Abstract
In a Genome Wide Association Study (GWAS) of individuals of European ancestry afflicted with Systemic Lupus Erythematosus (SLE) the extensive utilization of imputation, stepwise multiple regression, lasso regularization, and increasing study power by utilizing False Discovery Rate (FDR) instead of a Bonferroni multiple test correction enabled us to identify 13 novel non-human leukocyte antigen (HLA) genes and confirmed the association of 4 genes previously reported to be associated. Novel genes associated with SLE susceptibility included two transcription factors (EHF, and MED1), two components of the NFκB pathway (RASSF2 and RNF114), one gene involved in adhesion and endothelial migration (CNTN6), and two genes involved in antigen presentation (BIN1 and SEC61G). In addition, the strongly significant association of multiple single nucleotide polymorphisms (SNPs) in the HLA region was assigned to HLA alleles and serotypes and deconvoluted into four primary signals. The novel SLE-associated genes point to new directions for both the diagnosis and treatment of this debilitating autoimmune disease.
Introduction
Systemic Lupus Erythematosus (SLE) (OMIM 152700) is a debilitating autoimmune disease which affects multiple organs and is characterized by a loss of tolerance to self-antigens, inflammation, and dysregulated immune responses, resulting in significant morbidity and mortality. Although many new loci which contribute to the pathogenesis of SLE have been identified by Genome Wide Association Studies (GWAS) and other association studies, they collectively do not explain all the risk contributed by heritable factors [1], indicating that other, as-of-yet unidentified genes are likely to be involv1ed.
GWAS are commonly used to identify gene–phenotype associations. The power of GWAS is in their agnostic approach, which does not require prior knowledge of any of the genetics or cellular mechanisms underlying a phenotypic trait. This comes at the cost of testing a large number of single nucleotide polymorphisms (SNPs), thereby increasing the multiple-testing correction, and ignoring (often decades of) prior research into a phenotypic trait which may increase the power of a genetic study [2].
In the present study we report the discovery of 13 novel genes outside of the human leukocyte antigen (HLA) region (4 Family-Wise Error Rate (FWER) significant) and confirmation of 4 genes (4 FWER significant) which were reported as significant in previously published studies (Table 1).
Table 1.
Association of SLE with non-HLA genes. SNPs are assigned to the nearest gene; the ideogram is given in the loc column. The p value, FDR, and rsid correspond to the most significant SNP found within a gene. Previous Association columns indicate whether a gene has previously been associated with SLE and if so, p values and an rsid are given for a representative association. The Dependent On column indicates whether a genes significance can be explained by another gene or region.
| Gene | Loc | p | FDR | rsid | Previous Association | Dependent On | ||
|---|---|---|---|---|---|---|---|---|
| Assoc. | p | rsid | ||||||
| EDEM3 | 1q25.3 | 2.3×10−13 | 2.3×10−8 | rs10911628 | No | |||
| BIN1 | 2q14.3 | 4×10−6 | 4.7×10−2 | rs12993006 | No | |||
| KCNJ3 | 2q24.1 | 2×10−6 | 3×10−2 | rs4544377 | No | |||
| STAT4 | 2q32.2 | 5.1×10−9 | 1.9×10−4 | rs7574865 | Yes [33] | 8.2×10−14 | rs7574865 | |
| CNTN6 | 3p26.3 | 9.8×10−8 | 2.4×10−3 | rs4684256 | No | |||
| SEC61G | 7p11.2 | 1.8×10−6 | 2.9×10−2 | rs6946131 | No | |||
| IRF5 | 7q32.1 | 7.1×10−10 | 3×10−5 | rs4728142 | Yes [34] | 4.4×10−7 | rs2004640 | |
| TNPO3 | 7q32.1 | 1.5×10−13 | 2.1×10−8 | rs10488631 | Yes [35] | 6.4×10−13 | rs12531711 | |
| MTG1 | 10q26.3 | 3.3×10−6 | 4.5×10−2 | rs10857712 | No | |||
| EHF | 11p13 | 1.9×10−7 | 4×10−3 | rs10466455 | No | |||
| FAM98B | 15q14 | 9.9×10−15 | 2.9×10−9 | rs11073328 | No | |||
| TYRO3 | 15q15.1 | 3.4×10−6 | 4.5×10−2 | rs12259 | No3 [36] | FAM98B | ||
| SPATA8 | 15q26.2 | 1.2×10−8 | 4×10−4 | rs8023715 | No | |||
| ITGAM | 16p11.2 | 4.3×10−11 | 2.1×10−6 | rs9888739 | Yes [37] | 6.9×10−22 | rs1143679 | |
| MED1 | 17q12 | 6.7×10−7 | 1.2×10−2 | rs11655550 | No | |||
| RASSF2 | 20p13 | 2.1×10−6 | 3×10−2 | rs6084875 | No | |||
| RNF114 | 20q13.13 | 1.4×10−11 | 8×10−7 | rs11697848 | No | |||
Essential for the prevention of lupus-like autoimmunity via innate inflammatory responses
Furthermore, we have imputed all associated regions using the largest panels publicly available and used bioinformatics tools to test for deleterious effects of non-synonymous variants. Imputation can be used to deconvolute multiple signals in regions which have complex interdependent signals, such as the HLA region. Imputation (HLA*IMP) has also been used in this study to assign samples to HLA alleles and then to HLA serotypes, connecting modern GWAS based association results to earlier genetic and serotypic association results.
