Significance
Enteropathogenic bacteria, exemplified by Escherichia coli strain O157:H7, rely on elaborate acid-resistance systems (ARs) to survive the extremely acidic environment of the stomach. The amino acid antiporter AdiC plays a key role in AR3 and is activated only under acidic pH conditions. Despite elucidation of crystal structures, the molecular mechanism by which AdiC senses acidic pH remains unknown. In this study, we have identified a single aromatic amino acid, Tyr74, as a pH sensor in AdiC. The pH-sensing mechanism appears to be mediated by cation–π interactions between proton and the aromatic side chain of Tyr74. The identification of the pH sensor and the proposed mechanism of pH sensing contribute to mechanistic understanding of the pH-dependent acid resistance in bacteria.
Keywords: membrane transporter; pH sensing; amino acid, polyamine, and organocation superfamily
Abstract
Enteropathogenic bacteria, exemplified by Escherichia coli, rely on acid-resistance systems (ARs) to survive the acidic environment of the stomach. AR3 consumes intracellular protons through decarboxylation of arginine (Arg) in the cytoplasm and exchange of the reaction product agmatine (Agm) with extracellular Arg. The latter process is mediated by the Arg:Agm antiporter AdiC, which is activated in response to acidic pH and remains fully active at pH 6.0 and below. Despite our knowledge of structural information, the molecular mechanism by which AdiC senses acidic pH remains completely unknown. Relying on alanine-scanning mutagenesis and an in vitro proteoliposome-based transport assay, we have identified Tyr74 as a critical pH sensor in AdiC. The AdiC variant Y74A exhibited robust transport activity at all pH values examined while maintaining stringent substrate specificity for Arg:Agm. Replacement of Tyr74 by Phe, but not by any other amino acid, led to the maintenance of pH-dependent substrate transport. These observations, in conjunction with structural information, identify a working model for pH-induced activation of AdiC in which a closed conformation is disrupted by cation–π interactions between proton and the aromatic side chain of Tyr74.
Enteropathogenic bacteria, such as Escherichia coli, Salmonella enterica, and Yersinia pestis, have developed sophisticated acid-resistance systems (ARs) to survive the extremely acidic environment in the stomach (1). In E. coli, AR2 and AR3 each use two molecular components, a membrane-embedded amino acid antiporter and a cytosolic decarboxylase, to consume and expel intracellular protons (1–3). AR3 consists of the antiporter AdiC, which exchanges extracellular l-arginine (Arg) with intracellular agmatine (Agm), and the decarboxylase AdiA, which converts Arg to Agm by removing a carbon dioxide molecule from the α-carboxylate group of Arg and thus absorbing an intracellular proton (2–6). Similarly, AR2 comprises an l-glutamate (Glu):γ-aminobutyric acid (GABA) antiporter GadC and two Glu decarboxylases, GadA and GadB, which convert Glu to GABA (7, 8). Each cycle of transport and decarboxylation by AR2 or AR3 results in the consumption and expelling of one intracellular proton, effectively increasing intracellular pH (2).
The transport activity of the amino acid antiporter AdiC or GadC is strictly pH-dependent (6, 9). Both transporters display robust transport activity only at pH values of 6.0 or lower. At neutral pH or higher, neither AdiC nor GadC exhibits significant transport activity. In addition, expression of some of the AR components is regulated by pH (10, 11); for example, the transcription factor AdiY enhances the expression level of AdiA in response to acidic pH (12). The underlying molecular mechanisms of these pH-dependent functions remain largely unknown.
Structural and biochemical characterization of AdiC and GadC has provided important insights into substrate recognition and transport for these conserved amino acid antiporters (9, 13–16). Considerable effort has focused on identifying pH sensors—presumably one or more amino acids—in these membrane transporters (9). Because robust transport activities of AdiC or GadC are turned on at pH values of 5.5–6.5 (6, 9), the amino acid histidine (His), which has a pKa of 6.2, became the prime suspect. Unfortunately, however, despite intense scrutiny, no His residue in GadC or AdiC has been reported to constitute a pH sensor.
In this paper, we report the identification of a critical pH-sensing amino acid, Tyr74, in AdiC. Replacement of Tyr74 by any other amino acid (except Phe) resulted in loss of pH dependence. Importantly, substrate binding and selectivity are unaffected by these mutations. Our experimental evidence unambiguously demonstrates that Tyr74 is the long sought-after pH sensor in AdiC.
Results
Rationale for the Search for a pH Sensor.
Identification of a pH sensor is central to mechanistic understanding of the pH-dependent transport activity of AdiC. By definition, a pH sensor must have the ability to undergo pH-induced changes—either to mediate new interactions or to abolish existing interactions—so as to regulate substrate transport. Such pH-induced changes may involve protonation and deprotonation of the sensor or direct interaction between protons and the sensor. The former category may involve pH-induced protonation and deprotonation of the amino acids His, Glu, and Asp, whereas the latter category is exemplified by cation–π interactions between protons and an aromatic amino acid (17).
