SUMMARY
Mitochondrial complex I (CI) deficiency is associated with multiple neurological and metabolic disorders. However, its effect on innate immunity and bone remodeling is unclear. Using deletion of the essential CI subunit Ndufs4 as a model for mitochondrial dysfunction, we report that mitochondria suppress macrophage activation and inflammation while promoting osteoclast differentiation and bone resorption via both cell-autonomous and systemic regulation. Global Ndufs4 deletion causes systemic inflammation and osteopetrosis. Hematopoietic Ndufs4 deletion causes an intrinsic lineage shift from osteoclast to macrophage. Liver Ndufs4 deletion causes a metabolic shift from fatty acid oxidation to glycolysis, accumulating fatty acids and lactate (FA/LAC) in circulation. FA/LAC further activates Ndufs4−/− macrophages via ROS induction, and diminishes osteoclast lineage commitment in Ndufs4−/− progenitors; both inflammation and osteopetrosis in Ndufs4−/− mice are attenuated by TLR4/2 deletion. Together, these findings reveal mitochondrial CI as a critical rheostat of innate immunity and skeletal homeostasis.
INTRODUCTION
The nuclear-encoded protein NADH: ubiquinone oxidoreductase iron-sulfur protein 4 (Ndufs4) is critical for mitochondrial CI assembly (Karamanlidis et al., 2013; Kruse et al., 2008) and identified as a hotspot for mutations (Tucker et al., 2011). CI is the largest complex in mitochondrial respiratory chain. Its dysfunction is frequently linked with neurological diseases such as Alzheimer, Parkinson and Leigh syndrome (Coskun et al., 2012), as well as metabolic defects such as impairment of oxidative phosphorylation (OXPHOS) (Kirby et al., 1999). CI is also the site where the respiratory chain generates reactive oxygen species (ROS) (Kushnareva et al., 2002). Physiological changes in ROS production are associated with processes such as autophagy, differentiation, metabolic adaption and immune cell activation (Sena and Chandel, 2012). However, ROS deregulation is associated with many malfunctions (James et al., 2012).
Macrophages are essential components of the innate immunity and play a key role in inflammation. Emerging evidence reveal a tightly controlled crosstalk between metabolism and inflammation (Tschopp, 2011). Toll-like receptors (TLRs) not only recognize lipopolysaccharide (LPS) in bacterial cell wall but also sense nutritional signals such as fatty acids (Baker et al., 2011; Hotamisligil and Erbay, 2008; Kawai and Akira, 2007). Inflammation is also increasingly recognized as a key factor in the development of metabolic diseases including obesity, insulin resistance, type 2 diabetes and atherosclerosis (Rocha and Libby, 2009; Schenk et al., 2008). Furthermore, our recent studies reveal that there is also a delicate balance of metabolism and inflammation during lactation: maternal genetic defects such as PPARγ or VLDL receptor deletion, as well as maternal dietary defects such as high-fat-diet can cause metabolic defects during lactation that lead to the production of “toxic milk”, which triggers a systemic inflammation in wild-type (WT) nursing neonates that is manifested as a transient alopecia (Du et al., 2012a; Du et al., 2012b; Wan et al., 2007b). We hypothesize that genetic programs also exist in the offspring that allow the suckling neonates to cope with the lipid- and energy-rich milk produced by normal lactation and to prevent inflammation. Using alopecia as readout for neonatal inflammation, here we have identified the mitochondrial Ndufs4 as part of a critical anti-inflammatory genetic program in the offspring.
Osteoclasts are also derived from the monocyte-macrophage lineage. Upon binding of RANKL (receptor activator of nuclear factor kappa-B ligand) to its receptor RANK, macrophage precursors undergo differentiation into mature osteoclasts that are multinucleated, specialized, bone-resorbing cells. This process can be enhanced by other signaling pathways and small molecules such as the nuclear receptor PPARγ and its agonist rosiglitazone, a widely used diabetes drug (Lazarenko et al., 2007; Sottile et al., 2004; Wan, 2010; Wan et al., 2007a; Zinman et al., 2010). Osteoclasts are essential for physiological bone remodeling, deficiency of which can cause osteopetrosis. However, excessive osteoclasts can cause osteoporosis (Zaidi, 2007). Nuclear receptor ERRα and the transcription coactivator PGC1β are critical for osteoclast function, implicating that osteoclastogenesis demands high energy and intact mitochondria (Brown and Breton, 1996; Ishii et al., 2009; Wan, 2010; Wei et al., 2010). Here we have identified Ndufs4 as a key mitochondrial component required for osteoclastogenesis.
To enhance our understanding of the complex disorders associated with mitochondrial deficiency, it is pivotal to identify new mechanisms for how mitochondria modulate cellular and physiological processes. In light of the critical roles of macrophage and osteoclast in many aspects of physiology and disease, it is also important to delineate new mechanisms underlying the control of their lineage specification, differentiation and function. Using Ndufs4 deletion as a model for mitochondrial CI dysfunction, here we show that normal mitochondrial function suppresses macrophage activation and inflammation while promoting osteoclast differentiation and bone resorption via dual mechanisms of both cell-autonomous and systemic regulation.
RESULTS
Global Ndufs4 deletion causes systemic inflammation manifested as alopecia
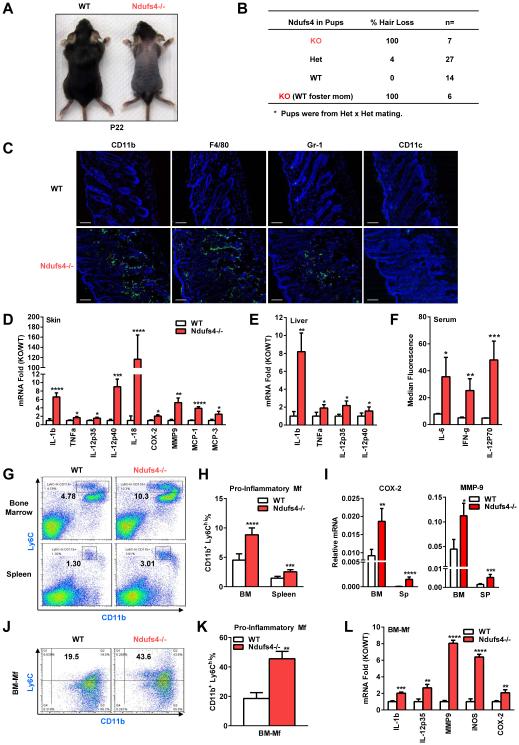
Global Ndufs4 loss in the Ndufs4−/− mice results in mitochondrial CI deficiency and encephalomyopathy around postnatal day 30 (P30), leading to lethality at ~7 weeks of age (Kruse et al., 2008). Interestingly, the Ndufs4−/− pups also exhibited a transient alopecia phenotype (Kruse et al., 2008). The first pelage appeared normal until ~P16 when precocious hair loss occurred; the hair loss peaked ~P20-P24 (Figure 1A), and then recovered after weaning (~P35) when a new coat of hair grew back. To determine whether the alopecia was dependent on offspring or maternal genotype, we performed the following experiments. First, when Ndufs4+/− females were bred with Ndufs4+/− males, 100% of the Ndufs4−/− pups and 4% of Ndufs4+/− pups developed hair loss, whereas all the wild-type (WT) control pups were normal (Figure 1B). Second, hair loss could not be rescued when Ndufs4−/− pups were fostered by WT lactating dams (Figure 1B). These results indicate that the alopecia was caused by the defects in the Ndufs4−/− pups rather than in the Ndufs4+/− dams.
Figure 1. Global Ndufs4 deletion causes systemic inflammation manifested as alopecia.
(A-B) Ndufs4−/− pups exhibit alopecia. (A) A representative image on postnatal day 22 (P22). (B) Alopecia could not be rescued by WT foster dams.
(C) Skin of Ndufs4−/− pups were infiltrated with leukocytes such as macrophages (detected by anti-CD11b, anti-F4/80 and anti-Gr-1) but not dendritic cells (detected by anti-CD11c) on P22. Scale bars, 25 μm.
(D-E) Expression of inflammatory genes in the skin (D) and liver (E) on P22 (n=6).
(F) Cytokine levels in the serum on P22 (n=6).
(G-H) Percentage of CD11b+Ly6Chi pro-inflammatory macrophages in bone marrow (BM) and spleen on P22. (G) FACS 2D dot plots. (H) Quantification (n=5).
