Abstract
The mucosal immune system – consisting of adaptive and innate immune cells as well as the epithelium – is profoundly influenced by its microbial environment. There is now growing evidence that the converse is also true, that the immune system shapes the composition of the intestinal microbiome. During conditions of health, this bidirectional interaction achieves a homeostasis in which inappropriate immune responses to nonpathogenic microbes are averted and immune activity suppresses blooms of potentially pathogenic microbes (pathobionts). Genetic alteration in immune/epithelial function can affect host gardening of the intestinal microbiome, contributing to the diversity of intestinal microbiota within a population and in some cases allowing for unfavorable microbial ecologies (dysbiosis) that confer disease susceptibility.
Introduction
The small intestine and colon house a complex bacterial ecosystem consisting of an estimated 1014 cells in humans, 10 fold greater than the number of human cells [1]. Many have beneficial functions such as harvesting energy from otherwise indigestible plant polysaccharides (transferred to the host via short chain fatty acids), triggering formation of an intestinal mucus barrier, promoting intestinal vascularization, metabolizing xenobiotics, and preventing colonization by pathogens [2–6]. The microbiome also has profound effects on the immune system, the subject of other reviews in this issue. The beneficial properties of the intestinal microbiome confer an evolutionary advantage to animals that can regulate the microbial communities in their digestive tracts. Intestinal microbes also benefit from the survival and reproduction of their hosts. It has been hypothesized that in response to evolutionary pressure favoring mutual survival, hosts and intestinal microbes have co-evolved traits that foster symbiosis [7,8].
The existence of genetically encoded mechanisms for regulating microbial composition is suggested by the observation that across mammals, the intestinal microbiome clusters by taxonomic order rather than geography [9]. This could also reflect similarities in diet, which has been shown across mammals and within human populations to strongly influence the intestinal microbiome [10–12]. However, the microbiome of taxonomic groups such as bears/pandas with highly variable diets (carnivore, omnivore, herbivore) most resembles the microbiome of other members of the same taxonomic group rather than taxonomically unrelated mammals with a similar diet [9]. An alternate non-genetic explanation for taxonomic clustering of microbiota is a founder effect due to initial colonization by microbes of parental origin, allowing for stable transmission of a species-specific microbiome without active host involvement. Human neonates are known to be initially colonized by maternal vaginal or skin bacteria depending on mode of delivery and founder effects have been demonstrated for specific microbes such as H. pylori [13,14]. However, longitudinal studies of human infants and ex-germ-free mice have demonstrated wide fluctuations in microbial community structure – possibly reflecting environmental or stochastic factors – before stabilization into an adult microbiota distinct from (though still influenced by) the founding microbiota [15–17].
Since diet and founder effects, while important, do not alone explain taxonomic specificity of microbial composition, it is apparent that genetically encoded mechanisms exist to ensure that early fluctuations in intestinal microbiota result in the emergence of a stable, species-appropriate microbiota. Host factors that could potentially be utilized to regulate the microbiota include immune activity, intestinal motility, attachment surfaces, and secreted products that are metabolized by bacteria. The existence of host gardening of the intestinal microbiome has largely been probed by comparing the microbiota of mice deficient in a putative gardening gene to littermate controls using 16S ribosomal RNA sequencing. The vast majority of gardening genes identified in this manner are involved in innate (including epithelial) and adaptive mucosal immunity (Table 1).
Table 1.
Genes that have been implicated in microbial gardening in mouse and/or human studies. Targets of microbial gardening are more abundant in KO mice except where noted.
| Gardening gene | Putative microbial targets of gardening | Phenotype of Knockout Mice | Disease Association (for human genetic variants) |
|---|---|---|---|
| Pattern recognition receptors | |||
| MyD88 [20] | Rikenellaceae, Porphyromonadaceae | ||
| MyD88, epithelium [21] | TM7, Lactobacillus (D), Klebsiella pneumonia (I) | Exacerbated DSS colitis | |
| TLR5 [23,25] | Escherichia coli (I), Prevotellaceae (A), Lachnospiraceae (A), Alistipes (A), Bacteroides (A) | * Obesity, spontaneous colitis | |
| # TLR2 [26] | Alistipes, Lactobacillus, Clostridium, Eubacterium | Protection from diet-induced metabolic syndrome | |
| NOD2 [31–34] | Proteobacteria, Bacillus, Clostridium group IV | * Exacerbated DSS colitis | Crohn’s disease, Blau syndrome |
| # RIP2 [31] | * Exacerbated DSS colitis | ||
| # NLRP6 [38] | Prevotellaceae, TM7 | * Exacerbated DSS colitis | |
| # ASC [38] | Prevotellaceae, TM7 | * Exacerbated DSS colitis | |
| # Caspase-1 [38] | Prevotellaceae, TM7 | * Exacerbated DSS colitis | |
| Epithelium | |||
| # IL-18 [38] | Prevotellaceae, TM7 | * Exacerbated DSS colitis | |
| Alpha defensins [46] | SFB, Bacteroides (D) | ||
| ACE2 [48] | Limibacter, Paludibacter | * Exacerbated DSS colitis | |
| Vitamin D receptor [53] | Desulfovibrionaceae, Bacteroidaceae, Erysipelotrichaceae | Exacerbated DSS colitis | |
| Cyp27B1 [53] | Desulfovibrionaceae, Bacteroidaceae, Erysipelotrichaceae | Exacerbated DSS colitis | |
| RegIIIγ [22] | SFB, Eubacterium rectale | Increased intestinal Th1 cells and IgA production | |
| RELMβ [68] | Bacteriodetes (A), Firmicutes (A), Proteobacteria (A) | Resistance to diet- induced obesity | |
| FUT2 [77–80] | Various (see text) | Crohn’s disease, primary sclerosing cholangitis, psoriasis, type 1 diabetes | |
| Innate lymphoid cells | |||
| IL-22 [92,95] | SFB, Alcaligenes (I) | ||
| # T-bet, with Rag [98,99] | Klebsiella pneumonia, Proteus mirabilis, Helicobacter typhlonius | * Spontaneous colitis | |
| Adaptive immunity | |||
| μMT (i.e. no B cells in KO) [106] | Paracoccus, Lactococcus, Clostridiaceae (D) | Lipid malabsorption | |
| AID [107–109] | SFB, culturable anaerobes | Susceptibility to Y. enterocolitica | |
| iNOS and TNFα, B cells [110] | SFB (D) | Susceptibility to C. rodentium | |
| # PD1 [114] | Erysipelotrichaceae, Prevotellaceae, Alcaligenaceae, TM7 | Arthritis, glomerulonephritis, cardiomyopathy | |
| CNS1 [116] | TM7, Alistipes | Enteritis, gastritis | |
| CD1d [122] | SFB, E. coli, Pseudomonas aeruginosa | ||
| Other | |||
| IRGM [135] | Prevotella | Crohn’s disease | |
| # Sialyltransferase [84] | Ruminococcaceae (D) | * Attenuated DSS colitis | |
| # Apolipoprotein A1 [85] | Rikenellaceae, Lachnospiraceae, Erysipelotrichaceae (D) | Impaired glucose tolerance, increased body fat | |
D=depleted, I=mucosal invasion, A=altered abundance that was not further described (or phylotypes within the taxonomic group were both increased and decreased).