In this study, we searched for a sensor that responds to pH changes in the weakly acidic range of pH 5.5–6.5. We first assessed the possibility that the substrate molecule Arg or Agm may serve as a pH sensor for AdiC. Given the pKa values of 12–13 for the guanidinium groups of Arg and Agm, neither substrate molecule can rely on its side chain to sense acidic pH. The α-amino groups of Arg and Agm have pKa values of 9–10, whereas the α-carboxylate of Arg has a pKa of ∼2.2. Consistent with the conclusions of a previous study (18), our analysis effectively rules out Arg or Agm as a potential pH sensor.
We next focused on specific amino acids in the primary sequences of AdiC (Fig. 1A). Three classes of amino acids have the potential to sense pH changes. The first class includes three types of amino acids, Asp, Glu, and His, all of which have acidic pKa on their side chains and may undergo pH-induced protonation and deprotonation at a pH range of 5.5–6.5 within the folded environment of AdiC. The second class comprises the aromatic amino acids Phe, Tyr, and Trp. These aromatic residues may sense protons through cation–π interactions, which are known to be considerably stronger than hydrogen bonds (19–21). The third class also includes three types of amino acids, Ser, Thr, and Cys, which have slightly basic pKa but theoretically might still serve as a pH sensor in certain circumstances. None of the other 11 types of amino acids can serve as a pH sensor.
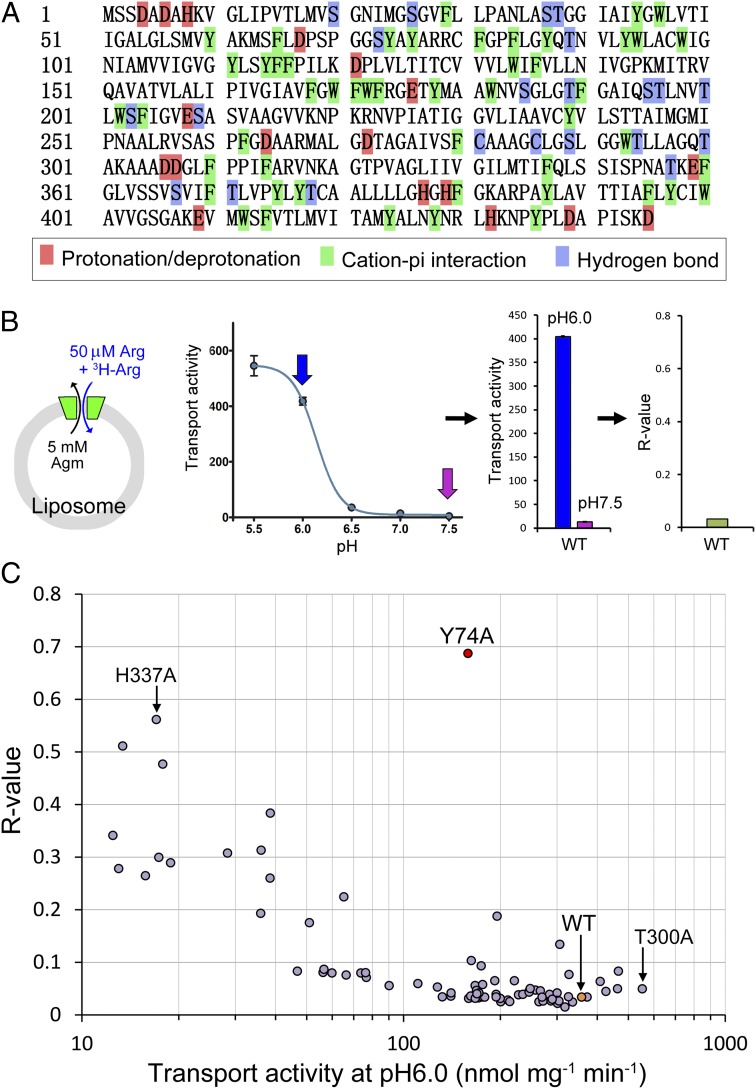
Fig. 1.
Identification of Tyr74 as a critical pH sensor in the amino acid antiporter AdiC. (A) The primary amino acid sequence AdiC. Three types of residues—Asp, Glu, and His—which may undergo pH-induced protonation and deprotonation, are highlighted in red. Aromatic amino acids, in green, may sense protons through cation–π interactions. A portion of the Ser, Thr, and Cys residues are shown in blue; these residues are located along the transport path or on the surface. (B) Definition of the R value. Initial rates of substrate transport were measured in a proteoliposome-based assay (Left) at different pH values. The ratio of the initial transport rate at pH 7.5 to that at pH 6.0 is defined as an R value (Center and Right). (C) Identification of Tyr74 as a critical pH sensor in AdiC. Shown is a plot of R values against initial transport rates at pH 6.0.
