Abstract
Porphyromonas gingivalis is a bacterial pathogen that produces the polyproteins RgpA and Kgp, which are proteolytically processed into proteinases and adhesins. We have demonstrated that the RgpA and Kgp proteinases and adhesins are C terminally processed by carboxypeptidase CPG70 by sequencing C-terminal peptides from both the wild type and an isogenic CPG70 mutant, using ion trap mass spectrometry.
Porphyromonas gingivalis is a gram-negative, anaerobic bacterium that has been associated with the onset and progression of human periodontitis, an inflammatory disease of the supporting tissues of the teeth (6). P. gingivalis produces three cysteine proteinases, namely, RgpA, RgpB, and Kgp, that are associated with the cell surface and/or secreted depending on the bacterial strain and environmental conditions (4). These proteinases make a significant contribution to the pathogenesis of P. gingivalis by indirectly causing periodontal tissue destruction through the activation of host matrix metalloproteinases and by degrading key proteins and peptides of the immune system (5). RgpA and Kgp are synthesized as polyproteins. Each is proteolytically processed to yield an N-terminal proteinase domain and several C-terminal adhesins (Fig. 1). N-terminal sequence analysis of each domain revealed that the processing occurred at either Arg-X or Lys-X peptide bonds, consistent with the activities of RgpA and Kgp, respectively (1). A peptide mass fingerprinting study using purified proteins from wild-type P. gingivalis W50 and isogenic mutants lacking either functional Kgp or RgpA/B confirmed that the N-terminal processing at Arg-X peptide bonds was dependent on the presence of functional RgpA or RgpB (7). In addition, the study demonstrated that each of the domains is C terminally processed at an X-Lys peptide bond (Fig. 1). The C-terminal processing of one of these domains was shown to be dependent on the presence of Kgp. Given that Kgp cleaves on the C-terminal side of Lys residues, the C-terminal processing was predicted to involve a Lys-specific carboxypeptidase in addition to Kgp. Recently, CPG70, an Arg/Lys-specific carboxypeptidase, was purified from the culture fluid of P. gingivalis (3). The protein shares C-terminal sequence similarity to the cysteine proteinases, suggesting a common mechanism for cell surface attachment and secretion (7). A W50 isogenic mutant lacking functional CPG70 (W50CPG) was found to be avirulent in a murine model of infection, indicating an important role for this enzyme in virulence (3). In this study, we employed tandem mass spectrometry (MS/MS) sequencing of C-terminal tryptic peptides of several RgpA and Kgp domains derived from wild-type W50 and its isogenic mutant strain W50CPG to demonstrate the involvement of CPG70 in the maturation of these polyproteins.
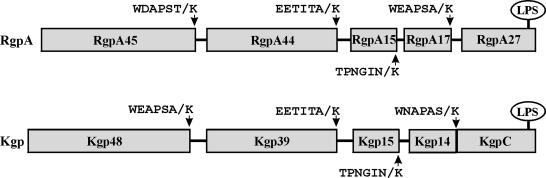
FIG. 1.
Domain structure of the RgpA and Kgp polyproteins, showing C-terminal processing sites. Eight domains are shown to be processed C terminally at an X-Lys peptide bond. The mature C terminus of RgpA27 has not been determined, and the C-terminal domain(s) of Kgp (KgpC) has not yet been identified. There is extensive sequence similarity (28 to 100% identity) between each RgpA domain and its equivalent domain in Kgp. RgpA15 is identical to Kgp15. The figure is derived from the work of Veith et al. (7). LPS, lipopolysaccharide.
