Abstract
Haemophilus ducreyi, the causative agent of chancroid, is highly resistant to the complement-mediated bactericidal activity of normal human serum (NHS). Previously, we identified DsrA (for ducreyi serum resistance A), a major factor required for expression of the serum resistance phenotype in H. ducreyi. We describe here a second outer membrane protein, DltA (for ducreyi lectin A), which also contributes to serum resistance in H. ducreyi. Isogenic dltA mutants, constructed in 35000HP wild-type and FX517 dsrA backgrounds, were more susceptible to the bactericidal effects of NHS than each respective parent, demonstrating the additive effect of the mutations. Furthermore, expression of dltA in H. influenzae strain Rd rendered this highly susceptible strain partially resistant to 5% NHS compared to a vector-control strain. Although primary basic local alignment search tool analysis of the dltA open reading frame revealed no close bacterial homologue, similarity to the β-chain of the eukaryotic lectin ricin was noted. DltA shares highly conserved structural motifs with the ricin β chain, such as cysteines and lectin-binding domains. To determine whether dltA was a lectin, ligand blots and affinity chromatography experiments were performed. DltA was affinity purified on immobilized lactose and N-acetylgalactosamine, and N-glycosylated but not glycosidase-treated model glycoproteins bound DltA. These data indicate that DltA is a lectin with specificity for lactose-related carbohydrates (CHO) and is important for H. ducreyi serum resistance.
Chancroid is a sexually transmitted genital ulcer disease (reviewed in references 34, 39, 65, and 75) characterized by painful ulcers and suppurative lymphadenopathy. Although rare in the United States (www.cdc.gov/std/stats00/slides/otherstds/sld002.htm) and Canada (http://cythera.ic.gc.ca/dsol/ndis/disease2/chan_e.html), chancroid is common in developing, resource-poor countries worldwide (65). Interest in the study of chancroid pathogenesis has been renewed since the discovery that genital ulcer diseases, including chancroid, are important independent cofactors in the heterosexual transmission of the human immunodeficiency virus (7, 9, 29, 40, 50). In fact, chancroid incidence is highest in countries where human immunodeficiency virus is epidemic (68).
Chancroid is caused by the gram-negative pleomorphic bacterium Haemophilus ducreyi (reviewed in reference 1). This fastidious microorganism is a strict human pathogen that localizes to the genital epithelia and does not cause systemic infection (39, 65, 75). Although H. ducreyi was first identified by Auguste Ducrey in 1889, the mechanisms responsible for the pathogenesis of chancroid remain poorly understood.
Several microorganisms, including H. ducreyi, can survive in the presence of fresh normal human serum (NHS), which contains immunoglobulins and complement (C′). C′ is a complex system of proteins that promotes killing of pathogens by opsonophagocytosis and lysis (38, 57), among other functions. Capsules of both gram-positive and gram-negative bacteria (12, 57), the O antigen on lipopolysaccharides of gram-negative bacteria (57), sialic acids on lipooligosaccharides (LOS) (54, 57), and several membrane proteins in a variety of bacterial species (16, 54, 57) have been shown to be involved in providing protection from the activity of C′. These bacterial components affect C′ activity by a variety of ways, such as spatially hindering the binding of C′ proteins (49) or antibodies (15) to the bacterial membrane, by binding “blocking” antibodies (58), by facilitating the inactivation of the C′ cascade through the binding of negative regulators of C′ (2, 37, 52, 53, 55, 56), by preventing the productive insertion of the membrane attack complex into the outer membrane (OM) (24, 30), or by interfering with opsonophagocytosis (71).
Numerous studies have documented the serum resistance of H. ducreyi (16, 19, 25, 33, 41-43). Since H. ducreyi is unencapsulated, the serum resistance phenotype is not due to the presence of a capsule (65). Furthermore, the ability to sialylated LOS does not render H. ducreyi strains resistant to NHS (25). Surprisingly, isogenic mutants of 35000HP lacking the dd-heptose and all subsequent terminal lactose-related carbohydrates (CHO) remain fully resistant to NHS (25). However, mutations affecting the ability to express the 30-kDa OM protein (OMP) DsrA substantially affect serum resistance (16), although minor effects were also seen in mutants lacking the major OMP (MOMP) (25). Earlier reports by Odumeru et al., who used nonisogenic strains expressing truncated LOS (41, 42) and containing cryptic mutations in dsrA, suggested that LOS composition might affect serum susceptibility in H. ducreyi. However, expression of dsrA restored serum resistance in these serum-susceptible strains without affecting LOS electrophoretic mobility (16).
Lectins are CHO-binding proteins found ubiquitously in nature, in eukaryotes and prokaryotes, and from single-cell to multicellular organisms (10). Lectins bind CHO through hydrophobic interactions (31) and can be grouped according to their CHO-recognition domains (CRD) (10). The lectin ricin, whose β chain binds terminal galactose and GalNAc residues, is an R-type (ricin-like) lectin. R-type CRDs are the only types of CRDs found in bacterial lectins (10). Lectins are important in cell adhesion and recognition because they bind CHO rapidly and in a reversible fashion (62). Twenty-five years ago, Ofek et al. showed that surface-exposed bacterial lectins were required for initiation of infection (44). Since then, several bacterial lectins have been shown to be involved in the attachment of host cell glycoconjugates, such as the P pili and type 1 fimbriae of Escherichia coli (78), the filamentous hemagglutinin of Bordetella pertussis (63), several adhesins of nontypeable Haemophilus influenzae (70), and type IV pili of Pseudomonas aeruginosa (20).
Thus far, no lectin has been identified or shown to be involved in the pathogenesis of chancroid. We demonstrate here that DltA is a lectin involved in the serum resistance of H. ducreyi.
(Portions of this study were presented at the 7th International Symposium on Haemophilus ducreyi Pathogenesis and Chancroid in June 2003 in Ottawa, Ontario, Canada.)
MATERIALS AND METHODS
Strains and media.
Strains used in the present study are described in Table 1. H. ducreyi strains were routinely maintained by subculture in 5% CO2 at 34.5°C on chocolate agar plates (CAP) containing 1× GGC (0.1% glucose, 0.01% glutamine, 0.026% cysteine) (74) and 5% fetal bovine serum (FBS) (Sigma, St. Louis, Mo.). For broth cultures, H. ducreyi was grown in GC broth (Difco, Detroit, Mich.) containing 5% FBS and 1% IsoVitalex (BBL, Cockeysville, Md.) at 34.5°C with 5% CO2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| H. ducreyi | ||
| 35000HPa | Human-passaged variant of strain 35000 (21) | S. M. Spinola et al. (67) |
| FX517 | 35000HPdsrA::CATc | 16 |
| FX533 | 35000HPdltA::Kan | This study |
| FX534 | 35000HPdsrA::CAT dltA::Kan | This study |
| H. influenzae | ||
| Rd | Wild type | ATCCb |
| Rd/p1286 | Strain Rd containing pUNCH1286 | This study |
| Rd/pLSKS | Strain Rd containing pLSKS | This study |
| E. coli | ||
| DH5α | recA gyrB | Bethesda Research Labs |
| BL21(DE3)/pLYSs | Cmr | Invitrogen |
| Plasmids | ||
| pCR2.1-TOPO | PCR cloning vector; Kanr Ampr | Invitrogen |
| pCRT7/NT-TOPO | Expression vector; Ampr | Invitrogen |
| pLSKS | H. ducreyi shuttle vector; Smr | 77 |
| pRSM1791 | Mutagenesis plasmid; β-Gal+ Ampr | 5 |
| pUNCH1284 | dltA expression clone in pCRT7/NT-TOPO; used for rDltA purification | This study |
| pUNCH1285 | dltA PCR clone in pCR2.1-TOPO | This study |
| pUNCH1286 | dltA subclone in pLSKS | This study |
| pUNCH1287 | Mutagenized dltA; pUNCH1286 mutagenized with Kan cassette in pLSKS; Kanr Smr | This study |
| pUNCH1289 | Mutagenized dltA subclone in pRSM1791 | This study |
| pUNCH1295 | NcaA expression clone | This study |
35000HP was isolated from a pustular lesion of a human volunteer 13 days after inoculation.
ATCC, American Type Culture Collection.
CAT, chloramphenicol acetyltransferase.
E. coli strains were grown either on Luria-Bertani (LB) agar or in LB broth. H. influenzae strains were grown on CAP supplemented with 1% IsoVitalex. The following concentrations of antibiotics were used: 100 μg of ampicillin (Amp)/ml, 30 μg of chloramphenicol (Cm)/ml, 30 μg of kanamycin (Kan)/ml, and 100 μg of streptomycin (Sm)/ml.
DIG labeling of FN, Tf, fH, and BSA.
Proteins were labeled by using digoxigenin-3-0-succinyl-ɛ-aminocaproic acid-N-hydroxysuccinimide ester (DIG-NHS) according to the manufacturer's instructions (Roche Diagnostics GmbH, Mannheim, Germany). Briefly, 1 mg of fibronectin (FN; kindly supplied by Kenneth Ingram, American Red Cross), 500 μg of transferrin (Tf; Sigma, St-Louis, MO) and factor H (fH; Advanced Research Technologies, San Diego, Calif.), and 1 mg of bovine serum albumin (BSA; Sigma) were exchanged in phosphate-buffered saline (PBS) by using a Centricon 50 concentrator, and then 10.6 μl of DIG-NHS (20 mg/ml in dimethyl sulfoxide [DMSO]) was added to the proteins, followed by incubation at room temperature for 2 h with gentle mixing. The reaction was quenched with 100 μl of 1 M Tris (pH. 8.0). DIG-labeled proteins were dialyzed overnight against PBS (pH 8.0) at 4°C to remove any unbound DIG. The activity of the probes was assessed by titration in a ligand blot as described below.