The full-length AdiC (residues 1–445) contains 18 residues in the first class of amino acids and 50 residues in the second class that have the potential to sense pH changes in the 5.5–6.5 range. In addition, 22 residues of the third class are located in the transport path or on the surface of AdiC and theoretically may serve as pH sensors. In total, we had 90 candidate residues for pH sensing. We decided to take the brute force approach of individually mutating all 90 amino acids and examining the transport activity of all 90 AdiC variants.
Identification of Tyr74 as a Critical pH Sensor.
Using a PCR-based method, we performed Ala-scanning mutagenesis on AdiC, targeting the aforementioned 90 residues. These AdiC variants were individually purified to homogeneity and examined for their transport activity in an in vitro proteoliposome-based assay (Fig. 1B). In this assay, each of the 90 different proteoliposomes, with a distinct AdiC variant incorporated in the membrane and 5 mM unlabeled Agm inside, was incubated in a buffer containing 50 μM unlabeled Arg and 0.21 μM 3H-labeled Arg at varying pH values. The initial rate of transport, calculated by measuring the first 15 s of transport on incubation, was plotted against pH values. As reported previously (9), the initial transport rates for the WT AdiC protein were quite robust at pH values of 5.5 and 6.0, but became barely detectable at pH 7.0 and 7.5 (Fig. 1B). To quantify the pH-dependent transport activity of AdiC, we defined an R value, the ratio of the initial transport rate at pH 7.5 over that at pH 6.0 (Fig. 1B). The R value is ∼0.03 for WT AdiC.
R values should be within the range of 0–1.0. A smaller R value indicates a larger difference of transport rates between pH 6.0 and pH 7.5, or a stronger dependence on acidic pH for transport. An R value of 1.0 suggests a transport process that is completely independent of pH. Given the influence of pH on other transport-related residues, an R value of 1.0 seemed rather unlikely. We measured the R values for all 90 AdiC variants and plotted these values against the initial rates of transport at pH 6.0 (Fig. 1C). The Y74A variant, involving replacement of Tyr74 by Ala, clearly stood out, exhibiting the highest R value of 0.69 and ∼60% of the transport activity compared with the WT protein (Fig. 1C). None of the other variants came close to Y74A. The H337A variant had a high R value of 0.56 but severely compromised transport, with only ∼5% of the WT transport activity (Fig. 1C). In contrast, the T300A variant exhibited higher transport activity than the WT protein, but had an R value of only 0.05 (Fig. 1C). This analysis unambiguously identifies Tyr74 as a critical pH sensor.
Confirmation of Tyr74 as a pH Sensor.
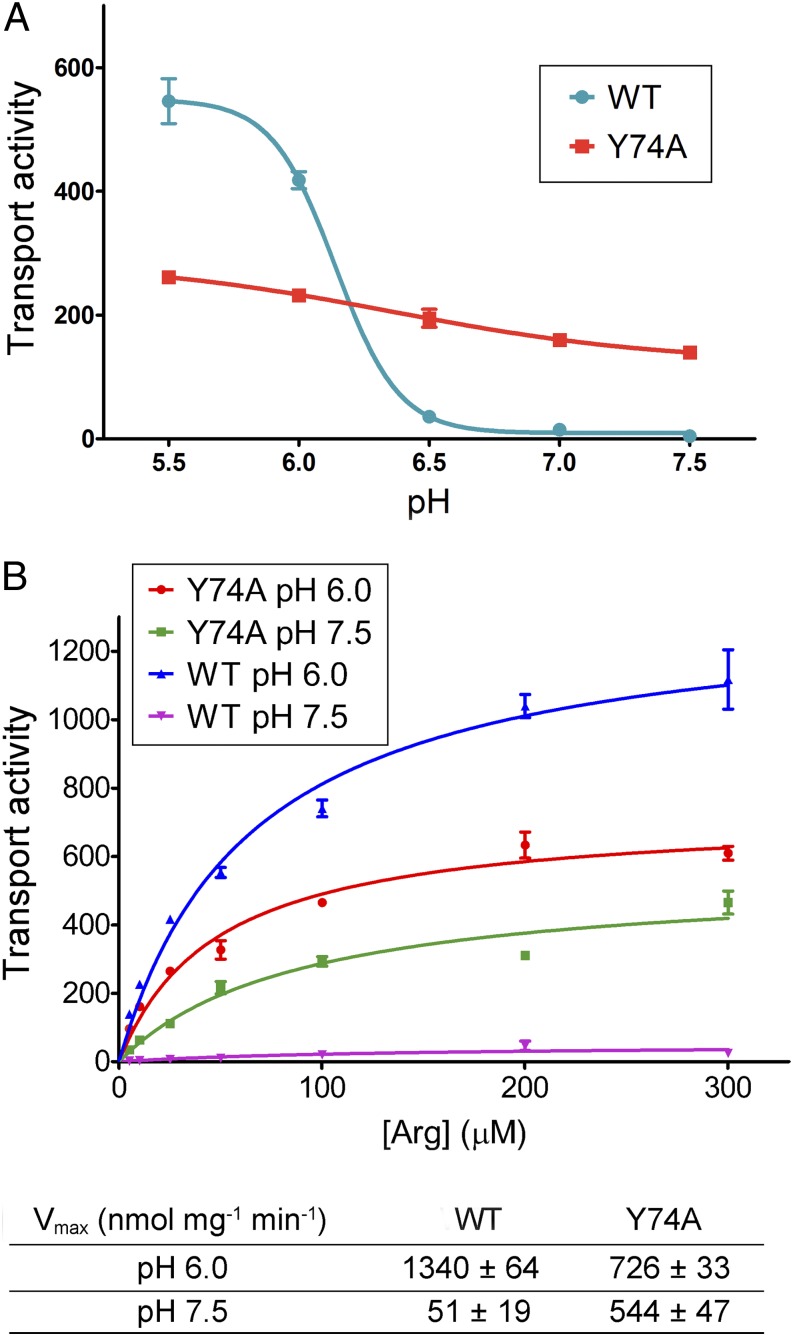