Many SNPs are associated with a disease because they are in Linkage Disequilibrium (LD) with another SNP which is the primary signal in that region. To disambiguate between independent and dependent signals, we used conditional regression analysis and lasso regularization to eliminate signals whose association was due to correlation with another primary signal. This enabled us to identify the minimum number of signals required to explain the association results present in every associated region, an approach pioneered by Raychaudhuri et al.[3], and adapted here for SLE.
This study has also assigned HLA alleles and serotypes to each subject and has refined the association signals within the HLA and other regions.
Results
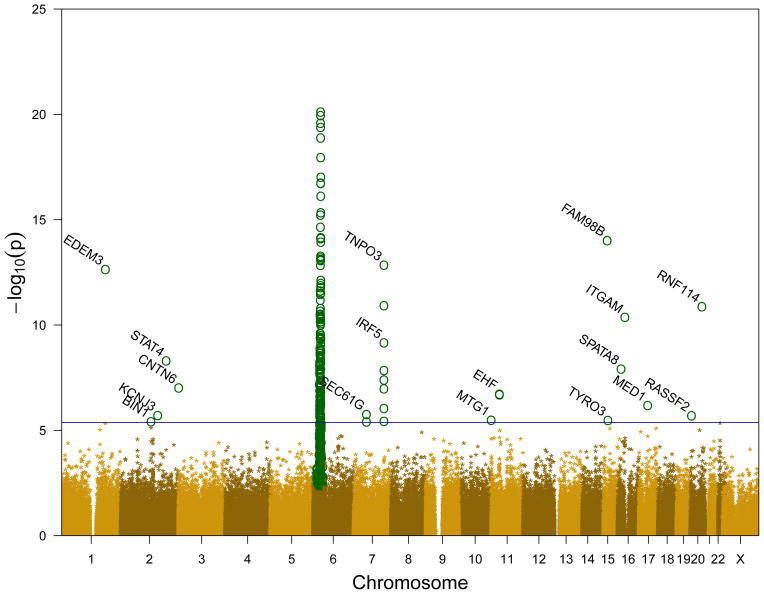
We detected 430 significant SNPs (False Discovery Rate (FDR) ≤ 0.05) in 160 genes. 405 SNPs were in the HLA region (Supplemental Table S1 and S3), and 25 SNPs were in 17 genes in other regions (Table 1, Supplemental Table S3). An additional 7 genes were found to be consistent with previously published results, although they did not meet the non-HLA FDR level of this study (Supplemental Table S2). Figure 1 shows a Manhattan plot of the genome wide association data showing the p values of all tested SNPs. As this figure shows, the HLA region (6p23-6p21) is dramatically enriched in significant SNPs. Because of these results, coupled with the well-known association of the HLA region with SLE and the extensive LD in the HLA region, we have separated the analysis and discussion of HLA genes from non-HLA genes.
Figure 1.
Manhattan plot of genome wide association data. Significant SNPs (FDR ≤ 0.05) are shown as circles in dark green (
 ), non-significant SNPs (FDR > 0.05) are shown as goldenrod asterisks (
), non-significant SNPs (FDR > 0.05) are shown as goldenrod asterisks (
 ). The blue horizontal line is an estimate of the p value where FDR = 0.05 for non-HLA regions.
). The blue horizontal line is an estimate of the p value where FDR = 0.05 for non-HLA regions.
Non-HLA regions
25 significant SNPs within 16 regions corresponding to 17 genes were detected in non-HLA regions. Of these genes, 13 have not been previously associated with SLE (Previous Association columns in Table 1). Two of the 17 genes are transcription factors (EHF and MED1), and two are involved in NFκB signaling (RASSF2 and RNF114). Two genes are involved in antigen presentation (BIN1 and SEC61G), and one in cell adhesion/tissue remodeling CNTN6. All of the significant genes which have mRNA expression information available are expressed in at least one relevant immune cell type (Figure S2).
The presence of multiple significant SNPs in a region can be due to LD or multiple independent signals. To distinguish between these possibilities, we performed multiple regression analysis on the significant SNPs in each of the 4 regions which contained multiple significant SNPs. We found SNPs in 1 gene which were dependent on other genes or interdependent on each other where significance of either gene could be explained by the other gene (Table 1 “Dependent On” column).