Ligand blots with DIG-labeled proteins.
Proteins from H. ducreyi cell pellets (∼107 CFU/lane) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (32) under nonreducing and reducing conditions (as indicated) and transferred to a 0.45-μm-pore-size nitrocellulose membrane by using a Mini-Protean II transfer apparatus (Bio-Rad, Hercules, Calif.). Membranes were blocked for a minimum of 1.5 h in 0.5% Tween 20 in PBS (block), incubated at room temperature for 1 h with DIG-labeled proteins (1 μg/ml in block), and washed three times in 0.05% Tween 20 in PBS. Membranes were thereafter incubated with anti-DIG alkaline phosphatase (anti-DIG-AP) antibody (Roche, Indianapolis, Ind.) (1:5,000 in block) for 1 h at room temperature and then washed, and detection was performed with a Lumi-Phos WB chemiluminescence substrate (Pierce, Rockford, Ill.).
Sequencing DltA and preliminary identification of dltA in the H. ducreyi genome.
Because N-terminal Edman degradation failed twice, we performed in situ trypsin digestion and mass spectrometry (MS) to sequence DltA. Sarkosyl-insoluble (SI) OM preparations were subjected to preparative SDS-PAGE and Coomassie blue staining, and the putative DltA band was cut out. To verify that we had extracted the correct band from the gel, part of the gel slice submitted for trypsin studies was electroeluted. The eluted protein, but not a control protein migrating just above DltA, bound FN in a ligand blot (data not shown). Internal tryptic peptide mapping of the gel slice was done, followed by matrix-assisted laser desorption ionization-time-of-flight MS, and MS/MS data were generated at the UNC Microchemical Core. We used the resulting protein sequence to search the H. ducreyi genome databases (www.stdgen.lanl.gov/ or http://www.ncbi.nlm.nih.gov/genomes/chrom.cgi?gi=309&db=G). The databases kindly supplied by Robert Munson (Ohio State University).
Globomycin treatment of H. ducreyi.
A 20-ml liquid culture of H. ducreyi inoculated with 2.4 × 106 CFU/ml was allowed to grow for 4 h at 34.5°C with 5% CO2. At that point, the culture was split into two 10-ml cultures. Globomycin (final concentration, 15 μg/ml) was added to one culture, and DMSO alone (1.5%) was added to the other (negative control). At 4 h after the addition of DMSO or globomycin, 1 ml of the culture (optical density at 600 nm [OD600] of 0.2 [∼108 CFU]) was harvested and then pelleted, and the cells were resuspended in 100 μl of Laemmli sample buffer with β2-mercaptoethanol (βME) (i.e., reducing conditions). The samples (10 μl) were thereafter subjected to SDS-PAGE and Western blotting with anti-recombinant DltA (anti-rDltA), anti-DsrA, and anti-Hlp antibodies.
Cloning and mutagenesis of the dltA gene.
The dltA gene was amplified by PCR from whole cells of H. ducreyi strain 35000HP as the template and oligonucleotides DLTA.01 (5′-CGC TTG TAC AAG CGG GC-3′) and DLTA.02 (5′-CAG CTT ACA AAA TGA TGG GC-3′) as primers. These oligonucleotides were based on the H. ducreyi genome sequence and flank the dltA open reading frame (ORF). An 815-bp product was predicted from PCR. The conditions for PCR were as follows: a single denaturing step was performed for 5 min at 95°C, followed by 35 cycles of denaturation for 1 min at 95°C, annealing at 50°C for 1 min, and elongation at 72°C for 1 min. A single product of ∼800 bp was observed and was gel purified by using the Qiagen PCR purification kit (Valencia, Calif.). The PCR product was cloned into the pCR2.1-TOPO according to the manufacturer's instructions to form pUNCH1285. To create a plasmid for complementation of dltA mutants, pUNCH1286 was constructed. The insert in pUNCH1285 was digested with EcoRI, Klenow treated, and gel purified. The vector pLSKS was digested with EcoRV and ligated to the insert containing dltA from pUNCH1285 to form pUNCH1286. The dltA ORF in pUNCH1286 was mutagenized by insertion of a Kan cassette. pUNCH1286 was made linear by digestion with Eco47III and ligated to EcoRI- and Klenow-treated Kan cassette from pUC4K to form pUNCH1287. The mutagenized dltA was then moved into suicide vector pRSM1791 (5) by digesting pUNCH1287 with XbaI and KpnI and filling the overhangs with the T4 DNA polymerase and deoxynucleoside triphosphates. Plasmid pRSM1791 was digested with NotI, treated with Klenow, and ligated to the mutagenized dltA insert from pUNCH1287 to create pUNCH1289. pUNCH1289 was electroporated into H. ducreyi 35000HP, and transformants were isolated on CAP containing Kan. Kan-resistant transformants (putative cointegrates) were streaked onto Kan-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) CAP to isolate mutants that had lost the wild-type locus (5). Mutants were confirmed by PCR amplification with whole cells of the mutants as DNA template and the oligonucleotides DLTA.01 and DLTA.02 as primers under the conditions described above. As expected, a 2-kb band was amplified in the mutants (data not shown).
Complementation of dltA mutants (FX533 and FX534) and expression of dltA in H. influenzae.
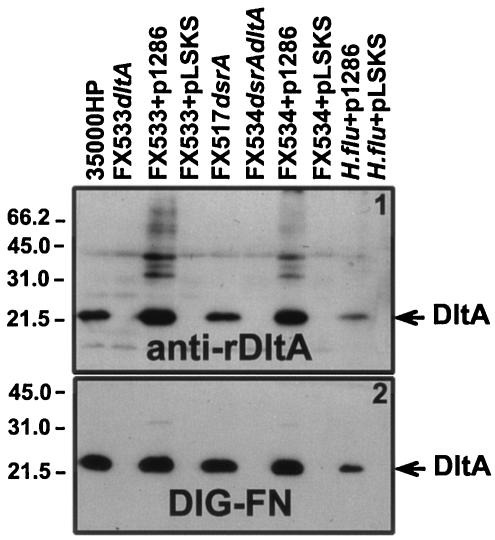
Plasmid pUNCH1286dltA was introduced into H. ducreyi strains FX533 and FX534 (Table 1) and in H. influenzae strain Rd by electroporation. Expression of dltA from pUNCH1286 was assessed by Western and DIG-FN ligand blots (Fig. 4).
FIG. 4.
Characterization of DltA expression in mutants. Proteins from solubilized H. ducreyi (∼107 CFU/lane) were subjected to SDS-PAGE under nonreducing conditions and Western blotting with anti-rDltA (panel 1) and DIG-FN (panel 2). Lanes: 35000HP, H. ducreyi 35000HP parental strain; FX533dltA, 35000HP dltA::Kan; FX533+p1286, FX533 complemented in trans with pUNCH1286dltA; FX533+pLSKS, FX533 complemented in trans with the empty vector pLSKS; FX517dsrA, 35000HP dsrA::CAT; FX534dsrAdltA, 35000HP dltA::Kan dsrA::CAT; FX534+p1286, FX534 complemented in trans with pUNCH1286dltA; FX534+pLSKS, FX534 complemented in trans with the empty vector pLSKS; H. flu+p1286, H. influenzae strain Rd containing pUNCH1286dltA; H. flu+pLSKS, H. influenzae strain Rd containing pLSKS.
Cloning, expression, and purification of rDltA.
The dltA gene was amplified from 35000HP whole cells by using oligonucleotides DLTA.03 (5′-TGT GTA TAT GCA CCT CCC G-3′) and DLTA.02. The PCR product was ligated to pCRT7/NT-TOPO to form pUNCH1284 and transformed into E. coli BL21(DE3)/pLysS (Invitrogen, Carlsbad, Calif.). This construct eliminated the putative lipoprotein N-terminal signal sequence and replaced it with an N-terminal fusion tag. After induction, the transformants were examined by SDS-PAGE and Coomassie blue staining for the presence of an abundant novel protein compared to the vector control. We observed several clones that contained an ∼30-kDa protein that bound DIG-FN in ligand blots. One clone was chosen for further study and designated pUNCH1284 (Table 1). The insert from pUNCH1284 was sequenced on both strands at the University of North Carolina at Chapel Hill Sequencing Facility utilizing Taq terminator chemistry.
Using the method of Qi et al. (51), rDltA was purified as previously described (17, 73). Inclusion bodies containing the recombinant proteins were subsequently purified by four centrifugations (10,000 × g for 20 min). Using SDS-PAGE and Coomassie blue staining, it was determined that rDltA was >95% pure (data not shown).
Generation of anti-DltA antisera.
Elite New Zealand White rabbits were immunized four times, at 2-week intervals, with 200 μg of rDltA in complete Freund adjuvant for the first immunization and in incomplete Freund adjuvant for the last three immunizations at Covance Research Products, Inc. (Denver, Pa.). Eight weeks after the first immunization, rabbits were exsanguinated, and sera were collected and stored at −70°C for later use.
Affinity chromatography of DltA with immobilized CHO.
