Significance
Hybridization in animals has commonly been considered rare. In contrast, recent studies suggest hybridization to play a central role in both speciation and adaptation. This work highlights a complex process resulting from hybridization; exchange of genomic regions between species in Formica wood ants is at the same time selected for and against, leading to an intragenomic conflict and extremely strong natural selection. The results suggest that the genomic regions acquired through hybridization harbor both incompatibilities and adaptive genetic variation. Our study has implications for the genomic architecture of speciation and argues for incorporating adaptive introgression into the current models of genomic divergence.
Keywords: adaptive introgression, haplodiploid, Hymenoptera, Haldane's rule, speciation continuum
Abstract
Hybridization is not a mere reproductive dead end but has been suggested to play a central role in speciation, for example, by introducing adaptive genetic variation. Our previous study uncovered a unique consequence of hybridization in Formica ants. In a population including two isolated but partially introgressed genetic groups, the females have an apparent hybrid background, whereas the males do not. This situation results in large-scale differences between male and female genomes that are stable throughout generations. Here, we compare genotypes from different developmental stages to investigate how sex-specific introgression and genetic differences between sexes are maintained. We show that strong selection rather than sex-dependent transmission maintains the genetic differences between sexes. All genotype combinations are produced and observed in the eggs of both sexes, but the alleles acquired through hybridization disappear from the haploid males during development from egg to adult as their frequencies drop toward zero. However, the same introgressed alleles increase in frequency and are favored when heterozygous in the females. Genotypes eliminated from males most likely represent incompatibilities arising from hybridization. Our results show an unusual situation of opposite selection, where introgression is favored in diploid females but selected against in haploid males. This finding suggests that introgressed genomic regions harbor both fitness-enhancing and -reducing elements. Our work highlights the complex consequences of hybridization and provides a rare opportunity to observe natural selection in real time in nature.
Recent studies suggest hybridization in animals to be more common than previously thought and play a role in both speciation and adaptation (1–3). Toward the end of the speciation process, hybridization commonly leads to inviability and reduced fertility because of incompatible epistatic interactions between genes from different species (4–6). Alternatively, it can result in adaptive introgression of genetic material across the species boundary, where fitness-enhancing gene complexes are transferred from one taxon into another (2, 3).
Ants offer unique opportunities to study the process of speciation. Because of sociality and haplodiploidy, some of the consequences of hybridization in ants differ from those in other species. Ants are haplodiploid, as the females (queens and workers) develop through normal sexual reproduction and are diploid, whereas the males develop from unfertilized eggs and are haploid. Therefore, even if a female mates with a male of another species, her haploid sons are not hybrids (7). Furthermore, haploid sons of a hybrid female reveal both recessive and dominant hybrid incompatibilities genome-wide. Haploid male hybrids lack the intact genome of both parental species and thus, face the incompatibility problems seen in F2 hybrids or backcrossed individuals of a diploid species (8, 9). These findings are in accordance with Haldane’s rule, which suggests that the effect of hybrid incompatibilities is strongest in not only the heterogametic but also, the hemizygous (haploid) sex (10). In previously described cases of ant hybridization, the workers are hybrids, but reproductive individuals are produced by mating within a pure line or a species as, for example, in the Pogonomyrmex harvester ants (11) and Solenopsis fire ants (12). Another system (dubbed social hybridogenesis) has been described in Cataglyphis desert ants, where new queens are produced parthenogenetically, whereas workers are hybrids between two genetic lineages (13).
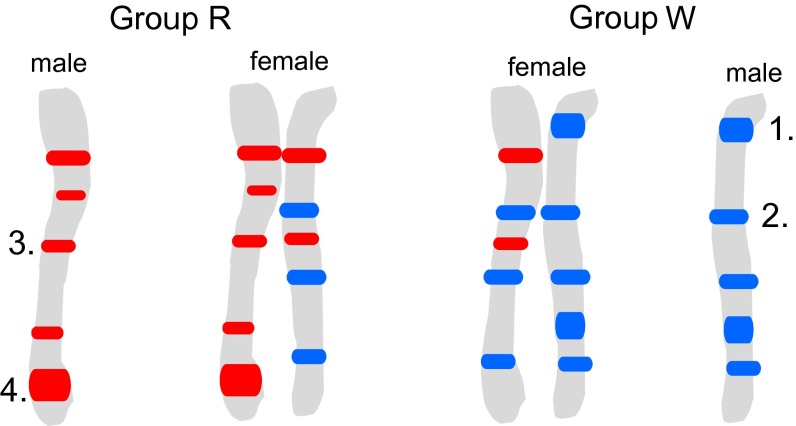
Our earlier study based on a population sample of adults revealed yet another exceptional example of hybridization in ants. The population has a history of hybridization between the mound-building wood ants Formica aquilonia and Formica polyctena (9), which has resulted in two genetic groups (R and W) that occupy the same nests and both have diagnostic alleles of their own (Fig. 1). The past history of hybridization has led to introgression of alleles between the groups in such a way that alleles present in one genetic group are also found in the diploid individuals (i.e., females) but not the haploid males of the other genetic group (Fig. 1). In other words, the females (both workers and queens) are apparent hybrids, but the males are not.
Fig. 1.
Schematic representation of the male (haploid) and the female (diploid) genomes in the two genetic groups (R and W) of the hybrid ant population. Red rectangles represent genome areas originating from the R group, and blue rectangles represents genome areas originating from the W group. Allele categories are (1) allele diagnostic to W, (2) allele introgressed from W to R, (3) allele introgressed from R to W, and (4) allele diagnostic to R. Introgression is apparent in the females of both groups but not in the males.