We measured the initial rates of transport for the Y74A variant at five different pH values and plotted these rates against pH (Fig. 2A). In sharp contrast to WT AdiC, the initial rate of transport for the Y74A variant showed only a slight decrease from pH 5.5 to pH 7.5. Consequently, the initial rate of transport for AdiC Y74A was ∼60% of that for WT AdiC at pH 6.0, but more than 20-fold greater than that for WT AdiC at pH 7.5 (Fig. 2A).
Fig. 2.
Confirmation of Tyr74 as a critical pH sensor in AdiC. (A) The AdiC variant Y74A exhibits pH-independent substrate transport. The initial rates of transport are plotted against five pH values for both WT AdiC and the Y74A variant. Only WT AdiC, and not the Y74A variant, exhibited pH-dependent transport activity. (B) The maximal rates of transport by AdiC Y74A are similar at pH 7.5 and pH 6.0. Vmax values were determined for both WT AdiC and the Y74A variant at pH 7.5 and pH 6.0.
Considering that the initial rate of transport represents only one aspect of the transport process, we sought to determine the maximal rate of transport (Vmax) for both WT AdiC and the Y74A variant at pH 6.0 and pH 7.5 (Fig. 2B). The initial rates of transport, measured at seven different Arg concentrations, were plotted against substrate concentration. Fitting of the curve gave rise to Vmax. The Vmax value for the Y74A variant at pH 6.0 was 726 ± 33 nmol mg−1 min−1, ∼54% of that for the WT AdiC protein (Fig. 2B). In contrast, the Vmax value for AdiC Y74A at pH 7.5 was 544 ± 47 nmol mg−1 min−1, more than 10-fold that for the WT protein. Importantly, for AdiC Y74A, the Vmax value at pH 7.5 was 75% of that at pH 6.0, whereas for WT AdiC, the Vmax value at pH 7.5 was <4% of that at pH 6.0.
Substrate Selectivity of AdiC Y74A.
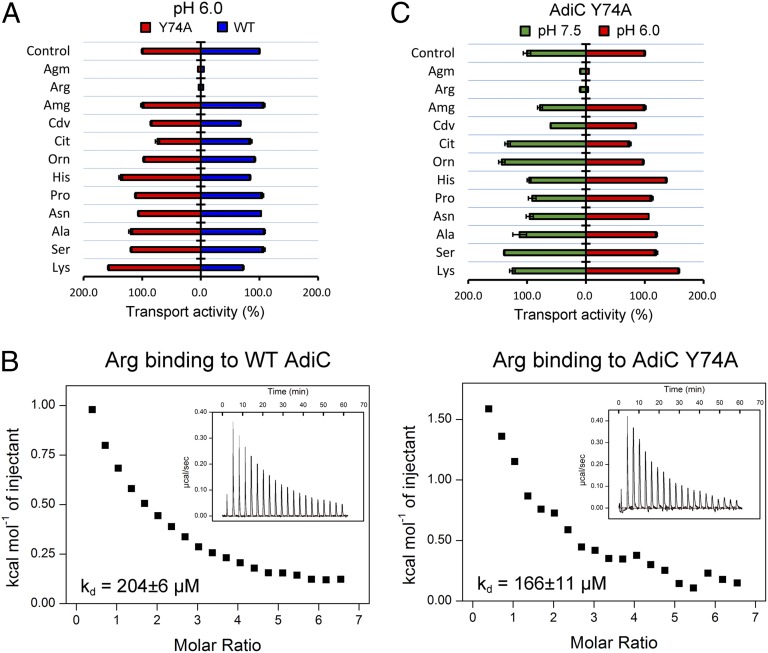
Our experimental evidence strongly identifies Tyr74 as a critical pH sensor that may underlie the pH-dependent transport activity of AdiC. An important question is whether the Y74A mutation alters substrate selectivity by causing nonspecific structural changes to AdiC. To address this question, we reconstituted a competition assay by conducting the transport assay in the presence of 5 mM unlabeled substrate molecules at pH 6.0. As anticipated, 5 mM unlabeled Arg or Agm, which is 100-fold greater than the usual amount of unlabeled substrate Arg, overwhelmed the WT AdiC, resulting in a sharp reduction in measured transport activity (Fig. 3A). Importantly, the same result was obtained for the Y74A variant. In contrast, 5 mM of other control amino acids, such as His and Lys, had little impact on the transport activity of WT AdiC or the Y74A variant, indicating a similar level of substrate selectivity in WT and Y74A. Most notably, aminoguanidine (Amg), cadaverine (Cdv), citrulline (Cit), and ornithine (Orn) are chemically similar to Arg/Agm, yet none of these molecules was able to compete with Arg/Agm for substrate transport by WT AdiC or the Y74A variant (Fig. 3A). Consistent with unaltered substrate selectivity, WT AdiC and the Y74A variant exhibited similar binding affinities toward Arg (Fig. 3B).