Denotes genes implicated in gardening by knockout mouse studies that did not use littermate controls from heterozygote breeding.
Denotes phenotypes that can be attributed to the altered intestinal microbiota based upon co-housing experiments or reconstitution of germ-free mice.
SFB=segmented filamentous bacteria
Microbial sensing via pattern recognition receptors
The immune system is equipped with an intricate system of germline encoded pattern recognition receptors (PRRs) that recognize microbial molecular patterns. Many of the genes found to affect the microbiome are PRRs, suggesting that gardening is an active process in which mucosal cells adjust their gardening activity in response to microbial composition.
Toll-like receptors (TLRs)
TLRs are transmembrane PRRs that exist either on the cell surface - where they recognize external components of bacteria, mycoplasma, fungi, and viruses - or in the endolysosomal compartment, where they recognize microbial nucleic acids [18]. The cell surface TLRs include TLR1 (which recognizes lipoproteins), TLR2 (lipoproteins), TLR4 (lipopolysaccharide), TLR5 (flagellin), and TLR6 (lipoproteins). The endolysosomal TLRs include TLR3 (double-stranded RNA), TLR7 (single stranded RNA), TLR 8 (single stranded RNA), and TLR9 (unmethylated DNA with CpG repeats). TLR signaling pathways are dependent upon two adaptor molecules, MyD88 (all TLRs except TLR3) and TRIF (TLR3, TLR4). The critical role of TLRs in recognizing microbial pathogens is revealed by the phenotype of MyD88-deficient humans, who develop recurrent pyogenic bacterial infections [19].
A number of studies have suggested a role for TLRs in immune gardening of the microbiome. An early study found that MyD88−/− mice had increased Rikenellaceae and Porphyromonadaceae in cecal contents [20]. Deletion of MyD88 specifically in intestinal epithelial cells resulted in increased TM7 and decreased Lactobacillus in feces [21]. These mice also developed overgrowth of colonic and ileal mucosa-associated bacteria, which was associated with invasion of bacteria such as Klebsiella pneumoniae into mesenteric lymph nodes [21,22]. This indicates that epithelial MyD88-dependent gardening mechanisms exist that modulate microbial composition and restrain invasive pathobionts. The specific TLRs involved are largely unknown, but strong evidence exists that TLR5 has a gardening effect. TLR5−/− mice developed an altered microbiome which caused spontaneous colitis or, after rederivation into a new facility, obesity that could be transmitted to co-housed wild-type mice [23,24]. Obese TLR5−/− mice had altered abundance of 116 phylotypes and colitic TLR5−/− mice were found to have a bloom of Enterobacteriaceae, in particular Escherichia coli, penetrating the colonic mucus layer [25]. This suggests that colitis can be precipitated by deficient gardening of invasive pathobionts. TLR2 has also been reported to affect microbial composition, with 22 differentially abundant phylotypes in knockout (KO) mice [26].
A recent systematic study of the microbiome in TLR-deficient mice raised questions about these findings [27]. No statistically significant differences in microbial composition were observed between individually caged TLR2, TLR4, TRL5, TLR9, and MyD88 KO mice compared to littermate controls. The reason for the discrepancy with prior studies is unknown. The earlier TLR2 data may have been misleading as the TLR2−/− and control mice were bred separately, making it possible that differences in microbial composition reflected environmental factors [26]. However, the earlier MyD88 and TLR5 studies used heterozygote breeders and the MyD88 study employed individual caging [20,23]. The lack of effect of TLR genotype in the recent study may have resulted from the mice being housed in a facility with autoclaved cages, irradiated food, and acidified water whereas other studies utilized standard SPF facilities [27]. This raises the point that gardening, while genetically encoded, acts on environmental input and so could potentially be mitigated in a clean environment.
NOD1/2
The NOD-like receptor (NLR) family consists of at least 23 cytoplasmic proteins, many with specificity for bacterial products [28]. The best studied are NOD1, NOD2, and the NLR components of the inflammasome. NOD1 and NOD2 recognize moieties from peptidoglycan – meso-diaminopimelic acid (DAP) and muramyl dipeptide (MDP), respectively – which are found on many bacterial and mycobacterial pathogens. NOD1 is expressed widely while NOD2 is expressed primarily in immune cells and epithelial cells. Following microbial sensing, NOD1 and NOD2 translocate to the plasma membrane and initiate downstream signaling pathways via RIP2 (also known as RICK) [29]. NOD1 and NOD2 are also able to induce the formation of autophagosomes by recruiting Atg16L1 to the plasma membrane, resulting in engulfment of invading bacteria [30].
NOD2 has drawn considerable attention due to the strong link between NOD2 genetic polymorphisms and Crohn’s disease. NOD2−/− mice had altered microbial composition in the ileum, colon, and feces as well as an increased number of ileal mucosa-associated microbes [31–33]. NOD2−/− mice treated with DSS (dextran sulfate sodium) and AOM (azoxymethane) to induce colitis-associated malignancy had increased colonic Rikenella and Paludibacter compared to controls [31]. The microbiome of NOD2−/− and RIP2−/− mice conferred susceptibility to DSS colitis that was transmissible to co-housed wild-type mice [31]. In humans, a study of the ileal microbiota of normal, colitis, and Crohn’s disease patients demonstrated that Crohn’s-associated NOD2 polymorphisms are associated with altered ileal composition independently of disease phenotype [34]. Specifically, NOD2 was found to influence the abundance of Proteobacteria, Bacillus, and Clostridium group IV by 16S sequencing and the Clostridium Coccoides – Eubacterium Rectales group by quantitative RT-PCR. Human patients homozygous for NOD2 polymorphisms have increased levels of both Firmicutes and Bacteroides in ileal tissue, paralleling results in mice [32].
We are unaware of any studies that have used 16S sequencing to evaluate the intestinal microbiome in NOD1−/− mice, though one paper using quantitative RT-PCR did not demonstrate any changes in abundance of a selected panel of bacterial groups [35].
Inflammasome
Inflammasomes are cytosolic protein complexes consisting of a pattern recognition receptor (generally a NLR), the adaptor protein ASC, and procaspase 1. The primary function of an inflammasome is to cleave caspase 1 into its active form, which can then activate the inflammatory cytokines IL-1β and IL-18 by proteolysis. NLRP1, NLRP3, NLRP6, and NLRC4 have been documented to form inflammasomes in response to diverse stimuli including MDP, flagellin, PrgJ-like proteins from Gram negative bacteria, and reactive oxygen species generated by mitochondrial stress [36,37]. NLRP6 is found predominantly in epithelial cells, but its ligand remains unknown [38]. Mice deficient in NLRP6, ASC, or caspase-1 developed an abnormal microbiota that conferred transmissible susceptibility to DSS colitis and fatty liver disease [38,39]. These mice were found to have increased Prevotellaceae and TM7 in feces and expansion of a crypt base microbe containing electron dense intracellular material (a feature seen in some Prevotella). This demonstrates that intracellular sensing of invasive pathobionts such as Prevotella by NLRP6 can activate gardening to prevent disease-promoting dysbiosis. Interestingly, stress was recently shown to downregulate NLRP6 via corticotropin-releasing hormone, causing alteration of the small intestinal microbiome and enteritis that was transmissible to co-housed mice [40].