The parental species of this hybrid population belong to the recently speciated Formica rufa group. At the European scale, F. polyctena is a southern species, whereas F. aquilonia has a boreoalpine distribution occurring in northern and mountainous areas. Their distributions overlap in southern Finland, where our study population is also located. Hybridization may readily take place in their sympatric populations, and the ants are taxonomically difficult to identify (14). Therefore, finding pure parental populations for comparative purposes is difficult. Furthermore, both F. polyctena and F. aquilonia form large supercolonies, a single nest containing many and up to hundreds of queens. The parental species can show large genetic differences within a species even locally because of founder effects and restricted dispersal (15).
Introgression into females but not into males is compatible with Haldane’s rule and has resulted in large-scale genetic differences between sexes within the two groups, R and W, of the hybrid population. Mating and sexual reproduction within a group should wipe out such genetic differences within a few generations. The intriguing question thus remains: how are genetic differences between the sexes within the genetic groups created and maintained? Here, we will test two hypotheses that were proposed to explain the situation (9).
The selection hypothesis predicts that the genetic differences between male and female genomes result from strong postzygotic selection eliminating incompatible genotype combinations. Strong selection against hybrid males could create large-scale differentiation between sexes, because recessive incompatibilities can be masked in diploid heterozygous females but not haploid males. Consequently, the hybrid males die, but females can survive. The observed genotype frequencies in the adult ants suggest that selection should be very strong and eliminate even most of the offspring genotypes depending on the genetic group and the sex (9).
The segregation hypothesis proposes that hybridization has led to the formation of two independently segregating allelic sets, one of which is always transmitted from the mother queens to their sons and the other one is always transmitted to daughters (as if females would be of type XY and males would be of type Y). When the allelic set transmitted to future daughters (X) is fertilized by sperm carrying the paternal complement (Y), the females remain hybrids, and differentiation between sexes is maintained. Two independently segregating allelic sets could be created by chromosomal rearrangements, which can contribute to hybrid incompatibilities and reproductive isolation (16, 17).
Results
Alleles Segregate to Eggs at Random.
To test the segregation hypothesis, we established 70 laboratory colonies, each headed by a single old queen (i.e., egg-laying queen from a colony) collected from the field in early spring. The queens were later genotyped; 44 queens belonged to the group W, and 24 queens belonged to group R. Two queens could not be included in either group, because they lacked diagnostic alleles (Table S1). We further genotyped the spermathecal content of each queen and 10–15 eggs per queen to study mating patterns and the segregation of the alleles from heterozygous queens. Under the segregation hypothesis, we expected that some alleles enter preferentially into unfertilized haploid (male) and others enter preferentially into fertilized diploid (female) eggs (as specified in Table S2). In detail, the alleles that are present in males and only as heterozygotes in adult females of the same genetic group were expected to be transmitted to haploid male eggs. Conversely, the alleles that were found as heterozygotes in females but not at all (or only exceptionally) in adult males were expected to be transmitted to diploid eggs (details in ref. 9).
Specifically, 19 alleles (6 alleles in group W, 10 alleles in group R, and 3 alleles in both groups) should segregate differentially into the haploid and diploid eggs based on the adult genotypes of our previous study (9). We tested whether segregation of these alleles to eggs departs from random by using a one-sided test, where the counterhypothesis is the sex-specific distortion indicated in Table S2. The probabilities combined over families showed no significant overall departure from random segregation for any of the alleles (all P values > 0.1) (Table S2). In fact, the observed bias, if any, was opposite to that predicted by the segregation hypothesis (Table S3). We also combined the probabilities over loci for each family to see if any of the queens showed a systematic segregation distortion (Table S4). Two significant departures from random segregation (at the level P < 0.05) were observed for 59 queens with informative brood, but none of the tests remained significant when correcting for multiple testing. Thus, a queen transmits her alleles at equal probabilities to both diploid (female) and haploid (male) eggs.
The family data of double heterozygous queens were also used to test for linkage of the loci. After correcting for multiple tests, none of the pairwise locus comparisons showed a significant departure from random combination of alleles (Table S5). The estimated proportion of putative recombinant eggs under the set assumption was close to 50%, which was expected under the null hypothesis (Table S5). Thus, the loci can be considered to be unlinked and segregate to eggs independently of each other. Based on these results, we can reject the segregation hypothesis.
Two Genetic Groups and Gene Flow Between Them.
Next, we inferred the mating type for each of 68 old queens (i.e., whether mating is within or between the two genetic groups). Combining the information from the spermathecae and eggs indicated that at least 19 queens had mated with two males. We detected only one clear case of mating between the two genetic groups, where all 13 diploid eggs of an R queen had been fertilized by a single W male and alleles in the spermatheca matched those in the eggs (SI Text). In conclusion, only 1 of 68 queens had mated with a male from the other genetic group, or 1 of 87 matings was between the groups W and R. Thus, these data indicate less potential gene flow between the groups than previously estimated (9).
The population samples included some diploid individuals that carried diagnostic alleles of both genetic groups and were likely hybrids produced by mating between the groups R and W. They were mostly found in one laboratory culture that had at least 20 old queens collected from a single nest, with the frequency of putative hybrids among the offspring being 87.5% (42 of 48 worker larvae and pupae). Otherwise, the proportion of such hybrids in the field samples was 7.3% (7 of 96) among the adult workers sampled in 1996 and 1.5% (13 of 863; 6 larvae, 5 pupae, and 2 adult workers) in the diploid females sampled between 2004 and 2011, but none of them were adult queens. No haploid males carried diagnostic alleles from both groups, meaning that we did not observe any males that would have been produced by a queen that is an F1 hybrid between the two groups. In conclusion, although the groups R and W have a hybrid history, our data suggest that current gene flow between the groups is low.
Genotypic Selection Within a Genetic Group.
