Significance
Tropical rainforest hunter-gatherer populations worldwide share the pygmy phenotype, or small human body size. The evolutionary history of this phenotype is largely unknown. Here we studied DNA from the Batwa, a rainforest hunter-gatherer population from east central Africa, to identify regions of the Batwa genome that underlie the pygmy phenotype. We then performed population genomic analyses to study the evolution of these regions, including comparisons with the Baka, a west central African rainforest hunter-gatherer population. We conclude that the pygmy phenotype likely arose due to positive natural selection and that it arose possibly multiple times within Africa. These results support longstanding anthropological hypotheses that small body size confers an important selective advantage for human rainforest hunter-gatherers.
Keywords: human evolutionary ecology, human hunter-gatherers, population genomics, convergent evolution
Abstract
The evolutionary history of the human pygmy phenotype (small body size), a characteristic of African and Southeast Asian rainforest hunter-gatherers, is largely unknown. Here we use a genome-wide admixture mapping analysis to identify 16 genomic regions that are significantly associated with the pygmy phenotype in the Batwa, a rainforest hunter-gatherer population from Uganda (east central Africa). The identified genomic regions have multiple attributes that provide supporting evidence of genuine association with the pygmy phenotype, including enrichments for SNPs previously associated with stature variation in Europeans and for genes with growth hormone receptor and regulation functions. To test adaptive evolutionary hypotheses, we computed the haplotype-based integrated haplotype score (iHS) statistic and the level of population differentiation (FST) between the Batwa and their agricultural neighbors, the Bakiga, for each genomic SNP. Both |iHS| and FST values were significantly higher for SNPs within the Batwa pygmy phenotype-associated regions than the remainder of the genome, a signature of polygenic adaptation. In contrast, when we expanded our analysis to include Baka rainforest hunter-gatherers from Cameroon and Gabon (west central Africa) and Nzebi and Nzime neighboring agriculturalists, we did not observe elevated |iHS| or FST values in these genomic regions. Together, these results suggest adaptive and at least partially convergent origins of the pygmy phenotype even within Africa, supporting the hypothesis that small body size confers a selective advantage for tropical rainforest hunter-gatherers but raising questions about the antiquity of this behavior.
Small human body size, or the “pygmy” phenotype, is strongly associated with populations who have traditionally hunted and gathered for food in tropical rainforest habitats (1, 2). The phenotype appears to have evolved independently at least twice: in both Central Africa and Southeast Asia. The likely convergence has led anthropologists to hypothesize that small body size may confer direct or indirect fitness benefits in response to one or more common ecological challenges of the tropical rainforest: (i) food limitation (1, 3), (ii) high heat and humidity (4), (iii) forest structural density (5), (iv) high pathogen load (2, 6), or (v) high adult mortality (7). However, an adaptive basis for the pygmy phenotype has not been convincingly shown.
In this study, we used genome-wide SNP genotyping data and an admixture mapping design to identify genomic regions associated with the pygmy phenotype in the Batwa, a rainforest hunter-gatherer population from east central Africa. We then tested the adaptive hypothesis for the evolution of the pygmy phenotype by evaluating these regions for signatures of positive selection in the Batwa and their agriculturalist neighbors, the Bakiga. Finally, we compared SNPs from the phenotype-associated regions of the genome to those from a sample of Baka rainforest hunter-gatherers from west central Africa (Fig. 1) to develop a comprehensive model for the evolution of the pygmy phenotype in Africa.
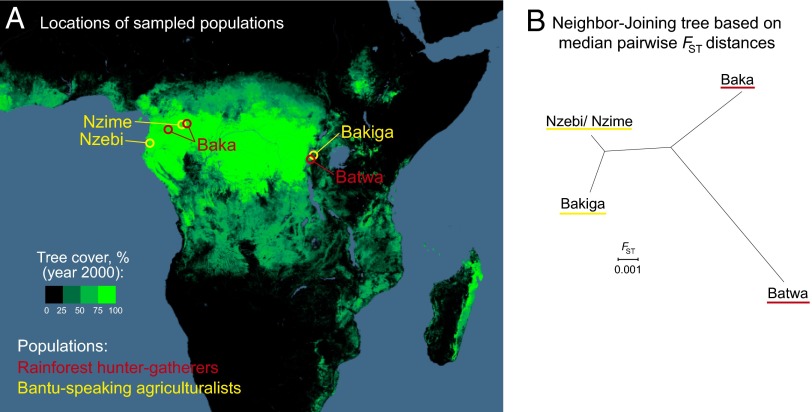
Fig. 1.
Rainforest hunter-gatherer and agriculturalist study populations. (A) Sampling locations for the east central and west central African rainforest hunter-gatherer and Bantu-speaking agriculturalist populations included in the study. The tree cover map is based on Landsat data and modified from visualization tools provided by Hansen et al. (67). (B) Neighbor-joining tree based on the median FST value for genome-wide autosomal SNPs. The Batwa and Baka population samples for this analysis excluded individuals with >10% Bakiga and Nzebi/Nzime ancestry, respectively, as estimated with ADMIXTURE (10). Pairwise median FST values: Batwa-Baka = 0.0135; Batwa-Bakiga = 0.0140; Batwa-Nzebi/Nzime = 0.0137; Baka-Bakiga = 0.0105; Baka-Nzebi/Nzime = 0.0086; Bakiga-Nzebi/Nzime = 0.0033.
Results
Batwa Stature Is Positively Correlated with Genome-Wide Proportion of Bakiga Ancestry.
We collected anthropometric data and DNA samples from 169 Batwa and 61 Bakiga adults (Fig. 1 and Dataset S1). We then used the Illumina Omni platform to genotype 1 million (1M) genome-wide SNPs for each of these individuals, of which 903,823 autosomal SNPs were included in our analyses following quality control filtering.
