–C1565. doi: 10.1152/ajpcell.00595.2001. [DOI] [PubMed] [Google Scholar]
- 95.Vandenburgh H, Shansky J, Benesch-Lee F, Barbata V, Reid J, Thorrez L, Valentini R, Crawford G. Drug-screening platform based on the contractility of tissue-engineered muscle. Muscle Nerve. 2008;37:438–447. doi: 10.1002/mus.20931. [DOI] [PubMed] [Google Scholar]
- 96.Shimizu K, Sasaki H, Hida H, Fujita H, Obinata K, Shikida M, Nagamori E. Assembly of skeletal muscle cells on a Si-MEMS device and their generative force measurement. Biomed Microdevices. 2010;12:247–252. doi: 10.1007/s10544-009-9379-4. [DOI] [PubMed] [Google Scholar]
- 97.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 98.Fasano A, Shea-Donohue T. Mechanisms of disease: the role of intestinal barrier function in the pathogenesis of gastrointestinal autoimmune diseases. Nat Rev Gastroenterol Hepatol. 2005;2:416–422. doi: 10.1038/ncpgasthep0259. [DOI] [PubMed] [Google Scholar]
- 99.Hawkins BT, Davis TP. The blood-brain barrier/neurovascular unit in health and disease. Pharmacol Rev. 2005;57:173–185. doi: 10.1124/pr.57.2.4. [DOI] [PubMed] [Google Scholar]
- 100.Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 2010;468:562–566. doi: 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rubas W, Cromwell ME, Shahrokh Z, Villagran J, Nguyen TN, Wellton M, Nguyen TH, Mrsny RJ. Flux measurements across Caco-2 monolayers may predict transport in human large intestinal tissue. J Pharm Sci. 1996;85:165–169. doi: 10.1021/js950267+. [DOI] [PubMed] [Google Scholar]
- 102.Douville NJ, Tung YC, Li R, Wang JD, El-Sayed ME, Takayama S. Fabrication of two-layered channel system with embedded electrodes to measure resistance across epithelial and endothelial barriers. Anal Chem. 2010;82:2505–2511. doi: 10.1021/ac9029345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun T, Swindle EJ, Collins JE, Holloway JA, Davies DE, Morgan H. On-chip epithelial barrier function assays using electrical impedance spectroscopy. Lab Chip. 2010;10:1611–1617. doi: 10.1039/c000699h. [DOI] [PubMed] [Google Scholar]
- 104.Jimison LH, Tria SA, Khodagholy D, Gurfinkel M, Lanzarini E, Hama A, Malliaras GG, Owens RM. Measurement of barrier tissue integrity with an organic electrochemical transistor. Adv Mater. 2012;24:5919–5923. doi: 10.1002/adma.201202612. [DOI] [PubMed] [Google Scholar]
- 105.Johnstone AF, Gross GW, Weiss DG, Schroeder OH, Gramowski A, Shafer TJ. Microelectrode arrays: a physiologically based neurotoxicity testing platform for the 21st century. Neurotoxicology. 2010;31:331–350. doi: 10.1016/j.neuro.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 106.Jones IL, Livi P, Lewandowska MK, Fiscella M, Roscic B, Hierlemann A. The potential of microelectrode arrays and microelectronics for biomedical research and diagnostics. Anal Bioanal Chem. 2011;399:2313–2329. doi: 10.1007/s00216-010-3968-1. [DOI] [PubMed] [Google Scholar]
- 107.Asai Y, Tada M, Otsuji TG, Nakatsuji N. Combination of functional cardiomyocytes derived from human stem cells and a highly-efficient microelectrode array system: an ideal hybrid model assay for drug development. Curr Stem Cell Res Ther. 2010;5:227–232. doi: 10.2174/157488810791824502. [DOI] [PubMed] [Google Scholar]
- 108.Nam Y, Wheeler BC. In vitro microelectrode array technology and neural recordings. Crit Rev Biomed Eng. 2011;39:45–61. doi: 10.1615/critrevbiomedeng.v39.i1.40. [DOI] [PubMed] [Google Scholar]