(I) Expression of inflammatory genes in the primary bone marrow cells or splenocytes on P22 (n=5).
(J-K) Percentage of CD11b+Ly6Chi pro-inflammatory macrophages in BM-Mf cultures on day 6.
(J) FACS 2D dot plots. (K) Quantification (n=5).
(L) Expression of inflammatory genes in BM-Mf cultures on day 6 (n=5). Error bars, SD.
To examine whether the alopecia is associated with excessive recruitment of myeloid immune cells, we collected skin from Ndufs4−/− and WT littermate controls at P22 and performed immuno-fluorescence staining with antibodies for myeloid cell markers. The skin from Ndufs4−/− pups displayed an infiltration of CD11b+, Gr-1+ and F4/80+ cells, which are largely inflammatory monocytes and macrophages (Figure 1C). In contrast, there was no overt accumulation of CD11c+ dendritic cells (Figure 1C). Consistent with these observations, the mRNA levels for a set of inflammatory markers were elevated in the skin of Ndufs4−/− pups (Figure 1D). These results indicate that the alopecia was likely caused by an inflammatory response during suckling.
We next investigated whether the inflammation was systemic or skin-specific. First, expression of inflammatory markers was also elevated in the liver of Ndufs4−/− pups (Figure 1E). Second, serum levels of inflammatory cytokines including IL-6, IFNγ and IL-12p70 were higher in Ndufs4−/− pups (Figure 1F). Third, percentage of CD11b+Ly6Chi pro-inflammatory monocytes/macrophages was increased in the bone marrow and spleen of Ndufs4−/− pups (Figure 1G-H). Fourth, expression of inflammatory markers was also elevated in primary bone marrow cells and splenocytes of Ndufs4−/− pups (Figure 1I). These results indicate that the Ndufs4−/− pups suffered from systemic inflammation.
To examine if Ndufs4 deletion in the myeloid progenitors confers an intrinsic activation of macrophage, we compared bone marrow-derived macrophages (BM-Mfs) from Ndufs4−/− mice or WT controls that were differentiated with a defined concentration of cytokine. Both the percentage of CD11b+Ly6Chi macrophages and the expression of inflammatory genes were higher in the Ndufs4−/− cultures (Figure 1J-L). These results indicate that the systemic inflammation in Ndufs4−/− pups is likely attributed to, at least in part, cell-intrinsic defects.
Ndufs4−/− mice exhibited unaltered B cells and T cells, as well as Th17 and Treg subpopulations (Figure S1A-D). In addition to the increased pro-inflammatory M1 markers in the GMCSF-differentiated Ndufs4−/− macrophages and pup skin (Figure 1D, L), there were also decreased anti-inflammatory M2 markers in the MCSF-differentiated Ndufs4−/− macrophages and pup skin (Figure S1E). Serum corticosterone levels were normal (Figure S1F). Mitochondrial CI activity was reduced by >99% in Ndufs4−/− macrophages (Figure S1G). Furthermore, treatment with a CI inhibitor rotenone also elevated the expression of inflammatory genes in WT macrophages (Figure S1J). These results further indicate that CI deficiency promotes inflammatory activation of macrophages.
Global Ndufs4 deletion decreases bone resorption and increases bone mass
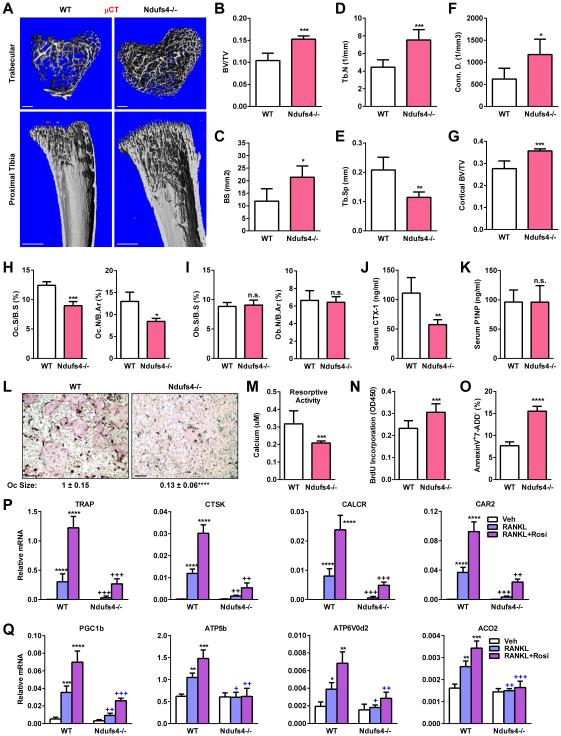
In light of the potential roles of mitochondria in osteoclast differentiation and bone remodeling, we investigated the skeletal phenotype in Ndufs4−/− mice. Micro-Computed Tomography (μCT) analysis of the proximal tibiae from P22 pups revealed that Ndufs4−/− mice exhibited a high-bone-mass phenotype (Figure 2A-G). Histomorphometry showed that osteoclast numbers and surface were decreased, whereas osteoblast numbers and surface were unaltered (Figure 2H-I). Consistently, serum bone resorption marker CTX-1 (C-terminal telopeptides of Type I collagen) was 48% lower (Figure 2J), whereas serum bone formation marker N-terminal propeptide of type I procollagen (P1NP) was normal (Figure 2K). These data indicate that the high-bone-mass in Ndufs4−/− mice was mainly caused by a compromised osteoclast function.
Figure 2. Global Ndufs4 deletion decreases bone resorption and increases bone mass.
(A-G) μCT analyses of tibiae on P22 (male, n=8). (A) Representative images of the trabecular bone of the tibial metaphysis (top) (scale bar, 10 μm) and the entire proximal tibia (bottom) (scale bar, 1 mm). (B-F) Quantification of trabecular bone volume and architecture. (B) BV/TV, bone volume/tissue volume ratio. (C) BS, bone surface. (D) Tb.N, trabecular number. (E) Tb.Sp, trabecular separation. (F) Conn.D., Connectivity Density. (G) Cortical BV/TV.
(H-I) Bone histomorphometry (P22, male, n=8). (H) Osteoclast number (Oc.N/B.Ar) and osteoclast surface (Oc.S/B.S) (n=8). (I) Osteoblast number (Ob.N/B.Ar) and osteoblast surface (Ob.S/B.S) (n=8). B.Ar, bone area.
(J) Serum CTX-1 (P22, male, n=8)
(K) Serum P1NP (P22, male, n=8)
(L-Q) Bone marrow osteoclast differentiation assay. (L) Representative images of differentiation cultures on day 12. Mature osteoclasts were multinucleated (>3 nuclei) TRAP+ (purple) cells. Scale bar, 25 μm. (M) Osteoclast activity (n=8). (N) Osteoclast precursor proliferation (n=8). (O) Osteoclast apoptosis (n=8). (P-Q) Expression of osteoclast differentiation markers (P) and osteoclast function genes (Q) (n=4). P-Q, * compares each treatment with vehicle (Veh) control, + compares Ndufs4−/− with WT control in the same treatment group. Error bars, SD.
Ex vivo bone marrow osteoclast differentiation assay using defined cytokine concentration showed that RANKL-mediated and rosiglitazone-stimulated osteoclastogenesis was severely impaired by Ndufs4 deletion. Ndufs4−/− cultures exhibited decreased number and size of multinucleated TRAP+ (tartrate-resistant acid phosphatase+) mature osteoclasts (Figure 2L), lower bone resorptive activity (Figure 2M), elevated precursor proliferation (Figure 2N) and increased apoptosis (Figure 2O). Moreover, induction of osteoclast differentiation markers, such as TRAP, CTSK, CALCR and CAR2, were diminished (Figure 2P); and induction of osteoclast function genes that promote mitochondria biogenesis and oxidative phosphorylation, such as PGC1β, ATP5b, ATP6V0d2 and ACO2, were severely blunted (Figure 2Q). CI activity was reduced by >99% in Ndufs4−/− osteoclasts (Figure S1G). Osteoclast and osteoblast co-culture showed that the osteoclastogenic defects were observed only when KO osteoclast progenitors were co-cultured with WT osteoblasts, but not when WT osteoclast progenitors were co-cultured with KO osteoblasts (Figure S2A-B). Moreover, osteoblast differentiation from Ndufs4−/− bone marrow mesenchymal stem cells was normal (Figure S2C). These results indicate that CI deficiency causes an intrinsic defect in osteoclastogenesis, leading to a decreased bone resorption and an increased bone mass.