Whole H. ducreyi cells (ca. 5 × 108 CFU) were solubilized in 2% Zwittergent 3,14 in Tris-EDTA-NaCl buffer for 1 h at room temperature and centrifuged for 20 min at 10,000 × g, and the pellet was discarded. A total of 900 μl of the supernatant was mixed with 100 μl of a 50% slurry of acrylamide-bound CHO (EY Laboratories, Inc., San Mateo, Calif.), followed by incubation at room temperature for 2 h. The pellet was washed four times, with a changing of the tubes between washes 3 and 4, and then 100 μl of 1× Laemmli buffer was added to the final pellet. Then, 20 μl of this suspension was subjected to SDS-PAGE and a ligand blot with DIG-FN (0.5 μg/ml) and anti-rDltA (1:30,000).
PNGase F treatment of DIG-labeled probes.
Deglycosylation of DIG-labeled probes with N-glycosidase F (PNGase F) was performed according to the manufacturer's instructions (New England Biolabs, Beverly, Mass.) as follows. DIG-labeled probes were boiled 10 min in denaturing buffer (5% SDS and 10% βME; New England Biolabs) and then incubated 1 h at 37°C in 1× G7 buffer (0.05 M sodium phosphate [pH 7.5]), 1% NP-40, and 250 U of PNGase F. Controls containing no PNGase F (−) were treated concurrently. Since boiling affected the integrity of DIG-fH, this probe was not subjected to denaturation prior to its treatment with PNGase F.
Fractionation of DltA.
OMs from H. ducreyi (grown in 1.5 liter of Columbia broth [Difco, Sparks, Md.] supplemented with 1% GGC and 1.5% FBS) were prepared as previously described (13), with several modifications. Whole H. ducreyi cells were passed through a French press and centrifuged at 10,000 × g at 4°C. The supernatant was further centrifuged at 100,000 × g for 1 h at 4°C. The pellet was rinsed three times with 10 mM HEPES and then solubilized in 1% N-lauroylsarcosine (Sarkosyl) in 10 mM HEPES (pH 8.0) for 30 min at 37°C. The suspension was then subjected to another 1 h of centrifugation at 100,000 × g. The SI pellet was resuspended in sterile PBS and stored at −20°C. Protein content was determined by using the BCA protein assay kit (Pierce), and 10 μg of each fraction was subjected to SDS-PAGE and Western blotting.
Protease cleavage of surface proteins of H. ducreyi.
Liquid media were inoculated with 2.4 × 106 CFU of H. ducreyi strains FX517 and FX533/ml. After 9 h of growth, the bacterial cells were harvested, washed three times in PBS plus Ca/Mg, and resuspended to an OD600 of 0.4 (ca. 2 × 108 CFU/ml). A 1-ml sample of the bacterial suspension from both strains was subjected to centrifugation, and the pellet was suspended in Laemmli sample buffer and labeled as “time zero.” To a 2.5-ml sample of bacterial suspension of both strains was added 12.5 μl of chymotrypsin at 1 mg/ml (final concentration, 5 μg/ml) or 5 μl of trypsin at 10 mg/ml (final concentration, 20 μg/ml). The suspensions were incubated at 35°C for 2 min and then centrifuged, and the pellets were resuspended in Laemmli buffer. The samples were subjected to SDS-PAGE and Western blotting with anti-rDltA, anti-rDsrA, anti-Hlp and anti-rHgbA antibodies. For simplicity sake, we only show protease cleavage of strain FX517 (positive control for cleavage) because we have evidence that DsrA shields some antigens on the surface of H. ducreyi (data not shown). Thus, DltA would more accessible to protease activity in the absence of DsrA. Protease cleavage in strain 35000HP is identical to that of FX517 (data not shown).
Surface iodination of whole H. ducreyi.
H. ducreyi strains 35000HP, FX517, FX533, and FX534 at an OD600 of 1.0 (ca. 5 × 108 CFU) were prepared. The cells were washed once and resuspended in PBS plus Ca/Mg. First, 1 mCi of [125I]Na was preactivated in Iodo-Gen tubes (Pierce), and then 0.25 mCi was added to each of the bacterial suspensions. The iodination process was allowed to proceed for 2 min to prevent lysis of bacteria and iodination of cytoplasmic proteins. The iodination reaction was quenched by adding cold GCB. The cells were washed three times with cold GCB, and the final pellet was suspended in Laemmli sample buffer plus βME. A sample containing 60,000 cpm was subjected to SDS-PAGE and Coomassie blue staining. The gel was dried before overnight exposure to −70°C with intensifying screens.
Bactericidal assay.
The resistance of H. ducreyi to NHS was determined as previously described (16). Briefly, bacteria (200 CFU) and NHS, the source of complement (5, 10, or 50%), were incubated for 45 min at 34.5°C in 5% CO2. Aliquots were plated onto CAP, and colonies were counted after incubation at 34.5°C in 5% CO2 for 48 h.
Analysis of LOS.
Whole cells from H. ducreyi (ca. 2 × 108 CFU) were suspended in Laemmli sample buffer (10% glycerol, 5% βME, SDS, 0.0025% bromophenol blue) and incubated with proteinase K (50 μg/ml final concentration) for 1 h at 56°C (27), boiled for 1 min, and stored at 4°C. Approximately 5 μl of this preparation was subjected to SDS-PAGE and silver stained according to the method of Tsai and Frasch (76) or subjected to Western blotting with monoclonal antibody 3E6 that reacts with purified LOS from H. ducreyi strain 35000HP (22, 69).
Statistical analysis.
Data from the bactericidal assays were compared with a paired t test by using the Excel (Microsoft) software. A P value of ≤0.05 was accepted as the level of statistical significance.
Nucleotide sequence.
The dltA gene corresponds to ORF Hd0746 of the H. ducreyi genome sequence (www.stdgen.lanl.gov/). The sequence of the H. ducreyi genome was released under GenBank accession no. AE017143.
RESULTS
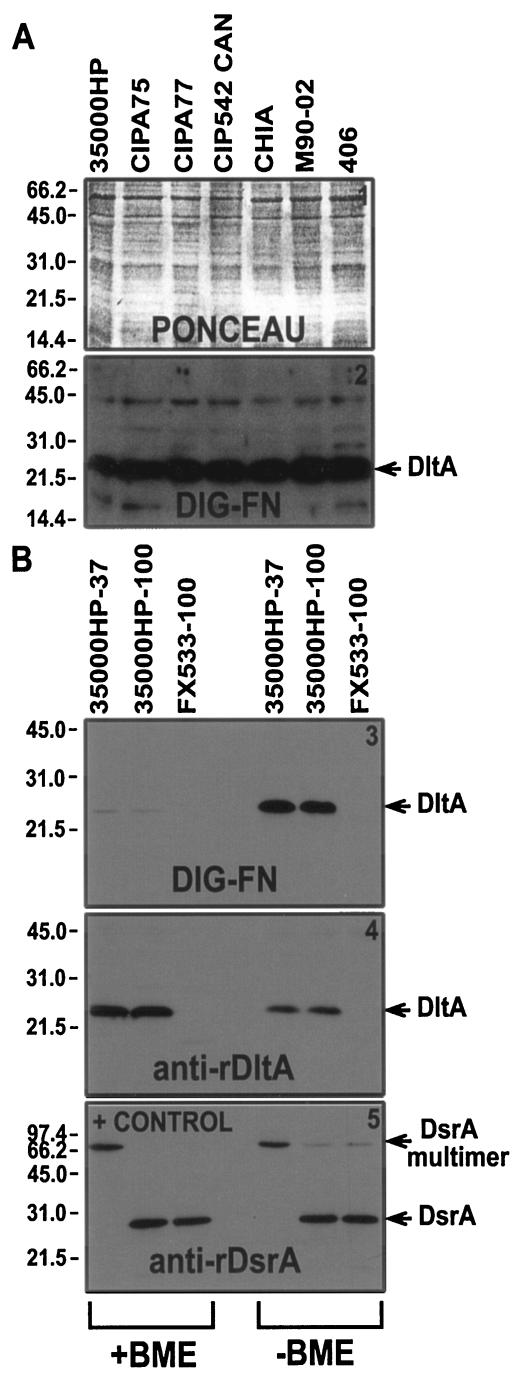
DltA is a 22-kDa H. ducreyi protein that binds FN. We have previously shown that DsrA binds keratinocytes and the extracellular matrix (ECM) protein vitronectin (8). In order to better understand ECM protein binding by H. ducreyi, we developed a ligand blot assay using DIG-labeled ECM proteins, including FN. We observed a 22-kDa low-abundance (by Coomassie blue staining) H. ducreyi protein that bound FN and that was expressed by all seven H. ducreyi strains tested (Fig. 1A and Table 2). Based on the functional characterization described below, this protein was named DltA (for ducreyi lectin A).
FIG. 1.
Identification of a novel 22-kDa H. ducreyi protein that binds FN. (A) Solubilized proteins from H. ducreyi (Table 2) (ca. 107 CFU/lane) were subjected to SDS-PAGE (reducing conditions) and ligand blotting with DIG-FN as a primary probe and anti-DIG-AP as a secondary probe and developed by using chemiluminescence. Panel 1 of this figure shows the Ponceau S stain of the membrane prior to the ligand blot shown on panel 2. (B) Proteins from H. ducreyi (∼107 CFU/lane) were solubilized at either 37 or 100°C, subjected to SDS-PAGE in reducing (+BME [left side]) and nonreducing conditions (−BME [right side]) and Western-ligand blotting with DIG-FN (panel 3), anti-rDltA (panel 4), or anti-rDsrA (panel 5) as primary probes and anti-DIG-AP or anti-rabbit-AP as secondary probes. Blots were developed by using chemiluminescence.