Our data indicate that mothers transmit alleles equally to both haploid male and diploid female eggs, thus rejecting the segregation hypothesis. According to the selection hypothesis, genetic differences between sexes could be maintained if the alleles that have been introgressed from the other genetic group are eliminated from males at a later developmental stage. If this elimination happened, adult males would not carry such introgressed alleles, and females could have them only as heterozygous. To test the selection hypothesis, we estimated allele and genotype frequencies in larvae (n = 252), pupae (n = 242), adults (i.e., workers, young queens, and males; n = 413), and actual reproductives (i.e., old queens and spermathecae; n = 155) to infer possible selective changes between the different developmental stages.
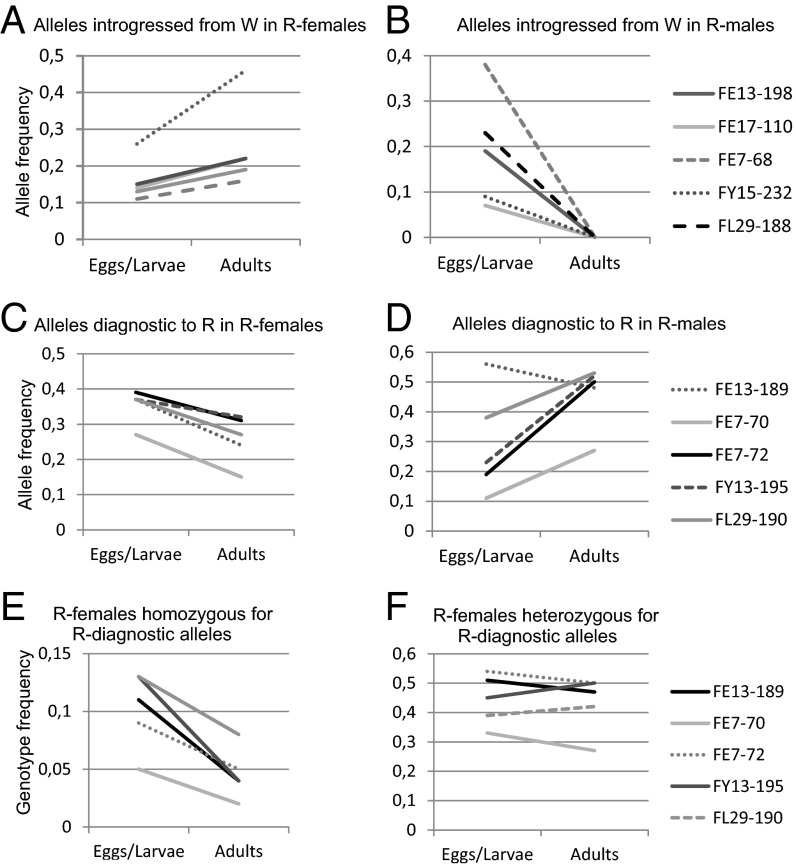
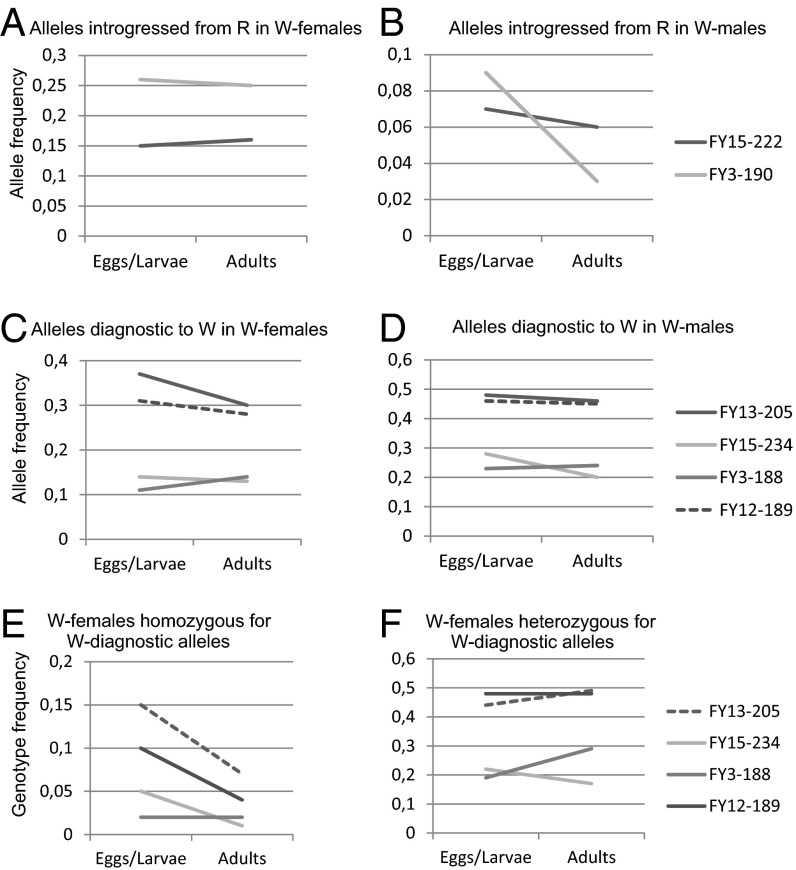
Haploid male eggs carried introgressed alleles with high frequencies, but they disappeared during development already at early stages (Figs. 2 and 3, Table 1, and Tables S6 and S7). The frequency of every introgressed allele clearly declined in males from egg/larval stage to adults (including the reproductives) from 0.09–0.42 to zero in R males (five alleles) (Fig. 2B and Table S6) or from 0.19–0.23 to close to zero in W males (two alleles) (Fig. 3B and Table S6). The number of introgressed alleles per haploid male dropped significantly from 0.77 in eggs to 0 in adult R males (Table 1) and from 0.33 to 0.05 in W males (Table S7). Variation among the developmental stages was significant in both genetic groups. Because the eggs laid by a single queen do not provide independent data points, the tests were done by selecting randomly one haploid egg from those genotyped from each queen and repeating the resampling 1,000 times. All of the tests were significant in both genetic groups at the 5% level.