The Batwa hunted and gathered in Bwindi Impenetrable Forest in southwest Uganda before it became a national park in 1991, and the Bakiga were and remain their Bantu-speaking agricultural neighbors. The oral histories of both populations suggest a long-term association, including the regular trade of forest products for cultivated goods and occasional intermarriage (8, 9). Indeed, results from a nonhierarchical clustering analysis of the SNP data implemented in the program ADMIXTURE (10) (Fig. 2A) revealed variable but considerable levels of Bakiga ancestry among the Batwa individuals (mean = 14.2%; range = 0–93%). Conversely, the estimated levels of Batwa ancestry among the Bakiga were much lower (mean = 5.3%; range = 0–10.4%), consistent with previous findings (11).
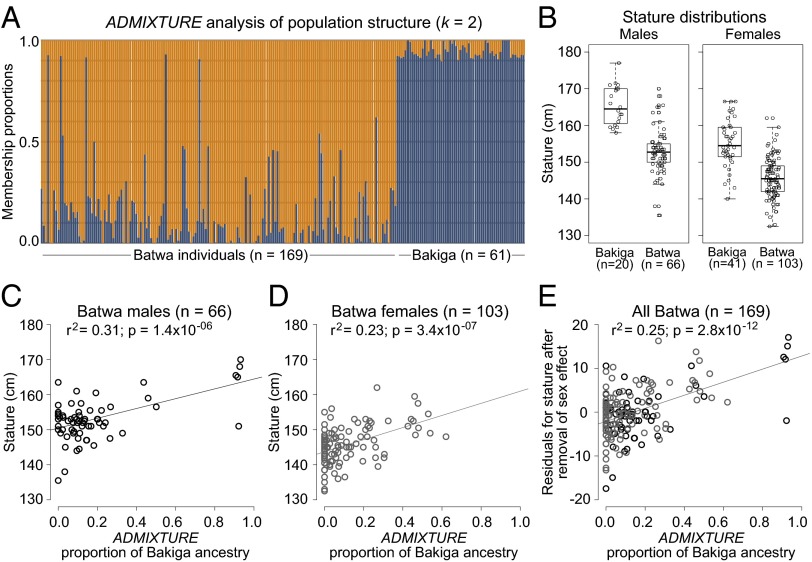
Fig. 2.
Batwa and Bakiga admixture and stature. (A) Population structure analysis based on autosomal SNPs. Each individual is represented as a vertical line, with population origins indicated below the lines. Cluster membership proportions are depicted in orange (inferred proportion of Batwa ancestry) and blue (inferred proportion of Bakiga ancestry). (B) Boxplots of Batwa and Bakiga male and female stature estimates. (C–E) Relationships between Bakiga ancestry and stature for (C) Batwa males, (D) Batwa females, and (E) all Batwa individuals after regressing out the sex effect from the stature estimate.
As expected, mean stature estimates for the Batwa (66 males, 152.9 cm; 103 females, 145.7 cm) were lower than those for the Bakiga (20 males, 165.4 cm; 41 females, 155.1 cm; Fig. 2B). Batwa stature is significantly positively correlated with the proportion of Bakiga admixture: for males, females, and for all samples combined after regressing out the sex effect (Fig. 2 C–E), confirming a genetic basis for the African pygmy phenotype (6, 12).
Admixture Mapping Analysis Identifies 16 Regions Associated with the Pygmy Phenotype.
We used HAPMIX (13) to estimate the number of chromosomes with Batwa and Bakiga ancestry for each autosomal SNP, for all Batwa individuals. The average ancestry proportions (across all SNPs) based on our final HAPMIX data were strongly correlated with those from the ADMIXTURE analysis (r = 0.98; Fig. S1 and Dataset S1). We obtained similar results using RFMix (14), an alternative method for local ancestry inference (Methods and Fig. S1).
We next used a linear model to perform an admixture mapping analysis, in which we identified regions of the genome for which Batwa stature (after accounting for sex and the genome-wide level of Bakiga ancestry) was significantly correlated with the level of local Bakiga ancestry (i.e., the HAPMIX estimated number of Bakiga chromosomes for a given SNP position). Although our sample size (n = 169) is far smaller than those of previous genome-wide association studies (GWAS) for stature (15–19), relatively fewer, larger effect loci may underlie the Batwa pygmy phenotype compared with that for heritable stature variation within the prior GWAS populations, which would translate to substantially more power to identify associated loci with a given sample size. Specifically, if small body size was subject to positive natural selection in rainforest hunter-gatherers, then larger-effect, short-stature alleles would more likely have been maintained and increased in frequency in the Batwa, relative to populations in which stature may have been less affected by natural selection. The substantial difference between the Batwa and Bakiga mean stature estimates (12.5 cm for males and 9.4 cm for females; Fig. 2B) also underscores the potential power of an admixture mapping approach.
Using the admixture mapping method, we identified 18 regions associated with the pygmy phenotype that passed the genome-wide level of significance at a false discovery rate (FDR) (20) of 0.25 (Fig. 3A and Dataset S2). Most of these regions were supported by a second association method, implemented in the program GEMMA (21), that specifically accounts for interindividual relatedness. We ranked the GEMMA P values for each genome-wide SNP from lowest to highest and found that, for 16 of our 18 admixture mapping-identified regions, at least one SNP had a GEMMA percentile rank >97% (Dataset S2). The remaining two regions (percentile ranks >90% and >92%) were excluded from most subsequent analyses.
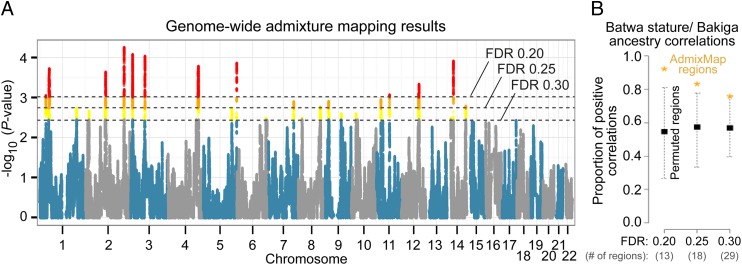
Fig. 3.