TABLE 2.
List of bacterial strains (Fig. 1)
| Strain | Other name | Site of isolation | Date of isolation (yr) | Source and reference |
|---|---|---|---|---|
| CIPA75 | HMC32 | Pasteur Institute | 1952 | P. Totten (11) |
| CIPA77 | HMC104 | Pasteur Institute | 1952 | P. Totten (11) |
| CIP542 Canada | France | 1954 | W. Albritton | |
| CHIA | HMC1 | CDCa | 1953 | P. Totten (11) |
| M90-02 | HMC48 | Bahamas | 1995 | P. Totten (11) |
| 406 | HMC49 | Jackson, Miss. | 1995 | P. Totten (11) |
CDC, Centers for Disease Control and Prevention.
To determine whether DltA could form multimers such as DsrA and whether it contained intradisulfide bonds, we incubated whole cells of H. ducreyi strain 35000HP at 37 and 100°C in nonreducing (−βME) and reducing (+βME) conditions. DIG-FN bound much better to DltA in nonreducing conditions (Fig. 1B3). Furthermore, DltA does not appear to form multimers (Fig. 1B4), as is the case for DsrA (Fig. 1B5).
DNA sequence and deduced amino acid sequence of the H. ducreyi 22-kDa FN-binding DltA protein from strain 35000HP.
DltA was subjected to trypsin digestion and MS as described in Materials and Methods. The protein sequence data were used to search the H. ducreyi genome and matched to a 540-bp ORF (H. ducreyi genome Hd0746). The G+C content of this ORF (42%) was comparable to the overall G+C content of this organism (38 to 39 mol%) (1). There were no obvious −35 (TGATAA) or −10 (TATATT) E. coli consensus sequences identified upstream of this ORF. The gene upstream of dltA, Hd0747 (nhaA), is in the same orientation as dltA and encodes a putative Na+/H+ antiporter. Downstream of the dltA ORF and in the opposite orientation is Hd0745, a gene encoding a protein of unknown function.
The nucleotide sequence of the dltA ORF predicts an immature lipoprotein of 21,016 Da containing 179 amino acids (Fig. 2). Cleavage of DltA at its putative signal peptidase II cleavage site would yield a mature protein of 164 amino acids. With the addition of a lipid component, this would be consistent with its electrophoretic mobility at 22 kDa, determined by ligand blotting (Fig. 1). DltA contains seven cysteines (Fig. 2), and the mature protein is predicted to have a pI of 8.30 (DNA STRIDER).
FIG. 2.
Predicted amino acid sequence of DltA and alignment of DltA with the ricin β chain. The putative signal peptidase II site, LSGC, is underlined. The arrow indicates the possible cleavage site. DltA contains seven cysteines shown in reverse highlight. The QXW “lectin repeats” patterns are indicated in boldface. Only the first module of the ricin β chain is shown. Identity of amino acid is shown by a dark gray highlight, and a light gray highlight indicates similarity.
A BLASTP search performed on the deduced amino sequence of full-length DltA revealed no significant similarities to any known protein. However, sections of DltA had similarity to various domains of a lipoprotein from Salmonella species, to xylanases and xylan-binding domains of proteins from Streptomyces species, and to parts of an N-acetylgalactosaminyltransferase from Homo sapiens (data not shown). In addition, putative conserved domains were found between DltA and the β chain of ricin, a well-studied R-type lectin, suggesting that DltA might be a lectin (Fig. 2). DltA shares three imperfect QRW lectin repeats with the ricin β chain, and the relative positions of 6 cysteines are conserved in both proteins (Fig. 2). Taken together, these results suggested that the FN-binding protein DltA identified above might be a lectin.
Globomycin treatment of H. ducreyi.
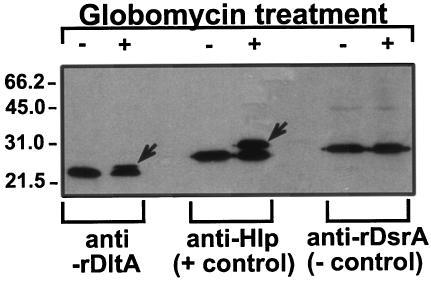
The data presented above suggest that DltA is a lipoprotein. To confirm this, we submitted the signal sequence of DltA to DOLOP (the Database of Bacterial Lipoproteins [http://www.mrc-lmb.cam.ac.uk/genomes/dolop/]), a website that predicts if a leader sequence is that of a lipoprotein. It revealed that DltA was a probable lipoprotein and that its lipobox was the sequence LSGC (underlined in Fig. 2). To further clarify this issue, whole cells of H. ducreyi were treated with globomycin, a known inhibitor of signal peptidase II, and subjected to SDS-PAGE and Western blotting with anti-rDtlA. Globomycin-treated H. ducreyi had an extra band above DltA that corresponded to the unprocessed precursor containing the signal sequence (Fig. 3). This phenomenon was also seen in Hlp, a surface-exposed lipoprotein of H. ducreyi, and was absent in DsrA, which is not a lipoprotein (Fig. 3). We therefore concluded that DltA is a lipoprotein.
FIG. 3.
Globomycin treatment of H. ducreyi. H. ducreyi cells were allowed to grow for 4 h before the addition of globomycin and DMSO. Four hours later, the cells were pelleted, solubilized in Laemmli sample buffer, and subjected to SDS-PAGE in reducing conditions (107 CFU/lane) and Western blotting with anti-rDltA, anti-Hlp, and anti-rDsrA antibodies. Lanes: +, addition of globomycin-DMSO in the culture; −, addition of DMSO to the culture. Note the additional band in globomycin-treated culture for DltA and Hlp.
Construction and characterization of H. ducreyi dltA mutant and dltA dsrA double mutant.
Mutants FX533 and FX534 lacking DltA were constructed in 35000HP wild-type and FX517dsrA backgrounds, respectively. Mutagenesis of the dltA locus in both strains was confirmed by PCR and Western blot analyses (data not shown). In PCR, products of ∼800 bp were amplified from the parent strains by using primers DLTA.01 and DLTA.02, a finding consistent with the DNA sequence. In contrast, 2-kb products were amplified from dltA mutant templates, a finding consistent with the insertion of the Kan cassette within the dltA locus.
In Western and FN-ligand blot analyses, both FX533 (35000HPdltA) and FX534 (FX517dsrAdltA) failed to express DltA (Fig. 4). DltA antibody binding and FN binding was restored in both mutants when DltA was expressed in trans from pUNCH1286 (Fig. 4) but not from a vector control. Furthermore, introduction of pUNCH1286dltA into the heterologous host H. influenzae strain Rd resulted in dltA expression, as indicated by DltA antibody and DIG-FN binding.
To examine the effect of the dltA mutation on the expression of other proteins, we subjected OMPs of both parents and mutants to SDS-PAGE and compared Coomassie-stained bands. The electrophoretic patterns of OMPs from strains FX533 and FX534 were indistinguishable from those of their parents (data not shown). We also examined the effect of the dltA mutation on LOS expression by SDS-PAGE and silver staining. No differences were observed. A Western blot probed with anti-LOS monoclonal antibody 3E6 (22, 69) revealed that there were no major differences in reactivity between the LOS moieties of parent and mutant strains (data not shown). There were also no differences in growth in rich broth between parent (35000HP and FX517) and mutant (FX533 and FX534) strains (data not shown).
Determination of the function of DltA.
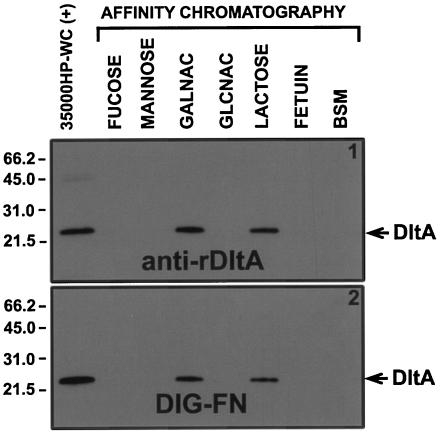
Similarities in the deduced amino acid sequence between DltA and the ricin β chain (Fig. 2 and text) suggested that DltA might be a lectin. To test this hypothesis, we examined the ability of DltA to bind to immobilized CHO and glycoproteins (Fig. 5). DltA was only recovered from the lactose and GalNAc affinity columns (Fig. 5). These data were confirmed in a similar affinity chromatography experiment with iodinated whole H. ducreyi cells and immobilized CHO. DltA was enriched, compared to other OMPs, only when mixed with immobilized lactose or GalNAc (data not shown).
FIG. 5.
DltA binding to immobilized CHO. Zwittergent 3,14-soluble total H. ducreyi proteins from ∼109 CFU were mixed with and allowed to bind to acrylamide beads containing the indicated covalently bound CHO or glycoprotein. After the unbound components were removed, the pellet was solubilized in Laemmli sample buffer and then subjected to SDS-PAGE under nonreducing conditions and Western blotting with anti-rDltA (panel 1) or DIG-FN (panel 2). Lanes: 35000HP-WC(+), Zwittergent 3,14-solubilized proteins from whole H. ducreyi cells (∼107 CFU/lane) used as positive control; GALNAC, N-acetylgalactosamine; GLCNAC, N-acetylglucosamine; BSM, bovine submaxillary mucin.