Fig. 3.
Substrate selectivity of the AdiC variant Y74A. (A) The Y74A variant maintains a similar level of substrate selectivity as the WT AdiC protein. The transport assay depicted in Fig. 1B was challenged individually by each of the 12 unlabeled substrate molecules at a final concentration of 5 mM. The measured transport activities relative to the control (no additional unlabeled substrate) are shown for both WT AdiC and the Y74A variant. (B) Arg exhibits a similar level of binding affinity for WT AdiC and the Y74A variant. The binding affinity was measured by ITC. (C) The Y74A variant exhibits a similar level of substrate selectivity at pH 6.0 and pH 7.5.
We next compared substrate selectivity of the Y74A variant at pH 6.0 and pH 7.5 (Fig. 3C). Similar to that at pH 6.0, a nearly identical pattern of substrate selectivity was observed at pH 7.5 for the Y74A variant. This result indicates that pH has no significant impact on the substrate selectivity of the AdiC variant Y74A, further validating Tyr74 as a critical pH sensor.
PH-Sensing Mechanism of Tyr74.
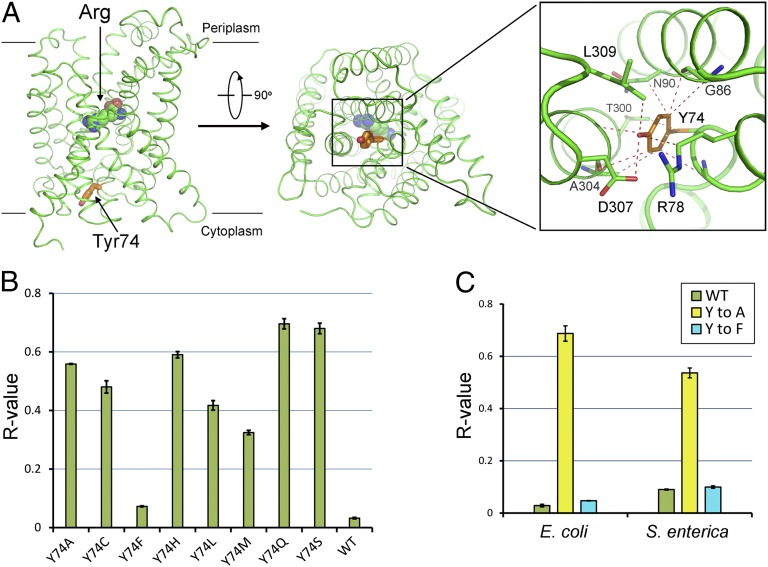
Our experimental evidence suggests that Tyr74 may serve as a pH-regulated gate. At pH 6.5 or higher, the gate closes and blocks substrate entry, whereas at pH 6.0 or lower, the gate opens and allows substrate passage. We speculate that at pH 6.5 or higher, Tyr74 may interact with surrounding amino acids to maintain a closed conformation. This speculation was confirmed by analysis of the AdiC crystal structure (14) (Fig. 4A). The crystals of AdiC were generated under the weakly basic condition, which does not support substrate transport. In the crystals, Tyr74 is located at the center of transport path and interacts with a number of amino acids on TM3 and TM8 to maintain the gate-closed conformation (14) (Fig. 4A).
Fig. 4.