Epithelial gardening
IL-18
The intestinal epithelium exists in close proximity to the intestinal microbiota and is a likely site of microbial sensing and gardening. The role of the epithelium in gardening is supported by the phenotype of mice with epithelial specific MyD88 deletion and further characterization of NLRP6-mediated gardening [21]. An analysis of IL-1β, IL-1R, and IL-18 KO mice revealed that only IL-18−/− mice had an altered microbiome conferring increased DSS colitis susceptibility [38]. While the microbiome of IL-18−/− mice was not identical to that of NLRP6 and ASC KO mice, it was characterized by the same increased abundance of Prevotellaceae and TM7. Bone marrow chimera experiments revealed that IL-18 expression was required in non-hematopoietic cells to prevent dysbiosis, strongly suggesting that the epithelium was the source of IL-18 processed by the NLRP6 inflammasome. The mechanism of IL-18 gardening is not known but is likely multifactorial as IL-18 has diverse effects on the immune system including promoting T helper responses (Th1, Th2, and Th17 depending on the cytokine milieu), increasing cytotoxic CD8+ T cell activity, activating NK cells, inducing macrophage inflammatory cytokine production, and recruiting neutrophils [41].
Antimicrobial products: defensins, cathelicidin, RegIIIγ
Intestinal epithelial cells are able to produce a variety of cationic peptides with broad antimicrobial activity including beta defensins, alpha defensins, and cathelicidins [42]. Beta defensins are both constitutively expressed by epithelial cells and induced by stimuli such as TLR ligands [43]. They act as direct antimicrobial agents and as chemoattracts for immune cells (including Th17 cells) via CCR6 [42,44]. While beta defensins are attractive candidates to garden the microbiota, this has not yet been studied to our knowledge. Alpha defensins are expressed by neutrophils and Paneth cells, a specialized epithelial cell type in the small intestine that releases granules containing defensins and other antimicrobial products in response to microbial stimuli [45]. Mice lacking an enzyme required to process alpha defensins have an altered small intestinal microbiome characterized by reduced Bacteroides [46]. Conversely, transgenic expression of an alpha defensin in Paneth cells resulted in increased Bacteroides and loss of segmented filamentous bacteria (SFB) colonization in the small intestine. Transgenic mice had fewer Th17 cells in the small intestine, consistent with reports that intestinal SFB colonization is critical for induction of Th17 responses in mice [46,47]. Alpha defensin-mediated gardening may be regulated by the nutrient composition of the diet. It was recently shown that dietary tryptophan deficiency or deletion of ACE2, an enzyme required for tryptophan transport into intestinal cells, caused reduced production of antimicrobial products including alpha defensins [48]. ACE2−/− mice had an altered ileal microbiome (including increased abundance of several members of the Limibacter and Paludibacter genera) and increased DSS colitis susceptibility that was transmissible to germ-free mice. The altered microbiome was reversible by treatment with nicotinamide (a product of tryptophan metabolism) or treatment with a dipeptide form of tryptophan which could be absorbed without ACE2.
Cathelicidin in its active peptide form, LL-37, has broad antimicrobial activity and acts as a chemoattract for neutrophils, monocytes, and T cells via the formyl peptide receptor-like 1 [42,49]. Expression of cathelicidin by epithelial cells and innate immune cells can be induced by short chain fatty acids derived from bacterial metabolism and by vitamin D [50–52]. Indeed, vitamin D deficiency resulting in inadequate cathelicidin production has been proposed as a mechanism of tuberculosis susceptibility in dark-skinned populations [52]. While we are unaware of any direct studies on microbial composition in cathelicidin deficient mice, one study has evaluated the microbiome of mice deficient in vitamin D receptor and Cyp27B1 (the enzyme that generates the active form of vitamin D, 1,25-dihydroxycholecalciferol) [53]. These mice had an altered microbiome characterized at the family level by higher abundance of Desulfovibrionaceae, Bacteroidaceae, and Erysipelotrichaceae and lower abundance of Lactobacillaceae, Lachnospiraceae, Ruminococcaceae, Streptococcaceae, and Deferribacteraceae. Intestinal expression of Cyp27B1 has recently been shown to be regulated by the microbiota, suggesting that vitamin D mediated gardening is modulated by PRR signaling [54]. Interestingly, deficiency of vitamin D receptor (VDR) or Cyp27B1 exacerbates DSS colitis [55,56]. This can be rescued by epithelial-specific transgenic expression of VDR, demonstrating the physiologic importance of vitamin D signaling for epithelial homeostasis [57]. However, vitamin D can enhance tight junctions and is involved in many immune processes including T cell homing to the intestine, maintenance of intraepithelial T lymphocytes, and development of natural killer T cells, so it is unclear if the microbial phenotype of these mice or their susceptibility to colitis is due to reduced cathelicidin expression [56,58–60].
Paneth cells and enterocytes also secrete RegIIIγ, an antimicrobial lectin produced predominantly in the ileum that has preferential activity against Gram positive bacteria [61]. It is minimally expressed in germ-free mice but rapidly induced upon conventionalization. This likely occurs through TLR-mediated microbial sensing by epithelial cells, as epithelial cell specific deletion of MyD88 greatly reduces RegIIIγ [22]. Loss of RegIIIγ expression after antibiotic treatment allowed for expansion of an antibiotic-resistant Gram positive pathogen, vancomycin-resistant Enterococcus [62]. RegIIIγ−/− mice have been found to have increased mucosa-associated microbiota in the ileum, characterized by overgrowth of at least two groups of Gram positive organisms - segmented filamentous bacteria (SFB) and Eubacterium rectale [22]. This corresponded to increased Th1 activity and IgA production in the small intestine, suggesting that the altered microbiome provoked adaptive immune responses. Increased intestinal Th17 activity was not observed in RegIIIγ−/− mice, but systemic Th17 responses were not assessed [22].
RELMβ
Intestinal goblet cells are capable of secreting not only mucus but also bioactive molecules including RELMβ, a product specific to goblet cells that is upregulated during inflammatory processes and parasite infection [63–65]. RELMβ is undetectable in germ-free mice but is induced within 48 hours of conventionalization, indicating that its expression is controlled by microbial factors [66]. It stimulates macrophages to secrete inflammatory cytokines including TNFα and IL-12, which then promotes IFNγ production by T cells [63,67]. KO mice show reduced DSS colitis severity, impaired responses to helminths, and resistance to diet induced obesity [63,65,67,68]. These mice were found to have an altered microbiome characterized by differential abundance of 15 Bacteriodetes, 15 Firmicutes, and 1 Proteobacteria lineages, indicating that RELMβ has a gardening effect [68]. However, it is unknown whether the phenotypes of these mice were due to alteration of the intestinal microbiota.