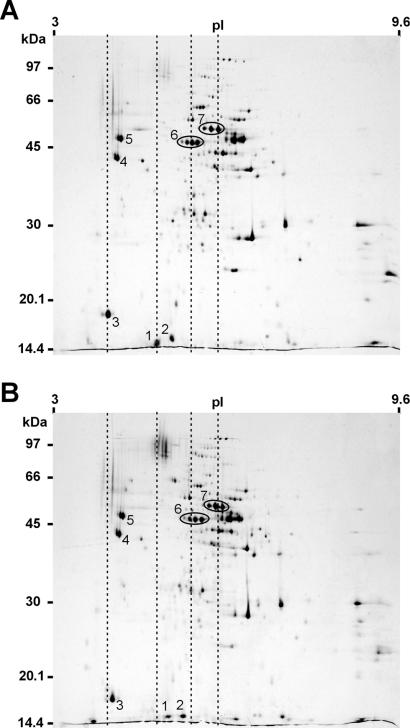
Surface protein extracts of P. gingivalis W50 and W50CPG were prepared by incubation of washed cells with 1% Triton X-114 (2) in a buffer containing 20 mM Tris-HCl (pH 7.4), 5 mM CaCl2, and 50 mM NaCl for 45 min at room temperature. After clarification of the extracts by centrifugation, protein was precipitated from the extract supernatant with 10% trichloroacetic acid and subjected to two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE), which was performed according to published methods (7) (Fig. 2). The gel spots corresponding to the processed domains of the Arg- and Lys-specific RgpA and Kgp polyproteins, respectively, are numbered according to their mobility on the gel, namely, Kgp14 (spot 1), RgpA17 (spot 2), RgpA15/Kgp15 (spot 3), Kgp39 (spot 4), RgpA44 (spot 5), RgpA45 (spot 6), and Kgp48 (spot 7). They were readily identified by comparison of the 2D gels obtained with the 2D gel pattern previously published (7). The intensities of these spots for the mutant (Fig. 2B) and for the wild type (Fig. 2A) were very similar, indicating that carboxypeptidase was not required for the initial processing of the polyproteins into discrete domains. However, when the gels were overlaid, it was apparent that the pIs of the domain spots generated from the mutant were higher (more basic) than those of the wild-type domains, consistent with the proposed role of CPG70 in the processing of C-terminal lysine residues (Fig. 2). To explore this difference further, each of the spots corresponding to the RgpA and Kgp domains was excised from the gel, subjected to in-gel digestion with trypsin, and subsequently analyzed by MS.
FIG. 2.
Two-dimensional gel electrophoresis of surface protein extract prepared from P. gingivalis W50 (A) and W50CPG (B). Protein extracts (300 μg) were subjected to 2D PAGE, and the gels were stained with colloidal Coomassie blue. The numbered spots were excised, digested, and identified by peptide mass fingerprinting. Spot 1, Kgp14; spot 2, RgpA17; spot 3, RgpA15/Kgp15; spot 4, Kgp39; spot 5, RgpA44; spot 6, RgpA45; spot 7, Kgp48. The dashed lines demonstrate that spots 1, 2, 3, 6 and 7 have moved to a higher pI in the gel from the mutant strain (B). Molecular mass markers are given on the left.
The identity of each domain was confirmed by peptide mass fingerprinting using a MALDI mass spectrometer (data not shown) (7). The mass spectrum obtained for RgpA15/Kgp15 from the wild type contained two major peaks, the one at m/z 2083 previously having been assigned to the processed C-terminal peptide (7). In the spectrum obtained for RgpA15/Kgp15 from the mutant, however, this peak was replaced by a peak 128 Da higher at m/z 2211, consistent with the presence of an additional Lys residue (data not shown). To provide direct evidence that these peaks at m/z 2083 and 2211 are the processed and unprocessed C-terminal peptides, respectively, the corresponding peptide digests were desalted and concentrated using μC18 Zip Tips (Millipore), following the manufacturer's instructions, and 2 μl of eluate was pipetted into a nanospray needle (Econo12 PicoTip; New Objective) and analyzed on an Esquire LC ion trap mass spectrometer fitted with a nanospray source (Bruker Daltonics, Bremen, Germany) (Fig. 3). The capillary voltage was set to 600 V, and the drying gas (N2) was set to 2 liters/min and 50°C. The trap drive, skim 1, and octopole voltages were optimized prior to each MS/MS analysis. Strong signals corresponding to the doubly charged forms of these peptides were observed and selected for MS/MS analyses. The identities of the peptides were determined by performing an MS/MS ion search against the National Center for Biotechnology Information database. A single, highly significant hit was obtained for both processed (Fig. 3A) and unprocessed (Fig. 3B) forms of the RgpA15/Kgp15 C-terminal peptide. The MS/MS spectra exhibited excellent sequence coverage, enabling the detection of a modified peptide in each spectrum. The modified peptides are the major forms present and contain a deamidated Asn residue, as indicated by “N*” in Fig. 3.
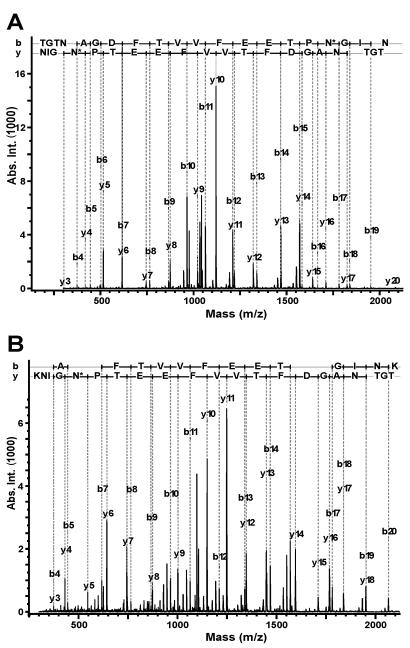
FIG. 3.