Hematopoietic Ndufs4 deletion enhances macrophage activation and inflammation
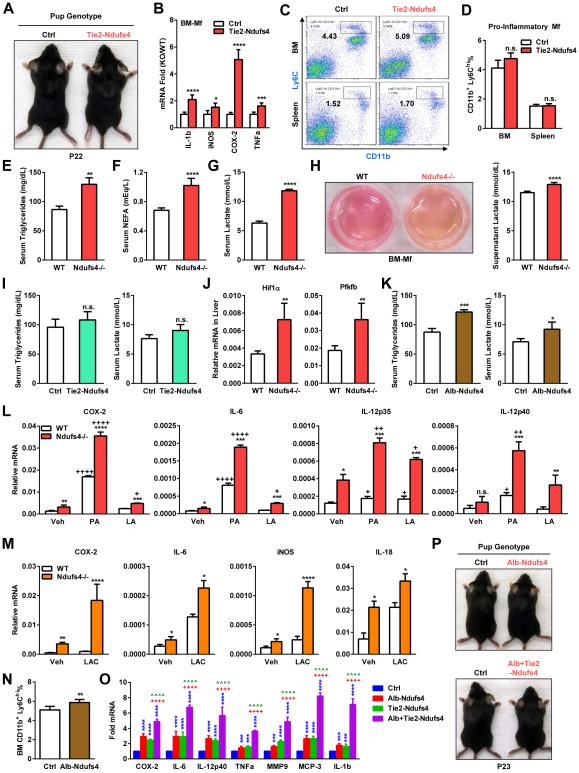
To further investigate whether CI plays a cell-autonomous role in macrophage activation and osteoclastogenesis, we generated hematopoietic Ndufs4 conditional knockout mice using Ndufs4flox/flox mice and Tie2-Cre mice. Tie2-Cre deletes flox’d alleles in all hematopoietic lineages but not in mesenchymal lineages; thus in macrophages and osteoclasts but not in osteoblasts (Wan et al., 2007a; Wan et al., 2007b; Wei et al., 2010). Tie2-Ndufs4 KO mice appeared healthy and survived to adulthood. Interestingly, Tie2-Ndufs4 KO pups did not exhibit alopecia (Figure 3A). Similar to Ndufs4−/− macrophages, Tie2-Ndufs4 KO macrophages also showed a >99% reduction in CI activity (Figure S1H), leading to a higher expression of inflammatory markers (Figure 3B). These results indicate that CI indeed plays a cell-autonomous role in macrophages to suppress inflammation.
Figure 3. Ndufs4 deletion shifts metabolism to potentiate macrophage activation.
(A) Representative image showing that Tie2-Ndufs4 KO mice had no alopecia.
(B) Expression of inflammatory genes in Tie2-Ndufs4 KO BM-Mfs was increased on day 6 (n=3). (C-D) Percentage of CD11b+Ly6Chi pro-inflammatory macrophages in the bone marrow (BM) and spleen was normal in Tie2-Ndufs4 KO pups on P22. (C) FACS 2D dot plots. (D) Quantification (n=5).
(E-G) Serum triglycerides (E), non-esterified fatty acid (NEFA) (F) and lactate (G) were increased in Ndufs4−/− pups (n=8).
(H) Ndufs4−/− macrophages secrets more lactic acid. Left, images showing the medium of Ndufs4−/− BM-Mf cultures was more acidic (yellow). Right, quantification of lactate in culture medium (n=6).
(I) Serum triglycerides (left) and lactate (right) (n=8).
(J) Expression of Hif1α and Pfkfb was higher in the liver of Ndufs4−/− mice (n=5).
(K) Serum triglycerides (left) and lactate (right) in Alb-Ndufs4 KO mice and controls (n=8).
(L-M) Fatty acids (L) and lactate (M) potentiate the activation of inflammatory genes in Ndufs4−/− macrophages (n=5). BM-Mfs were treated with palmitic acid (PA) or linoleic acid (LA) (400 μM) or lactate (15 mM) for 15 hr.
(N) Percentage of CD11b+Ly6Chi pro-inflammatory macrophages in the bone marrow (BM) was increased in Alb-Ndufs4 KO pups on P22 (n=5).
(O) Expression of inflammatory genes in the spleen was increased in Tie2-Ndufs4 KO and Alb-Ndufs4 KO, and further potentiated in Alb+Tie2-Ndufs4 DKO mice on P22 (n=5).
(P) Alb-Ndufs4 and Alb+Tie2-Ndufs4 DKO mice had no alopecia. Error bars, SD.
In contrast to the Ndufs4−/− global KO mice, the percentage of CD11b+Ly6Chi pro-inflammatory macrophages in the bone marrow and spleen of Tie2-Ndufs4 KO pups were unaltered (Figure 3C-D). This observation and the absence of the alopecia phenotype in Tie2-Ndufs4 KO mice indicate that other tissues outside of the hematopoietic lineages may also contribute to the systemic inflammation caused by global CI deficiency.
Ndufs4 deletion shifts metabolism from fatty acid oxidation to glycolysis
Mitochondrial CI plays a central role in energy metabolism by promoting fatty acid β-oxidation and TCA (tricarboxylic acid) cycle. Because milk provides the suckling neonates rich nutrients such as fat and sugar, we hypothesize that Ndufs4−/− pups also exhibit whole body metabolic defects. We found that serum levels of triglyceride and non-esterified fatty acid (NEFA) were both 50% higher in Ndufs4−/− pups at P22 (Figure 3E-F); at the same time, serum lactate was also 88% higher (Figure 3G). BM-Mfs from Ndufs4−/− mice also produced more lactate into the culture supernatant, leading to a more acidic environment (Figure 3H). These results (Figure 3E-H) indicate a metabolic shift in Ndufs4−/− pups from fatty acid oxidation to glycolysis, leading to decreased fatty acid catabolism and increased triglyceride accumulation, and at the same time, a compensatory elevated glucose catabolism and lactate accumulation.
Interestingly, serum levels of triglycerides and lactate were not significantly altered in Tie2-Ndufs4 KO pups on P22 (Figure 3I), suggesting that tissues other than the hematopoietic population were largely responsible for the global metabolic changes. Liver is a major metabolic tissue in suckling neonates, along with muscle and fat. We found that indeed expression of the pro-glycolysis genes Hif1α and Pfkfb2 were up-regulated in the liver of Ndufs4−/− pups on P22 (Figure 3J). Thus, we generated liver-specific Ndufs4 KO using Albumin-Cre mice. CI activity in the liver was reduced by 84% in Alb-Ndufs4 KO compared to controls (Figure S1I). Serum triglycerides and lactate were 39% and 30% higher, respectively, in Alb-Ndufs4 KO pups than controls on P22 (Figure 3K). Although the metabolic defects in Alb-Ndufs4 KO pups were less severe than in Ndufs4−/− pups, these results indicate that liver is a major tissue that mediates CI regulation of neonatal metabolism; nonetheless, additional tissues such as heart, skeletal muscle and fat may also contribute to the global phenotype.
Fatty acids and lactate potentiate the activation of Ndufs4−/− macrophage
We next asked whether the excessive fatty acids as the result of decreased β-oxidation could potentiate the pro-inflammatory phenotype of Ndufs4−/− macrophages. BM-Mfs from Ndufs4−/− mice or controls were treated for 15 hr with either palmitic acid (PA), a representative saturated fatty acid that is abundant in milk; or linoleic acid (LA), a representative unsaturated fatty acid. Expression of pro-inflammatory genes was stimulated by PA and to a lesser extent by LA in WT macrophages (Figure 3L). This effect was more pronounced in Ndufs4−/− macrophages, which already exhibited higher basal expression than WT macrophages (Figure 3L). Similarly, lactate also increased the expression of pro-inflammatory genes in WT macrophages, and more dramatically in Ndufs4−/− macrophages (Figure 3M).