Fig. 2.
(A–D) Changes in the allele frequencies in males and females in the group R during development from egg/larva (Nf = 364, Nm = 29) to adult (Nf = 173, Nm = 124) showing opposite trends between sexes. E and F show genotype frequencies for diagnostic alleles in females. Note that the scales of the allele frequency axis vary.
Fig. 3.
(A–D) Changes in the allele frequencies in males and females in the group W during development from egg/larva (Nf = 329, Nm = 358) to adult (Nf = 188, Nm = 186). E and F show genotype frequencies for diagnostic alleles in females.
Table 1.
Frequencies of different genotypes in genetic group R
| Genotype and developmental stage | N | No. of loci with the specified genotype per individual | Mean per individual | |||||
| 0 | 1 | 2 | 3 | 4 | 5 | |||
| Haploid males | ||||||||
| Introgressed alleles | ||||||||
| Eggs | 23 | 0.40 | 0.47 | 0.09 | 0.04 | 0 | 0 | 0.77 |
| Larvae | 6 | 0.50 | 0.17 | 0.33 | 0 | 0 | 0 | 0.83 |
| Pupae | 0 | — | — | — | — | — | — | — |
| Adults | 100 | 0.99 | 0.01 | 0 | 0 | 0 | 0 | 0.01 |
| Reproductive fathers | 27 | 1.00 | 0 | 0 | 0 | 0 | 0 | 0.00 |
| Diploid females | ||||||||
| Heterozygote for an introgressed allele | ||||||||
| Eggs | 215 | 0.21 | 0.49 | 0.24 | 0.05 | 0.01 | 0 | 1.14 |
| Worker larvae | 71 | 0.14 | 0.44 | 0.28 | 0.11 | 0.03 | 0 | 1.45 |
| Queen larvae | 78 | 0.12 | 0.44 | 0.31 | 0.12 | 0.03 | 0 | 1.50 |
| Worker pupae | 113 | 0.13 | 0.38 | 0.33 | 0.13 | 0.03 | 0 | 1.54 |
| Queen pupae | 36 | 0.17 | 0.39 | 0.28 | 0.08 | 0.08 | 0 | 1.53 |
| Adult workers | 123 | 0.12 | 0.19 | 0.40 | 0.16 | 0.09 | 0.03 | 2.00 |
| Young queens | 26 | 0 | 0.12 | 0.27 | 0.31 | 0.23 | 0.08 | 2.88 |
| Old queens | 24 | 0 | 0 | 0.59 | 0.18 | 0.23 | 0 | 2.64 |
| Homozygote for a diagnostic allele | ||||||||
| Eggs | 215 | 0.46 | 0.37 | 0.14 | 0.03 | 0 | 0 | 0.74 |
| Worker larvae | 71 | 0.54 | 0.39 | 0.06 | 0.01 | 0 | 0 | 0.54 |
| Queen larvae | 78 | 0.47 | 0.39 | 0.14 | 0 | 0 | 0 | 0.67 |
| Worker pupae | 113 | 0.56 | 0.35 | 0.09 | 0.01 | 0 | 0 | 0.56 |
| Queen pupae | 36 | 0.69 | 0.28 | 0.03 | 0 | 0 | 0 | 0.33 |
| Adult workers | 123 | 0.74 | 0.20 | 0.05 | 0.01 | 0 | 0 | 0.33 |
| Young queens | 26 | 0.96 | 0 | 0 | 0.04 | 0 | 0 | 0.12 |
| Old queens | 24 | 1.00 | 0 | 0 | 0 | 0 | 0 | 0.00 |
The fraction of haploid males carrying any introgressed alleles dropped from 60% in eggs (n = 23) to 1% in adult males (n = 100) and 0% in reproductive males (n = 27) in the R group and from 29% (n = 151) to 9% (n = 128) and 5% (n = 60) in the W group. Based on these frequencies, the introgressed alleles (or linked genomic sequences) are lethal in the R group, and more than one-half of the haploid eggs are apparently lost. Likewise, the fitness (w) of males with introgressed alleles is 0.13 in the W group (compared with fitness = 1 for other males) to cause a frequency change from x0 = 0.29 to x1 = 0.05 (w = [x1(1 − x0)]/[(1 − x1)x0]) (SI Text).
Diploid females also experienced selection, because genotypes present at the egg stage were absent in adults (9). Based on our earlier study, five alleles diagnostic for the R group are present only as heterozygotes in adult R females. Both R males and females transmit these alleles to diploid eggs, and homozygotes are formed. The number of such homozygous loci per diploid female showed significant variation among the developmental stages (Table 1), with all of the resampled (i.e., one diploid egg per mother) χ2 values being significant at least at the 5% level. R eggs had, on average, 0.74 loci homozygous for a diagnostic allele. The corresponding number was 0.12–0.33 in adults and 0.0 in old queens (Table 1), and developmental stages differed significantly from each other (χ2 = 49.7, df = 8, P < 0.0001) (SI Text). All five R-diagnostic alleles showed a decline in the frequency of homozygous diploid females from the egg/larval stage to adults (Fig. 2E and Tables S8 and S9) in the R group. Genotypes heterozygous for these same alleles did not show such declines (Fig. 2F), but the overall frequencies of diagnostic alleles in females declined (Fig. 2C). Thus, alleles diagnostic for the R group behave as recessive deleterious alleles, because homozygotes are progressively lost during development.