FUT2
Epithelial gardening involves not only antimicrobial and immunomodulatory molecules but also mucus glycoproteins, which serve as attachment sites and energy sources for microbes. Considerable evidence now exists that the intestinal microbiome is influenced by fucosylation – the addition of L-fucose to the terminal β-D-galactose residue of mucus glycoproteins by α1–2 fucosyltransferase (encoded by the FUT2 gene). Fucose is utilized as a nutrient source by some bacteria, most notably B. thetaiotaomicron, and can be scavenged by pathogens such as E. coli and Salmonella typhimurium early during infection [69–71]. Some intestinal microbes utilize fucose to synthesize capsular polysaccharides [72]. In addition, the H antigen created by FUT2 has been shown to serve as an attachment site by pathogens such as norovirus and Campylobacter jejuni [73,74]. Fucosylation is a microbiota-dependent process. In germ-free mice, fucosylation occurs immediately after birth but is lost over time [75]. Fucosylation was restored by conventionalization or colonization with B. thetaiotaomicron, which expresses an unidentified product in response to low fucose levels that induces fucosylation [76]. Germ-free FUT2−/− mice colonized with human feces had increased abundance of Parabacteroides, Eubacterium, Parasutterella, Bacteroides, and Lachnospiraceae and decreased abundance of an unclassified Clostridiales genus in their feces [77]. Interestingly, the microbiome of FUT2−/− mice fed a polysaccharide-deficient diet was unchanged from controls, demonstrating the interplay that exists between diet and host gardening. In humans, deficiency of FUT2 – which occurs in around 20% of individuals, termed “non-secretors – was associated with shifts in microbial composition in the colonic mucosa (increased Coprococcus and an unclassified Lachnospiraceae), feces (reduced Bifidobacteria), and bile (reduced Proteobacteria) [78–80]. The altered intestinal microbiome in non-secretors is suspected to account for their increased risk for inflammatory/autoimmune diseases including Crohn’s disease, primary sclerosing cholangitis, psoriasis, and type 1 diabetes [80–83].
FUT2 is not the only reported example of non-immune genetic gardening. Sialyltransferase is an enzyme that synthesizes sialyllactose in milk, which may act as a nutrient source for some microbes and affect bacterial adhesion. In its absence, knockout and WT co-fostered mice developed an altered microbiome characterized by decreased susceptibility to DSS colitis [84]. Apolipoprotein A1 is a component of high-density lipoprotein. Knockout mice have features of metabolic syndrome, in particular impaired glucose tolerance, and alterations in microbial composition [85]. In addition, other glycosylation enzymes besides FUT2 involved in mucus formation may also affect microbial composition. For instance, deficiency of enzymes necessary for the synthesis of core 1, core 2, and core 3 O-glycans present in mucus have been reported to increase DSS colitis susceptibility or induce a microbiota-dependent spontaneous colitis [86–88]. It is not yet known if these phenotypes were related to alterations in microbial composition, but this is a plausible mechanism.
Innate lymphoid cells (ILCs)
In recent years, groups of cells have been characterized that resemble subsets of T cells but lack a rearranged antigen receptor. These innate lymphoid cells include natural killer (NK) cells, GATA3-dependent cells with a Th2-like cytokine profile, and RORγt-dependent cells [89]. RORγt-dependent ILCs are the primary intestinal source of IL-22, a cytokine induced by IL-23 which is critical for epithelial production of antimicrobial proteins including RegIIIβ, RegIIIγ, S100A8, and S100A9 [90–92]. Mice lacking ILCs have minimal RegIIIγ expression by the intestinal epithelium [90,93]. IL-22 production appears to be regulated by the microbiota in a manner that depends on the route of exposure to microbial ligands. Colonization of germ-free mice reduced IL-22 production by ILCs, possibly via induction of epithelial IL-25 by microbial products [90]. In contrast, systemic administration of a bacterial product, flagellin, increased IL-22 production by triggering IL-23 release from CD103+ DCs via TLR5 [94].
The ability of ILCs to regulate antimicrobial production by epithelial cells has made them an attractive candidate gardening cell type in the mucosa. Indeed, antibody-mediated depletion of ILCs and anti-IL-22 treatment resulted in increased SFB levels in the feces [95]. It has not yet been reported whether ILCs influence the composition of the remainder of the microbiome in the intestinal lumen. ILC depletion and anti-IL-22 treatment has also been found to permit systemic dissemination of Alcaligenes xylosoxidians, an invasive Proteobacteria species that normally is contained to mouse Peyer’s patches and mesenteric lymph nodes [92]. This suggests that in addition to gardening luminal SFB, possibly via regulation of IL-22, ILCs also guard against invasive species that have penetrated the epithelial barrier.
A subset of RORγt-dependent ILCs has recently been described that expresses T-bet, lacks CCR6 expression, and differentiates into NKp46+ RORγt-dependent ILCs [96]. These cells are reduced in number in germ-free mice. A similar reduction is seen in MyD88−/−TRIF−/− mice, suggesting that TLR signaling controls this population. IFNγ-producing T-bet+CCR6-RORγt-dependent ILCs played a critical role in the intestinal response to Salmonella and may also be involved in microbial gardening. This ILC subset is absent in Rag−/−Tbet−/− mice, which develop an altered microbiota causing transmissible colitis [97,98]. These mice had differential abundance of 69 phylotypes in feces by 16S sequencing and increased abundance of Klebsiella pneumoniae and Proteus mirabilis (both Proteobacteria) by culture [99]. It was subsequently discovered that the colitis phenotype was attributable to colonization with Helicobacter typhlonius [98]. This invasive Proteobacteria triggered a response by an IL-17-producing CCR6+ subset of RORγt-dependent ILCs, previously shown to mediate colitis induced by Helicobacter hepaticus [98,100]. Of note, the emergence of a colitogenic microbiota in Rag−/−Tbet−/− mice was initially attributed to increased production of TNFα by dendritic cells (DCs) although it now appears to be most likely due to a deficit in T-bet-dependent ILCs [97]. Current evidence argues against a role for DCs in gardening, as mice deficient in the two CD103+ lamina propria DC populations had no alteration in intestinal microbiota [98]. However, the CD103-CD11b+ DC subset which expresses TNFα in Rag−/−Tbet−/− mice has not been studied [101].
Adaptive immune cells
B cells
Evidence for a role of the adaptive immune system in microbial gardening has come largely from studies of B cells. Intestinal B cells are present in the lamina propria, isolated lymphoid follicles (ILF), and Peyer’s patches (PP). They develop into IgA-producing plasma cells after undergoing class switching and affinity maturation in a T-cell-dependent manner in PP germinal centers or a T-cell-independent manner in ILFs and the lamina propria [102,103]. This process requires activation-induced cytidine deaminase (AID) and microbial triggers. Germ-free mice have few IgA-secreting intestinal B cells despite normal numbers of B cells [104]. IgA is rapidly induced upon bacterial colonization and persists even after disappearance of bacteria from the gut, suggesting that IgA-secreting B cells are imprinted by microbial exposure rather than requiring ongoing stimulation [105].