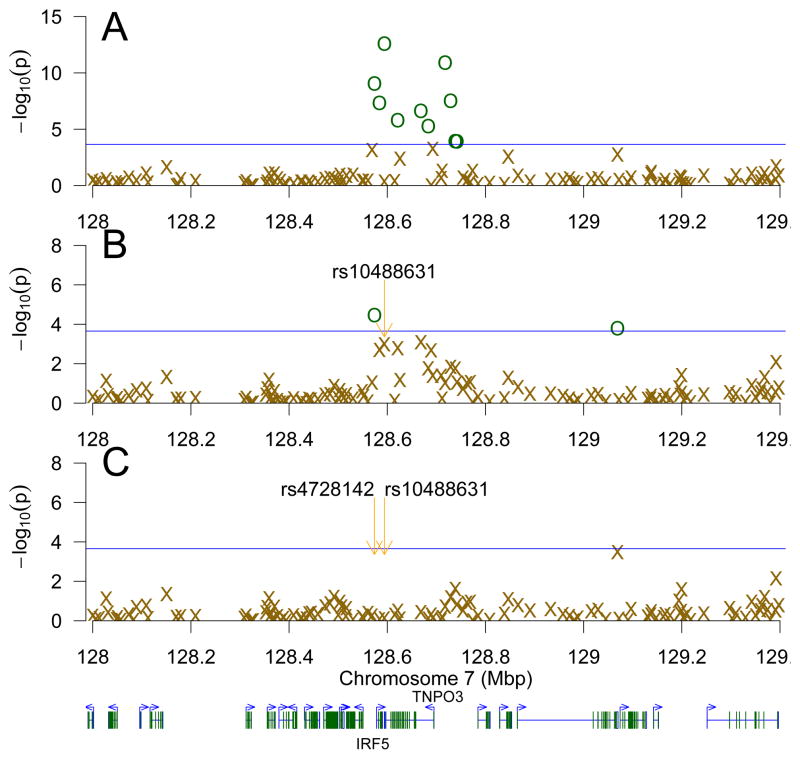
In the region including and surrounding IRF5 and TNPO3 (7q32.1) there are 8 significant SNPs (Figure 2 panel A, Supplemental Table S3). However, after accounting for rs10488631 in TNPO3 (Figure 2 panel B) and rs4728142 in IRF5 (Figure 2 panel C), the next most-significant SNP is rs1665105, whose p value (3.29×10−4) is greater than the IRF5/TNPO3 region multiple-family FDR (2.22×10−4) [4], and furthermore was not found to be significant in this study. This suggests that there are at least two causal variants in the IRF5 and TNPO3 region, and that IRF5 and TNPO3 are independently associated with SLE.
Figure 2.
Step-wise multiple regression of the IRF5 and TNPO3 region. Panel A shows all SNPs in the IRF5 and TNPO3 region. Each subsequent panel (B–C) shows the same SNPs after accounting for the most significant remaining SNP in the model, with SNPs more significant than the FDR (BH−Rq/m) threshold for this region (a blue horizontal line) shown in dark green circles (
 ); those less significant than the threshold are shown as goldenrod Xs (
); those less significant than the threshold are shown as goldenrod Xs (
 ). Panel B accounted for rs10488631 and panel C, for rs4728142 and rs10488631. Genes are depicted below the figure, with the starting positions of IRF5 and TNPO3 indicated.
). Panel B accounted for rs10488631 and panel C, for rs4728142 and rs10488631. Genes are depicted below the figure, with the starting positions of IRF5 and TNPO3 indicated.
In the 15q14 region, the significant SNP in FAM98B, rs11073328, accounts for the original significance of the SNP in TYRO3, rs12259 (pmult = 2.61×10−18, pmult = 8.30×10−2 in the multiple regression, respectively). This indicates that the association of TYRO3 is due to FAM98B; there is evidence for only one signal in the 15q14 region. The final two regions which have multiple SNPs are SEC61G and EHF; each of these regions has two significantly associated SNPs, and in both regions, the significantly associated SNPs are in complete LD with each other. Thus, there is only evidence for a single signal in SEC61G and another single signal in EHF.
Imputation was performed on all 16 non-HLA regions comprising over 2.9 million SNPs. 840 additional significant SNPs were found in these regions, 3 of these SNPs were non-synonymous. However, bioinformatic analyses of these non-synonymous SNPs using PolyPhen [5], SIFT [6], and PROVEAN [7] do not predict deleterious effects as a consequence of these mutations. Thus, it is unlikely that we have identified the causal mutations responsible for the association of these regions with SLE, and deep sequencing or other follow up studies of these regions are required.
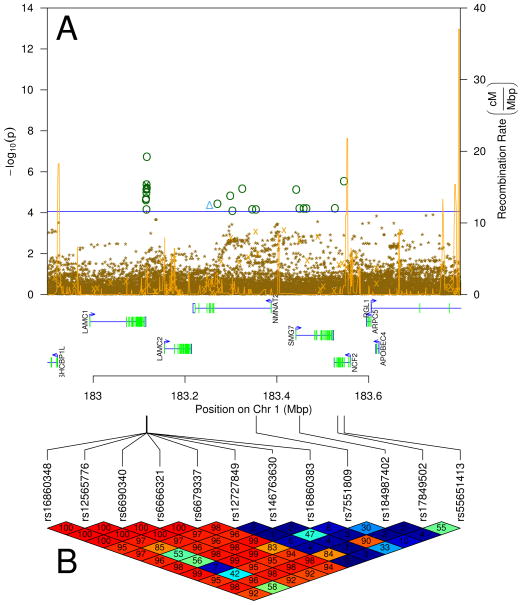
We have previously established that rs17849502 in NCF2 (1q25) is a causal mutation associated with SLE which produces a two-fold reduction in Fcγ receptor-mediated Vav1 NADPH oxidase response [8]. In this region there is are multiple significant and suggestive (FDR ≤ 0.1) SNPs by genotyping and imputation in each of EDEM3, LAMC1, and NCF2 genes as shown in Figure 3. Multiple regression indicates that the signals in EDEM3 and LAMC1 are independent of each other (data not shown). To determine if the significant association of EDEM3 and the suggestive association of LAMC1 was due to the presence of rs17849502 in NCF2, we examined the imputation results in this region. While rs17849502 was present in the imputation panels, it is rare, so the ability of the imputation panels to accurately impute this SNP is greatly diminished, leading to its non-significance upon imputation (IMPUTE v2 reports an “info” metric of 0.05 for this SNP; accurately imputed SNPs should have an “info” metric close to 1 [9]). However, rs17849502 and rs12565776 are in strong linkage disequilibrium (D′ = 0.96, Figure 3 Panel B), so it remains possible that the significance of rs12565776 is due to rs1789502.