- 109.Morin F, Nishimura N, Griscom L, Lepioufle B, Fujita H, Takamura Y, Tamiya E. Constraining the connectivity of neuronal networks cultured on microelectrode arrays with microfluidic techniques: a step towards neuron-based functional chips. Biosens Bioelectron. 2006;21:1093–1100. doi: 10.1016/j.bios.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 110.Kanagasabapathi TT, Ciliberti D, Martinoia S, Wadman WJ, Decre MM. Dual-compartment neurofluidic system for electrophysiological measurements in physically segregated and functionally connected neuronal cell culture. Front Neuroeng. 2011;4:13. doi: 10.3389/fneng.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Braam SR, Tertoolen L, van de Stolpe A, Meyer T, Passier R, Mummery CL. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4:107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 112.Caspi O, Itzhaki I, Kehat I, Gepstein A, Arbel G, Huber I, Satin J, Gepstein L. In vitro electrophysiological drug testing using human embryonic stem cell derived cardiomyocytes. Stem Cells Dev. 2009;18:161–172. doi: 10.1089/scd.2007.0280. [DOI] [PubMed] [Google Scholar]
- 113.Varghese K, Molnar P, Das M, Bhargava N, Lambert S, Kindy MS, Hickman JJ. A new target for amyloid beta toxicity validated by standard and high-throughput electrophysiology. PloS One. 2010;5:e8643. doi: 10.1371/journal.pone.0008643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.He M, Herr AE. Microfluidic polyacrylamide gel electrophoresis with in situ immunoblotting for native protein analysis. Anal Chem. 2009;81:8177–8184. doi: 10.1021/ac901392u. [DOI] [PubMed] [Google Scholar]
- 115.Zhang C, Xu J, Ma W, Zheng W. PCR microfluidic devices for DNA amplification. Biotechnol Adv. 2006;24:243–284. doi: 10.1016/j.biotechadv.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 116.Roux A, Lison D, Junot C, Heilier JF. Applications of liquid chromatography coupled to mass spectrometry-based metabolomics in clinical chemistry and toxicology: A review. Clin Biochem. 2011;44:119–135. doi: 10.1016/j.clinbiochem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 117.Wikswo JP, Block FE, 3rd, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, McLean DL, McLean JA, McKenzie JR, Reiserer RS, Samson PC, Schaffer DK, Seale KT, Sherrod SD. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans Biomed Eng. 2013;60:682–690. doi: 10.1109/TBME.2013.2244891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Enders JR, Marasco CC, Kole A, Nguyen B, Sevugarajan S, Seale KT, Wikswo JP, McLean JA. Towards monitoring real-time cellular response using an integrated microfluidics-matrix assisted laser desorption ionisation/nanoelectrospray ionisation-ion mobility-mass spectrometry platform. IET Syst Biol. 2010;4:416–427. doi: 10.1049/iet-syb.2010.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu H, Alexander CM, Beebe DJ. A plate reader-compatible microchannel array for cell biology assays. Lab Chip. 2007;7:388–391. doi: 10.1039/b612358a. [DOI] [PubMed] [Google Scholar]
- 120.Shuler ML, Esch MB. Body-on-a chip: Using microfluidic systems to predict human responses to drugs. Pure Appl Chem. 2010;82:1635–1645. [Google Scholar]
- 121.Meyvantsson I, Warrick JW, Hayes S, Skoien A, Beebe DJ. Automated cell culture in high density tubeless microfluidic device arrays. Lab Chip. 2008;8:717–724. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 122.Faley SL, Copland M, Wlodkowic D, Kolch W, Seale KT, Wikswo JP, Cooper JM. Microfluidic single cell arrays to interrogate signalling dynamics of individual, patient-derived hematopoietic stem cells. Lab Chip. 2009;9:2659–2664. doi: 10.1039/b902083g. [DOI] [PubMed] [Google Scholar]
- 123.Simmons CS, Petzold BC, Pruitt BL. Microsystems for biomimetic stimulation of cardiac cells. Lab Chip. 2012;12:3235–3248. doi: 10.1039/c2lc40308k. [DOI] [PubMed] [Google Scholar]
- 124.Oh TI, Sung JH, Tatosian DA, Shuler ML, Kim D. Real-time fluorescence detection of multiple microscale cell culture analog devices in situ. Cytometry A. 2007;71:857–865. doi: 10.1002/cyto.a.20427. [DOI] [PubMed] [Google Scholar]