The percentage of CD11b+Ly6Chi pro-inflammatory macrophages in the bone marrow of Alb-Ndufs4 KO pups was higher than controls (Figure 3N). Moreover, the expression of inflammatory genes in the spleen was increased in Alb-Ndufs4 KO pups and Tie2-Ndufs4 KO pups, and further potentiated in Alb+Tie2-Ndufs4 double KO (DKO) pups (Figure 3O). These results indicate that the combination of the intrinsic defects in Ndufs4−/− macrophages and the systemic metabolic defects of FA/LAC accumulation in the circulation of the Ndufs4−/− pups lead to an exacerbated macrophage activation and systemic inflammation compared with WT macrophages in the normal environment. Alb-Ndufs4 KO pups or Alb+Tie2-Ndufs4 DKO pups also did not exhibit alopecia (Figure 3P), suggesting that Ndufs4 deletion in other metabolically active tissues, such as heart, skeletal muscle and fat, may be also required to replicate metabolic and inflammatory defects as severe as in the global Ndufs4−/− pups.
ROS production is elevated in Ndufs4−/− macrophages and exacerbated by fatty acids
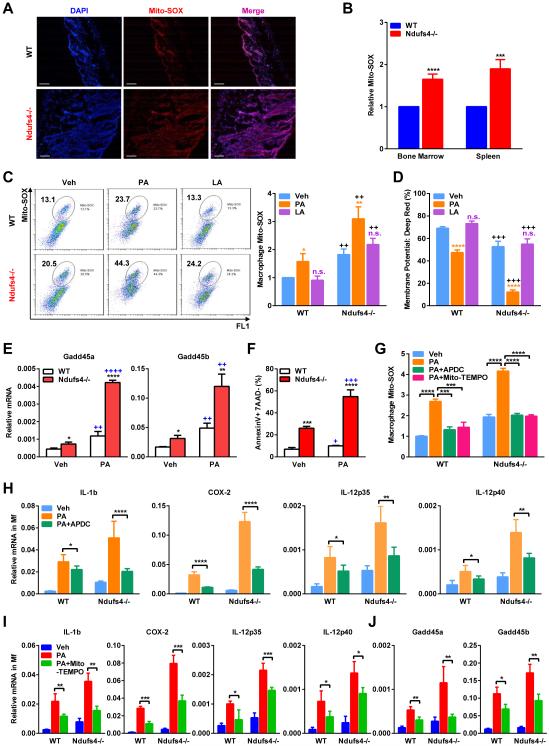
Mitochondria-derived ROS is critical for macrophage activation (West et al., 2011). Thus, we next investigated if the exacerbated inflammation by Ndufs4 deletion is contributed by excessive ROS production in vivo and in vitro. Quantified by MitoSOX staining (Misawa et al., 2013; Zhou et al., 2011), ROS was found to be more abundant in the skin, bone marrow and spleen of Ndufs4−/− pups compare with WT controls (Figure 4A-B). ROS level was much higher in Ndufs4−/− BM-Mfs, and was further increased by PA treatment (Figure 4C). Mitochondrial membrane potential was lower in Ndufs4−/− BM-Mfs and further decreased by PA (Figure 4D). In line with the increased ROS production and oxidative stress, the expression of the stress sensor genes Gadd45α/β, as well as apoptosis, was also higher in Ndufs4−/− macrophages and further potentiated by PA (Figure 4E-F). Importantly, PA induction of ROS (Figure 4G) and pro-inflammatory genes (Figure 4H) was attenuated by the mitochondrial ROS inhibitor 2R,4R-APDC (2R,4R-aminopyrrolidine dicarboxylate). Mito-TEMPO, another mitochondria-targeted antioxidant and a specific scavenger of mitochondrial ROS, also attenuated PA induction of ROS (Figure 4G), inflammatory genes (Figure 4I) and stress sensor genes (Figure 4J). These findings indicate that increased mitochondrial ROS production represents an important mechanism underlying the systemic inflammation in Ndufs4−/− suckling neonates.
Figure 4. ROS level is elevated in Ndufs4−/− macrophages and exacerbated by fatty acids.
(A-B) Representative images of Mito-SOX staining of skin on P22. Scale bars, 25 μm.
(B) FACS quantification of Mito-SOX levels in bone marrow and spleen on P22 (n=5).
(C) Mito-SOX levels in BM-Mfs treated with PA or LA (400 μM). Left, FACS 2D dot plots. Right, Quantification (n=5).
(D) Mitochondrial membrane potential was decreased in Ndufs4−/− macrophages and further reduced by PA, but not LA (n=5), quantified by percentage of MitoTracker Deep Red via FACS. C-D, * and n.s. compare treatment with vehicle control in the same genotype; + compares Ndufs4−/− with WT in the same treatment group.
(E-F) Expression of the stress sensor genes Gadd45α/β (E) and apoptosis (F) were higher in Ndufs4−/− macrophages and further potentiated by PA (n=3). * compares Ndufs4−/− with WT under the same treatment; + compares PA with vehicle in the same genotype.
(G) PA induction of mitochondrial ROS in macrophages, measured by Mito-SOX, was abolished by ROS inhibitors 2R,4R-APDC (50 μM) or Mito-TEMPO (50 μM).
(H-I) ROS inhibitors 2R,4R-APDC (H) or Mito-TEMPO (I) partially rescued the PA-activated inflammatory gene expression in WT and Ndufs4−/− macrophages. (J) Mito-TEMPO partially rescued the PA-activated stress sensor gene expression in WT and Ndufs4−/− macrophages. BM-Mfs were treated with PA (400 μM), with or without APDC or Mito-TEMPO (50 μM). Error bars, SD.
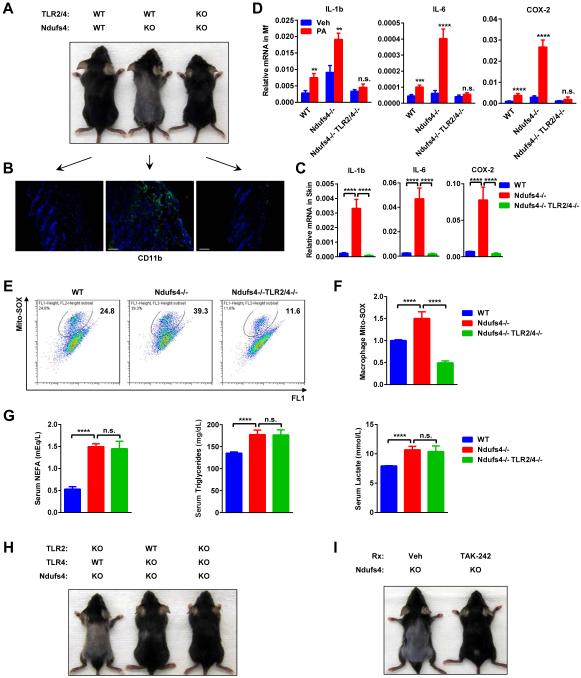
Inflammation and alopecia in Ndufs4−/− pups can be rescued by TLR4/2 deletion
Fatty acids and lactate induce inflammation and insulin resistance via TLR4/2 (Samuvel et al., 2009; Senn, 2006; Shi et al., 2006). To further investigate the significance of the excessive FA/LAC in the systemic inflammation in Ndufs4−/− pups in vivo, we examined whether the alopecia could be rescued by TLR4/2 deletion. Ndufs4/TLR2/TLR4 triple KO (TKO) mice were completely resistant to hair loss (Figure 5A). Consistent with this observation, the excessive macrophage infiltration (Figure 5B) and pro-inflammatory gene expression (Figure 5C) in the skin of Ndufs4−/− pups were ameliorated in Ndufs4/TLR2/TLR4 TKO pups. Moreover, PA-stimulated inflammatory gene expression (Figure 5D) and ROS production (Figure 5E-F) in Ndufs4−/− macrophages was also dampened by TLR2/4 deletion. Serum NEFA, triglyceride and lactate were unaffected by TLR2/4 deletion as they were elevated to a similar extent in both Ndufs4-KO and Ndufs4/TLR2/4 TKO (Figure 5G), indicating that the metabolic defects remained, but the response to this metabolic shift was abolished by TLR2/4 deletion.
Figure 5. Inflammation and alopecia in Ndufs4−/− pups can be rescued by TLR2/4 deletion.
(A-F) Comparison of Ndufs4/TLR4/TLR2 triple KO, Ndufs4−/− and WT controls on P22 (n=4).
(A) Representative images showing alopecia phenotype.
(B) CD11b immuno-staining of skin. Scale bars, 25 μm.
(C) Expression of inflammatory genes in the skin.
(D) Expression of inflammatory genes in BM-Mfs treated with PA or vehicle (Veh).
(E-F) Mito-SOX levels in BM-Mfs treated with PA (400 μM). (E) FACS 2D dot plots. (F) Quantification (n=5).
(G) TLR2/4 deletion did not alter the metabolic defects in Ndufs4−/− mice (n=5).