Figure 3.
Imputation in the LAMC1, EDEM3, and NCF2 region. Panel A shows the imputation results. Significant imputed SNPs are shown as dark green circles (
 ), significant genotyped SNPs are shown as cyan triangles (
), significant genotyped SNPs are shown as cyan triangles (
 ), non-significant SNPs (FDR > 0.05) are shown as goldenrod asterisks (
), non-significant SNPs (FDR > 0.05) are shown as goldenrod asterisks (
 ) The blue horizontal line is an estimate of the p value where FDR = 0.05. The spiky gold line depicts the recombination rate in centimorgans per megabase. Panel B shows the LD (D′) of significant SNPs and rs17849502. SNPs in LD (D′ near 100) are red; those not in LD (D′ near 0) are blue, intermediate LD are on the red-orange-yellow-green-blue color continuum.
) The blue horizontal line is an estimate of the p value where FDR = 0.05. The spiky gold line depicts the recombination rate in centimorgans per megabase. Panel B shows the LD (D′) of significant SNPs and rs17849502. SNPs in LD (D′ near 100) are red; those not in LD (D′ near 0) are blue, intermediate LD are on the red-orange-yellow-green-blue color continuum.
HLA region
The HLA region has by far the largest number and most highly significant SNPs, reaching a minimum p value of 7.76×10−21 (FDR=2.56×10−17) (Figure 1, Supplemental Table S1).
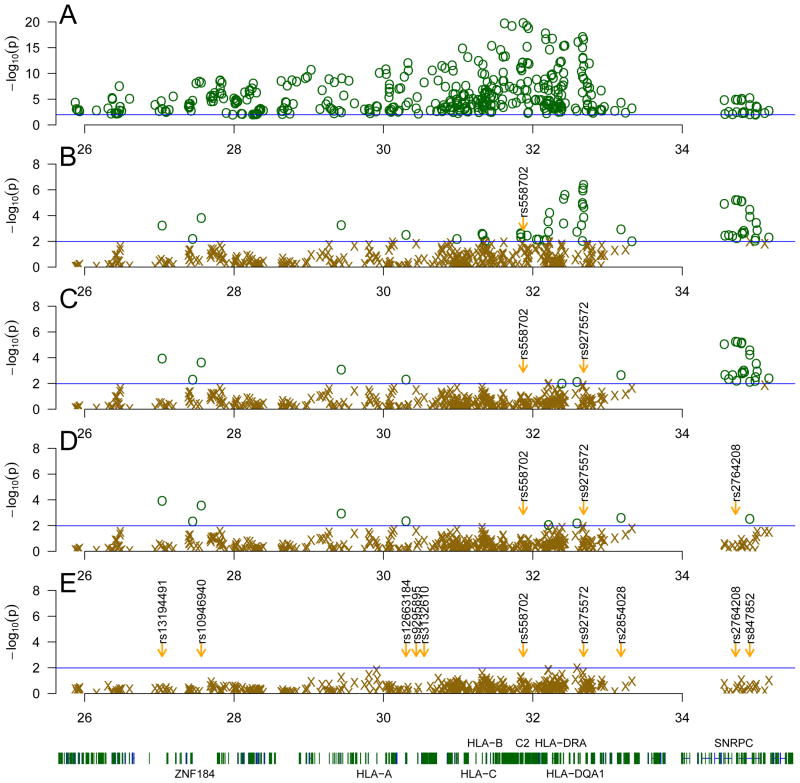
The 6p21-22 region, which includes the classical HLA genes, contains many alleles which are in linkage disequilibrium with each other. Thus, many of the significant results seen could be due to a smaller number of primary signals. To determine the minimum number of signals capable of accounting for the significance seen in the 6p21-22 region we performed a step-wise multiple regression analysis, accounting for the most significant SNP in the model at each step, and repeating until no additional terms had p values less than the p value corresponding to an FDR of 0.05 in this study. As shown in Figure 4, the most significant SNP (from 405 significant SNPs out of 4,748 genotyped SNPs in this region) was rs558702 (p = 7.76 × 10−21) (Figure 4 Panel A). After accounting for rs558702, the most significant unaccounted SNP is rs9275572 (p = 1.94 × 10−6, Figure 4 Panel B). Subsequently, after accounting for both of these SNPs, rs2764208 was the most significant SNP unaccounted for (p = 3.61×10−6, Figure 4 Panel C). Finally, rs10946940 was most significant SNP after accounting for all three of these SNPs (p = 2.37×10−4, Figure 4 Panel D). These four SNPs accounted for the vast majority of the significant SNPs seen in the 6p21-22 region, although 6 additional weak signals remained (Figure 4 Panel E). Using lasso regularization [10] across all significant SNPs in this region, both rs558702 and rs9275572 were the components in the model with the largest coefficients, further indicating their importance (data not shown). Thus, two signals in the classical HLA are able to account for the vast majority of the significant associations seen in HLA, and two additional signals in ZNF184 and SNRPC explain most significance seen in the periphery of the HLA region (Table 2 and Figure 4). ZNF184 is a transcription factor with no known gene targets. The other signal is in SNRPC which is involved in the formation of the spliceosome which is often a target of autoantibodies in SLE [11]. Thus, the majority of the association of classical HLA with SLE in this study can be explained by two primary signals.