Mice deficient in all B cells were found to have alterations in the jejunal microbiome including increased Paracoccus, increased Lactococcus, and decreased Clostridiaceae [106]. The absence of non-IgM antibodies and somatic hypermutation due to AID deficiency caused increased culturable anaerobes and SFB in the small intestine [107,108]. This could be reversed by surgically connecting the circulatory system of AID knockouts to wild-type mice, indicating that serum factors (presumably immunoglobins) mediated the phenotype [108]. Similar microbial changes were observed in Rag−/− mice (i.e. T and B cell deficient), which could be reversed by bone marrow transplantation from WT but not AID−/− mice. The importance of somatic hypermutation in IgA gardening was recently shown using AID knock-in mice carrying a mutation that allowed for normal class switching but disrupted somatic hypermutation [109]. These mice had alterations in their small intestinal microbiome characterized by increased culturable anaerobes and increased abundance of Lachnospiraceae, Bacteroidales, and Lactobacillus. Recently, a population of small intestinal IgA-producing plasma cells was discovered that expresses inducible nitric oxide synthase (iNOS) and TNFα in a microbiota-dependent manner [110]. Absence of expression of both iNOS and TNFα in B cells resulted in deficiency of IgA-producing cells in the lamina propria. Interestingly, this was accompanied by reduced small intestinal SFB, suggesting that B cells can garden in a TNFα/iNOS manner that favors SFB whereas IgA gardens in a manner that limits SFB.
T cells
T cells, in particular T follicular helper (TFH) cells, have been implicated in microbial gardening through their regulation of B cell class switching and affinity maturation in PP germinal centers [111]. TFH cells have characteristically high expression of the inhibitory receptor, programmed cell death-1 (PD1). In the absence of PD1, mice develop antibody-mediated arthritis, glomerulonephritis, and cardiomyopathy [112,113]. PD1−/− mice also have an altered microbiome, including increased culturable anaerobic bacteria in the small intestine and increased Erysipelotrichaceae, Prevotellaceae, Alcaligenaceae, and TM7 in the cecum [114]. This was attributed to poor affinity maturation of the IgA repertoire due to aberrant B cell selection in PP germinal centers [115]. Possible additional evidence for T cell regulation of B-cell-mediated gardening comes from a study of mice lacking TGFβ-induced FoxP3-expressing regulatory T cells due to CNS1 deficiency. These mice had increased numbers of germinal center B cells and produced autoantibodies reacting against intestinal antigens [116]. They developed a B-cell-mediated enteritis characterized by plasma cell expansion and Th2 cytokine expression. These mice had increased TM7 and Alistipes in their feces. However, it is unclear if this was due to altered gardening (perhaps secondary to heightened B cell activity) or was a secondary consequence of intestinal inflammation.
CD1d-restricted T cells
Natural killer T (NKT) cells are a class of T cells that express NK markers and respond to lipid antigens presented by CD1d as well as to cytokine stimulation by IL-12, IL-18, and type 1 interferons [117]. They have potent effector activity through their release of cytokines including IFNγ, IL-4, IL-17, and TNFα. A subset of these cells, termed invariant NKT (iNKT) cells, has a restricted TCR repertoire and has been shown to be strongly regulated by microbial signals. In germ-free mice, iNKTs are increased in the colonic lamina propria in a CXCL16 dependent manner but decreased in the liver and spleen, which could be reversed by colonization with Sphingomonas (which expresses ligands recognized by iNKT cells) [118–120]. iNKTs respond to a wide range of pathogens and may also be involved in gardening [121]. One study of CD1d−/− mice observed alterations in the fecal microbiota, increased ileal SFB, and increased small intestinal colonization by pathogens including E. coli and Pseudomonas aeruginosa [122]. NKTs may be involved in gardening downstream of NLRP6 and IL-18, but this has not yet been evaluated.
γδ intraepithelial lymphocytes (IELs)
A diverse collection of T cells reside within the intestinal epithelium, of which about 60% express TCRγδ. This IEL subset secretes growth factors that promote epithelial growth/repair and can combat enteric pathogens through IFNγ production and direct cytotoxic activity [123]. After epithelial injury, γδ IELs prevent mucosal penetration of bacteria by secreting growth factors, chemokines, and antimicrobial products in a partially MyD88-dependent manner [124]. γδ IELs may also play a role in gardening invasive pathobionts in the absence of epithelial injury. Colonization of germ-free mice induces expression of antimicrobials by γδ IELs including RegIIIγ and RegIIIβ [125]. This could be reproduced by colonization with a single endogenous mucosa-invasive E. coli species or wild-type Salmonella (but not a non-invasive mutant), suggesting that γδ T cells respond to microbes that penetrate the mucus barrier. Absence of γδ T cells resulted in an inability to control new invasive species early after exposure. WT mice co-housed with TCRγδ−/− mice showed increased RegIIIγ expression by γδ T IELs, suggesting that these mice were exposed to mucosa invasive bacteria which arose in TCRγδ−/− mice due to deficient gardening. However, it has not yet been directly demonstrated to our knowledge that loss of γδ T cells alters the composition of the microbiome.
Inter-individual variation in microbial composition due to genetic variation in gardening
The existence of multiple mechanisms for gardening the intestinal microbiome raises the possibility that variation in gardening genes can contribute to microbial diversity within populations. This has been supported by multiple mouse studies. Comparison of 8 inbred strains revealed that each had a distinct microbial composition which was not eliminated by cohabitation, though founder effects could not be excluded [126]. Two quantitative trait loci (QTL) analyses of interbred mouse lines have identified multiple genetic factors influencing the abundance of microbes at various taxonomic levels [127,128]. The first study identified thirteen QTLs affecting microbial composition, including one mapping to a region containing IL-22, IFNγ, and Irak3 (an inhibitor of TLR signaling) that affected Lactococcus and Coriobacteriaceae abundance [127]. The second study identified five QTLs, of which three contained candidate genes affecting immune activity (consistent with the immune system playing a major role in gardening) [128]. This included type 1 interferons, Irak4 (a signaling molecule utilized by some TLRs), and Tgfb3, which influenced the abundance of Bacteroides, Rikenellaceae, and Prevotellaceae, respectively. Host genetics can also interact with diet, as was shown in a study reporting wide variation among 52 strains of mice in the effect of a high-fat, high-sucrose diet on the microbiome [129]. The authors identified a single nucleotide polymorphism (SNP) near 3 amylase genes which was associated with enrichment of Enterobacteriaceae on a high fat, high-sucrose diet.