Batwa pygmy phenotype-associated genomic regions. (A) Manhattan plot of results for admixture mapping analysis of Batwa stature. SNPs passing the genome-wide level of significance at FDR = 0.30 (29 genomic regions), FDR = 0.25 (18 regions), and FDR 0.20 (13 regions) are highlighted in yellow, orange, and red, respectively. (B) Proportion of positive correlations between Batwa stature (corrected for genome-wide Bakiga ancestry) and local level of Bakiga ancestry for pygmy phenotype-associated regions identified by the admixture mapping analysis at the three FDR cutoffs. Observed results are indicated by orange asterisks; the proportions of positive correlations are 0.92, 0.83, and 0.76 for FDR = 0.20, 0.25, and 0.30, respectively. Black squares and dashed lines indicate mean and 95% CI results, respectively, from a permutation analysis in which admixture mapping was performed 100 times after permuting the stature estimates among individuals. Genomic regions significantly associated with stature in each permutation (always fewer than observed in the original data analysis) were stored and ultimately pooled across all of the permutations (separately for each of the three FDR cutoffs). From these pools, we drew 100 random samples of significant regions for each cutoff to match the observed number of significant regions from the original analysis and computed the proportion of regions from each random sample with positive stature–ancestry correlations. The proportion of observed regions with positive stature–ancestry correlations is significantly greater than expected based on the permutation results for the two most stringent FDR cutoffs (P < 0.01, P = 0.01, and P = 0.07 for FDR = 0.20, 0.25, and 0.30, respectively).
In addition to the admixture mapping signal, these regions have three other attributes that provide supporting evidence of genuine association with the pygmy phenotype. First, by chance, we would expect to observe positive correlations between Batwa stature (ancestry and sex corrected) and the local level of Bakiga ancestry for ∼50% of associated genomic regions, because we tested for both positive and negative associations. However, these values would be positively correlated for regions truly associated with the pygmy phenotype because we expect taller individuals (after genome-wide ancestry and sex correction) to have relatively higher levels of local Bakiga ancestry. Indeed, we observed positive correlations for 15 of the 18 regions (83%) detected by our admixture mapping approach at an FDR = 0.25, a significantly higher proportion than expected based on a permutation analysis (mean = 57%; P = 0.01), 14 of the 16 regions (88%) supported by the GEMMA analysis, and a pattern of greater positive correlation enrichment among more strongly associated regions (Fig. 3B).
Second, 4 of the 16 GEMMA-supported regions (25%) overlapped at least one SNP previously associated with stature variation in populations of European descent (n = 433 GWAS SNPs; Dataset S2; www.genome.gov/gwastudies/), whereas only an average of 1.34 regions were expected to overlap stature GWAS SNPs by chance alone (permutation analysis, P = 0.031; Fig. S2). The same variants are not necessarily involved in both European stature variation and the pygmy phenotype, but this result demonstrates the stature and growth functional potential of the identified regions. In contrast, similar enrichments for the 16 regions were not observed with SNPs associated with other highly polygenic phenotypes in Europeans, for example, when considering the GWAS SNP sets for IgG glycosylation (n = 699; P = 0.89), Crohn’s disease (n = 210; P = 1.0), type 2 diabetes (n = 207; P = 0.44), and multiple sclerosis (n = 175; P = 1.0; Fig. S2). Third, 4 of the 16 regions (25%), including 3 regions that also overlapped stature GWAS SNPs, overlapped one or more genes with growth hormone receptor signaling or regulation of multicellular growth Gene Ontology (GO) (22) functions, which is significantly more than expected by chance (P = 0.003; Fig. S2 and Dataset S2).
Signature of Polygenic Adaptation for Pygmy Phenotype-Associated Genomic Regions.
We computed three statistics to identify potential signatures of natural selection from the Batwa and Bakiga 1M SNP data, after restricting the Batwa dataset to the 95 individuals with ≤10% overall Bakiga ancestry: (i) BayeScan q value estimates to identify SNPs with levels of Batwa-Bakiga population differentiation that exceed that expected under neutrality (23–25), (ii) standard FST-based estimates of the level of Batwa-Bakiga population differentiation for each autosomal SNP (26), and (iii) integrated haplotype score (iHS), a widely used haplotype-based method (27).
We first used a sliding window approach to identify 105 Batwa and 180 Bakiga genomic regions with average absolute iHS values that exceeded the 99th percentile of genomic windows, and 56 genomic regions significantly differentiated between Batwa and Bakiga as identified by BayeScan (Dataset S3 and Fig. S3). We then used Genetrail (28) to identify overrepresented Gene Ontology functions or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways among the genes that overlap these regions (Dataset S3). Only for the Batwa |iHS| windows did we observe any enrichment for genes with an obvious functional category connection to body size variation (GO:0005026; TGFβ receptor activity; P = 0.0006). However, this result was only marginally significant after accounting for multiple tests (FDR = 0.089). The most significant gene functional enrichments for the Batwa-Bakiga BayeScan highly differentiated regions were related to olfactory perception and taste transduction, similar to previous reports for other African rainforest hunter-gatherer populations (29).
We next compared our results from the admixture mapping analysis to the BayeScan and iHS data. None of the 16 regions overlapped any of the 56 Batwa-Bakiga BayeScan candidate regions. Only 1 of the 16 (6.3%) pygmy phenotype-associated regions that was also supported by the GEMMA analysis overlapped any of the 105 Batwa iHS candidate regions, which is consistent with chance expectations (permutation analysis, P = 0.50). Thus, these results provide no indication that genomic regions underlying the pygmy phenotype have been subject to the type of recent, strong positive selection that is best identified by these extreme outlier approaches (i.e., a selective sweep).
However, when we simply considered the FST and iHS values for each individual SNP, rather than considering only the most extreme outlying values, we found that Batwa-Bakiga FST values for SNPs within the 16 pygmy phenotype-associated regions were significantly higher than those from the remainder of the genome (one-tailed Wilcoxon test, P = 1.26 × 10−11; Fig. 4A; permutation analysis, P = 0.031; Fig. S2). Similarly, Batwa |iHS| values were also significantly higher for SNPs within the 16 regions vs. outside them (Wilcoxon test, P = 1.14 × 10−23; Fig. 4A; permutation analysis, P = 0.062; Fig. S2). Simulations and further analysis of the genome-wide FST and |iHS| relationship show that concomitant FST and |iHS| increases to the extent observed in our data are unexpected under neutrality (Figs. S4 and S5). Therefore, the FST and iHS results combine to strongly support a history of positive selection for alleles contained within the 16 pygmy phenotype-associated regions.