MS/MS spectra of RgpA15 C-terminal peptide from W50 (A) and W50CPG (B). An in-gel digest of RgpA15 from W50 and W50CPG was micropurified by Zip Tips and analyzed by nanospray ion trap MS. (A) The MS/MS spectrum of the m/z 1042.7 (2+) parent ion from W50 was found to match the sequence of the processed C-terminal peptide. The sequence is TGTNAGDFTVVFEETPN*GIN, with * denoting deamidation of the Asn residue. (B) The MS/MS spectrum of the m/z 1106.5 (2+) parent ion from the mutant strain W50CPG was found to match the unprocessed form of the same peptide. The sequence is TGTNAGDFTVVFEETPN*GINK. Only b- and y-type ions are labeled in both spectra. N*, deamidated Asn residue; Abs. Int., absolute intensity.
Using the same technique, the processed and unprocessed forms of the C-terminal peptides of RgpA45, Kgp48, RgpA17, and Kgp14 were identified (Table 1), indicating that the same carboxypeptidase, CPG70, was responsible for the removal of C-terminal Lys in each domain. The C-terminal peptides of RgpA44 and Kgp39 are identical and have average masses (MH+) of 4,619 and 4,747 Da for the processed and unprocessed forms, respectively. The large size of these made them difficult to analyze by ion trap MS; however, peaks at these masses were observed by matrix-assisted laser desorption ionization MS and were specific to domains derived from the wild-type and mutant strains, respectively (data not shown).
TABLE 1.
MS/MS data for C-terminal peptides of RgpA45, Kgp48, RgpA17, and Kgp14
| Domain | W50 sequence | Parent iona | MS/MS ionsb | W50CPG sequence | Parent ionc | MS/MS ionsb |
|---|---|---|---|---|---|---|
| RgpA45 | WDAPST | 676.3 | b2-b6, y2-y4 | WDAPSTK | 402.8 (2+) | b1-b3, y1-y6 |
| Kgp48 | WEAPSA | 660.3 | b1-b6, y3-y4 | WEAPSAK | 394.8 (2+) | b1,2,5,6, y1-y6 |
| RgpA17 | WEAPSA | 660.3 | b1-b6, y3-y4 | WEAPSAK | 394.8 (2+) | b1,2,5,6, y1-y6 |
| Kgp14 | WNAPAS | 645.6 | b1-b6, y3-y5 | WNAPASK | 387.3 (2+) | b1-3,5,6, y1-y7 |
Singly charged form of peptide dominant due to lack of Arg/Lys.
All observed b- and y-type ions in the MS/MS spectrum.
Doubly charged form of peptide dominant due to presence of Lys.
Having shown that CPG70 is involved in processing of RgpA and Kgp, an important question that arises is the relevance of this processing to virulence. In addition to the possibility that CPG70 has a direct role in virulence by processing host factors, it is also possible that the role of CPG70 in virulence is linked to Kgp in their dual role of C-terminal processing. One possibility is that the removal of C-terminal lysine is required to enable the various adhesin and proteolytic domains of both RgpA and Kgp to correctly associate and form a virulent conformation. Processed RgpA15/Kgp15 has a theoretical mass of 12.8 kDa. This adhesin migrated on the 2D-PAGE gel, however, at approximately 18 kDa (Fig. 2A, spot 3). In comparison, under the same experimental conditions, unprocessed RgpA15/Kgp15 was found to migrate to a lower molecular mass (Fig. 2B, spot 3), suggesting that the presence of C-terminal Lys has an effect on the structure of this domain. Whether the complex of RgpA and Kgp domains with unprocessed C-terminal Lys residues is less virulent, however, remains to be established. In conclusion, we have shown that CPG70 is involved in the C-terminal processing of the RgpA and Kgp polyproteins.
Editor: A. D. O'Brien
REFERENCES
- 1.Bhogal, P. S., N. Slakeski, and E. C. Reynolds. 1997. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology 143:2485-2495. [DOI] [PubMed] [Google Scholar]
- 2.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 3.Chen, Y. Y., K. J. Cross, R. A. Paolini, J. E. Fielding, N. Slakeski, and E. C. Reynolds. 2002. CPG70 is a novel basic metallocarboxypeptidase with C-terminal polycystic kidney disease domains from Porphyromonas gingivalis. J. Biol. Chem. 277:23433-23440. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffman, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potempa, J., A. Banbula, and J. Travis. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol. 2000 24:153-192. [DOI] [PubMed] [Google Scholar]
- 6.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 7.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]