In humans, twin studies have been used to evaluate the global impact of genetic variation on microbial composition. The intestinal microbiome has consistently been shown to be more similar between pairs of monozygotic twins than between pairs of unrelated individuals [130–134]. However, monozygotic twins share many environmental factors including maternal colonizers, early life environment, and childhood diet. This can be controlled by comparing monozygotic twins to dizygotic twins. Significantly increased microbial similarity between monozygotic twins compared to dizygotic twins has been reported in two studies using the Sorensen index to analyze data from 16S Sanger sequencing and temporal temperature gradient gel electrophoresis [130,133]. In contrast, two studies using high throughput 16S sequencing and a phylogenetic similarity measure, Unifrac, reported no difference in microbial similarity between monozygotic twins compared to dizygotic twins [131,132]. However, even if this is correct, the global similarity analysis used in these studies may not reveal shifts in the abundance of specific microbial groups due to genetic variation.
This review has already mentioned two genes, FUT2 and NOD2, with genetic variants that affect microbial composition [34,78–80]. Recently, a microbiome association study using 30 Crohn’s associated SNPs identified a genetic variant, the rs11747270a SNP, that correlates with Prevotella abundance in sigmoid colon biopsies [135]. This SNP maps to the locus for immunity-related GTPase family M (IRGM) [136]. This protein regulates autophagy, a process triggered by PRRs such as NOD2 that is important for clearance of intracellular bacteria including Salmonella, Mycobacterium, and invasive Escherichia coli [137]. A second Crohn’s associated IRGM polymorphism has been reported to reduce the effectiveness of autophagy in response to intracellular bacteria [138]. Interestingly, Prevotella species including P. intermedia and P. bivia have been reported to invade epithelial cell lines [139,140]. It is conceivable that the elevated abundance of Prevotella in colonic biopsies was due to increased survival of invasive organisms due to deficient IRGM activity. As further genome wide association studies are performed for microbial composition, it is likely that many additional examples of genetic polymorphism in gardening genes will be identified in human populations.
Acquired immune deficits can influence the microbiota
The critical role of mucosal immunity in shaping the intestinal microbiome suggests that not only genetic but also acquired deficits in immune function could affect the microbiota. A common cause of acquired immunodeficiency in humans in recent decades has been HIV. It is now appreciated that HIV transmission, replication, and persistence occur largely in mucosal tissues [141]. Rapid depletion of mucosal memory CD4+ T cells occurs during acute infection and is sustained throughout the chronic phase of disease [142]. This is accompanied by a decrease in the ratio of Th17:Tregs, increased CD8+ T cells, and decreased NK cells. As a consequence of these immune disturbances, HIV patients have increased epithelial permeability, systemic microbial translocation, and elevated serum inflammatory markers consistent with a chronic inflammatory state [143]. In most patients, these mucosal immune disturbances are not reversed completely by highly active anti-retroviral therapy [144]. In light of changes in mucosal immunity induced by HIV, it has been predicted that HIV infection would impact the intestinal microbiome. A longitudinal study of chimpanzees before and after SIV infection observed increased abundance of Sarcina, Staphylococcus, and Selenomonas in the feces [145]. Three recent papers evaluating the mucosal and fecal microbiome of untreated and treated HIV patients have confirmed altered microbial communities in HIV [146–148]. All three identified a reduction in Alistepes and two identified a reduction in Bacteroides. However, they each reported enrichment of different microbial groups in HIV patients. One identified enrichment of multiple members of the Proteobacteria phylum (including pathobionts such as Pseudomonas), another Prevotella, and the third Fusobacterium (a genus which includes pathobionts that can invade epithelial cells) [149]. The discrepancies may reflect different sampling sites in each of the studies or stochastic overgrowth of pathobionts in the face of disrupted immune gardening. Acquired deficits in immune function can also be seen in patients treated with immunosuppressive medication to prevent rejection of solid organ transplants. While we are not aware of published reports on the intestinal microbiome of this population, at least one study has examined the oral microbiome. Transplant patients were found to have altered microbial composition including increased abundance of pathobionts such as Klebsiella, Acenitobacter, and Pseudomonas [150]. More broadly, medications that affect the immune system – such as immunomodulators and biologics (not including antibiotics) used to treat inflammatory bowel disease, rheumatic diseases, and psoriasis – could also potentially affect microbial gardening and result in disease-promoting dysbiosis. There is minimal data at present on this topic, though the existence of medication-specific microbial profiles was suggested in the oral microbiome study and one of the HIV studies [148,150].
Conclusion
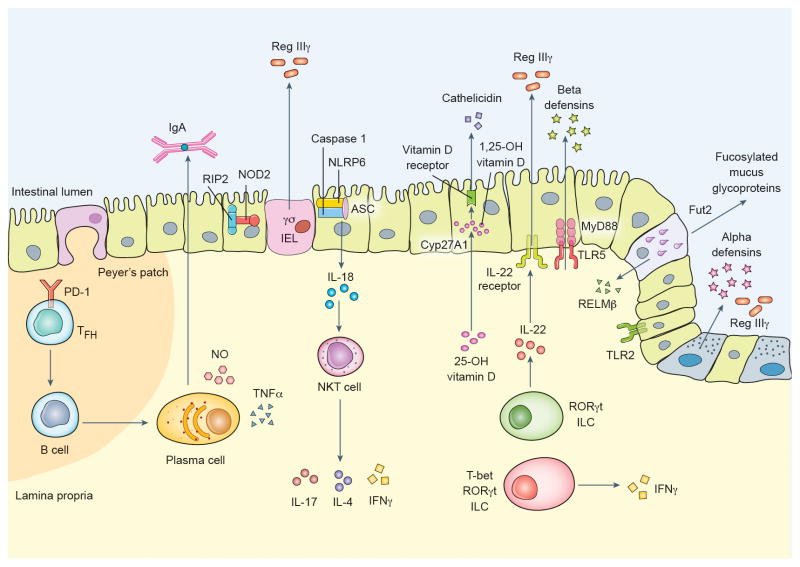
There is now extensive evidence that genetically encoded mechanisms exist for gardening the intestinal microbiome. These pathways pair microbial sensing by PRRs to antimicrobial effector mechanisms such as defensins, RegIIIγ, and IgA (Figure 1). However, many questions remain. At present, the gardening activity of specific PRRs has been characterized in terms of shifts at the genus or higher taxonomic level in knockout mice. It is unknown what changes occur at the species or strain level, whether PRRs have overlapping activity, and which microbial features trigger gardening pathways dependent upon a single PRR (e.g. NLRP6-mediated gardening of Prevotella, which was unaffected by loss of any other inflammasome-forming NLR) [38]. The effector mechanisms that allow for targeted gardening of specific microbes are also unknown (other than IgA), as host antimicrobial products generally have a wide spectrum of activity. While many immune cell types have been implicated in gardening, the relationships between each immune cell type and specific microbial taxa are largely uncharacterized with exceptions such as SFB gardening by B cells and NK T cells.
Figure 1.
Putative mechanisms for immune gardening of the intestinal microbiome.