Proposed mechanism of pH-sensing by Tyr74. (A) Tyr74 blocks the transport path by interacting with various surrounding residues in AdiC. Tyr74 (orange) serves as a gate at the cytosolic side of AdiC. Van der Waals contacts within 4 Å are represented by red dashed lines. The bound Arg is shown in spheres. (B) Replacement of Tyr74 by Phe, but not by any other amino acid, resulted in abrogation of pH-dependent substrate transport. The R values for eight variants are compared with that for the WT AdiC protein. (C) Tyr74 likely serves as the pH sensor in AdiC from S. enterica. The R value for WT AdiC from S. enterica is similar to that for the Y74F variant. In contrast, the R value for the Y74A variant from S. enterica is ∼0.54, sixfold higher than that for WT AdiC from S. enterica.
How does Tyr74 sense acidic pH? This action is likely mediated by cation–π interactions between protons and the aromatic side chain of Tyr74. In the crystal structure of the gate-closed AdiC at basic pH (14), there is no space to accommodate a proton atom. Under acidic pH, an increased proton concentration allows the formation of cation–π interactions between the protons and Tyr74, causing local structural rearrangement and consequent gate opening. Among the other 19 amino acids, only Phe meets the two criteria required for pH sensing: maintenance of interactions with neighboring residues to close the gate and interactions with protons through cation–π interactions to open the gate. This analysis predicts that replacement of Tyr74 by Phe, but not by any other amino acid, may allow retention of the pH-sensing mechanism. Confirming this prediction, the Y74F variant retained the ability to sense pH (Fig. 4B). In contrast, replacement of Tyr74 by Ala, Cys, His, Leu, Met, Gln, or Ser led to a loss of pH-dependent substrate transport.
pH-dependent transport is also observed in AdiC from other virulent enteric pathogens. Because Tyr74 is conserved in various other AdiC orthologs, we suspect that the corresponding Tyr may play a similar role in other bacteria. To examine this possibility, we cloned the AdiC ortholog from Salmonella enterica, generated variants Y74A and Y74F, and assessed their transport activities. In complete agreement with our analysis, Y74A, but not Y74F, in S. enterica abrogated pH-dependent substrate transport (Fig. 4C). This result suggests that Tyr74 may be a conserved pH sensor in multiple bacterial species.
Discussion
Similar to AdiC, the amino acid antiporter GadC is activated only under acidic pH conditions (9). Considerable investigative efforts have focused on a potential role of the substrate molecules Glu and GABA, because the pKa values of their carboxylate side chains are close to the pH range in which GadC undergoes activation transition. Gratifyingly, GadC selectively transports Glu and GABA with their carboxylate side chains fully protonated (22, 23). Because protonation of the carboxylate is favored by decreasing the pH value from 6.5 to 5.5 or lower, Glu and GABA clearly play a role in their pH-dependent transport. It is important to note that pH-dependent protonation of Glu and GABA does not fully explain pH-dependent substrate transport by GadC. For example, deletion of a C-terminal sequence (known as the C-plug) in GadC results in a shift of pH-dependence to higher pH by 0.5 pH unit (9), suggesting a role for the C-plug in this process. A C-plug truncated variant of GadC could transport Gln, a mimic of Glu with no net charge in the side chain, even at pH 8.0, although the transport activity is much lower than that at pH 5.5 (22). These observations suggest the existence of an as-yet unidentified pH sensor in GadC.
Similar to GadC, the charged states of Arg were found to affect the transport activity of AdiC, with Arg+ clearly favored over Arg2+ (18). However, unlike the transport of Glu and GABA by GadC, the substrate molecule Arg does not play a role in its pH-dependent transport by AdiC. Arg exists in an equilibrium between Arg+ and Arg2+; at pH 2.0 or below, the equilibrium is shifted toward Arg2+. The pH range of 2.0 and below is very different from the pH range of 5.5–6.5, where transition for pH-dependent transport occurs. Thus, before this study, little was known about the mechanism of pH-dependent transport by AdiC.
In this study, we took a brute force approach by individually mutating each potentially important residue in AdiC and assessing its impact on pH-dependent transport. To facilitate functional assessment, we defined an R value, the ratio of transport activity at pH 7.5 over that at pH 6.0. Notably, although we used pH 6.0 as the activated pH for calculating R values, a lower pH value, such as 5.5 or even 5.0, works equally well for AdiC. Our systematic effort led to the identification of a single amino acid, Tyr74 on the cytosolic side of AdiC, as a dominant pH sensor. Subsequent biochemical characterization unequivocally confirmed this finding. How can an aromatic residue sense acidic pH? We postulate that this action is mediated by cation–π interactions between a proton and the aromatic side chain of Tyr74. Such interactions are thought to open the intracellular gate of AdiC by causing local structural rearrangement. Scrutiny of this model may have to await information of the AdiC structure at acidic pH or in the activated conformation.