In W females, the frequencies of diagnostic alleles were lower than in R. Thus, there were fewer homozygous eggs to begin with and also less elimination. There is also significant variation among the developmental stages in W group (SI Text), and three of four diagnostic alleles show a decline in the frequency of homozygotes from the egg/larval stage to adults (Fig. 3E and Tables S8 and S9), although the decline is not as steep as in group R. In both R and W groups, the frequencies of diagnostic alleles decrease in both workers and queens (Table 1 and Tables S7 and S10).
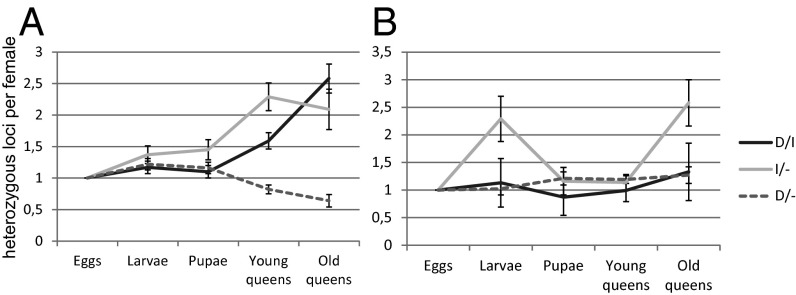
While diagnostic homozygotes decline, the proportion of heterozygous females increases. These changes did not, however, affect all heterozygous genotypes equally. The number of loci heterozygous for introgressed alleles per female increased in the R group (Fig. 4A, Table 1, and Tables S11 and S12), with the developmental stages differing significantly from each other (χ2 = 102.0, df = 12, P < 0.0001). All old queens (n = 24) and 72% of the other diploid females (n = 149) in group R had at least two loci heterozygous for introgressed alleles. The proportion of such individuals in the R eggs (n = 215) was 30%. Thus, selection seems to favor females heterozygous for foreign alleles (i.e., hybrid females). The trends in W females were not as strong as in R females (Fig. 4B). However, the average number of homozygous diagnostic loci per W female dropped from eggs (0.39) to adults (0.17 in workers and 0.30 in young queens) and further to old queens (0.02) (Table S7). Likewise, the number of loci heterozygous for introgressed alleles increased in W females from 0.47 in eggs to 1.02 in old queens (Tables S8 and S9).
Fig. 4.
Frequency changes of heterozygous genotypes through developmental stages in (A) R females and (B) W females. The values are scaled by setting the frequency in eggs equal to one, and error bars (1 SE) are calculated by bootstrapping. The different heterozygotes are D/I (one diagnostic and one introgressed allele), I/− (one introgressed allele and any other allele that is not diagnostic), and D/− (one diagnostic allele and any other allele that is neither diagnostic nor introgressed). The increase of heterozygotes with introgressed alleles indicates that hybrid females are favored in group R.
In summary, the females as well as the males experience strong selection. In the R group, only 16% of the eggs, 27% of the larvae, 29% of the pupae, and 65% of the adults (Tables S8 and S9) represent genotypes that are present in the old egg-laying queens (combining the elimination of the diagnostic homozygotes and the increase in the number of introgressed alleles as heterozygotes). These results indicate that 71–84% of the genotypes present in early developmental stages do not enter the reproductive gene pool. In the W group, 98% of the reproductive queens had no diagnostic alleles as homozygotes. The corresponding frequency was 80% in other adult ants, 57% in pupae, 73% in larvae, and 65% in eggs (Tables S8 and S9). If the change from eggs to adults and reproductive queens is caused by selection, the relative fitness of individuals carrying such homozygous loci should be less than 0.5, and about one-third of the eggs belong to that category.
Discussion
Our earlier study (9) found extraordinary genetic patterns of introgression in a hybrid wood ant population with two coexisting genetic groups (R and W). These two groups are not only both hybrids, but also, each can be considered to be a distinct hybrid species, because a total of 19 (35%) alleles were diagnostic to either group and 27 (49%) alleles were diagnostic comparing only the males of the two groups (17 microsatellite loci; total of 55 alleles). Past hybridization has resulted in mutual introgression of some alleles to the females but not to the males of the other species. As a result, females carry alleles not detected in conspecific adult males. In addition, adult females also lack homozygous genotypes for many alleles that are diagnostic for their own genetic group, although such homozygotes would be expected from random mating with conspecifics and were here detected in eggs. These departures from the expected genotype frequencies lead to strong differences between male and female genomes, which were hypothesized to result from either prezygotic sex-specific segregation distortion or postzygotic selection among the genotypes (9). These results clearly support the selection hypothesis, because some genotypes are lost during the individual development, whereas some are favored. Interestingly, selection is opposite in males and females in such a way that introgression is favored in the diploid females but selected against in the haploid males. Differential selection in haploid and diploid genomes suggests that the introgressed genomic regions harbor both fitness-enhancing and -reducing elements.
No Support for Segregation Hypothesis.
The segregation hypothesis predicts that the queen should transfer her paternally and maternally derived haploid genome complements to haploid and diploid eggs, respectively, without recombination. Cytological mechanisms allowing the segregation hypothesis exist (16), but our family data refute the hypothesis in the case of Formica hybrids, because the alleles from heterozygous queens segregate randomly to both haploid and diploid eggs. The egg genotypes, both haploid and diploid, in our Formica data are compatible with Mendelian segregation from their mother, also ruling out clonal reproduction such as that maintaining genetic differentiation between the sexes in the little fire ant Wasmannia auropunctata (18).
Opposite Selection in Males and Females.