Figure 4.
Step-wise multiple regression of the HLA region. Panel A shows all significant SNPs in the HLA region. Each subsequent panel (B–E) shows the same SNPs after accounting for of the most significant remaining SNP in the model, with SNPs more significant than the genome-wide FDR threshold for this study shown in dark green circles (
 ); those less significant than the threshold are shown as goldenrod Xs (
); those less significant than the threshold are shown as goldenrod Xs (
 ). Panel B accounted for rs558702, panel C accounted for rs9275572 and rs558702, panel D accounted for rs2764208, rs9275572 and rs558702. After accounting for rs10946940, rs2764208, rs9275572 and rs558702 (panel E), there were no SNPs meeting the genome-wide FDR threshold.
). Panel B accounted for rs558702, panel C accounted for rs9275572 and rs558702, panel D accounted for rs2764208, rs9275572 and rs558702. After accounting for rs10946940, rs2764208, rs9275572 and rs558702 (panel E), there were no SNPs meeting the genome-wide FDR threshold.
Table 2.
Significance of four SNP model terms in the seven term model, showing the need for at least four separate signals in the HLA region to account for all of the significant associations with SLE seen. p is the probability of obtaining the results given that the coefficient of the term is equal to zero.
| rsID | Gene | p value |
|---|---|---|
| rs558702 | C2 | 4.40 × 10−7 |
| rs9275572 | HLA-DQA2 | 2.82 × 10−6 |
| rs2764208 | SNRPC | 4.10 × 10−6 |
| rs10946940 | ZNF184 | 2.37 × 10−4 |
To reconcile our SNP findings with the established HLA allele and serotype association with SLE, we imputed HLA alleles using HLA*IMP [12–14] which assigned an HLA haplotype to each sample on the basis of genotyped SNPs in the HLA region. The alleles of HLA-A, B, C, DPB1, DQA1, DQB1, and DRB1 were imputed at four digit accuracy and HLA-DRB3, DRB4, and DRB5 were imputed at two digit accuracy. Table 3 shows the alleles which were significantly associated with SLE. Notably, HLA-DQA1*05:01 and HLADQB1* 02:01, which have previously been shown to be significantly associated with SLE and are part of the DR17/DQ2 extended haplotype [15], were found to be highly associated in this study. We did not detect any significant associations with any of the alleles of HLA-A, C, DPB1, DRB4, or DRB5. Finally, multiple regression with HLA-DQB1 and HLA-DRB1 indicated that HLA-DQB1 and HLA-DRB1 have an independent association with SLE, and the association of one is not merely due to linkage with the other (Table S5). Furthermore, the association of HLA-B is largely explained by HLA-DRB1, but not HLA-DQB1 (Table S5). A regression analysis with the significant alleles of HLA-DQB1 and HLA-DRB1 with the two significant primary SNPs signals in classical HLA (rs558702 and rs9275572) indicates that HLA-DQB1 and HLA-DRB1 are strongly correlated with these SNPs. The SNPs outside of the HLA region (rs2764208 and rs10946940) were not accounted for by HLA-DQB1 or HLA-DRB1, indicating that their association is independent of the classical HLA signal.
Table 3.
HLA allele association with SLE. Gene*Allele is the gene and four (two for DRB3-5) digit HLA allele assignment. OR with ±95% CI in parentheses and p values of association with SLE with three PCA axes. Only alleles with p ≤ 10−3 are shown.
| Gene*Allele | OR ± 95% CI | p | FDR |
|---|---|---|---|
| HLA-B*08:01 | 1.802 (1.346, 2.432) | 9.35 × 10−5 | 2.36 × 10−3 |
| HLA-DQA1*05:01 | 1.890 (1.534, 2.335) | 2.80 × 10−9 | 4.96 × 10−7 |
| HLA-DQB1*02:01 | 1.755 (1.381, 2.238) | 4.85 × 10−6 | 2.39 × 10−4 |
| HLA-DQB1*03:01 | 0.643 (0.502, 0.825) | 4.95 × 10−4 | 7.80 × 10−3 |
| HLA-DRB1*03:01 | 1.934 (1.507, 2.494) | 2.88 × 10−7 | 2.55 × 10−5 |
| HLA-DRB3*01 | 1.791 (1.296, 2.507) | 5.28 × 10−4 | 7.80 × 10−3 |
| HLA-DRB3*02 | 1.791 (1.296, 2.507) | 5.28 × 10−4 | 7.80 × 10−3 |
The HLA alleles were then used to assign each sample to an HLA serotype. Table 4 shows the antigens which were significantly associated with SLE. DQ7 (broad antigen DQ3) had a protective effect, whereas DR17 and B8 were associated with disease. We did not see any significant associations with the HLA Class I A or C antigens, nor did we see any significant associations with alleles of the extended DR/DQ haplotype.