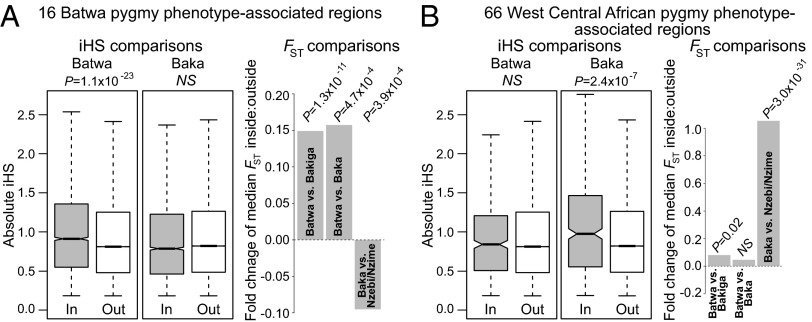
Fig. 4.
Cross-population analysis of selection signatures for pygmy phenotype-associated genomic regions. (A) Boxplot and fold-change comparisons of |iHS| and median FST values for SNPs located inside (In) vs. outside (Out) the 16 Batwa pygmy phenotype-associated regions identified by admixture mapping and supported by GEMMA (21). P values were computed with Wilcoxon tests. Fold-change = (median In − median Out)/median In. (B) Similar comparisons of |iHS| and median FST values for SNPs located inside versus outside 66 genomic regions previously reported to be associated with stature variation among west central African rainforest hunter-gatherers (6, 29). The Batwa are east central African rainforest hunter-gatherers and the Baka are west central African rainforest hunter-gatherers. The Bakiga and Nzebi/Nzime, respectively, are their Bantu-speaking agricultural neighbors.
Collectively, these results are consistent with a potential pattern of polygenic adaptation (30–33) on the pygmy phenotype, involving relatively small frequency shifts across multiple phenotype-associated alleles in aggregate rather than major selective sweeps on one or a few major-effect loci. We note that we use polygenic to reference multiple regions of the genome generally (30) rather than necessarily implicate the involvement of any gene coding changes in the phenotype.
Batwa-Baka Comparison Suggests Pygmy Phenotype Convergence Within Africa.
The pygmy phenotype is a characteristic of both east central and west central African rainforest hunter-gatherers, two groups that diverged ∼20–30 kya (34–37). It is unknown whether the pygmy phenotype evolved (i) once in Africa, in a common ancestor of east central and west central rainforest hunter-gatherers or (ii) multiple times independently within Africa, as proposed by Bahuchet based on the inference of Pleistocene rainforest refugia from paleoclimatological data (38).
To test this hypothesis, we expanded our analysis to include 1M SNP genotype data from 74 Baka rainforest hunter-gatherers from west central Africa (Gabon and Cameroon) and 73 Nzebi and Nzime, the Bantu-speaking agriculturalist neighbors of the Baka (data originally reported in ref. 11). We computed iHS and pairwise FST values for all populations (after excluding Baka individuals with ≥10% overall Nzebi/Nzime ancestry, to match our Batwa dataset; Fig. S6). We also considered our 1M SNP genotype data against stature association results from two previous genomic studies of west central African rainforest hunter-gatherers (6, 29) and with a set of candidate height SNPs for individuals of African descent (39).
If the pygmy phenotype is ancestral to all African rainforest hunter-gatherers and maintains the same genetic basis in both the Batwa and Baka populations, then we might expect a similar pattern of relatively high Baka |iHS| values in the 16 Batwa pygmy phenotype-associated regions, as we had observed for the Batwa (Fig. 4A). However, the Baka |iHS| values were not higher for SNPs within vs. outside the 16 regions (one-tailed Wilcoxon test, P = 0.99; Fig. 4A; permutation analysis, P = 0.82; Fig. S2). The expanded FST comparisons also fail to support a single-origin, ancestral model for the African pygmy phenotype. Specifically, Batwa-Baka FST values for SNPs within the 16 regions were significantly higher than the genomic background (Wilcoxon test, P = 0.0005; Fig. 4A; permutation analysis, P = 0.04; Fig. S2), whereas we may have expected relatively lower levels of differentiation in the 16 regions under the ancestral phenotype model (i.e., if ancestral phenotype-associated alleles had been maintained at similar frequencies in the Batwa and Baka, following population divergence). Additionally, the Baka-Nzebi/Nzime FST values were lower, rather than higher, inside the 16 regions compared with the remainder of the genome (Wilcoxon test, P = 0.0004; Fig. 4A; permutation analysis, P = 0.81; Fig. S6), in the opposite direction of the Batwa-Bakiga result.
Our comparative analyses thus far have been one-sided, involving the study of pygmy phenotype-associated regions ascertained in an east central African population (the Batwa). To perform a complementary analysis, we considered data from Jarvis et al. (6) and Lachance et al. (29), who performed genotype/stature association analyses in a sample of 57 total Baka, Bakola, and Bedzan west central African rainforest hunter-gatherers and 38–39 total Tikar and Ngumba Bantu-speaking agriculturalists. They focused on candidate SNPs from genomic regions highlighted by their initial genome-wide scans for selection and reported significant genotype/stature associations for 130 SNPs from 66 genomic regions (we assigned SNPs ≤ 50kb apart to the same region; Dataset S4). These data are not equivalent to our Batwa pygmy phenotype-associated regions, which we identified without reference to signatures of selection. Additionally, their association analyses were performed on combined rainforest hunter-gatherer and agriculturalist population samples, a study design in which population structure may be difficult to effectively control. However, if the associations reported in Jarvis et al. (6) and Lachance et al. (29) are at least enriched for alleles that do underlie the west central African pygmy phenotype, then we can still examine these regions under our comparative population framework to further assess the ancestral vs. convergent models for African pygmy phenotype origins.