According to the selection hypothesis, the genetic differences between the sexes within a genetic group and the departures of genotype frequencies from those expected under random mating arise from strong selection among the genotypes, some genotypes being practically lethal. Significant directional changes in genotype frequencies, as a function of developmental stage in both genetic groups R and W, clearly support the selection hypothesis. We could not follow these changes within a single cohort of individuals, but pooling genotypes from different nests and time points makes the data genetically representative at the population level. Each nest may have a large number, even hundreds, of reproductive queens, and therefore, the within-nest relatedness is close to zero (19). Thus, although queens were here sampled from relatively few nests, individuals collected from the same nest are likely to represent many different families.
Alleles introgressed from the other genetic group (Fig. 1) (i.e., have a hybrid origin) are strongly selected against in both W and R males. Selection is consistent at different loci, and eight alleles (five introgressed alleles and three other alleles) are completely lost from the R males. The ultimate reason for the elimination of these genotypes could either be intrinsic, leading to inviability or sterility, or extrinsic, where the hybrids suffer lower fitness for ecological reasons (20). Frequency changes at individual loci show that males carrying introgressed alleles mainly disappear early in development (larval stages) in the R group, whereas in the W group, the frequencies decrease gradually (Table S7). This result suggests inviability of hybrid males, which especially in the R group, could be linked to developmental difficulties. Inviability but also behavioral and spermatogenic sterility of hybrid males have been shown in the haplodiploid genus Nasonia (8). Examples of hybrid inviability outside haplodiploids are numerous and intensely studied, for example, in Drosophila (21, 22).
Interestingly, diploid females heterozygous for the introgressed alleles are favored in the R group, because the proportion of loci heterozygous for introgressed alleles per female increased during development (Fig. 4A). Apparently, the more hybrid-like the queen, the better its fitness. Furthermore, these results suggest that the same alleles or genomic regions that are selected against in males have apparent heterotic effects in females. Heterosis is a common consequence of hybridization (23, 24), but this type of antagonistic situation, where the same alleles or genomic regions have opposite effects in different sexes or between haploid and diploid genomes, is unique. Selection for introgression heterozygotes among W females is not as strong as among R females (Fig. 4B), suggesting that the consequences of hybridization are asymmetric.
Another type of selection in females takes place by elimination of individuals homozygous for alleles diagnostic to their own group. Selection against such putatively parental alleles is surprising but could be caused by the facts that the diploid females have high heterozygosities and their genomes are historically of hybrid type. When females in both groups have a strong hybrid background, it could create incompatibilities with loci homozygous for one of the parental gene pools (i.e., loci homozygous for diagnostic alleles). These data show that there is no single developmental stage where the homozygotes especially suffer, but their frequencies decline gradually and consistently at different loci. Selection in the females, especially in queens but also workers, seems, thus, as strong as in the males, although for different reasons.
Possible Long-Term Consequences of Selection.
Our results indicate a potential benefit of extreme polygyny or supercolonialism for hybrid individuals, because the hybrid population persists despite extremely strong selection. A likely explanation is that the presence of several hundred long-lived queens in a nest produces a reproductive excess, where more offspring are born than necessary for colony survival. Selection is stronger in the R group, and it also suffers more problems in worker production. Indeed, the R group is rarer in the population, being present in 19% of the nests (9), whereas the W group is present in all of the nests. The survival of the R group in this population may, thus, depend on its ability to share nests with the W group and use W workers for colony maintenance. There is also a possibility that the observed selection depends, at least partly, on the social context in such a way that worker ants selectively remove developing brood based on some genetically determined cues. If that is the case, the lower frequency of the R group could lead to stronger selection among R females, with W workers favoring those with introgressed alleles and removing those that are homozygous for R-diagnostic alleles. It is, however, hard to see how that would cause selection in the opposite direction in R males.
Haldane’s Rule and the Underlying Causes.
The above results are compatible with the extended Haldane’s rule (10, 25), according to which incompatibilities arising from hybridization are stronger in the heterogametic or hemizygous (i.e., haploid) sex. Several genetic scenarios can lead to Haldane’s rule, the most supported ones being the dominance theory and the faster male theory (26). Our results support the dominance theory. The faster male hypothesis predicts that male traits evolve faster than female traits because of strong sexual selection in males (20, 27). Thus, hybrid females are not expected to suffer from incompatibilities at all, because male-expressed genes are not expected to affect females (20). This expectation contrasts our data, where we also see selection in hybrid females. Furthermore, the faster male theory predicts sterility of males rather than inviability (10, 20), but our data show inviability of hybrid males. The dominance theory remains a plausible explanation, because our data show selection against recessive incompatibilities in haploid males.
Factors Underlying Speciation.
What do these data reveal about factors restricting introgression and contributing to speciation? The parental species of this hybrid population are F. aquilonia and F. polyctena (9), which probably diverged during the last 0.5 My (28). During this time, these species have not completely lost the ability to produce viable hybrids, but they have developed both pre- and postzygotic isolation. The segregation analysis shows that individual marker loci affected by selection are not linked. This observation means that the loci underlying incompatibilities and creating the postzygotic isolation are scattered in the genome and not organized in a few genomic islands of speciation, which is found in Anopheles gambiae strains (29, 30) and Heliconius butterflies (31).
In addition to postzygotic genetic incompatibilities, the two genetic groups of Formica also showed strong prezygotic isolation, because the fraction of intergroup mating is 1–2%. It should be emphasized that opportunities for mixed mating exist, because the two groups share nests, and sexuals of both groups are produced in the same nests and have been collected on the same day. Interestingly, in one of our laboratory cultures, nearly 90% of the offspring were new diploid hybrids produced from intergroup matings. The presence of intergroup hybrids in laboratory conditions but the lack of them in natural conditions could suggest that environmental factors play a role in hybrid female survival.