Table 4.
HLA antigen association with SLE. In the case of split antigens, the broad antigen is given in the Broad column. OR with ±95% CI in parentheses and p values of association with SLE with three PCA axes. Only antigens with p < 10−3 are shown.
| Antigen | Broad | OR ± 95% CI | p |
|---|---|---|---|
| B8 | 1.803 (1.347, 2.434) | 9.12 × 10 −5 | |
| DQ7 | DQ3 | 0.643 (0.501, 0.825) | 4.98 × 10 −4 |
| DR17 | DR3 | 1.962 (1.589, 2.426) | 4.15 × 10 −10 |
Discussion
In the present study we report the discovery of novel non-HLA genes not previously associated with SLE and the confirmation of genes which were reported as significant in previously published studies (Table 1). The novel discoveries were made in part due to the increase in statistical power of the study through the use of a FDR multiple testing correction instead of a FWER multiple testing correction. SLE is largely a disease of aberrant immunoregulation affecting most types of immune cells. It is therefore expected that genes involved in regulation of various immune cells and their functions would be an important part of the genetic susceptibility architecture of SLE. Indeed, among the novel genes associated with SLE susceptibility found in the present study, two genes code for transcription factors (EHF and MED1), two are components of the NFκB pathway (RASSF2 and RNF114), two are involved in antigen presentation (BIN1 and SEC61G), and one is involved in adhesion and endothelial migration (CNTN6).
Some of the proteins encoded by these susceptibility genes have been previously considered to be involved in the pathogenesis of the disease, but this is the first study to demonstrate that they are actually associated with genetic susceptibility to SLE.
The Ets family of transcription factors and especially Ets-1, have been previously associated with SLE[16]. Here we identified EHF, another member of the Ets family as associated with SLE. EHF is particularly important in the differentiation of dendritic cells (DC), an important antigen presenting cell and producer of cytokines, including type I IFNs that are essential for disease pathogenesis [17]. Interestingly, the cytokine TGFA, specifically involved in DC differentiation [17], is suggestively associated with SLE (p = 2.62 × 10−5, FDR = 1.60×10−1). Furthermore, MAFB, a repressor of Ets-mediated transcription, is also suggestively associated with SLE (p = 9.88×10−6, FDR = 8.83×10−2). The Ets pathway exemplifies the recurring motif of genes at multiple steps of a disease-relevant pathway being found to be associated with that disease.
The transcription factor MED1 is essential for the intrathymic development of iNKT cells, a subgroup of immune cells found at abnormal levels in SLE subjects[18].
Components of the NFκB pathway, the master transcriptional regulator of inflammation, have been associated with increased SLE susceptibility (IRAK1, TNFAIP3, UBE2L3, PRKCB)[1]. The present study adds two new genes involved in NFκB signaling (RASSF2 and RNF114), highlighting the importance of this pathway for SLE genetic susceptibility and pathogenesis. RASSF2 downregulates NFκB signaling via association with IKK. RNF114, an ubiquitin binding protein, regulates a positive feedback loop that enhances dsRNA-induced production of type I IFN through the activation of the IRF3 and NFκB transcription factors, previously shown to be associated with the immune-mediated skin disease psoriasis[19].
Genetic variants related to adhesion and endothelial migration of the various immune cell types have been associated with SLE susceptibility (ITGAM, ICAM1, SELP)[1, 20]. In the present study we have identified an additional gene involved in cell adhesion and tissue remodeling (CNTN6).
Although the primary auto-antigens driving SLE pathogenesis are quite elusive, it is clear that abnormal antigen processing and presentation play an important part[1]. In the current study we identified two new susceptibility genes involved in antigen processing and presentation (BIN1 and SEC61G). These susceptibility genes accompany and supplement the MHC genes within the HLA region. Classical MHC class I and class II genes on the surface of antigen presenting cells are responsible for antigen presentation to the T cell receptor on T lymphocytes. MHC loci were the first reported genetic association with SLE and remain the strongest. Understanding the genetic risk of this region is therefore critical to the understanding of the pathogenesis of the disease. However, the region is extremely gene-dense with long range LD and hundreds of immunologically active genes, which makes identification of the true causal loci very difficult.
Following the approach of Raychaudhuri et al. [3] using conditional logistic regression, we showed that two signals inside of the classical HLA and two additional in the periphery can account for the vast majority of the significant SNPs in the entire HLA region. We confirm the findings of Morris et al.[21] by showing the involvement of HLA-DRB1*03:01 and the involvement of SNPs in addition to HLA alleles. We extend their findings by showing the association of SLE with serotypes, confirm the number of primary signals using lasso regularization, and provide evidence for the requirement of HLA-DQB1*02:01 to explain association of HLA with SLE.
Our use of HLA imputation has also enabled us to deconvolute the large number of associated SNPs in the HLA region and reconcile these findings with decades-old work showing the association of serotypes and HLA alleles with SLE.
The new SLE-associated genes and genetic regions discovered in this study expand our understanding of SLE and provide new putative targets for diagnostics and treatment of this important autoimmune disease.