(H) Alopecia in Ndufs4−/− pups was rescued by TLR4 deletion but not TLR2 deletion (P22).
(I) Alopecia in Ndufs4−/− pups was rescued by TLR4 inhibitor TAK-242 (P22). Ndufs4−/− pups were gavaged with TAK-242 (10mg/kg/day) or vehicle control starting P1. Error bars, SD.
Further genetic dissection revealed that although TLR2/4 double deletion has the strongest rescue effects, TLR4 deletion alone, but not TLR2 deletion alone, also conferred significant attenuation of the alopecia, indicating that TLR4 is the major mediator of the inflammation in the Ndufs4−/− neonates (Figure 5H).
This genetic evidence promoted us to investigate whether pharmacological inhibition of TLR4 can ameliorate the inflammation in Ndufs4−/− neonates, using alopecia as a convenient readout. The results showed that Ndufs4−/− pups treated with the TLR4 inhibitor TAK-242 were spared from hair loss (Figure 5I). These findings uncover TLR4 as an essential mediator of the systemic inflammation in the Ndufs4−/− pups, and highlight TLR4 inhibitors as an effective treatment of this mitochondrial disorder.
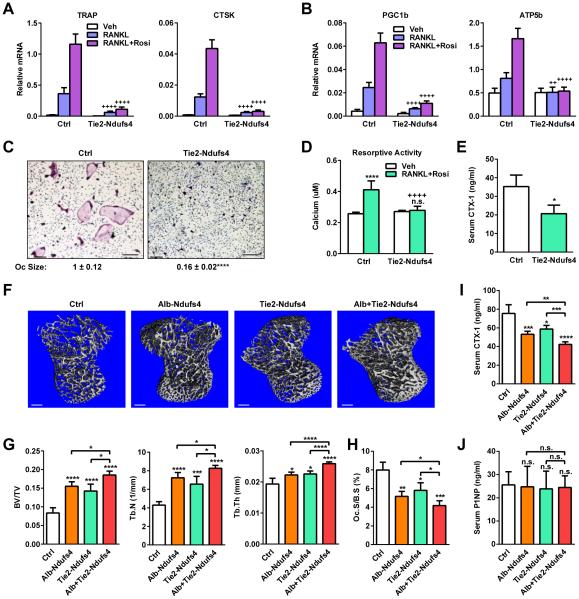
Hematopoietic Ndufs4 deletion impairs osteoclastogenesis and bone resorption
We next examined the hematopoietic-intrinsic regulation of osteoclastogenesis by CI using the Tie2-Ndufs4 KO mice. Osteoclast differentiation and function from Tie2-Ndufs4 KO bone marrow were severely impaired (Figure 6A-D), causing a 41% lower serum CTX-1 (Figure 6E) and a higher bone mass (Figure 6F-G). We have also generated myeloid-specific Ndufs4 KO mice using the Lysozyme-Cre mice (Lyz-Ndufs4). A time course analysis of osteoclast differentiation revealed that Ndufs4 deletion was later and less complete in Lyz-Ndufs4 cultures than Tie2-Ndufs4 cultures, leading to a less severe osteoclast differentiation defect (Figure S5A-E). Nonetheless, Lyz-Ndufs4 KO mice also showed a similar phenotype as Tie2-Ndufs4 KO mice with lower bone resorption, higher bone mass, and elevated inflammatory gene expression in spleen (Figure S5F-M). These results indicate that CI plays a cell-intrinsic role in the osteoclast lineage to promote osteoclastogenesis and decrease bone mass.
Figure 6. Both hematopoietic and liver Ndufs4 deletion inhibits bone resorption.
(A-D) Ex vivo bone marrow osteoclast differentiation assay of Tie2-Ndufs4 KO mice. (A) Expression of osteoclast differentiation markers (n=4). (B) Expression of osteoclast function genes (n=4). (C) Images of differentiation cultures on day 12. Scale bar, 25 μm. (D) Osteoclast activity (n=8).
(E) Serum CTX-1 (3 month, male, n=6).
(F-J) Comparison of Alb-Ndufs4 KO, Tie2-Ndufs4 KO, Alb+Tie2-Ndufs4 DKO pups or controls (P22, male, n=6). (F-G) μCT. (F) Images of the trabecular bone of the tibial metaphysis (scale bar, 10 μm). (G) Trabecular bone volume and architecture. (H) Osteoclast surface. (I) Serum CTX-1. (J) Serum P1NP. Error bars, SD.
Interestingly, Ndufs4 deletion in the liver also significantly elevated the bone mass in the Alb-Ndufs4 KO mice (Figure 6F-G). Moreover, Ndufs4 deletion in both osteoclast and liver as in the Alb+Tie2-Ndufs4 DKO mice further increased bone mass compare to individual single KO (Figure 6F-G). Osteoclast surface (Figure 6H) and serum CTX-1 (Figure 6I) were also lower in Alb-Ndufs4 KO mice and further decreased in Alb+Tie2-Ndufs4 DKO mice; whereas serum P1NP was unaltered (Figure 6J). These results indicate that CI promotes osteoclastogenesis and bone resorption through both cell-intrinsic and systemic metabolic mechanisms.
In light of the dual regulation of bone resorption and inflammation by CI, we next examined an LPS-induced inflammatory arthritis model (Chang et al., 2008; Jimi et al., 2004; Takayanagi et al., 2000; Zhao et al., 2009). LPS induction of bone resorption and bone loss was abolished in Alb-Ndufs4 KO, Tie2-Ndufs4 KO and Alb+Tie2-Ndufs4 DKO mice (Figure S3A-C), despite the exacerbated inflammation (Figure S3D). This indicates that osteoclastogenic defects are dominant over inflammatory defects in the context of inflammatory arthritis, and mitochondrial CI inhibition may confer resistance to inflammation-induced bone loss.
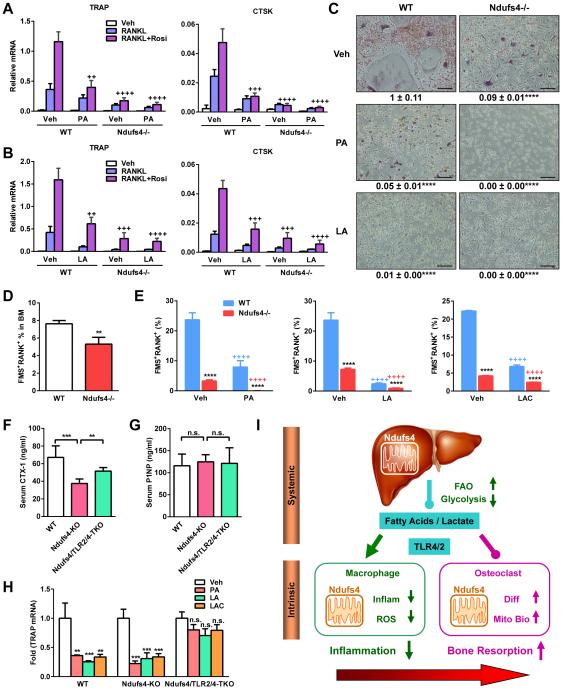
Fatty acids and lactate exacerbate the osteoclastogenic defects in Ndufs4−/− cells
Macrophage activation and osteoclast maturation represents two divergent and competing differentiation outcomes for monocyte precursors. The findings that the excessive FA/LAC in Ndufs4−/− mice potentiate macrophage activation, and liver Ndufs4 deletion also decrease bone resorption, prompted us to investigate whether FA/LAC also suppress osteoclast differentiation and lineage allocation. Osteoclast differentiation from WT bone marrow was suppressed by fatty acids such as PA and LA (Figure 7A-C); in contrast, osteoblast differentiation was unaffected (Figure S2D). Moreover, fatty acids exacerbated the osteoclastogenic defects in Ndufs4−/− cells, leading to a further reduction in osteoclast differentiation (Figure 7A-C). Lactate had similar effects (not shown). RANK and FMS are two receptors that define osteoclast precursors and are functionally required for osteoclastogenesis in response to RANKL and MCSF. The percentage of FMS+RANK+ osteoclast precursors was lower in the bone marrow of Ndufs4−/− mice and Alb-Ndufs4 KO mice (Figure 7D and S6A). Similarly, the percentage of FMS+RANK+ osteoclast precursors was also reduced in Ndufs4−/− osteoclast differentiation cultures (Figure 7E). The osteoclastogenic defects in Ndufs4−/− cultures could not be rescued by higher RANKL concentrations (Figure S4A), further supporting the severely compromised RANK level. Decreased FMS level in Ndufs4−/− cultures was only observed during osteoclast differentiation after RANKL treatment but not during proliferation (Figure S4B), in agreement of the specific impairment of osteoclast differentiation but not precursor proliferation (Figure 2L-N). Because the osteoclastogenic defects in Ndufs4−/− cultures were associated with reduced expression of not only FMS and RANK but also NFATc1 and c-fos, two key transcription factors for osteoclast differentiation, we examined the importance of each factor in this regulation. The results show that over-expression of FMS, RANK and NFATc1, but not c-fos, was able to partially rescue the osteoclastogenic defects (Figure S4E-L).