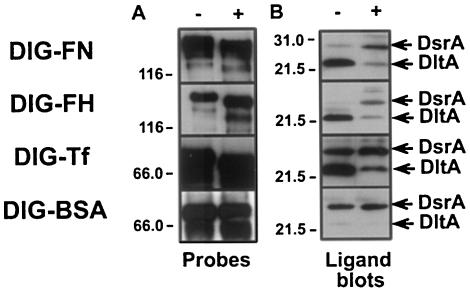
Additional evidence that DltA is a lectin was obtained from deglycosylation experiments (36). DIG-labeled glycoproteins (FN, Tf, and fH) that bound DltA in preliminary experiments were deglycosylated with PNGase F, which removes N-linked CHO by cleaving the entire glycose moiety at the first residue (36, 72). The deglycosylation state of glycoproteins was monitored by Western blots with an AP-conjugated anti-DIG antibody (Fig. 6A). Increased electrophoretic mobility of a probe was interpreted as removal of the CHO moiety (Fig. 6A). DIG-fH was efficiently deglycosylated in nondenaturing conditions (Fig. 6A), but Tf and FN required denaturing conditions to be completely deglycosylated. BSA was not affected by glycosidase treatment, as expected for a nonglycosylated protein (Fig. 6A).
FIG. 6.
Effect of PNGase F treatment of DIG-labeled glycoproteins on DltA binding. (A) Before (−) and after (+) treatment with glycosidase (PNGase F), each DIG-labeled glycoprotein was subjected to SDS-PAGE under nonreducing conditions and Western blotting with an anti-DIG probe to determine the effect of PNGase F treatment on the electrophoretic mobility of the glycoproteins. Deglycosylation was carried out under denaturing conditions for all probes, except for DIG-FH. (B) Untreated (−) and PNGase F-treated (+) DIG-labeled glycoproteins were tested for their ability to bind DltA in ligand blots of whole-cell preparations of strain 35000HP (nonreducing conditions).
Ligand blots of total H. ducreyi proteins (strain 35000HP) probed with glycosidase-treated glycoproteins are shown in Fig. 6B. Glycosidase treatment dramatically reduced the binding of DIG-FN, DIG-fH, and DIG-Tf to DltA compared to nontreated probes (Fig. 6B). DIG-BSA binding to DltA was minimally affected after treatment, since DIG-BSA bound very poorly to DltA prior to treatment (Fig. 6B). However, DIG-BSA did bind well to the very abundant protein DsrA, the protein in the 30-kDa range in Fig. 6B. Although glycosidase treatment did not affect the binding of DIG-BSA to DsrA, it did increase the binding of DIG-FN, DIG-fH, and DIG-Tf to this protein (Fig. 6B). We conclude that glycosidase treatment affected the mobility of glycoproteins by removing CHO. Furthermore, deglycosylation substantially reduced binding to DltA. These data suggest that DltA is a lectin that binds lactose-related residues.
Localization of DltA.
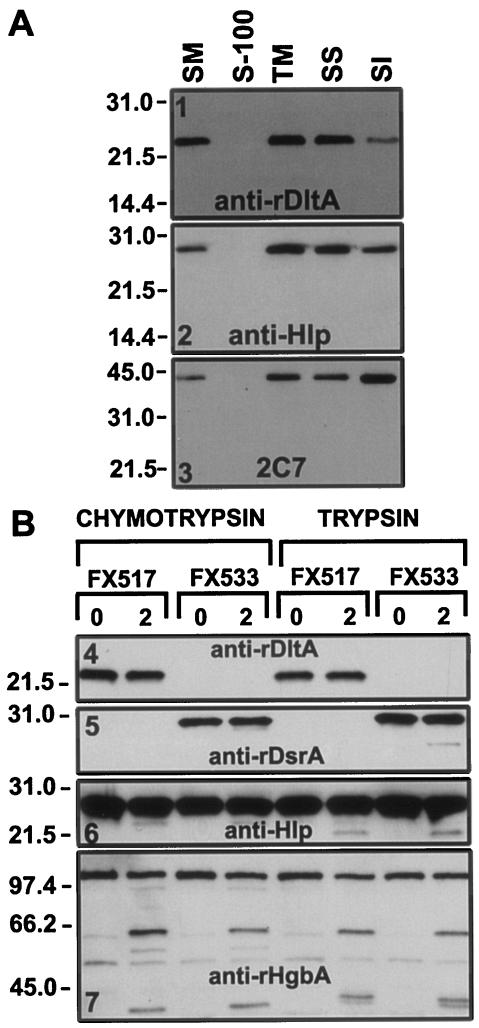
DltA appeared to be localized in the OM since it was initially isolated from an OMP preparation. Fractionation of bacterial membrane proteins can be achieved with certain detergents (18). With this technique, the more detergent-soluble inner membrane proteins generally partition to the Sarkosyl-soluble (SS) fraction, whereas most integral OMPs fractionate in the SI pellet. To examine the OM localization of DltA, cell fractionation was done. DltA fractionated with the cell envelope (Fig. 7A, TM in panel 1) and was found primarily in the SS fraction (Fig. 7A, SS in panel 1). Hlp, another H. ducreyi lipoprotein previously shown to be surface exposed (26), fractionated similarly to DltA (Fig. 7A, panel 2). As another control, we examined the fractionation of the MOMP, an integral OMP (66). MOMP was found predominantly in the SI fraction (Fig. 7A, panel 3). Although these data do not show conclusively that DltA is present in the OM, they clearly show that it is present in the cell envelope. Furthermore, these data do not exclude the possibility that DltA is surface exposed since Hlp, a known surface-exposed lipoprotein, also partially fractionated in the SS fraction (Fig. 7A, panel 2).
FIG. 7.
Cell fractionation and protease cleavage of H. ducreyi. (A) Cell fractionation. Whole cells of H. ducreyi strain 35000HP were subjected to detergent fractionation as described in the Materials and Methods. Then, 10 μg of each preparation was subjected to SDS-PAGE and Western blotting with anti-rDltA, anti-Hlp, and 2C7 antibodies as primary probes and with anti-rabbit-AP or anti-mouse-AP (2C7) as secondary probes. The blots were developed by using chemiluminescence. Lanes: SM, starting material; S-100, supernatant from a centrifugation at 100,000 × g; TM, total membranes; SS, SS supernatant; SI, SI pellet. (B) Protease cleavage. H. ducreyi cells from strains FX517 and FX533 (ca. 2 × 108 CFU/ml) were subjected to protease cleavage with either chymotrypsin (5 μg/ml) or trypsin (20 μg/ml) for 2 min at 35°C. Cells were pelleted, solubilized in Laemmli sample buffer, and subjected to SDS-PAGE (∼107 CFU/lane) in reducing conditions and Western blotting. Lanes: 0, before protease treatment; 2, after 2 min of protease treatment.
To determine whether DltA is surface exposed, whole cells of H. ducreyi were treated with trypsin and chymotrypsin (Fig. 7B). Although we were unable to detect degradation products of DltA after this treatment (Fig. 7B4), both DsrA and Hlp were also minimally affected by this procedure, whereas HgbA was readily cleaved (Fig. 7B7).
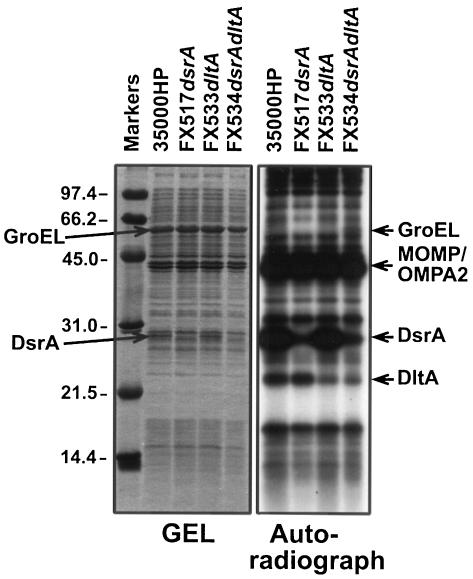
Surface exposure of a protein can also be shown by gentle iodination of intact cells, whereby only surface-exposed OMPs such as MOMP, OMPA2, DsrA, and HgbA are iodinated (13, 14). When this was performed in wild-type and in dltA dsrA single and double mutants of H. ducreyi, dltA-expressing strains showed a more intense band at 22-kDa compared to dltA mutants (Fig. 8). Intracellular contamination with 125I was minimal since GroEL, the major H. ducreyi protein in a cytoplasmic location (6, 47, 48), was not labeled. These iodination data suggest that DltA is surface exposed, although more experiments are needed.
FIG. 8.
Cell surface iodination of whole H. ducreyi cells. Whole cells of H. ducreyi (ca. 5 × 108 CFU) were surface labeled with 125I and subjected to SDS-PAGE in reducing conditions. The gel was stained with Coomassie blue stain, destained, and dried. The left panel of the figure shows the Coomassie blue-stained gel, and the right panel shows the autoradiograph.
The anti-rDltA antiserum was used in whole-cell binding assays to determine the surface exposure of DltA. However, in two experiments, there was no difference in binding between the wild type and the mutants (data not shown). These data suggest either that this antibody does not bind conformational epitopes, that the immunodominant epitopes are not exposed, or that DltA is not surface exposed. Consistent with the notion that the anti-rDltA antibody binds linear epitopes, it bound to DltA much better under reducing conditions in Western blot (Fig. 1B4). Thus, the lack of surface binding by anti-rDltA does not rule out that DltA is surface exposed.