Materials and Methods
All subjects provided written consent using IRB approved consent forms. This study was approved by the IRB of University of Southern California School of Medicine, and by the IRB of the institutions participating in the study, namely, University of California Los Angeles, University of California San Francisco, Oklahoma Medical research Foundation, University of Oklahoma Health Sciences Center, University of Alabama at Birmingham, Cincinnati Children’s Hospital Medical Center, and the IRB of King’s College, London. 735 unrelated self-identified females of European ancestry with SLE (satisfying the revised criterion for SLE from the American College of Rheumatology [22, 23]) were collected (along with 480 unrelated female controls) by SLEGEN members. Details of their collection are reported in Harley et al.[24] In addition, 2057 female controls were obtained from the Illumina iControlDB resource1 giving 2537 total controls. Genotyping was performed using the Illumina Infinium HumanHap300 genotyping bead chip as previously described [24].
In order to avoid including samples with genetic background not representative of the overall study, a principle component analysis (PCA) analysis was performed which identified 109 non-representative samples which were removed from further analysis.
To eliminate duplicate and cryptic relationships, a set of 500 SNPs with minor allele frequency (MAF) ≥ 0.4 were selected. 98 sets of duplicates (three cases, two SLEGEN controls, 93 Illumina controls) which matched at 99.9% of the SNPs and 44 sets of related samples (16 cases, three SLEGEN controls and 25 Illumina controls) had all but one duplicate/related removed. In order to address population structure which can be a source of confounders [25, 26], we performed PCA on a subset of SNPs which are informative for ancestry (Ancestry Informative Markers (AIMs)) [27]. In Figure S1, a PCA biplot of the samples collected for this study is shown. The vast majority of samples are present in a homogeneous grouping, with no clear delineations between case and control samples. A prominent exception to this are the samples in orange which are found in the lower left of panel A and lower right of panel B. These 109 samples (10 cases, 6 SLEGEN controls, and 93 Illumina controls) are not representative of the ancestral background of the other samples, and therefore have been excluded from further analysis.
To control for any remaining population structure, the same first three PCA rotation axes were used as cofactors in the association analysis. After correcting for these three factors and eliminating the outlier samples, the remaining dispersion of cases and controls is even and symmetrical (Figure S1 panels C and D).
The significance of association was calculated using logistic regression with SLE status as the dependent variable and the additive dosage of the SNP and the three first PCA axes as independent variables with custom routines written in R [28]. p values obtained were corrected for multiple testing by estimating the FDR using the Benjamini and Hochberg method [29]. SNPs with FDR ≤ 0.05 were considered to be significant.
Since the vast majority of significant SNPs are located in the HLA region, calculating the FDR study-wide would result in an under-estimation of the FDR in the non-HLA region. Thus, the FDR was calculated separately for the HLA regions and non HLA regions. Because each of these regions contains significant results, there is no need for additional correction due to splitting into multiple families [4].
Imputation on regions containing significantly associated SNPs was performed using Impute (v2.3.0) [30] with the 1k genomes [31] and hapmap [32] panels, covering over 3.1 million SNPs. Significance of association of imputed SNPs was calculated in the same manner as for genotyped SNPs, with multiple-testing corrections being applied within the region instead of study-wide.
Multiple logistic regression was performed by adding the most significant SNP in a region as covariates to the logistic regression model, and repeating the regression on the remaining SNPs in the region. This procedure was repeated until no SNPs had a multiple-testing corrected FDR less than the per-family FDR cutoff of 2.68×10−3 (q = 0.05, m = 298, R = 16 as defined in [4] using the (BH − q, BH − Rq/m) procedure). Verification was performed by starting the regression procedure from random significant starting positions. Lasso regularization path regression was performed using glmnet [10].
HLA haplotype imputation was performed using HLA*IMP v2.0 [12]. The best guess haplotype was used in logistic regression with three PCA axes as was done for genotyped values. HLA haplotypes were assigned to serotypes using the WMDA directory release 3.12.02, and logistic regression with three PCA axes was performed on the resultant serotypes.
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Institutes of Health grants: [R01AR043814 to BPT, R01AR057172 to COJ, R01AR043274 to KMS, R56AI063274, P20GM103456 to PMG, N01AR62277, R37AI024717, R01AR042460, and P20RR020143 to JBH, P01AI083194 to JBH, KMS, RPK, LAC, TJV, MEA-R, COJ, BPT, and PMG, P01AR49084 to RPK, JCE, EEB, RR-G, LMV, and MAP, R01AR33062 to RPK, R01CA141700 and RC1AR058621 to MEA-R, P60AR053308, R01AR052300, and UL1TR000004 to LAC]; the Lupus Research Institute grant to BPT; the Alliance for Lupus Research grants to BPT, LAC, and COJ; the Arthritis Research UK to TJV; the Arthritis Foundation to PMG; the Kirkland Scholar Award to LAC; Wake Forest University Health Sciences Center for Public Health Genomics to CDL; Swedish Research Council to MEA-R; and Instituto de Salud Carlos III, co-financed by FEDER funds of the European Union to MEA-R. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would also like to specifically acknowledge the helpful suggestions of Reviewer 2 to improve the analysis and paper.
Footnotes
Conflict of Interest
The authors declare that there are no competing financial interests in the publication of this work.