Figure 7. Fatty acids and lactate exacerbate osteoclastogenic defects in Ndufs4−/− cells.
(A-C) Bone marrow osteoclast differentiation cultures from WT or Ndufs4−/− mice were treated with PA or LA (400 μM). (A-B) Expression of osteoclast differentiation markers (n=3). + compares with WT cells treated with vehicle (Veh). (C) Images of differentiation cultures on day 12. Scale bar, 25 μm.
(D-E) Percentage of FMS+RANK+ osteoclast precursors in the bone marrow (D) (n=6) and osteoclast differentiation cultures 48 hr after RANKL treatment, with or without PA, LA (400 μM) or lactate (15 mM) (E) (n=6).
(F-H) TLR2/4 deletion partially restored the osteoclastogenic defect in Ndufs4−/− mice (P22, male, n=4). (F) Serum CTX-1. (G) Serum P1NP. (H) TRAP expression in osteoclast differentiation cultures treated with PA, LA (400 μM) or lactate (15 mM).
(I) A simplified model for how Ndufs4 and mitochondrial CI shift macrophage-osteoclast polarization to inhibit inflammation and stimulate bone resorption. FAO, fatty acid oxidation; Inflam, inflammation; Diff, differentiation; Mito Bio, mitochondria biogenesis. Error bars, SD.
Interestingly, the percentage of FMS+RANK+ osteoclast precursors in WT and Ndufs4−/− cultures was further diminished by PA, LA or lactate (Figure 7E). The anti-osteoclastogenic effects of PA, LA or lactate remained when the treatment was restricted to only the first 3 days after RANKL treatment and then removed (Figure S4C), further supporting that the regulation resides mainly in the precursor stage. In line with this in vitro finding, WT mice treated with PA and LA for 2 weeks displayed a decreased bone resorption and an increased bone mass (Figure S4D). TLR2 and TLR4 expression in Ndufs4−/− macrophage precursors was normal (Figure S6B). TLR2/4 deletion partially rescued the bone resorption defects in Ndufs4−/− mice both in vivo (Figure 7F) and in vitro (Figure 7H), while bone formation was unaffected (Figure 7G). Consequently, the high-bone-mass in Ndufs4−/− mice was attenuated in Ndufs4/TLR2/4-TKO mice (Figure S6C). These results provide critical mechanisms for the low bone resorption and high bone mass in mitochondrial dysfunction: CI deficiency reduces bone resorption by inhibiting both osteoclast differentiation and osteoclast lineage allocation via both cell-intrinsic defects and systematic metabolic defects.
DISCUSSION
Our study uncovers a critical yet previously unrecognized role of mitochondrial CI in macrophage-osteoclast polarization to suppress systemic inflammation but enhance bone resorption. This regulation involves mechanisms at both cell-intrinsic and systemic levels (Fig. 7I). First, CI in the myeloid precursors impedes inflammatory responses in macrophages but activates osteoclast differentiation by functioning in a cell-autonomous manner. Second, CI in metabolically active tissues such as liver shifts metabolism from glycolysis to fatty acid oxidation, leading to less accumulation of fatty acids and lactate in circulation, which in turn further dampen macrophage activation and enhance osteoclast differentiation via TLR4/2 signaling. Furthermore, CI favors osteoclastogenesis over macrophage activation at several stages by modulating lineage allocation, differentiation and activity. Therefore, mitochondrial CI orchestrates an intricate regulatory program to synergistically control physiology and disease.
Using primary bone marrow osteoclast differentiation assays, we found that fatty acids, such as PA and LA, suppress osteoclastogenesis by inhibiting osteoclast differentiation and diminishing FMS and RANK expression in osteoclast precursors. The anti-osteoclastogenic effects of fatty acids have also been reported in other studies (Cornish et al., 2008). Importantly, our ex vivo observations were also supported by in vivo findings that the increased serum levels of fatty acids in the Ndufs4−/− and Alb-Ndufs4-KO mice, as well as PA+LA treated mice, led to lower bone resorption and higher bone mass. Using RAW264.7 macrophage cell line, a recent report shows that PA may enhance osteoclast differentiation by increasing TNFα (Drosatos-Tampakaki et al., 2014). This discrepancy may be explained by the differences in the culture systems, for example, PA treatment of an immortalized cell line may not reflect the full effects of PA on the entire osteoclast differentiation process.
Inflammation-induced bone erosion such as in arthritis is a highly prevalent disorder. In line with the report that rotenone treatment attenuates LPS-induced bone loss (Kwak et al., 2010), our genetic dissection reveal that Ndufs4 deletion confers resistance to inflammation-induced bone erosion via both osteoclast-intrinsic and metabolic/systemic regulation. These findings highlight the exciting therapeutic potential of mitochondrial CI inhibition in treating bone degenerative diseases.
In our previous studies, we identified maternal genetic and dietary factors as key regulators of lactation, milk quality and neonatal inflammation (Du et al., 2012a; Du et al., 2012b; Wan et al., 2007b). In this study, we identified mitochondrial CI as a critical neonatal genetic program that is essential for the postnatal metabolic adaptation to prevent inflammation and ensure normal bone remodeling. Ndufs4 deletion in mice causes early postnatal lethality with metabolic defects including increased serum triglycerides and lactic acid; similarly, human mitochondrial CI deficiency also leads to disorders with early childhood onset (Distelmaier et al., 2009) that are manifested as metabolic abnormalities including fatal infantile lactic acidosis (FILA) (Loeffen et al., 2000; Smeitink et al., 2004). Similar to the alopecia in Ndufs4−/− mice, human patients with mitochondrial mutation also often have skin or hair problems, which are already accepted as an indicator of mitochondrial defects in clinic (Bodemer et al., 1999; Kubota et al., 1999; Silengo et al., 2003). This is the first in vivo study that reveals systemic inflammation as a key etiology for mitochondrial diseases in a unique natural metabolic context of lactation and suckling neonates. Several prevalent and devastating infantile disorders, such as necrotizing enterocolitis (NEC), still have no known causes or effective treatments. Future clinical investigations will further examine whether inflammation and mitochondrial dysfunction contribute to these newborn diseases. Importantly, our findings not only elucidate the mechanisms underlying the neonatal defects in systemic inflammation and low bone resorption caused by mitochondrial CI deficiency, but also uncover TLR4 inhibition as a potential treatment of infantile disorders and mitochondrial diseases.
EXPERIMENTAL PROCEDURES
Mice
Ndufs4−/− and Ndufs4flox/flox mice (Kruse et al., 2008) (backcrossed to C57BL/6 mice for > 15 and 7 generations, respectively) were provided by Dr. Richard Palmiter (University of Washington). Tie2-Cre (Kisanuki et al., 2001), Lysozyme-Cre and Albumin-Cre (Jackson Laboratory) transgenic mice were maintained on a pure C57BL/6 background. To generate hematopoietic-, myeloid- and liver-specific Ndufs4 KO mice, Ndufs4flox/flox mice were bred with Tie2-Cre+/−, Lyz-Cre+/− or Alb-Cre+/− mice, respectively; the Ndufs4flox/+Cre+/− F1 mice were bred with Ndufs4flox/flox mice to obtain Ndufs4flox/floxCre+/− F2 mice, which were bred with Ndufs4flox/flox mice to generate Ndufs4flox/floxCre+/− mice and littermate Ndufs4flox/floxCre−/− controls. TLR2/4 DKO mice on a pure C57BL/6 background (Hoshino et al., 1999; Takeuchi et al., 1999) were bred with Ndufs4+/− mice to generate Ndufs4/TLR4/TLR2 triple KO mice, Ndufs4/TLR4 or Ndufs4/TLR2 double KO mice, and Ndufs4 KO controls. All experiments were conducted using littermates. Mice were fed ad libitum with standard chow (Harlan Laboratories). Serum triglyceride and lactate levels were measured using a Vitros-250 chemistry analyzer at UTSW Metabolic Phenotyping Core. Serum non-esterified fatty acids were measured using the NEFA-HR assay (Wako). The same panels of cytokines were analyzed for expression, and only the ones with significant changes are shown to be concise. All mRNA expression was normalized by L19. Serum cytokines were measured using Cytometric Bead Array Mouse Inflammation Kits (BD Biosciences), and the ones that were detectable and significantly different are reported. Sample size estimate was based on power analyses performed using SAS 9.3 TS X64_7PRO platform at the UTSW Biostatistics Core. With the observed group differences and the relatively small variation of the in vivo measurements, n=4 and n=3 will provide >90% and >80% power at type I error rate of 0.05 (two-sided test), respectively. All protocols for mouse experiments were approved by the Institutional Animal Care and Use Committee of UTSW.