Unsurprisingly, given the partially iHS-based ascertainment schemes of Jarvis et al. (6) and Lachance et al. (29), we found that our Baka |iHS| values were significantly higher for SNPs located within the 66 west central Africa stature-association regions compared with the remainder of the genome (one-tailed Wilcoxon test, P = 2.4 × 10−7; Fig. 4B; permutation analysis, P = 0.01; Fig. S2). In contrast, and contrary to expectations from the ancestry pygmy phenotype hypothesis, the Batwa |iHS| values within these 66 regions were similar to those across the genome (Wilcoxon test, P = 0.41; Fig. 4B; permutation analysis, P = 0.41; Fig. S2). The median Baka-Nzebi/Nzime FST value for SNPs within the 66 regions was more than double that of the remainder of the genome (Wilcoxon test, P = 2.91 × 10−32; Fig. 4B; permutation analysis, P < 0.01; Fig. S2). Although Batwa-Bakiga FST values were also relatively high in these regions depending on the statistical test, the difference was not nearly of the same magnitude (Wilcoxon test, P = 0.016; Fig. 4B; permutation analysis, P = 0.41; Fig. S2). The higher Batwa-Bakiga FST values within these 66 regions are primarily accounted for by one large potential ancestral adaptation signal on chromosome 3 (Fig. S7) that had been previously identified by Lachance et al. (29). Excluding this single locus, the overall pattern of iHS and FST results for the Jarvis et al. (6) and Lachance et al. (29) regions is in accordance with the convergent origins model for the African pygmy phenotype (Fig. S7). In other words, regions identified as associated with the pygmy phenotype in west central and east central African rainforest hunter-gatherers are enriched for signatures of natural selection predominantly in west central African and east central African rainforest hunter-gatherers, respectively, as opposed to an ancestral signal shared by both populations.
Finally, we considered a set of 30 candidate height-associated SNPs in individuals of African descent (primarily African Americans) (39) that were also included on our 1M SNP array (Methods). We tested whether these SNPs were enriched for relatively high iHS values (absolute iHS > 2) relative to the genomic background. |iHS| values were > 2 in either the Batwa or Baka for 5 of 27 SNPs (18.5%) variable in both of these populations, more than double the proportion for SNPs across the remainder of the genome (8.3%; Fisher’s exact test, P = 0.069; Fig. S8). In contrast, similar proportions of Bakiga or Nzebi/Nzime high |iHS| values were observed for the stature associated SNPs (10.3%) and all other SNPs (7.4%; P = 0.47). Strikingly, none of the five Batwa or Baka high |iHS| values for the African stature associated SNPs were shared across populations (three were observed in the Batwa and two in the Baka). Thus, these results further support a model of polygenic adaptation and at least partial convergence for the pygmy phenotype, consistent with our cross-population analyses of the east central and west central African pygmy phenotype-associated regions.
Discussion
We can draw four primary conclusions from our analyses. (i) The African pygmy phenotype has a genetic basis, rather than a solely environmental one, based on the positive correlation between stature and Bakiga admixture for Batwa individuals raised in Batwa communities (Fig. 2 C–E). These results confirm those obtained from other African rainforest hunter-gatherer populations by Becker et al. (12) and Jarvis et al. (6) and are consistent with individual case observations from Cavalli-Sforza (4). (ii) The Batwa pygmy phenotype is likely polygenic in nature, as multiple genomic regions were highlighted by our admixture mapping analysis (Fig. 3A). (iii) The Batwa pygmy phenotype was likely adaptive in origin, based on the elevated levels of population differentiation (FST) and haplotype-based signatures of selection (iHS) for SNPs within the regions identified by our admixture mapping analysis (Fig. 4). Finally, the disparate FST and iHS patterns between the Batwa and the Baka in these regions, along with generally converse observations for the regions reportedly associated with stature in west central African rainforest hunter-gatherers (Fig. 4), suggest that (iv) the pygmy phenotype evolved convergently within Africa, at least in part. The possibility of an additional, ancestral component cannot be excluded, as we have certainly not yet identified the full genetic basis of the Batwa pygmy phenotype, and one of the genomic regions we did identify does have a potential ancestral adaptation signature (Fig. S7).
These findings advance our broader understanding of rainforest hunter-gatherer evolutionary history and ecology. First, adaptive hypotheses explaining small body size have been at least partly motivated by past assumptions that the pygmy phenotype evolved convergently among both African and Southeast Asian rainforest hunter-gatherers (1). Our study suggests that the pygmy phenotype likely evolved convergently even within Africa. This result parallels a recent inference from Migliano et al. (2) that the pygmy phenotype may have also evolved at least twice in Southeast Asia based on their conclusion that small-bodied populations from the Philippines and Papua New Guinea are each genetically related more closely to neighboring agriculturalists than they are to each other.
The new support for an expanded model of convergent evolution, along with our discovery of signatures of polygenic adaptation for genomic regions associated with the Batwa pygmy phenotype, should justify and encourage expanded hypothesis testing to understand the specific selective advantage(s) of small body size in the tropical rainforest habitat. Although our results alone cannot identify the specific ecological factor(s) linked to the evolution of the pygmy phenotype, future association and evolutionary population genetic studies of metabolism, thermoregulation, mobility, immunity, or other traits could theoretically establish the contemporaneous presence of selection pressures relevant to the various adaptive origin hypotheses.
Moreover, if our inference of within-Africa convergent evolution is correct, then this means that the pygmy phenotype evolved (at least in part) more recently than ∼20–30 kya, which is the estimated time of divergence between east central and west central African rainforest hunter-gatherers (11, 34). This insight potentially informs competing hypotheses concerning the antiquity of rainforest hunter-gatherer behavior (40, 41). Specifically, although many anthropologists envision a long-term history of tropical rainforest hunter-gatherer behavior (42–49), the severe seasonal food limitations of this habitat (50) have led others to question whether full-time human rainforest occupation is even possible without the ability to trade for agricultural food (50–53). Ancestors of the modern Batwa and Baka might have originally inhabited ecotonal rainforest edge environments that harbor more stable food resources for hunter-gatherers, before being displaced to deeper forest habitats by the farming populations with whom they now trade (50, 52). The absence of preserved skeletal remains of appreciable antiquity in the rainforest (54) has made it difficult to test this hypothesis.