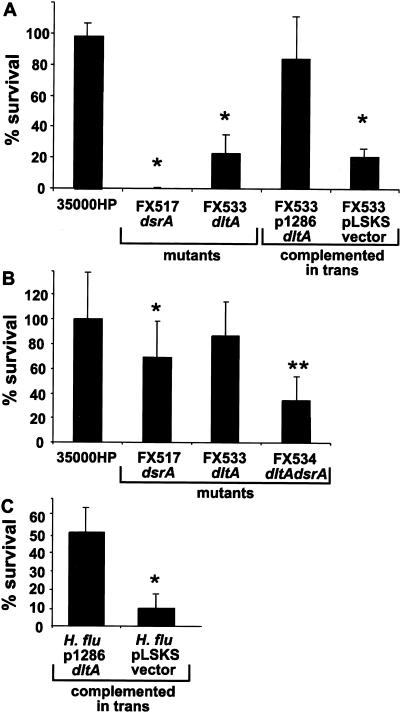
Serum susceptibility of the dltA mutants.
We tested H. ducreyi strains for sensitivity to NHS. FX533, the single dltA mutant, was significantly more susceptible to 50% NHS (23% survival) than the parent strain 35000HP (98% survival) (P = 0.001) (n = 6) (Fig. 9A). The serum resistance of FX533 was partially restored when dltA was expressed from pUNCH1286 (84% survival; P = 0.205) but not with the vector pLSKS alone (21% survival [P = 0.0001 compared to 35000HP]). In 50% NHS, strain FX534dltAdsrA was as susceptible as strain FX517dsrA (0% survival; Fig. 9A). However, in 10% NHS (Fig. 9B), the double mutant was significantly more susceptible (35% survival) than the dsrA single mutant strain (69% survival) (n = 10) (P = 0.012).
FIG. 9.
Serum susceptibility of single dltA and double dltA dsrA H. ducreyi mutants in 50% NHS (A) and 10% NHS (B) and the serum susceptibility of H. influenzae strain Rd expressing dltA in 5% NHS (C). Strains were mixed with either heated NHS (ΔNHS) or fresh (fNHS) for 45 min at 35°C, and viable counts were determined by plating. The data are expressed as percent survival in fNHS compared to survival in ΔNHS [i.e., (the number of CFU in fNHS/the number of CFU in ΔNHS) × 100]. The data are compiled from separate experiments performed on at least three different days. The asterisk denotes statistically different survival rates between the indicated strain and the parent strain (35000HP for panels A and B and H. influenzae strain Rd complemented with pLSKS for panel C). The double asterisk denotes statistically different survival rates between FX534 and FX517 (in panel B).
When dltA was expressed in H. influenzae strain Rd from pUNCH1286, it rendered this highly susceptible strain partially resistant to 5% NHS (51% survival) compared to the parent strain (10% survival) (n = 3) (P = 0.01). These data indicate that DltA is required for expression of a full serum resistance phenotype in H. ducreyi.
DltA expression and conservation.
We first examined the expression of DltA at different stages of growth in broth and found that there were no obvious differences in the amount of the DltA expressed (data not shown). DltA expression was also examined in a variety of temporally and geographically diverse strains (38 strains were tested). All strains studied expressed a 22-kDa protein that reacted with the polyclonal anti-rDltA generated from strain 35000HP (data not shown).
DISCUSSION
We provide here functional and structural evidence that DltA from H. ducreyi is a lectin. Functionally, we showed two lines of evidence that DltA binds CHO. First, we were able to affinity purify native DltA from Zwittergent-soluble total cellular protein from H. ducreyi by using immobilized lactose and GalNAc. Second, glycosidase treatment of glycoproteins reduced their binding to DltA in ligand blots. We also provided structural evidence that DltA is a lectin by showing that it has homology to the β chain of ricin. Both proteins contain QxW lectin repeats at the primary amino acid level, a common hallmark of R-type lectins of similarity (23). Furthermore, the number and position of cysteines are also conserved. Cysteines are important in ricin to form the α, β, and γ secondary structure domains and for binding CHO (23). Thus, we conclude that DltA is a lectin.
The ricin β chain has two lectin domains each capable of binding a lactose molecule (45, 59). DltA has a single lectin domain, and it is most similar to the first domain of ricin. Each ricin domain can be divided in three subdomains (α, β, and γ), each containing a QxW internal lectin repeat (23). We identified three subdomains in DltA (Fig. 2), although in two of the three QxW repeats, the “W” was conservatively replaced with an “F.” This substitution has also been found in BoNT hemagglutinin domains 1 and 2, mosquitocidal toxin domains 1 and 2 and in bovine GalNAc transferase (23).
Protein sequence comparison also revealed that the cysteines from the ricin β chain are conserved in DltA (Fig. 2). Cysteines in the ricin β chain and in many QxW repeat proteins form conserved disulfide bonds and are functionally important (23). By analogy to other R-type lectins that have been more intensively studied (23, 59), we speculate that cysteines 2 and 3, cysteines 4 and 5, and cysteines 6 and 7 are involved in intrapeptide disulfide bonds in DltA. Intrapeptide bonds were critical to the function of DltA because the addition of a reducing agent (βME) for a ligand blot significantly reduced DIG-FN binding to DltA (Fig. 1B). The presence of three disulfide bonds in such a small protein also suggests that DltA exhibits a small compact structure. We predict that the C-terminal two-thirds of DltA containing the lectin module is exposed to the external face of the OM, and the lipid tail, affixed to the N-terminal one-third, is associated with the OM.
In the present study, we conclude that DltA is a lectin because it binds to lactose and GalNAc in affinity chromatography experiments (Fig. 5) and to several N-glycosylated glycoproteins in ligand blots (Fig. 6). Lectins from many pathogenic organisms have been shown to be important adhesion molecules involved in the initiation of infection (62). In the case of chancroid, H. ducreyi is thought to enter the skin through microabrasions (65). However, very little is known about how H. ducreyi binds to cells and the ECM to cause disease. In the human model of chancroid, H. ducreyi has been shown to associate with fibrin, collagen, macrophages, and polymorphonuclear leukocytes (3, 65). Since these ECM proteins and eukaryotic cells are glycosylated, DltA may play a role in adhesion of H. ducreyi to components of the skin and initiation of infection. Future studies to determine the role of DltA in the pathogenesis of chancroid include testing the dltA mutant in the pig and human models of chancroid and studying the binding phenotype of this mutant in cell culture.
Several lines of evidence suggest that DltA is localized to the OM. First, DltA was originally purified and sequenced from a SI OMP preparation. Second, most serum-susceptible mutants are the result of mutation in a gene encoding OM constituents, including the previously described DsrA and MOMP of H. ducreyi (16, 25). Third, the sequence of DltA predicts its export to the OM. Indeed, type II signal sequences contain the consensus sequence LXXC (termed a lipobox), where C is the first amino acid of the mature protein and is acylated (35). The residues of the lipobox are numbered l−3X−2X−1C+1. The lipobox affords a protein containing this sequence lipid modification and localization to the membrane (46). However, it is the amino acid in position “+2” (X+2), the amino acid immediately after the acylated cysteine “C+1” contained in the lipobox that determines whether the protein is inserted in the cytoplasmic membrane or the OM (61). An aspartate residue in “X+2” position predicts retention of the protein in the cytoplasmic membrane, whereas any other amino acid in this position sends the protein to the OM (61). DltA possesses a valine residue at position “X+2” (Fig. 2), suggesting that it is translocated to the OM. This is also true of two other H. ducreyi lipoproteins, Hlp and PAL. Hlp has an aspartic acid (26) at position “X+2” and PAL, a serine (64), which is consistent with the notion of surface exposure, at least for Hlp (26). Finally, the other data that suggest that DltA may be present at the surface of the cell comes from iodination experiments shown in the present study. Indeed, we demonstrate that dltA-expressing strains show a more intense signal in the 22-kDa range after surface labeling compared to dltA mutants (Fig. 8). Overall, these data are consistent with the presence of DltA in the OM and suggest that DltA is surface exposed.
Some data imply that DltA may not be surface accessible. FN and fH that bound to DltA in a ligand blot did not bind DltA expressed in the membrane of whole H. ducreyi cells (data not shown). Although the glycoproteins used here bind DltA in a ligand blot, there is no evidence that they are the natural ligands. In addition, trypsin and chymotrypsin treatment of intact H. ducreyi cells did not result in DltA cleavage. Conceivably, the small compact structure proposed for DltA may account for the lack of surface accessibility. Conversely, DltA may be resistant to protease cleavage, which may to be the case for DsrA and Hlp (Fig. 7B). More experiments are needed to prove that DltA is surface exposed.
Some bacterial heme acquisition systems, such as in Neisseria spp., contain lipoproteins as part of the receptor complex required for hemoglobin utilization (60). Apparently, DltA does not function as part of the hemoglobin utilization system of H. ducreyi since the dltA mutant (FX533) grew as well as the parent strain 35000HP on medium containing hemoglobin as the sole source of heme (data not shown).
An H. ducreyi dltA mutant exhibited a moderate degree of serum susceptibility. However, the profound susceptibility of the double dltA dsrA mutant indicates that these mutations are additive, if not synergistic. The finding of a second locus, dltA, important for the expression of serum resistance is not surprising, given the redundant nature of many bacterial systems for essential properties such as iron acquisition (60) and serum resistance (54, 57). In earlier studies, we found that DsrA was an important determinant for expression of serum resistance. Along with the previously described momp (ompA1) locus (25), this brings to three the number of loci important for expression of complete serum resistance phenotype in H. ducreyi.