- 125.Sung JH, Choi JR, Kim D, Shuler ML. Fluorescence optical detection in situ for real-time monitoring of cytochrome P450 enzymatic activity of liver cells in multiple microfluidic devices. Biotechnol Bioeng. 2009;104:516–525. doi: 10.1002/bit.22413. [DOI] [PubMed] [Google Scholar]
- 126.Lee SR, Lee YT, Sawada K, Takao H, Ishida M. Development of a disposable glucose biosensor using electroless-plated Au/Ni/copper low electrical resistance electrodes. Biosens Bioelectron. 2008;24:410–414. doi: 10.1016/j.bios.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 127.Luo X, Davis JJ. Electrical biosensors and the label free detection of protein disease biomarkers. Chem Soc Rev. 2013;42:5944–5962. doi: 10.1039/c3cs60077g. [DOI] [PubMed] [Google Scholar]
- 128.Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- 129.Choucha-Snouber L, Aninat C, Grsicom L, Madalinski G, Brochot C, Poleni PE, Razan F, Guillouzo CG, Legallais C, Corlu A, Leclerc E. Investigation of ifosfamide nephrotoxicity induced in a liver-kidney co-culture biochip. Biotechnol Bioeng. 2013;110:597–608. doi: 10.1002/bit.24707. [DOI] [PubMed] [Google Scholar]
- 130.van der Meer AD, van den Berg A. Organs-on-chips: breaking the in vitro impasse. Integr Biol (Camb) 2012;4:461–470. doi: 10.1039/c2ib00176d. [DOI] [PubMed] [Google Scholar]
- 131.van der Valk J, Brunner D, De Smet K, Fex Svenningsen A, Honegger P, Knudsen LE, Lindl T, Noraberg J, Price A, Scarino ML, Gstraunthaler G. Optimization of chemically defined cell culture media--replacing fetal bovine serum in mammalian in vitro methods. Toxicol In Vitro. 2010;24:1053–1063. doi: 10.1016/j.tiv.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 132.Schaffner AE, Barker JL, Stenger DA, Hickman JJ. Investigation of the factors necessary for growth of hippocampal neurons in a defined system. J Neurosci Methods. 1995;62:111–119. doi: 10.1016/0165-0270(95)00063-1. [DOI] [PubMed] [Google Scholar]
- 133.Das M, Molnar P, Devaraj H, Poeta M, Hickman JJ. Electrophysiological and morphological characterization of rat embryonic motoneurons in a defined system. Biotechnol Prog. 2003;19:1756–1761. doi: 10.1021/bp034076l. [DOI] [PubMed] [Google Scholar]
- 134.Guo X, Das M, Rumsey J, Gonzalez M, Stancescu M, Hickman J. Neuromuscular junction formation between human stem-cell-derived motoneurons and rat skeletal muscle in a defined system. Tissue Eng Part C Methods. 2010;16:1347–1355. doi: 10.1089/ten.tec.2010.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smith AST, Long CJ, Pirozzi K, Hickman JJ. A functional system for high-content screening of neuromuscular junctions in vitro. Technology. 2013:1–12. doi: 10.1142/S2339547813500015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guo X, Gonzalez M, Stancescu M, Vandenburgh HH, Hickman JJ. Neuromuscular junction formation between human stem cell-derived motoneurons and human skeletal muscle in a defined system. Biomaterials. 2011;32:9602–9611. doi: 10.1016/j.biomaterials.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Davis H, Gonzalez M, Bhargava N, Stancescu M, Hickman JJ, Lambert S. Rat Cortical Oligodendrocyte-Embryonic Motoneuron Co-Culture: An Axon-Oligodendrocyte Interaction Model. J Biomater Tissue Eng. 2012;2:206–214. doi: 10.1166/jbt.2012.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Levenberg S, Huang NF, Lavik E, Rogers AB, Itskovitz-Eldor J, Langer R. Differentiation of human embryonic stem cells on three-dimensional polymer scaffolds. Proc Natl Acad Sci U S A. 2003;100:12741–12746. doi: 10.1073/pnas.1735463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Page H, Flood P, Reynaud E. Three-dimensional tissue cultures: current trends and beyond. Cell Tissue Res. 2013;352:123–131. doi: 10.1007/s00441-012-1441-5. [DOI] [PubMed] [Google Scholar]