Because the evolution of small body size may not necessarily have been restricted to full-time rainforest occupation, our results cannot directly resolve this debate. However, a relatively recent rather than ancient origin of the pygmy phenotype is at the least not inconsistent with the agriculture-dependency model for rainforest hunter-gatherer behavior, as agriculture originated in west central Africa within the last 5,000 y and then spread eastward and southward across sub-Saharan Africa (55). Such a scenario may be perceived as unlikely, because it would require major, convergent body size reductions over only several thousand years. However, recent nonhuman studies have shown that body size may undergo substantial evolutionary change in relatively short timeframes (56, 57). The case of rainforest hunter-gatherers could be a similar human example of this phenomenon. Combined with fine-mapping analyses to identify the specific mutations that underlie the pygmy phenotype, advances in statistical methods to precisely estimate the timing of polygenic adaptation events may let us determine whether selection for small body size alleles occurred concomitantly with the timing of the spread of agriculture across the continent. These and other studies of the evolutionary history of the pygmy phenotype would help to further our understanding of rainforest hunter-gatherer evolutionary ecology.
Methods
Population Samples.
We collected anthropometric data and DNA samples from 169 Batwa (rainforest hunter-gatherer) and 61 Bakiga (Bantu-speaking agriculturalist) adults (Dataset S1). These samples were collected during one field season in 2010. The precise ages of rainforest hunter-gatherers are not always known. To ensure that only adult Batwa participated in our study, we only sampled individuals who were born before the 1991 formation of Bwindi Impenetrable Forest National Park, a time point known well to the Batwa. The DNA samples were collected from either whole blood or saliva. DNA from whole blood was extracted using the PureGene DNA extraction kit (Gentra Systems), and DNA from saliva was extracted using the Oragene DNA sample collection and extraction kit. The Batwa samples were collected in six settlements located in the surroundings of the Bwindi Impenetrable Forest in southwest Uganda. To avoid potential confusion, we note that the Batwa east central African rainforest hunter-gatherers reside in Uganda and Rwanda. These groups are distinct from the west central African hunter-gatherer population also referred to as the Batwa, from the Lake Ntomba and Lake Ekonda region of Western Democratic Republic of Congo. Our sample of west central Africans, described recently by Patin et al. (11), included 74 Baka rainforest hunter-gatherers from Gabon and Cameron and 73 Nzebi and Nzime, the Bantu-speaking agriculturalist neighbors of the Baka. The Batwa and Bakiga individuals were collected under informed consent (Institutional Review Board protocols 2009-137 from Makerere University, Uganda, and 16986A from the University of Chicago). The project was also approved by the Uganda National Council for Science and Technology (HS617). The genetic analyses of the west central African samples were approved by the Institutional Review Boards of Institut Pasteur, France (2008-06 and 2011-54/IRB/2).
Genome-Wide Genotyping.
All samples were genotyped on the Illumina HumanOmni1-Quad genotyping array (Illumina). The 1M SNP genotype data for the Baka, Nzebi, and Nzime and for 40 Batwa and 40 Bakiga individuals were reported in Patin et al. (11). Genotypes of 928,705 SNPs were called in all samples using the Illumina Genome Studio v2010. SNPs were excluded if they had a call rate <95% across all samples or if they showed a significant deviation from Hardy–Weinberg equilibrium (P < 0.001) in any of the individual populations. In total, 903,823 autosomal SNPs passed quality-control filters. For these SNPs, the average genotype concordance rate across four pairs of duplicated samples was 99.93%.
Estimation of Genome-Wide Admixture Levels.
We calculated the levels of admixture of each of the hunter-gatherer individuals with their neighboring agriculturalist population using the unsupervised clustering algorithm ADMIXTURE (10). We ran the analyses separately for east central African (i.e., Batwa and Bakiga) and west central African populations (i.e., Baka and Nzebi/Nzime) for a range of ancestral clusters: K = 2, 3, and 4. K = 2 led to the lowest mean cross-validation error rates in both cases. Because the method implemented in ADMIXTURE does not explicitly model linkage disequilibrium (LD), we tested the extent to which LD between SNPs could affect our estimates. To do so, we ran similar analyses but after pruning SNP pairs with r2 values above predefined cutoffs ranging from 0.1 to 0.8 (by 0.1). All results were strongly correlated with r > 0.99. We therefore decided to use the results with the largest number of SNPs.
Local Ancestry Estimations.
We used HAPMIX (13) to estimate the number of chromosomes with Batwa and Bakiga ancestry for each autosomal SNP for all Batwa individuals. For optimal performance, HAPMIX requires knowledge of allele frequencies from both parental populations. In our sample, most of the Batwa individuals show some degree of Bakiga ancestry, with some individuals substantially admixed (Fig. 2). Initially, we thought that the Mbuti (another east central African rainforest hunter-gatherer population, from the Ituri forest of the Democratic Republic of Congo, with genome-wide SNP genotype data available for a small population sample of individuals) (58) could serve as a good parental population for the Batwa in the HAPMIX analysis. However, the Batwa and the Mbuti are actually significantly differentiated genetically (FST = 0.036) (11), despite their geographic proximity. Such differentiation could lead to spurious local ancestry assignments. Indeed, when using the Mbuti as a reference population in the HAPMIX analysis, the lowest proportion of Bakiga ancestry assigned to any Batwa individual was 38%, a result in clear disaccord with the ADMIXTURE results. Thus, in the absence of a true parental population for the Batwa, we performed 100 different HAPMIX analyses, each with a randomly drawn set of 20 Batwa from among the individuals with ≤10% Bakiga ancestry based on the ADMIXTURE estimate as the rainforest hunter-gatherer parental population, and computed the mean ancestry estimate for each SNP per individual. Our final estimates of the number of chromosomes with Batwa and Bakiga ancestry for each SNP were the average from the 100 analyses (the estimates for individuals with ≤10% Bakiga ancestry were based on <100 HAPMIX analyses, because those individuals were included in the parental population group in a subset of the analyses, as described above). The average ancestry proportions (across all SNPs) based on our final HAPMIX data were strongly correlated with those from the ADMIXTURE analysis (r = 0.98; Fig. S1 and Dataset S1). To further validate the accuracy of our local ancestry estimation, we used RFMix (14), an alternative method for local ancestry inference that does not require previous knowledge of the parental population (although the method still requires a learning step from a group of putatively nonadmixed individuals). Specifically, we performed 50 different RFMix analyses, each with a randomly drawn set of 20 Batwa from among the individuals with ≤10% Bakiga ancestry. RFMix can correct phasing assignments based on inferred ancestry tracts. We performed such a phasing correction before proceeding to the forward–backward inference. Afterward, we used RFMix to predict the forward–backward estimations. The expectation–maximization algorithm was iterated four times with the reference panels dropped after the first iteration. The average ancestry proportions calculated using RFMix were strongly correlated with those from the ADMIXTURE analysis (r = 0.97), as well as with those obtained using HAPMIX (r = 0.99; Fig. S1), and the local ancestry estimates (per SNP) from HAPMIX and RFMix were also strongly correlated (the mean and median Pearson correlation coefficients for all individuals were 0.8 and 0.83, respectively), demonstrating that our local ancestry estimates are largely robust to the method used.