The H. ducreyi genome is relatively small (28) (www.ncbi.nlm.nih.gov/genomes/chrom.cgi?gi = 309&db = G and http://www.stdgen.lanl.gov/), and one way of utilizing this limited genetic information is to evolve or acquire proteins with multiple functions. DsrA is multifunctional in that it mediates serum resistance (16) and attachment to keratinocytes and vitronectin (8) and is required for virulence in the experimental model of human chancroid infection (4). We do not know the natural ligands for DltA, but it is possible there are several, given the widespread nature of glycosylation. In ligand blots, where the surrounding membrane and other OMPs do not shield DltA, it binds a number of glycoproteins. The relative inaccessibility of DltA in the context of intact cells may confer specificity for its natural ligand(s). DltA may also influence serum susceptibility by interfering with the complement cascade, perhaps by binding a negative regulator of C′. An alternative or additional function is suggested by the similarity of DltA to the ricin β chain; DltA may mediate binding to host cell membrane glycoproteins. Studies are now under way to answer these important questions.
Acknowledgments
We thank Annice Rountree and Bonnie Olsen for expert technical assistance. We are grateful to Patricia Totten, Josef Bogaerts, and William Albritton for H. ducreyi strains; Stanley Spinola for monoclonal antibodies 5C9 and 2C7; and Kenneth Ingram for FN. We thank Robert S. Munson for providing sequence of H. ducreyi strain 35000, Chris Thomas for help with the figures, and Marcia Hobbs, Fred Sparling, Gour Biswas, and Galyna Afonina for careful review of the manuscript.
The study presented was supported by grant AI31496 to C.E.
Editor: D. L. Burns
REFERENCES
- 1.Albritton, W. L. 1989. Biology of Haemophilus ducreyi. Microbiol. Rev. 53:377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alitalo, A., T. Meri, H. Lankinen, I. Seppala, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, M. E., M. P. Goheen, C. A. Townsend, and S. M. Spinola. 2001. Haemophilus ducreyi associates with phagocytes, collagen, and fibrin and remains extracellular throughout infection of human volunteers. Infect. Immun. 69:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bong, C. T., R. E. Throm, K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2001. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect. Immun. 69:1488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bozue, J. A., L. Tarantino, and R. S. Munson, Jr. 1998. Facile construction of mutations in Haemophilus ducreyi using lacZ as a counter-selectable marker. FEMS Microbiol. Lett. 164:269-273. [DOI] [PubMed] [Google Scholar]
- 6.Brown, T. J., J. Jardine, and C. A. Ison. 1993. Antibodies directed against Haemophilus ducreyi heat shock proteins. Microb. Pathog. 15:131-139. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, M. 1998. Sexually transmitted diseases enhance HIV transmission: no longer a hypothesis. Lancet 351:5-7. [DOI] [PubMed] [Google Scholar]
- 8.Cole, L. E., T. H. Kawula, K. L. Toeffer, and C. Elkins. 2002. The Haemophilus ducreyi serum resistance antigen, DsrA, binds vitronectin and confers attachment to human keratinocytes. Infect. Immun. 70:6158-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickerson, M. C., J. Johnston, T. E. Delea, A. White, and E. Andrews. 1996. The causal role for genital ulcer disease as a risk factor for transmission of human immunodeficiency virus: an application of the Bradford Hill criteria. Sex. Transm. Dis. 23:429-440. [DOI] [PubMed] [Google Scholar]
- 10.Dodd, R. B., and K. Drickamer. 2001. Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology 11:71R-9R. [DOI] [PubMed] [Google Scholar]
- 11.Dutro, S. M., G. Wood, and P. Totten. 1999. Prevalence of, antibody response to, and immunity induced by Haemophilus ducreyi hemolysin. Infect. Immun. 67:3317-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, M. S., D. L. Kasper, H. J. Jennings, C. J. Baker, and A. Nicholson-Weller. 1982. Capsular sialic acid prevents activation of the alternative complement pathway by type III, group B streptococci. J. Immunol. 128:1278-1283. [PubMed] [Google Scholar]
- 13.Elkins, C. 1995. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect. Immun. 63:1241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkins, C., K. B. Barkley, N. H. Carbonetti, A. J. Coimbre, and P. F. Sparling. 1994. Immunobiology of purified recombinant outer membrane porin protein I of Neisseria gonorrhoeae. Mol. Microbiol. 14:1059-1075. [DOI] [PubMed] [Google Scholar]
- 15.Elkins, C., N. H. Carbonetti, V. A. Varela, D. Stirewalt, D. G. Klapper, and P. F. Sparling. 1992. Antibodies to N-terminal peptides of gonococcal porin are bactericidal when gonococcal lipopolysaccharide is not sialylated. Mol. Microbiol. 5:2617-2628. [DOI] [PubMed] [Google Scholar]
- 16.Elkins, C., K. J. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkins, C., K. Yi, B. Olsen, C. Thomas, K. Thomas, and S. Morse. 2000. Development of a serological test for Haemophilus ducreyi for seroprevalence studies. J. Clin. Microbiol. 38:1520-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frisk, A., H. J. Ahmed, E. Van Dyck, and T. Lagergard. 1998. Antibodies specific to surface antigens are not effective in complement-mediated killing of Haemophilus ducreyi. Microb. Pathog. 25:67-75. [DOI] [PubMed] [Google Scholar]
- 20.Hahn, H. P. 1997. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa: a review. Gene 192:99-108. [DOI] [PubMed] [Google Scholar]
- 21.Hammond, G. W., C. J. Lian, J. C. Wilt, W. L. Albritton, and A. R. Ronald. 1978. Determination of the hemin requirement of Haemophilus ducreyi: evaluation of the porphyrin test and media used in the satellite growth test. J. Clin. Microbiol. 7:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen, E. J., S. R. Lumbley, H. Saxen, K. Kern, L. D. Cope, and J. D. Radolf. 1995. Detection of Haemophilus ducreyi lipooligosaccharide by means of an immunolimulus assay. J. Immunol. Methods 185:225-235. [DOI] [PubMed] [Google Scholar]
- 23.Hazes, B. 1996. The (QxW)3 domain: a flexible lectin scaffold. Protein Sci. 5:1490-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heffernan, E. J., L. Wu, J. Louie, S. Okamoto, J. Fierer, and D. G. Guiney. 1994. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect. Immun. 62:5183-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiltke, T. J., M. E. Bauer, J. Klesney-Tait, E. J. Hansen, R. S. Munson, Jr., and S. M. Spinola. 1999. Effect of normal and immune sera on Haemophilus ducreyi 35000HP and its isogenic MOMP and LOS mutants. Microb. Pathog. 26:93-102. [DOI] [PubMed] [Google Scholar]
- 26.Hiltke, T. J., A. A. Campagnari, and S. M. Spinola. 1996. Characterization of a novel lipoprotein expressed by Haemophilus ducreyi. Infect. Immun. 64:5047-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobbs, M. M., M. J. Leonardi, F. R. Zaretzky, T. H. Wang, and T. H. Kawula. 1996. Organization of the Haemophilus ducreyi 35000 chromosome. Microbiology 142(Pt. 9):2587-2594. [DOI] [PubMed] [Google Scholar]
- 29.Jessamine, P. G., F. A. Plummer, A. J. O. Ndinya, M. A. Wainberg, I. Wamola, L. J. D'Costa, D. W. Cameron, J. N. Simonsen, P. Plourde, and A. R. Ronald. 1990. Human immunodeficiency virus, genital ulcers and the male foreskin: synergism in HIV-1 transmission. Scand. J. Infect. Dis. Suppl. 69:181-186. [PubMed] [Google Scholar]
- 30.Joiner, K. A., K. A. Warren, C. Hammer, and M. M. Frank. 1985. Bactericidal but not nonbactericidal C5b-9 is associated with distinctive outer membrane proteins in Neisseria gonorrhoeae. J. Immunol. 134:1920-1925. [PubMed] [Google Scholar]
- 31.Kennedy, J. F., P. M. G. Palva, M. T. S. Corella, M. S. M. Cavalcanti, and L. C. B. B. Coelho. 1995. Lectins, versatile proteins of recognition: a review. Carbohydr. Polymers 26:219-230. [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lagergard, T., A. Frisk, M. Purven, and L. A. Nilsson. 1995. Serum bactericidal activity and phagocytosis in host defense against Haemophilus ducreyi. Microb. Pathol. 18:37-51. [PubMed] [Google Scholar]
- 34.Lewis, D. A. 2000. Chancroid: from clinical practice to basic science. AIDS Patient Care STDS 14:19-36. [DOI] [PubMed] [Google Scholar]
- 35.Madan Babu, M., and K. Sankaran. 2002. DOLOP: database of bacterial lipoproteins. Bioinformatics 18:641-643. [DOI] [PubMed] [Google Scholar]
- 36.Maley, F., R. B. Trimble, A. L. Tarentino, and T. H. Plummer, Jr. 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180:195-204. [DOI] [PubMed] [Google Scholar]
- 37.McDowell, J. V., J. Wolfgang, E. Tran, M. S. Metts, D. Hamilton, and R. T. Marconi. 2003. Comprehensive analysis of the factor h binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor h binding proteins. Infect. Immun. 71:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan, B. P., and C. L. Harris. 1999. Complement regulatory proteins. Academic Press, London, England.