Despite the many unknowns, it is clear that gardening of the intestinal microbiome is critical for health. In many studies, disruption of gardening resulted in dysbiosis which conferred susceptibility to diseases including colitis, obesity, and fatty liver disease. This often corresponded to a bloom of pathobionts, especially Proteobacteria such as Escherichia coli in TLR5−/− mice, Prevotella in NLRP6−/− mice, Helicobacter in Rag−/−Tbet−/− mice, and Alcaligenes in ILC-deficient mice. The three human genetic variants associated with gardening are all also risk factors for Crohn’s disease, possibly due to dysbiosis. Gardening can be affected not only by genetic variation but also by diet (as shown for tryptophan/defensins), stress (as shown for NLRP6), and acquired immune deficits (including, potentially, due to immunosuppresive medication). Disease-promoting dysbiosis arising from aberrant microbial gardening could prove to be a common theme in medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Braun J. Unsettling facts of life: bacterial commensalism, epithelial adherence, and inflammatory bowel disease. Gastroenterology. 2002;122:228–30. doi: 10.1053/gast.2002.31109. [DOI] [PubMed] [Google Scholar]
- 2.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–84. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersson J, et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G327–33. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinhardt C, Bergentall M, Greiner TU, Schaffner F, Ostergren-Lunden G, Petersen LC, Ruf W, Backhed F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483:627–31. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swann J, et al. Gut microbiome modulates the toxicity of hydrazine: a metabonomic study. Mol Biosyst. 2009;5:351–5. doi: 10.1039/b811468d. [DOI] [PubMed] [Google Scholar]
- 6.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claesson MJ, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 12.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moodley Y, et al. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527–30. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCafferty J, Muhlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, Fodor AA. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116–25. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 19.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321:691–6. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frantz AL, Rogier EW, Weber CR, Shen L, Cohen DA, Fenton LA, Bruno ME, Kaetzel CS. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 2012;5:501–12. doi: 10.1038/mi.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–8. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vijay-Kumar M, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–21. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho FA, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–52. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellermayer R, et al. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 2011;25:1449–60. doi: 10.1096/fj.10-172205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, Khanin R, Pamer EG. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–56. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–28. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–90. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 31.Couturier-Maillard A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123:700–11. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, Schreiber S, Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–62. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 33.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009;106:15813–8. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li E, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One. 2012;7:e26284. doi: 10.1371/journal.pone.0026284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson SJ, Zhou JY, Geddes K, Rubino SJ, Cho JH, Girardin SE, Philpott DJ. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 2013;4:222–31. doi: 10.4161/gmic.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012;13:325–32. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 38.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, et al. Stress-induced corticotropin-releasing hormone-mediated NLRP6 inflammasome inhibition and transmissible enteritis in mice. Gastroenterology. 2013;144:1478–87. 1487 e1–8. doi: 10.1053/j.gastro.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 42.Cederlund A, Gudmundsson GH, Agerberth B. Antimicrobial peptides important in innate immunity. FEBS J. 2011;278:3942–51. doi: 10.1111/j.1742-4658.2011.08302.x. [DOI] [PubMed] [Google Scholar]
- 43.Vora P, et al. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J Immunol. 2004;173:5398–405. doi: 10.4049/jimmunol.173.9.5398. [DOI] [PubMed] [Google Scholar]
- 44.Ghannam S, et al. CCL20 and beta-defensin-2 induce arrest of human Th17 cells on inflamed endothelium in vitro under flow conditions. J Immunol. 2011;186:1411–20. doi: 10.4049/jimmunol.1000597. [DOI] [PubMed] [Google Scholar]
- 45.Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- 46.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto T, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–81. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Y, Chen Q, Schmidt AP, Anderson GM, Wang JM, Wooters J, Oppenheim JJ, Chertov O. LL-37, the neutrophil granule- and epithelial cell-derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J Exp Med. 2000;192:1069–74. doi: 10.1084/jem.192.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schauber J, et al. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52:735–41. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raqib R, et al. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc Natl Acad Sci U S A. 2006;103:9178–83. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu PT, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–3. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 53.Ooi JH, Li Y, Rogers CJ, Cantorna MT. Vitamin D regulates the gut microbiome and protects mice from dextran sodium sulfate-induced colitis. J Nutr. 2013;143:1679–86. doi: 10.3945/jn.113.180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–27. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 55.Liu N, et al. Altered endocrine and autocrine metabolism of vitamin D in a mouse model of gastrointestinal inflammation. Endocrinology. 2008;149:4799–808. doi: 10.1210/en.2008-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong J, Zhang Z, Musch MW, Ning G, Sun J, Hart J, Bissonnette M, Li YC. Novel role of the vitamin D receptor in maintaining the integrity of the intestinal mucosal barrier. Am J Physiol Gastrointest Liver Physiol. 2008;294:G208–16. doi: 10.1152/ajpgi.00398.2007. [DOI] [PubMed] [Google Scholar]
- 57.Liu W, et al. Intestinal epithelial vitamin D receptor signaling inhibits experimental colitis. J Clin Invest. 2013;123:3983–96. doi: 10.1172/JCI65842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bruce D, Cantorna MT. Intrinsic requirement for the vitamin D receptor in the development of CD8alphaalpha-expressing T cells. J Immunol. 2011;186:2819–25. doi: 10.4049/jimmunol.1003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu S, Bruce D, Froicu M, Weaver V, Cantorna MT. Failure of T cell homing, reduced CD4/CD8alphaalpha intraepithelial lymphocytes, and inflammation in the gut of vitamin D receptor KO mice. Proc Natl Acad Sci U S A. 2008;105:20834–9. doi: 10.1073/pnas.0808700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu S, Cantorna MT. The vitamin D receptor is required for iNKT cell development. Proc Natl Acad Sci U S A. 2008;105:5207–12. doi: 10.1073/pnas.0711558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–7. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McVay LD, et al. Absence of bacterially induced RELMbeta reduces injury in the dextran sodium sulfate model of colitis. J Clin Invest. 2006;116:2914–23. doi: 10.1172/JCI28121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barnes SL, Vidrich A, Wang ML, Wu GD, Cominelli F, Rivera-Nieves J, Bamias G, Cohn SM. Resistin-like molecule beta (RELMbeta/FIZZ2) is highly expressed in the ileum of SAMP1/YitFc mice and is associated with initiation of ileitis. J Immunol. 2007;179:7012–20. doi: 10.4049/jimmunol.179.10.7012. [DOI] [PubMed] [Google Scholar]
- 65.Herbert DR, et al. Intestinal epithelial cell secretion of RELM-beta protects against gastrointestinal worm infection. J Exp Med. 2009;206:2947–57. doi: 10.1084/jem.20091268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He W, et al. Bacterial colonization leads to the colonic secretion of RELMbeta/FIZZ2, a novel goblet cell-specific protein. Gastroenterology. 2003;125:1388–97. doi: 10.1016/j.gastro.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 67.Nair MG, et al. Goblet cell-derived resistin-like molecule beta augments CD4+ T cell production of IFN-gamma and infection-induced intestinal inflammation. J Immunol. 2008;181:4709–15. doi: 10.4049/jimmunol.181.7.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hildebrandt MA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–24. e1–2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martens EC, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–7. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–9. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–81. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 73.Lindesmith L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9:548–53. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 74.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–20. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 75.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–3. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 76.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci U S A. 1999;96:9833–8. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kashyap PC, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A. 2013;110:17059–64. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rausch P, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A. 2011;108:19030–5. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wacklin P, et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One. 2011;6:e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Folseraas T, et al. Extended analysis of a genome-wide association study in primary sclerosing cholangitis detects multiple novel risk loci. J Hepatol. 2012;57:366–75. doi: 10.1016/j.jhep.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGovern DP, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum Mol Genet. 2010;19:3468–76. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ellinghaus D, et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am J Hum Genet. 2012;90:636–47. doi: 10.1016/j.ajhg.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smyth DJ, et al. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. 2011;60:3081–4. doi: 10.2337/db11-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fuhrer A, Sprenger N, Kurakevich E, Borsig L, Chassard C, Hennet T. Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J Exp Med. 2010;207:2843–54. doi: 10.1084/jem.20101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang C, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–41. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 86.Fu J, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest. 2011;121:1657–66. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stone EL, et al. Glycosyltransferase function in core 2-type protein O glycosylation. Mol Cell Biol. 2009;29:3770–82. doi: 10.1128/MCB.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD, Braun J, Xia L. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204:1417–29. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–10. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sawa S, et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–6. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 91.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–9. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 92.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–5. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sanos SL, Vonarbourg C, Mortha A, Diefenbach A. Control of epithelial cell function by interleukin-22-producing RORgammat+ innate lymphoid cells. Immunology. 2011;132:453–65. doi: 10.1111/j.1365-2567.2011.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kinnebrew MA, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–87. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–99. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klose CS, et al. A T-bet gradient controls the fate and function of CCR6(−)RORgammat(+) innate lymphoid cells. Nature. 2013 doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 97.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Powell N, et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37:674–84. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–5. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Welty NE, Staley C, Ghilardi N, Sadowsky MJ, Igyarto BZ, Kaplan DH. Intestinal lamina propria dendritic cells maintain T cell homeostasis but do not affect commensalism. J Exp Med. 2013;210:2011–24. doi: 10.1084/jem.20130728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. 2010;28:243–73. doi: 10.1146/annurev-immunol-030409-101314. [DOI] [PubMed] [Google Scholar]
- 103.Tsuji M, et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–71. doi: 10.1016/j.immuni.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 104.Hansson J, et al. Influence of gut microbiota on mouse B2 B cell ontogeny and function. Mol Immunol. 2011;48:1091–101. doi: 10.1016/j.molimm.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 105.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–9. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shulzhenko N, et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med. 2011;17:1585–93. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–7. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 108.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–6. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–70. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- 110.Fritz JH, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kato LM, Kawamoto S, Maruya M, Fagarasan S. Gut T and IgA: key players for regulation of bacterial communities and immune homeostasis. Immunol Cell Biol. 2013 doi: 10.1038/icb.2013.54. [DOI] [PubMed] [Google Scholar]
- 112.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–51. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 113.Nishimura H, et al. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291:319–22. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- 114.Maruya M, Kawamoto S, Kato LM, Fagarasan S. Impaired selection of IgA and intestinal dysbiosis associated with PD-1-deficiency. Gut Microbes. 2013;4:165–71. doi: 10.4161/gmic.23595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, Kato LM, Fagarasan S. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–9. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 116.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–9. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–17. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 118.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wingender G, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–28. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wei B, et al. Commensal microbiota and CD8+ T cells shape the formation of invariant NKT cells. J Immunol. 2010;184:1218–26. doi: 10.4049/jimmunol.0902620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tupin E, Kinjo Y, Kronenberg M. The unique role of natural killer T cells in the response to microorganisms. Nat Rev Microbiol. 2007;5:405–17. doi: 10.1038/nrmicro1657. [DOI] [PubMed] [Google Scholar]
- 122.Nieuwenhuis EE, et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest. 2009;119:1241–50. doi: 10.1172/JCI36509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–56. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ismail AS, Behrendt CL, Hooper LV. Reciprocal interactions between commensal bacteria and gamma delta intraepithelial lymphocytes during mucosal injury. J Immunol. 2009;182:3047–54. doi: 10.4049/jimmunol.0802705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ismail AS, et al. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci U S A. 2011;108:8743–8. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Campbell JH, et al. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 2012;6:2033–44. doi: 10.1038/ismej.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107:18933–8. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McKnite AM, et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One. 2012;7:e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Parks BW, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17:141–52. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stewart JA, Chadwick VS, Murray A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol. 2005;54:1239–42. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- 131.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lepage P, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–36. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 134.Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, de Vos WM, Zoetendal EG. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013;7:707–17. doi: 10.1038/ismej.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Quince C, et al. The impact of Crohn’s disease genes on healthy human gut microbiota: a pilot study. Gut. 2013;62:952–4. doi: 10.1136/gutjnl-2012-304214. [DOI] [PubMed] [Google Scholar]
- 136.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–37. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brest P, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–5. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 139.Dorn BR, Leung KL, Progulske-Fox A. Invasion of human oral epithelial cells by Prevotella intermedia. Infect Immun. 1998;66:6054–7. doi: 10.1128/iai.66.12.6054-6057.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Strombeck L, Sandros J, Holst E, Madianos P, Nannmark U, Papapanou P, Mattsby-Baltzer I. Prevotella bivia can invade human cervix epithelial (HeLa) cells. APMIS. 2007;115:241–51. doi: 10.1111/j.1600-0463.2007.apm_512.x. [DOI] [PubMed] [Google Scholar]
- 141.Veazey RS, Lackner AA. HIV swiftly guts the immune system. Nat Med. 2005;11:469–70. doi: 10.1038/nm0505-469. [DOI] [PubMed] [Google Scholar]
- 142.Costiniuk CT, Angel JB. Human immunodeficiency virus and the gastrointestinal immune system: does highly active antiretroviral therapy restore gut immunity? Mucosal Immunol. 2012;5:596–604. doi: 10.1038/mi.2012.82. [DOI] [PubMed] [Google Scholar]
- 143.Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol. 2012;10:655–66. doi: 10.1038/nrmicro2848. [DOI] [PubMed] [Google Scholar]
- 144.Mehandru S, et al. Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. PLoS Med. 2006;3:e484. doi: 10.1371/journal.pmed.0030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moeller AH, et al. SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe. 2013;14:340–5. doi: 10.1016/j.chom.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vujkovic-Cvijin I, et al. Dysbiosis of the gut microbiota is associated with hiv disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra91. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lozupone CA, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–39. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McHardy IHLXTMRPJJBJAPBJ. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Castellarin M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Diaz PI, et al. Transplantation-associated long-term immunosuppression promotes oral colonization by potentially opportunistic pathogens without impacting other members of the salivary bacteriome. Clin Vaccine Immunol. 2013;20:920–30. doi: 10.1128/CVI.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]