The pH sensor Tyr74 is located on the cytosolic side, not the periplasmic side. This location safeguards against inadvertent environmental pH disturbances and ensures effective responses to true acidic stress. Anecdotal pH changes in the surrounding environment may not cause a pH decrease within the E. coli cell, where AdiC maintains an inactive conformation. Under true acidic stress, the cytoplasmic pH of E. coli drops precipitously to pH 6.5 or below, resulting in pH sensing by Tyr74 and consequent activation of antiporter activity by AdiC. The efficient ARs in E. coli allow maintenance of cytoplasmic pH ∼2.5 pH units higher than that in the periplasm.
Although AdiC and GadC share considerable sequence homology, they use clearly different mechanisms for pH-dependent substrate transport. Although Tyr74 is conserved in AdiC orthologs from a few bacterial species, it is not conserved in GadC. A diversity of pH-sensing mechanisms may represent a survival advantage for bacteria, given that any single environmental factor is unlikely to simultaneously shut down both AR2 and AR3.
Methods
Generation of AdiC Variants.
The ORF of AdiC from E. coli O157:H7 was cloned into pET15b (Novagen) and expressed in E. coli BL21 (DE3). Site-directed mutagenesis was carried out using a standard protocol involving two PCR steps. All AdiC variants (more than 100) were confirmed by double-stranded plasmid sequencing. The sequences of the primers used for mutagenesis are not provided herein, but are available from the authors on request.
Protein Preparation.
All AdiC variants were overexpressed and purified as described previously (13). Cells were grown at 37 °C. The transformed cells was induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at an OD600 of 1.5. The cells were harvested, homogenized in a lysis buffer containing 25 mM Tris pH 8.0 and 150 mM NaCl, and disrupted by sonication. Cell debris was removed by centrifugation at 23,000 × g for 10 min. The membrane fraction was collected by ultracentrifugation at 150,000 × g for 1 h and then incubated with 2% (wt/vol) dodecyl-β-d-maltopyranoside (DDM; Anatrace) at 4 °C. Protein was purified with Ni2+-NTA affinity resin (Qiagen). The protein was released from the affinity resin by thrombin cleavage. The protein was concentrated to 1.5 mL and applied to a Superdex-200 column (GE Healthcare) in a buffer containing 25 mM Tris pH 8.0, 300 mM NaCl, and 0.04% DDM. The peak fraction was collected and concentrated for transport assays.
Preparation of Proteoliposomes.
Proteoliposomes were prepared following published protocols (6, 24). E. coli polar lipid extract (Avanti Polar Lipids) was solubilized in CM buffer [chloroform:methanol, 3:1 (vol/vol)] to a final concentration of 50 mg/mL, then dried under a stream of nitrogen to remove the solvent and to obtain a thin layer dry lipids in a glass tube. The dried lipids were resuspended in PA Buffer (containing 30 mM KH2PO4/K2HPO4,pH 7.5, 150 mM KCl, and 5 mM Agm) by vortexing for 20 min, to yield a final lipid concentration of 20 mg/mL After 10 cycles of quick freezing and thawing, the liposomes were extruded at least 21 times in an Avestin extruder through a 400-nm polycarbonate filter (Avanti) to obtain unilamellar vesicles of a homogeneous size. Protein concentration was determined using Bio-Rad reagents, and the liposomes were mixed with purified protein at a concentration of 5 μg/mg lipids. To destabilize the liposomes, β-d-octyl glucoside (OG) was added to a final concentration of 1.25%, followed by incubation at 4 °C for 2 h. DDM and OG were removed by incubating with 300 mg/mL Bio-Beads (Bio-Rad) overnight and then with 100 mg/mL Bio-Beads for 2 h. After one cycle of quick freezing and thawing, the proteoliposomes were extruded 21 times in the extruder through a 400-nm polycarbonate filter. Finally, the proteoliposomes were ultracentrifuged at 100,000 × g at 4 °C for 1 h, and the resulting pellet was resuspended in 150 mM KCl to 50 mg/mL.
Transport Assay.
Each reaction system had 4 μL volume of proteoliposomes and 100 μL reaction buffer (30 mM KH2PO4/K2HPO4,with pH adjusted to 5.5–7.5, 0.21 μM [3H]-Arg, and 50 μM unlabeled Arg). The initial rate of transport was measured at 15 s after incubation of the proteoliposomes with reaction buffer, by following the uptake of [3H]-Arg into the proteoliposomes at 25 °C. The reaction was quickly placed into a 0.4-μm filter and washed in 2 mL of lysis buffer. Then the filter was placed into a 24-hole plate for liquid scintillation counting. To perform the substrate selectivity assay, 5 mM candidate substrate was added to the reaction buffer (9). As for the Vmax assay, the concentration of nonradioactive Arg ranged between 5 and 300 μM (9). All of the transport assays were repeated at least three times.
Isothermal Titration Calorimetry.