The buildup of genomic divergence along the speciation continuum is a topic of current studies (32–34). These studies have emphasized the need for a genome-wide perspective, combining the effects of multiple loci (32). However, the models of divergence and genomic hitchhiking emphasize only negatively and neutrally selected genome areas (32, 33). Our results show that the same genomic regions enhance fitness when heterozygous (heterosis) but are virtually lethal when hemizygous (recessive hybrid incompatibilities). This finding suggests that some loci are selected both positively and negatively depending on context or perhaps more likely, that positively and negatively selected loci are linked within the genome. When hybridization results in the introgression of large chromosome blocks, these blocks likely harbor incompatibilities as well as adaptive variation. Such selection is likely in diplodiploid organisms but would be more difficult to observe.
These discoveries have implications for the current discussions on the source of adaptive genetic variation. Adaptation has mainly been thought to arise from standing genetic variation or new mutations (35). However, a third alternative exists: adaptive introgression (2, 3, 36). Our results show a strong case of adaptive introgression, where we can document the ancestry of the alleles and their fitness effects throughout different developmental stages. In taxa, where hybridization is common, introgression may serve as an important source of adaptive genetic variation (1, 36). Future studies within the recently speciated F. rufa group with several hybridizing species will show if adaptive introgression is a common process within this taxon.
Materials and Methods
Sampling.
To study the transmission of maternal alleles, old egg-laying queens were collected in the spring from a hybrid population situated in Tvärminne, southern Finland from two nests in 2007 (n = 23) and seven nests in 2008 (n = 50). In the laboratory, one queen and several workers with nest material were put into each nest box. Ten to fifteen eggs or very young embryos (0–3 d old) were collected and genotyped per each queen. Eggs could be collected from 70 queens. Also, the queen and its spermathecal content were genotyped. We have been unsuccessful in raising males and queens in laboratory conditions. Thus, to study the viability of different genotypes, individuals from different developmental stages were collected from the field between 2008 and 2011, including sexual larvae (n = 166), sexual pupae (n = 127), worker larvae (n = 89), worker pupae (n = 149), adult females (n = 235), adult males (n = 224), and adult workers (n = 307). All of the samples used in this study are listed in Table S1.
Genotyping and Data Analysis.
DNA was extracted with the DNAeasy Tissue Kit (Qiagen) using the manufacturer’s protocol designed for insects. The samples were genotyped in the nine most informative microsatellite loci (i.e., loci that had the largest numbers of alleles and most diagnostic and introgressed alleles and thus, showed the differences between the two genetic groups most clearly). Microsatellite loci used were FE7, FE13, FE19, FE17 (37), and FL29 (38) as well as FY12, FY13, FY15, and FY3 (39) (Table S13). Individuals were assigned into the groups R and W on the basis of diagnostic alleles (i.e., alleles that exist only in one of the groups). The basic analyses (e.g., allele frequencies) of the microsatellite data were done by the program GENEPOP (40), and additional analyses were done with programs written specially for this project (Dataset S1). Random segregation of maternal alleles from heterozygous queens was tested with a one-sided test, where the alternative hypothesis is the sex-specific distortion indicated in Table S2. The linkage between loci was tested under the segregation hypothesis. The linkage phase in double heterozygous queens is unknown. Therefore, we assumed that the linkage phase followed the segregation hypothesis and combined the data from different families. The probability of random segregation was calculated separately for each locus and family, and these probabilities were combined using the Fisher method (χ2 = −2ΣlogP). We also combined the probabilities by using the z transformation (41). The problem of multiple testing was controlled by adjusting the significance level using Bonferroni correction (0.05/ntests). More details on methods are in SI Text.
Supplementary Material
Acknowledgments
We thank R. Jokela, T. Mölläri, and E. Kulmuni for technical assistance and H. Helanterä, H. Johansson, the editor, and two anonymous referees for valuable comments on the manuscript. This work was supported by the Biocenter Oulu Graduate School (J.K.), Center of Excellence in Biological Interactions, Academy of Finland Grant 122210 (to P.P.), and the Finnish Society of Sciences and Letters (P.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.M. is a guest editor invited by the Editorial Board.
Data deposition: The genotype data reported in this paper have been deposited in the Dryad database, http://datadryad.org (doi:10.5061/dryad.64fs5).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323045111/-/DCSupplemental.