Admixture Mapping.
We examined associations between stature and local ancestry estimates at every autosomal SNP by using a linear regression model in which height was regressed against local ancestry estimates while taking into account the sex and the genome-wide level of Bakiga ancestry of each of the 169 Batwa individuals. All regressions were performed using the R statistical package. Specifically, we used the following model:
Here, corresponds to the intercept, corresponds to the effect of local proportion of Bakiga ancestry of individual i at locus j, corresponds to the sex of individual i, represents the genome-wide average of Bakiga ancestry for individual i, and are the residual of the model that are assumed to follow a normal distribution with mean 0 and variance . The admixture mapping model above does not explicitly take into account the levels of relatedness among individuals, an inherent feature of small population groups such as the Batwa. Thus, to validate that the results from the model above were robust to individuals’ relatedness, we performed a stature association analysis with the linear mixed models approach implemented in GEMMA (21). For this analysis, we regressed out the sex effect and quantile transformed the residuals to a standard normal distribution to serve as the final phenotype. We then calculated the Balding-Nichols matrix using all autosomal SNPs to estimate the relatedness matrix (21, 59). Afterward, for each locus, we fitted a linear mixed model with both local and genome-wide average admixture estimates as fixed effects and individual relatedness as random effects and calculated the corresponding P value using the software GEMMA (21, 60). For both models, the FDR was calculated using the q value approach described by Storey et al. (20).
Permutation Analyses.
We used permutation analyses to assess the likelihood that a given number of our pygmy phenotype-associated genomic regions would overlap various GWAS SNP or functional gene datasets by chance alone. We first generated 10,000 sets of the same number of size-matched genomic regions as the actual pygmy phenotype-associated regions. To avoid centromeres and highly repetitive sequence, we masked regions of the genome with >100-kb gaps between SNPs on the 1M SNP array. Otherwise, the size-matched genomic regions for each of the 10,000 sets were randomly chosen from the autosomal genome. For each permuted set of regions, we then counted how many regions overlapped one or more of the GWAS SNPs or genes in question. The resulting distribution of the number of overlapping regions was compared with the observed number of overlaps to generate an empirical P value (Fig. S2). GWAS SNP data from height association studies conducted in European populations (15–17) were downloaded from www.genome.gov/gwastudies/ on August 1, 2013. GO functional annotations were obtained from www.geneontology.org on July 30, 2013. We used a similar permutation scheme to analyze differences between median FST and |iHS| values within pygmy phenotype-associated regions compared with the rest of the genome. Specifically, we created 1,000 sets of size-matched regions [separately for the 16 Batwa pygmy phenotype-associated regions identified by our admixture mapping approach and the 66 stature-associated regions for west central African rainforest hunter-gatherers reported by Jarvis et al. (6) and Lachance et al. (29)]. For each population and population comparison, we then determined FST and |iHS| values for SNPs within and outside each set of permuted regions. The resulting distributions of the inside-outside difference were then compared with the inside-outside differences for the actual regions to generate empirical P values (Fig. S2).
Signatures of Selection.
SNP genotypes were computationally phased using SHAPEIT2 (61), treating each chromosome independently. Following phase reconstruction, we used a modified version of the iHS statistic (62) that weighs the contribution of each individual based on its uniqueness in the entire sample. In other words, this modified iHS statistic corrects for levels of relatedness among individuals, leading to a significant increase in power to detect selective sweeps among all demographic scenarios tested (62). iHS was calculated for each population separately on all autosomal SNPs with a minor allele frequency of at least 5%. The genetic map used was that released by the HapMap Consortium (www.hapmap.org), which is averaged across the three original HapMap populations and thus is unlikely to be influenced by selection in any individual population. We determined the ancestral state of each SNP using the Ensembl EPO pipeline, which uses sequence information from five primate species to define the most likely ancestral state (63, 64). To identify genomic regions with extreme |iHS| values, we used a sliding-window approach. Specifically, we computed the average |iHS| on windows of the genome of 50 contiguous SNPs with an offset of 25 SNPs to the next window (62). The average length of each window was 203,901 and 182,659 bp for the Batwa and Bakiga, respectively, which is similar to the window sizes previously used to detect signatures of positive selection in African populations using iHS (65). In each window, we then converted the test statistic to an empirical P value based on the proportion of windows showing an average |iHS| value equal or larger to the one observed in the window being tested. We defined extreme regions as those showing P < 0.01. Extreme regions within 50 kb of each other were merged into a single region. We also used BayeScan v2.1 (23) to identify SNPs significantly more differentiated between the Batwa and the Bakiga than expected under genetic drift alone. We excluded SNPs with a global minor allele frequency below 5% and we used only Batwa individuals with ≤10% of Bakiga ancestry, as estimated with ADMIXTURE. BayeScan estimates the probability that a SNP has been under local positive selection, which is then used to calculate q values defined as the minimum FDR at which a SNP may become significant (66). We identified highly differentiated genomic regions by using a sliding windows approach. Specifically, we considered windows of 500 kb, with an offset of 25 kb to the next window. The average number of SNPs per window was 127 (SD = 69) after discarding any window containing less than 50 SNPs. We considered a window as a candidate target for selection if the 5% quantile of the q values in the window was lower than 0.1, and we merged overlapping significant windows into larger regions. Finally, for each pair of populations, we calculated Wright’s FST for each SNP (26), with Batwa and Baka population samples that only included individuals with ≤10% ancestry of their respective Bantu-speaking agriculturalist neighbors, as estimated with ADMIXTURE.