Immuno-fluorescence Staining and FACS
Frozen sections were stained with FITC-CD11b, FITC-Gr-1, FITC-CD11c (BD Biosciences) or FITC-F4/80 (Serotec), washed twice, and mounted with medium containing DAPI (Vector Laboratories). FACS was conducted with FACSCalibur using FITC-Ly6C, PE-CD11b (BD Biosciences), PE-RANK and APC-FMS (eBioscience).
Mitochondrial ROS, membrane potential and CI activity
To detect mitochondrial generated ROS by FACS, cells were detached from culture plates with Cell Dissociation Buffer (Life Technologies) and washed in cold PBS. Detached cells or primary bone marrow and spleen cells were incubated with Mito-SOX red mitochondrial superoxide indicator (Life Technologies) at 37°C for 20 minutes, washed twice with PBS and then analyzed by flow cytometry. To detect mitochondrial ROS in skin, frozen skin sections were incubated with Mito-SOX at 37°C for 20 minutes, washed and counter-stained with DAPI before images acquisition. To quantify mitochondrial membrane potential, cells were stained with MitoTracker Deep Red (a fluorescent probe sensitive to the membrane potential) and MitoTracker Green (a probe that stains mitochondrial membrane lipids independently of membrane potential), and percentage of Deep Red+ cells were quantified (Nakahira et al., 2011; Tal et al., 2009). CI enzyme activity was analyzed with a microplate assay (Abcam, ab109721) using mitochondria isolated from tissue (Abcam, ab110168) or cells (Abcam, ab110170).
Bone Analyses
μCT was performed using a Scanco μCT-35 instrument (SCANCO Medical) as described (Wei et al., 2010). Histomorphometry were performed as described (Wan et al., 2007a; Wei et al., 2011). Serum CTX-1 and P1NP were measured with RatLaps™ EIA kit and Rat/Mouse PINP EIA kit (Immunodiagnostic Systems). For LPS-induced bone loss, we i.p. injected mice at 6-8 weeks of age with PBS or LPS (5mg/kg) at day 0 and day 4, and sacrificed them at day 7.
Ex Vivo Macrophage and Osteoclast Differentiation
Macrophage and osteoclasts were differentiated from bone marrow cells as described (Wan et al., 2007a). For macrophages, the cells were differentiated with 20ng/ml of mouse GM-CSF (R&D Systems) in α-MEM containing 10% FBS for 6 days. For FA/LAC stimulation, cells were treated with 400 μM PA or LA (Sigma) (Wen et al., 2011) or 15 mM L−(+)-lactic acid (Sigma) (Samuvel et al., 2009) for 15 hr on day 6 unless indicated otherwise. To inhibit mitochondrial ROS, macrophages were treated with 50 μM 2R,4R-APDC or Mito-TEMPO (Sigma) 24 hrs before the addition of fatty acids. For osteoclasts, the cells were differentiated with 40 ng/ml of mouse M-CSF in α-MEM containing 10% FBS for 3 days (day 1-3), then with 40 ng/ml of mouse MCSF and 100 ng/ml of mouse RANKL (R&D Systems) for 3-9 days (day 4-12), with or without rosiglitazone (1 μM). Mature osteoclasts were identified as multinucleated (>3 nuclei) TRAP+ cells on day 12. Osteoclast differentiation was quantified by the RNA expression of osteoclast markers on day 6 using RT-QPCR analysis. Osteoclast precursor proliferation was quantified by BrdU incorporation (GE Healthcare). Osteoclast apoptosis was quantified by FACS analysis of AnnexinV+7-AAD− cells (BD Biosciences). To quantify osteoclast function, osteoclast differentiation was conducted on OsteoAssay bone plates (Lonza), and osteoclast activity was measured as calcium release using CalciFluo ELISA assay (Lonza).
Statistical Analyses
All statistical analyses were performed with Student's t-Test and represented as mean ± standard deviation (SD) unless noted otherwise. The p values were designated as: *, p<0.05; **, p<0.01; ***, p<0.005; ****, p<0.001; n.s. non-significant (p>0.05).
Supplementary Material
HIGHLIGHTS.
Mitochondrial dysfunction triggers macrophage activation and systemic inflammation
Mitochondrial dysfunction impairs osteoclast differentiation and bone resorption
Mitochondria play a cell-intrinsic role in osteoclast-macrophage lineage allocation
Mitochondria alter metabolism to modulate osteoclast-macrophage function
ACKNOWLEDGMENTS
We thank Dr. Richard Palmiter (University of Washington) for Ndufs4−/− and Ndufs4flox/flox mice; UT Southwestern Mouse Metabolic Phenotyping Core and Biostatistics Core for their assistance in our studies; Drs. Paul Dechow and Jerry Feng (Baylor College of Dentistry) for assistance with μCT and histomorphometry. Y. Wan is a Virginia Murchison Linthicum Scholar in Medical Research. This work was in part supported by March of Dimes (#6-FY13-137, YW), The Welch Foundation (I-1751, YW), NIH (R01 DK089113, YW), CPRIT (RP130145, YW), DOD BCRP Idea Award (BC122877, YW) and UTSW Endowed Scholar Startup Fund (YW). The authors declare that they have no financial conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- Baker RG, Hayden MS, Ghosh S. NF-kappaB, inflammation, and metabolic disease. Cell metabolism. 2011;13:11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodemer C, Rotig A, Rustin P, Cormier V, Niaudet P, Saudubray JM, Rabier D, Munnich A, de Prost Y. Hair and skin disorders as signs of mitochondrial disease. Pediatrics. 1999;103:428–433. doi: 10.1542/peds.103.2.428. [DOI] [PubMed] [Google Scholar]
- Brown D, Breton S. Mitochondria-rich, proton-secreting epithelial cells. J Exp Biol. 1996;199:2345–2358. doi: 10.1242/jeb.199.11.2345. [DOI] [PubMed] [Google Scholar]
- Chang EJ, Ha J, Oerlemans F, Lee YJ, Lee SW, Ryu J, Kim HJ, Lee Y, Kim HM, Choi JY, et al. Brain-type creatine kinase has a crucial role in osteoclast-mediated bone resorption. Nat Med. 2008;14:966–972. doi: 10.1038/nm.1860. [DOI] [PubMed] [Google Scholar]
- Cornish J, MacGibbon A, Lin JM, Watson M, Callon KE, Tong PC, Dunford JE, van der Does Y, Williams GA, Grey AB, et al. Modulation of osteoclastogenesis by fatty acids. Endocrinology. 2008;149:5688–5695. doi: 10.1210/en.2008-0111. [DOI] [PubMed] [Google Scholar]
- Coskun P, Wyrembak J, Schriner SE, Chen HW, Marciniack C, Laferla F, Wallace DC. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim Biophys Acta. 2012;1820:553–564. doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelmaier F, Koopman WJ, van den Heuvel LP, Rodenburg RJ, Mayatepek E, Willems PH, Smeitink JA. Mitochondrial complex I deficiency: from organelle dysfunction to clinical disease. Brain : a journal of neurology. 2009;132:833–842. doi: 10.1093/brain/awp058. [DOI] [PubMed] [Google Scholar]
- Drosatos-Tampakaki Z, Drosatos K, Siegelin Y, Gong S, Khan S, Van Dyke T, Goldberg IJ, Schulze PC, Schulze-Spate U. Palmitic Acid and DGAT1 Deficiency Enhance Osteoclastogenesis, while Oleic Acid-Induced Triglyceride Formation Prevents It. J Bone Miner Res. 2014;29:1183–1195. doi: 10.1002/jbmr.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Yang M, Lee S, Behrendt CL, Hooper LV, Saghatelian A, Wan Y. Maternal western diet causes inflammatory milk and TLR2/4-dependent neonatal toxicity. Genes Dev. 2012a;26:1306–1311. doi: 10.1101/gad.191031.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Yang M, Wei W, Huynh HD, Herz J, Saghatelian A, Wan Y. Macrophage VLDL receptor promotes PAFAH secretion in mother's milk and suppresses systemic inflammation in nursing neonates. Nat Commun. 2012b;3:1008. doi: 10.1038/ncomms2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nature reviews. Immunology. 2008;8:923–934. doi: 10.1038/nri2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KA, Fumoto T, Iwai K, Takeshita S, Ito M, Shimohata N, Aburatani H, Taketani S, Lelliott CJ, Vidal-Puig A, et al. Coordination of PGC-1beta and iron uptake in mitochondrial biogenesis and osteoclast activation. Nat Med. 2009;15:259–266. doi: 10.1038/nm.1910. [DOI] [PubMed] [Google Scholar]