References
- 1.Abbott R, et al. Hybridization and speciation. J Evol Biol. 2013;26(2):229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 2.The Heliconius Genome Consortium Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487(7405):94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song Y, et al. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr Biol. 2011;21(15):1296–1301. doi: 10.1016/j.cub.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateson W. In: Darwin and Modern Science. Seward AC, editor. Cambridge, United Kingdom: Cambridge Univ Press; 1909. pp. 85–101. [Google Scholar]
- 5.Dobzhansky T. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics. 1936;21(2):113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller HJ. Temperature, Evolution, Development, Biological Symposia. 1942. Isolating mechanisms, evolution, and temperature. ed Dobzhansky T (Jaques Cattell Press, Lancaster, PA), Vol 6, pp 71–125. [Google Scholar]
- 7.Feldhaar H, Foitzik S, Heinze J. Review. Lifelong commitment to the wrong partner: Hybridization in ants. Philos Trans R Soc Lond B Biol Sci. 2008;363(1505):2891–2899. doi: 10.1098/rstb.2008.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koevoets T, Niehuis O, van de Zande L, Beukeboom LW. Hybrid incompatibilities in the parasitic wasp genus Nasonia: Negative effects of hemizygosity and the identification of transmission ratio distortion loci. Heredity (Edinb) 2012;108(3):302–311. doi: 10.1038/hdy.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulmuni J, Seifert B, Pamilo P. Segregation distortion causes large-scale differences between male and female genomes in hybrid ants. Proc Natl Acad Sci USA. 2010;107(16):7371–7376. doi: 10.1073/pnas.0912409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koevoets T, Beukeboom LW. Genetics of postzygotic isolation and Haldane’s rule in haplodiploids. Heredity (Edinb) 2009;102(1):16–23. doi: 10.1038/hdy.2008.44. [DOI] [PubMed] [Google Scholar]
- 11.Helms Cahan S, et al. Extreme genetic differences between queens and workers in hybridizing Pogonomyrmex harvester ants. Proc Biol Sci. 2002;269:1871–1877. doi: 10.1098/rspb.2002.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helms Cahan S, Vinson SB. Reproductive division of labor between hybrid and nonhybrid offspring in a fire ant hybrid zone. Evolution. 2003;57(7):1562–1570. doi: 10.1111/j.0014-3820.2003.tb00364.x. [DOI] [PubMed] [Google Scholar]
- 13.Leniaud L, Darras H, Boulay R, Aron S. Social hybridogenesis in the clonal ant Cataglyphis hispanica. Curr Biol. 2012;22(13):1188–1193. doi: 10.1016/j.cub.2012.04.060. [DOI] [PubMed] [Google Scholar]
- 14.Seifert B, Goropashnaya A. Ideal phenotypes and mismatching haplotypes - errors of mtDNA treeing in ants (Hymenoptera: Formicidae) detected by standardized morphometry. Org Divers Evol. 2004;4(4):295–305. [Google Scholar]
- 15.Pamilo P, et al. Genetic patchwork of network-building wood ant populations. Ann Zool Fennici. 2005;42(3):179–187. [Google Scholar]
- 16.Holsinger KE, Ellstrand NC. The evolution and ecology of permanent translocation heterozygotes. Am Nat. 1984;124(1):48–71. [Google Scholar]
- 17.Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010;8(9):e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fournier D, et al. Clonal reproduction by males and females in the little fire ant. Nature. 2005;435(7046):1230–1234. doi: 10.1038/nature03705. [DOI] [PubMed] [Google Scholar]
- 19.Pamilo P. Polyandry and allele frequency differences between the sexes in the ant Formica aquilonia. Heredity. 1993;70:472–480. [Google Scholar]
- 20.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer; 2004. pp. 248–292. [Google Scholar]
- 21.Sturtevant AH. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics. 1920;5(5):488–500. doi: 10.1093/genetics/5.5.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Presgraves DC, Balagopalan L, Abmayr SM, Orr HA. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature. 2003;423(6941):715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- 23.Rieseberg LH, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301(5637):1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz D, Matta BM, Shakir-Botteri NL, McPheron BA. Host shift to an invasive plant triggers rapid animal hybrid speciation. Nature. 2005;436(7050):546–549. doi: 10.1038/nature03800. [DOI] [PubMed] [Google Scholar]
- 25.Haldane JBS. Sex ratio and unisexual sterility in hybrid animals. J Genet. 1922;12(2):101–109. [Google Scholar]
- 26.Schilthuizen M, Giesbers MCWG, Beukeboom LW. Haldane’s rule in the 21st century. Heredity (Edinb) 2011;107(2):95–102. doi: 10.1038/hdy.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu CI, Davis AW. Evolution of postmating reproductive isolation: The composite nature of Haldane’s rule and its genetic bases. Am Nat. 1993;142(2):187–212. doi: 10.1086/285534. [DOI] [PubMed] [Google Scholar]
- 28.Goropashnaya AV, Fedorov VB, Pamilo P. Recent speciation in the Formica rufa group ants (Hymenoptera, Formicidae): Inference from mitochondrial DNA phylogeny. Mol Phylogenet Evol. 2004;32(1):198–206. doi: 10.1016/j.ympev.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 29.Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3(9):e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahn MW, White BJ, Muir CD, Besansky NJ. No evidence for biased co-transmission of speciation islands in Anopheles gambiae. Philos Trans R Soc Lond B Biol Sci. 2012;367(1587):374–384. doi: 10.1098/rstb.2011.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadeau NJ, et al. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos Trans R Soc Lond B Biol Sci. 2012;367(1587):343–353. doi: 10.1098/rstb.2011.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nosil P, Feder JL. Genomic divergence during speciation: Causes and consequences. Philos Trans R Soc Lond B Biol Sci. 2012;367(1587):332–342. doi: 10.1098/rstb.2011.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feder JL, Gejji R, Yeaman S, Nosil P. Establishment of new mutations under divergence and genome hitchhiking. Philos Trans R Soc Lond B Biol Sci. 2012;367(1587):461–474. doi: 10.1098/rstb.2011.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feder JL, Flaxman SM, Egan SP, Nosil P. Hybridization and the build-up of genomic divergence during speciation. J Evol Biol. 2013;26(2):261–266. doi: 10.1111/jeb.12009. [DOI] [PubMed] [Google Scholar]
- 35.Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23(1):38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Hedrick PW. Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol. 2013;22(18):4606–4618. doi: 10.1111/mec.12415. [DOI] [PubMed] [Google Scholar]
- 37.Gyllenstrand N, Gertsch PJ, Pamilo P. Polymorphic microsatellite DNA markers in the ant Formica exsecta. Mol Ecol Notes. 2002;2(1):67–69. [Google Scholar]
- 38.Chapuisat M. Characterization of microsatellite loci in Formica lugubris B and their variability in other ant species. Mol Ecol. 1996;5(4):599–601. doi: 10.1111/j.1365-294x.1996.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 39.Hasegawa E, Imai S. Characterization of microsatellite loci in red wood ants Formica (s.str.) spp. and the related genus Polyergus. Mol Ecol Notes. 2004;4:200–203. [Google Scholar]
- 40.Raymond M, Rousset F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- 41.Whitlock MC. Combining probability from independent tests: The weighted Z-method is superior to Fisher’s approach. J Evol Biol. 2005;18(5):1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.