References
- 1.Rullo OJ, Tsao BP. Recent insights into the genetic basis of systemic lupus erythematosus. Ann Rheum Dis. 2013;72 (Suppl 2):56–61. doi: 10.1136/annrheumdis-2012-202351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong DL, Reiff A, Myones BL, Quismorio FP, Klein-Gitelman M, McCurdy D, et al. Identification of new SLE-associated genes with a two-step Bayesian study design. Genes Immun. 2009;10:446–456. doi: 10.1038/gene.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjamini Y, Bogomolov M. Selective inference on multiple families of hypotheses. J Royal Stat Soc B. 2014;76:297–318. doi: 10.1111/rssb.12028. [DOI] [Google Scholar]
- 5.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 7.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacob CO, Eisenstein M, Dinauer MC, Ming W, Liu Q, John S, et al. Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase. Proc Natl Acad Sci US A. 2012;109:59–67. doi: 10.1073/pnas.1113251108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 10.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 11.McClain MT, Ramsland PA, Kaufman KM, James JA. Anti-sm autoantibodies in systemic lupus target highly basic surface structures of complexed spliceosomal autoantigens. J Immunol. 2002;168:2054–2062. doi: 10.4049/jimmunol.168.4.2054. [DOI] [PubMed] [Google Scholar]
- 12.Dilthey A, Leslie S, Moutsianas L, Shen J, Cox C, Nelson MR, et al. Multi-population classical HLA type imputation. PLoS Comput Biol. 2013;9:1002877. doi: 10.1371/journal.pcbi.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dilthey AT, Moutsianas L, Leslie S, McVean G. HLA*IMP–an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics. 2011;27:968–972. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tjernström F, Hellmer G, Nived O, Truedsson L, Sturfelt G. Synergetic effect between interleukin-1 receptor antagonist allele (IL1RN*2) and MHC class II (DR17,DQ2) in determining susceptibility to systemic lupus erythematosus. Lupus. 1999;8:103– 108. doi: 10.1191/096120399678847560. [DOI] [PubMed] [Google Scholar]
- 16.Leng RX, Wang W, Cen H, Zhou M, Feng CC, Zhu Y, et al. Gene-gene and gene-sex epistatic interactions of MiR146a, IRF5, IKZF1, ETS1 and IL21 in systemic lupus erythematosus. PLoS ONE. 2012;7:51090. doi: 10.1371/journal.pone0051090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehtonen A, Ahlfors H, Veckman V, Miettinen M, Lahesmaa R, Julkunen I. Gene expression profiling during differentiation of human monocytes to macrophages or dendritic cells. J Leukoc Biol. 2007;82:710–720. doi: 10.1189/jlb.0307194. [DOI] [PubMed] [Google Scholar]
- 18.Gabriel L, Morley BJ, Rogers NJ. The role of iNKT cells in the immunopathology of systemic lupus erythematosus. Ann N Y Acad Sci. 2009;1173:435–441. doi: 10.1111/j.1749-6632.2009.04743.x. [DOI] [PubMed] [Google Scholar]
- 19.Bijlmakers MJ, Kanneganti SK, Barker JN, Trembath RC, Capon F. Functional analysis of the RNF114 psoriasis susceptibility gene implicates innate immune responses to double-stranded RNA in disease pathogenesis. Hum Mol Genet. 2011;20:3129–3137. doi: 10.1093/hmg/ddr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob CO, Reiff A, Armstrong DL, Myones BL, Silverman E, Klein-Gitelman M, et al. Identification of novel susceptibility genes in childhood-onset systemic lupus erythematosus using a uniquely designed candidate gene pathway platform. Arthritis Rheum. 2007;56:4164–4173. doi: 10.1002/art.23060. [DOI] [PubMed] [Google Scholar]
- 21.Morris DL, Taylor KE, Fernando MMA, Nititham J, Alarcón-Riquelme ME, Barcellos LF, et al. Unraveling multiple MHC gene associations with systemic lupus erythematosus: model choice indicates a role for HLA alleles and non-HLA genes in Europeans. Am J Hum Genet. 2012;91:778–793. doi: 10.1016/j.ajhg.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/1529-0131(199709)40:9<1725::AID-ART29>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 23.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 24.Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouaziz M, Ambroise C, Guedj M. Accounting for population stratification in practice: a comparison of the main strategies dedicated to genome-wide association studies. PLoS ONE. 2011;6:28845. doi: 10.1371/journal.pone.0028845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng HW. Population admixture may appear to mask, change or reverse genetic effects of genes underlying complex traits. Genetics. 2001;159:1319–1323. doi: 10.1093/genetics/159.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baye TM, Tiwari HK, Allison DB, Go RC. Database mining for selection of SNP markers useful in admixture mapping. BioData Min. 2009;2:1. doi: 10.1186/1756-0381-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 30.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3. 1 million SNPs. Nature. 2007;449:851– 861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, et al. Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PLoS Genet. 2008;4:1000084. doi: 10.1371/journal.pgen.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sigurdsson S, Nordmark G, Göring HHH, Lindroos K, Wiman AC, Sturfelt G, et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet. 2005;76:528– 537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep. 2012;39:10627–10635. doi: 10.1007/s11033-012-1952-x. [DOI] [PubMed] [Google Scholar]
- 36.Bauer T, Zagórska A, Jurkin J, Yasmin N, Köffel R, Richter S, et al. Identification of Axl as a downstream effector of TGF-β1 during Langerhans cell differentiation and epidermal homeostasis. J Exp Med. 2012;209:2033–2047. doi: 10.1084/jem.20120493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.