- James AM, Collins Y, Logan A, Murphy MP. Mitochondrial oxidative stress and the metabolic syndrome. Trends in endocrinology and metabolism: TEM. 2012;23:429–434. doi: 10.1016/j.tem.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Jimi E, Aoki K, Saito H, D'Acquisto F, May MJ, Nakamura I, Sudo T, Kojima T, Okamoto F, Fukushima H, et al. Selective inhibition of NF-kappa B blocks osteoclastogenesis and prevents inflammatory bone destruction in vivo. Nat Med. 2004;10:617–624. doi: 10.1038/nm1054. [DOI] [PubMed] [Google Scholar]
- Karamanlidis G, Lee CF, Garcia-Menendez L, Kolwicz SC, Jr., Suthammarak W, Gong G, Sedensky MM, Morgan PG, Wang W, Tian R. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell metabolism. 2013;18:239–250. doi: 10.1016/j.cmet.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Seminars in immunology. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, Thorburn DR. Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology. 1999;52:1255–1264. doi: 10.1212/wnl.52.6.1255. [DOI] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Kruse SE, Watt WC, Marcinek DJ, Kapur RP, Schenkman KA, Palmiter RD. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell metabolism. 2008;7:312–320. doi: 10.1016/j.cmet.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Ishii T, Sugihara H, Goto Y, Mizoguchi M. Skin manifestations of a patient with mitochondrial encephalomyopathy with lactic acidosis and strokelike episodes (MELAS syndrome) Journal of the American Academy of Dermatology. 1999;41:469–473. doi: 10.1016/s0190-9622(99)70123-4. [DOI] [PubMed] [Google Scholar]
- Kushnareva Y, Murphy AN, Andreyev A. Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. The Biochemical journal. 2002;368:545–553. doi: 10.1042/BJ20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak HB, Lee BK, Oh J, Yeon JT, Choi SW, Cho HJ, Lee MS, Kim JJ, Bae JM, Kim SH, et al. Inhibition of osteoclast differentiation and bone resorption by rotenone, through down-regulation of RANKL-induced c-Fos and NFATc1 expression. Bone. 2010;46:724–731. doi: 10.1016/j.bone.2009.10.042. [DOI] [PubMed] [Google Scholar]
- Lazarenko OP, Rzonca SO, Hogue WR, Swain FL, Suva LJ, Lecka-Czernik B. Rosiglitazone induces decreases in bone mass and strength that are reminiscent of aged bone. Endocrinology. 2007;148:2669–2680. doi: 10.1210/en.2006-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffen JL, Smeitink JA, Trijbels JM, Janssen AJ, Triepels RH, Sengers RC, van den Heuvel LP. Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Human mutation. 2000;15:123–134. doi: 10.1002/(SICI)1098-1004(200002)15:2<123::AID-HUMU1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha VZ, Libby P. Obesity, inflammation, and atherosclerosis. Nature reviews. Cardiology. 2009;6:399–409. doi: 10.1038/nrcardio.2009.55. [DOI] [PubMed] [Google Scholar]
- Samuvel DJ, Sundararaj KP, Nareika A, Lopes-Virella MF, Huang Y. Lactate boosts TLR4 signaling and NF-kappaB pathway-mediated gene transcription in macrophages via monocarboxylate transporters and MD-2 up-regulation. Journal of immunology. 2009;182:2476–2484. doi: 10.4049/jimmunol.0802059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk S, Saberi M, Olefsky JM. Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest. 2008;118:2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn JJ. Toll-like receptor-2 is essential for the development of palmitate-induced insulin resistance in myotubes. J Biol Chem. 2006;281:26865–26875. doi: 10.1074/jbc.M513304200. [DOI] [PubMed] [Google Scholar]
- Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006;116:3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silengo M, Valenzise M, Spada M, Ferrero GB, Ferraris S, Dassi P, Jarre L. Hair anomalies as a sign of mitochondrial disease. Eur J Pediatr. 2003;162:459–461. doi: 10.1007/s00431-003-1228-5. [DOI] [PubMed] [Google Scholar]
- Smeitink JA, van den Heuvel LW, Koopman WJ, Nijtmans LG, Ugalde C, Willems PH. Cell biological consequences of mitochondrial NADH: ubiquinone oxidoreductase deficiency. Current neurovascular research. 2004;1:29–40. doi: 10.2174/1567202043480224. [DOI] [PubMed] [Google Scholar]
- Sottile V, Seuwen K, Kneissel M. Enhanced marrow adipogenesis and bone resorption in estrogen-deprived rats treated with the PPARgamma agonist BRL49653 (rosiglitazone) Calcif Tissue Int. 2004;75:329–337. doi: 10.1007/s00223-004-0224-8. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Ogasawara K, Hida S, Chiba T, Murata S, Sato K, Takaoka A, Yokochi T, Oda H, Tanaka K, et al. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408:600–605. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J. Mitochondria: Sovereign of inflammation? European journal of immunology. 2011;41:1196–1202. doi: 10.1002/eji.201141436. [DOI] [PubMed] [Google Scholar]
- Tucker EJ, Compton AG, Calvo SE, Thorburn DR. The molecular basis of human complex I deficiency. IUBMB life. 2011;63:669–677. doi: 10.1002/iub.495. [DOI] [PubMed] [Google Scholar]
- Wan Y. PPARgamma in bone homeostasis. Trends in endocrinology and metabolism: TEM. 2010;21:722–728. doi: 10.1016/j.tem.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Wan Y, Chong LW, Evans RM. PPAR-gamma regulates osteoclastogenesis in mice. Nat Med. 2007a;13:1496–1503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
- Wan Y, Saghatelian A, Chong LW, Zhang CL, Cravatt BF, Evans RM. Maternal PPAR gamma protects nursing neonates by suppressing the production of inflammatory milk. Genes Dev. 2007b;21:1895–1908. doi: 10.1101/gad.1567207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Wang X, Yang M, Smith LC, Dechow PC, Sonoda J, Evans RM, Wan Y. PGC1beta mediates PPARgamma activation of osteoclastogenesis and rosiglitazone-induced bone loss. Cell Metab. 2010;11:503–516. doi: 10.1016/j.cmet.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Zeve D, Suh JM, Wang X, Du Y, Zerwekh JE, Dechow PC, Graff JM, Wan Y. Biphasic and dosage-dependent regulation of osteoclastogenesis by beta-catenin. Mol Cell Biol. 2011;31:4706–4719. doi: 10.1128/MCB.05980-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Gris D, Lei Y, Jha S, Zhang L, Huang MT, Brickey WJ, Ting JP. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nature immunology. 2011;12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi M. Skeletal remodeling in health and disease. Nat Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- Zhao B, Takami M, Yamada A, Wang X, Koga T, Hu X, Tamura T, Ozato K, Choi Y, Ivashkiv LB, et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med. 2009;15:1066–1071. doi: 10.1038/nm.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- Zinman B, Haffner SM, Herman WH, Holman RR, Lachin JM, Kravitz BG, Paul G, Jones NP, Aftring RP, Viberti G, et al. Effect of rosiglitazone, metformin, and glyburide on bone biomarkers in patients with type 2 diabetes. J Clin Endocrinol Metab. 2010;95:134–142. doi: 10.1210/jc.2009-0572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.