The isothermal titration calorimetry (ITC) assays were performed as described previously (13).
Supplementary Material
Acknowledgments
This work was funded by the Ministry of Science and Technology of China (Grant 2009CB918801) and the National Natural Science Foundation of China (Projects 30888001, 31021002, and 31130002).
Footnotes
The authors declare no conflict of interest.
References
- 1.Foster JW. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat Rev Microbiol. 2004;2(11):898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 2.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186(18):6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanjee U, Houry WA. Mechanisms of acid resistance in Escherichia coli. Annu Rev Microbiol. 2013;67:65–81. doi: 10.1146/annurev-micro-092412-155708. [DOI] [PubMed] [Google Scholar]
- 4.Iyer R, Williams C, Miller C. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol. 2003;185(22):6556–6561. doi: 10.1128/JB.185.22.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong S, Richard H, Foster JW. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol. 2003;185(15):4402–4409. doi: 10.1128/JB.185.15.4402-4409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Y, Kolmakova-Partensky L, Miller C. A bacterial arginine-agmatine exchange transporter involved in extreme acid resistance. J Biol Chem. 2007;282(1):176–182. doi: 10.1074/jbc.M610075200. [DOI] [PubMed] [Google Scholar]
- 7.Hersh BM, Farooq FT, Barstad DN, Blankenhorn DL, Slonczewski JL. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178(13):3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181(11):3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma D, et al. Structure and mechanism of a glutamate-GABA antiporter. Nature. 2012;483(7391):632–636. doi: 10.1038/nature10917. [DOI] [PubMed] [Google Scholar]
- 10.Blethen SL, Boeker EA, Snell EE. Argenine decarboxylase from Escherichia coli, I: Purification and specificity for substrates and coenzyme. J Biol Chem. 1968;243(8):1671–1677. [PubMed] [Google Scholar]
- 11.Kanjee U, Gutsche I, Ramachandran S, Houry WA. The enzymatic activities of the Escherichia coli basic aliphatic amino acid decarboxylases exhibit a pH zone of inhibition. Biochemistry. 2011;50(43):9388–9398. doi: 10.1021/bi201161k. [DOI] [PubMed] [Google Scholar]
- 12.Stim-Herndon KP, Flores TM, Bennett GN. Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology. 1996;142(Pt 5):1311–1320. doi: 10.1099/13500872-142-5-1311. [DOI] [PubMed] [Google Scholar]
- 13.Gao X, et al. Structure and mechanism of an amino acid antiporter. Science. 2009;324(5934):1565–1568. doi: 10.1126/science.1173654. [DOI] [PubMed] [Google Scholar]
- 14.Gao X, et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 2010;463(7282):828–832. doi: 10.1038/nature08741. [DOI] [PubMed] [Google Scholar]
- 15.Fang Y, et al. Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature. 2009;460(7258):1040–1043. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalczyk L, et al. Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc Natl Acad Sci USA. 2011;108(10):3935–3940. doi: 10.1073/pnas.1018081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arras E, Seitsonen AP, Klappenberger F, Barth JV. Nature of the attractive interaction between proton acceptors and organic ring systems. Phys Chem Chem Phys. 2012;14(46):15995–16001. doi: 10.1039/c2cp42293j. [DOI] [PubMed] [Google Scholar]
- 18.Tsai MF, Miller C. Substrate selectivity in arginine-dependent acid resistance in enteric bacteria. Proc Natl Acad Sci USA. 2013;110(15):5893–5897. doi: 10.1073/pnas.1301442110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallivan JP, Dougherty DA. Cation–pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96(17):9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dougherty DA. Cation–pi interactions involving aromatic amino acids. J Nutr. 2007;137(6 Suppl 1):1504S–1508S. doi: 10.1093/jn/137.6.1504S. discussion 1516S-1517S. [DOI] [PubMed] [Google Scholar]
- 21.Gallivan JP, Dougherty DA. A computational study of cation−π interactions vs salt bridges in aqueous media: Implications for protein engineering. J Am Chem Soc. 2000;122(5):870–874. [Google Scholar]
- 22.Ma D, Lu P, Shi Y. Substrate selectivity of the acid-activated glutamate/γ-aminobutyric acid (GABA) antiporter GadC from Escherichia coli. J Biol Chem. 2013;288(21):15148–15153. doi: 10.1074/jbc.M113.474502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai MF, McCarthy P, Miller C. Substrate selectivity in glutamate-dependent acid resistance in enteric bacteria. Proc Natl Acad Sci USA. 2013;110(15):5898–5902. doi: 10.1073/pnas.1301444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reig N, et al. Functional and structural characterization of the first prokaryotic member of the L-amino acid transporter (LAT) family: A model for APC transporters. J Biol Chem. 2007;282(18):13270–13281. doi: 10.1074/jbc.M610695200. [DOI] [PubMed] [Google Scholar]