- 39.Morse, S. A. 1989. Chancroid and Haemophilus ducreyi. Clin. Microbiol. Rev. 2:137-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Farrell, N. 2002. Genital ulcers, stigma, HIV, and STI control in sub-Saharan Africa. Sex. Transm. Infect. 78:143-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odumeru, J. A., G. M. Wiseman, and A. R. Ronald. 1987. Relationship between lipopolysaccharide composition and virulence of Haemophilus ducreyi. J. Med. Microbiol. 23:155-162. [DOI] [PubMed] [Google Scholar]
- 42.Odumeru, J. A., G. M. Wiseman, and A. R. Ronald. 1985. Role of lipopolysaccharide and complement in susceptibility of Haemophilus ducreyi to human serum. Infect. Immun. 50:495-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Odumeru, J. A., G. M. Wiseman, and A. R. Ronald. 1984. Virulence factors of Haemophilus ducreyi. Infect. Immun. 43:607-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ofek, I., E. H. Beachey, and N. Sharon. 1978. Suface sugars of animal cells as determinants of recognition in bacterial adherence. Trends Biochemical Sci. 3:159-160. [Google Scholar]
- 45.Olsnes, S., and J. V. Kozlov. 2001. Ricin. Toxicon 39:1723-1728. [DOI] [PubMed] [Google Scholar]
- 46.Oudega, B., D. Clark, F. Stegehuis, M. J. Majoor, and J. Luirink. 1993. A lipoprotein signal peptide plus a cysteine residue at the amino-terminal end of the periplasmic protein β-lactamase is sufficient for its lipid modification, processing, and membrane localization in Escherichia coli. FEMS Microbiol. Lett. 108:353-359. [DOI] [PubMed] [Google Scholar]
- 47.Parsons, L. M., R. J. Limberger, and M. Shayegani. 1997. Alterations in levels of DnaK and GroEL result in diminished survival and adherence of stressed Haemophilus ducreyi. Infect. Immun. 65:2413-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsons, L. M., A. L. Waring, and M. Shayegani. 1992. Molecular analysis of the Haemophilus ducreyi groE heat shock operon. Infect. Immun. 60:4111-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parsons, N. J., J. R. Andrade, P. V. Patel, J. A. Cole, and H. Smith. 1989. Sialylation of lipopolysaccharide and loss of absorption of bactericidal antibody during conversion of gonococci to serum resistance by cytidine 5′-monophospho-N-acetylneuraminic acid. Microb. Pathog. 7:63-72. [DOI] [PubMed] [Google Scholar]
- 50.Plummer, F. A., J. N. Simonsen, D. W. Cameron, J. O. Ndinya-Achola, J. K. Kreiss, M. N. Gakinya, P. Waiyaki, M. Cheang, P. Piot, A. R. Ronald, and E. N. Ngugi. 1991. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J. Infect. Dis. 163:233-239. [DOI] [PubMed] [Google Scholar]
- 51.Qi, H. L., J. Y. Tai, and M. S. Blake. 1994. Expression of large amounts of neisserial porin proteins in Escherichia coli and refolding of the proteins into native trimers. Infect. Immun. 62:2432-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ram, S., M. Cullinane, A. M. Blom, S. Gulati, D. P. McQuillen, R. Boden, B. G. Monks, C. O'Connell, C. Elkins, M. K. Pangburn, B. Dahlback, and P. A. Rice. 2001. C4bp binding to porin mediates stable serum resistance of Neisseria gonorrhoeae. Int. Immunopharmacol. 1:423-432. [DOI] [PubMed] [Google Scholar]
- 53.Ram, S., M. Cullinane, A. M. Blom, S. Gulati, D. P. McQuillen, B. G. Monks, C. O'Connell, R. Boden, C. Elkins, M. K. Pangburn, B. Dahlback, and P. A. Rice. 2001. Binding of C4b-binding protein to porin: a molecular mechanism of serum resistance of Neisseria gonorrhoeae. J. Exp. Med. 193:281-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ram, S., F. G. Mackinnon, S. Gulati, D. P. McQuillen, U. Vogel, M. Frosch, C. Elkins, H. K. Guttormsen, L. M. Wetzler, M. Oppermann, M. K. Pangburn, and P. A. Rice. 1999. The contrasting mechanisms of serum resistance of Neisseria gonorrhoeae and group B Neisseria meningitidis. Mol. Immunol. 36:915-928. [DOI] [PubMed] [Google Scholar]
- 55.Ram, S., D. P. McQuillen, S. Gulati, C. Elkins, M. K. Pangburn, and P. A. Rice. 1998. Binding of complement factor H to loop 5 of porin protein 1A: a molecular mechanism of serum resistance of nonsialylated Neisseria gonorrhoeae. J. Exp. Med. 188:671-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ram, S., A. K. Sharma, S. D. Simpson, S. Gulati, D. P. McQuillen, M. K. Pangburn, and P. A. Rice. 1998. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J. Exp. Med. 187:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rautemaa, R., and S. Meri. 1999. Complement-resistance mechanisms of bacteria. Microbes Infect. 1:785-794. [DOI] [PubMed] [Google Scholar]
- 58.Rice, P. A. 1989. Molecular basis for serum resistance in Neisseria gonorrhoeae. Clin. Microbiol. Rev. 2:S112-S117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rutenber, E., and J. D. Robertus. 1991. Structure of ricin B-chain at 2.5 Å resolution. Proteins 10:260-269. [DOI] [PubMed] [Google Scholar]
- 60.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 61.Seydel, A., P. Gounon, and A. P. Pugsley. 1999. Testing the “+2 rule” for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34:810-821. [DOI] [PubMed] [Google Scholar]
- 62.Sharon, N. 1996. Carbohydrate-lectin interactions in infectious disease. Adv. Exp. Med. Biol. 408:1-8. [PubMed] [Google Scholar]
- 63.Smith, A. M., C. A. Guzman, and M. J. Walker. 2001. The virulence factors of Bordetella pertussis: a matter of control. FEMS Microbiol. Rev. 25:309-333. [DOI] [PubMed] [Google Scholar]
- 64.Spinola, S., T. Hiltke, K. Fortney, and K. Shanks. 1996. The conserved 18,000-molecular-weight outer membrane protein of Haemophilus ducreyi has homology to PAL. Infect. Immun. 64:1950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spinola, S. M., M. E. Bauer, and R. S. Munson. 2002. Immunopathogenesis of Haemophilus ducreyi infection (chancroid). Infect. Immun. 70:1667-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spinola, S. M., G. E. Griffiths, K. L. Shanks, and M. S. Blake. 1993. The major outer membrane protein of Haemophilus ducreyi is a member of the OmpA family of proteins. Infect. Immun. 61:1346-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spinola, S. M., A. Orazi, J. N. Arno, K. Fortney, P. Kotylo, C. Y. Chen, A. A. Campagnari, and A. F. Hood. 1996. Haemophilus ducreyi elicits a cutaneous infiltrate of CD4 cells during experimental human infection. J. Infect. Dis. 173:394-402. [DOI] [PubMed] [Google Scholar]
- 68.Steen, R. 2001. Eradicating chancroid. Bull. W. H. O. 79:818-826. [PMC free article] [PubMed] [Google Scholar]
- 69.Stevens, M. K., L. D. Cope, J. D. Radolf, and E. J. Hansen. 1995. A system for generalized mutagenesis of Haemophilus ducreyi. Infect. Immun. 63:2976-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.St. Geme, J. W., III. 2002. Molecular and cellular determinants of non-typeable Haemophilus influenzae adherence and invasion. Cell Microbiol. 4:191-200. [DOI] [PubMed] [Google Scholar]
- 71.Takahashi, S., Y. Nagano, N. Nagano, O. Hayashi, F. Taguchi, and Y. Okuwaki. 1995. Role of C5a-ase in group B streptococcal resistance to opsonophagocytic killing. Infect. Immun. 63:4764-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tarentino, A. L., G. Quinones, A. Trumble, L. M. Changchien, B. Duceman, F. Maley, and T. H. Plummer, Jr. 1990. Molecular cloning and amino acid sequence of peptide-N4-(N-acetyl-β-d-glucosaminyl)asparagine amidase from Flavobacterium meningosepticum. J. Biol. Chem. 265:6961-6966. [PubMed] [Google Scholar]
- 73.Thomas, K., I. Leduc, B. Olsen, C. Thomas, W. Cameron, and C. Elkins. 2001. Cloning, overexpression, purification, and immunobiology of an 85-kilodalton outer membrane protein from Haemophilus ducreyi. Infect. Immun. 69:4438-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Totten, P. A., and W. E. Stamm. 1994. Clear broth and plate media for the culture of Haemophilus ducreyi. J. Clin. Microbiol. 32:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trees, D. L., and S. A. Morse. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8:357-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsai, C.-M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 155:115-119. [DOI] [PubMed] [Google Scholar]
- 77.Wood, G. E., S. M. Dutro, and P. A. Totten. 1999. Target cell range of the Haemophilus ducreyi hemolysin and its involvement in invasion of human epithelial cells. Infect. Immun. 67:3740-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wullt, B., G. Bergsten, M. Samuelsson, and C. Svanborg. 2002. The role of P fimbriae for Escherichia coli establishment and mucosal inflammation in the human urinary tract. Int. J. Antimicrob. Agents 19:522-538. [DOI] [PubMed] [Google Scholar]