Cross-Population Analyses of Selection and Convergent Evolution.
To complement the cross-population analysis of the Batwa pygmy phenotype-associated regions, we obtained and curated a list of 130 SNPs that Jarvis et al. (6) and Lachance et al. (29) reported to be statistically associated with stature (following correction for multiple tests) in a combined sample of west central African rainforest hunter-gatherers and Bantu-speaking agriculturalists, following their identification of candidate positive selection regions. We created a list of 66 regions from this list of 130 SNPs by grouping SNPs located within 50 kb of each other, and then adding 10 kb to each end of each resulting start and end position (e.g., such that an individual SNP not located within 50 kb of another significant SNP would be represented in a region of 20,001 bp centered around the significantly associated SNP; Dataset S4). We also analyzed a set of candidate African stature SNPs reported by N’Diaye et al. (39). The authors of this study began with a database of European GWAS height SNPs, but then identified the tag SNPs for each locus that best reflected linkage disequilibrium patterns in Africans. Each of these SNPs was then studied for association with stature in 20,427 individuals of African descent (predominantly African Americans). Of the 78 of these SNPs for which they reported significant associations (P < 0.05), 30 were present on our 1M SNP array and variable in at least one of our four study populations. These 30 SNPs were used in our analysis.
Supplementary Material
Acknowledgments
We thank the Batwa community and all of the Batwa and Bakiga individuals for interest and participation; Stephen “Blackie” Gonsalves (deceased), Scott Kellerman, Philip Rosenthal, Bryan Greenhouse, Moses Kamya, and Joseph Ochieng for enthusiasm and encouragement; and the Batwa Development Program, Byaruhanga Julius, Magambo Michael, Byamugisha Patrick, Twesigomwe Sabastian, Safari Joseph, and Busingye Levi for expert assistance during the sample collection process in Uganda. We also thank Nanyunja Sarah for technical laboratory assistance. We thank Emily Davenport for comments on an earlier version of the manuscript. Funding for this study was provided by David and Lucille Packard Foundation Fellowship 2007-31754 (to N.J.D.), Penn State University College of the Liberal Arts (G.H.P.), Human Frontiers Science Program Grant CDA00025-2012 (to L.B.B.), and the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS), a CNRS Maladies Infectieuses et Environment grant, and a Foundation Simone and Cino del Duca Research grant (all to L.Q.-M.). M.F. was supported by a Swiss National Science Foundation grant (to L.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.C. is a guest editor invited by the Editorial Board.
Data deposition: The 1M SNP genotype data for the Baka, Nzebi, and Nzemi and for 40 Batwa and 40 Bakiga individuals were reported in Patin et al. (11) and are available at the European Genome-Phenome archive, www.ebi.ac.uk/ega/ (accession no. EGAS00001000605). The 1M SNP genotype data for the remaining 129 Batwa and 21 Bakiga individuals have been deposited in the European Genome-Phenome archive (accession no. EGAS00001000908). The per-SNP and per-individual HAPMIX estimates of local ancestry, the per-SNP P values and q values (false-discovery rate) from the admixture mapping and GEMMA analyses, and the per-SNP pairwise FST values and population-specific iHS values have been deposited in the Dryad Digital Repository (doi:10.5061/dryad.ms8k7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402875111/-/DCSupplemental.
References
- 1.Perry GH, Dominy NJ. Evolution of the human pygmy phenotype. Trends Ecol Evol. 2009;24(4):218–225. doi: 10.1016/j.tree.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Migliano AB, et al. Evolution of the pygmy phenotype: Evidence of positive selection fro genome-wide scans in African, Asian, and Melanesian pygmies. Hum Biol. 2013;85(1-3):251–284. doi: 10.3378/027.085.0313. [DOI] [PubMed] [Google Scholar]
- 3.López Herráez D, et al. Genetic variation and recent positive selection in worldwide human populations: Evidence from nearly 1 million SNPs. PLoS ONE. 2009;4(11):e7888. doi: 10.1371/journal.pone.0007888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalli-Sforza LL. African pygmies: An evaluation of the state of research. In: Cavalli-Sforza LL, editor. African Pygmies. Orlando, FL: Academic Press; 1986. pp. 361–426. [Google Scholar]
- 5.Diamond JM. Anthropology. Why are pygmies small? Nature. 1991;354(6349):111–112. doi: 10.1038/354111a0. [DOI] [PubMed] [Google Scholar]
- 6.Jarvis JP, et al. Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genet. 2012;8(4):e1002641. doi: 10.1371/journal.pgen.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Migliano AB, Vinicius L, Lahr MM. Life history trade-offs explain the evolution of human pygmies. Proc Natl Acad Sci USA. 2007;104(51):20216–20219. doi: 10.1073/pnas.0708024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edel MM. The Chiga of Uganda. 2nd Ed. New Brunswick, New Jersey: Transaction Publishers; 1996. [Google Scholar]
- 9.Ngologoza P. Kigezi and Its People. Kampala, Uganda: Fountain Publishers; 1998. [Google Scholar]
- 10.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patin E, et al. The impact of agricultural emergence on the genetic history of African rainforest hunter-gatherers and agriculturalists. Nat Commun. 2014;5:3163. doi: 10.1038/ncomms4163. [DOI] [PubMed] [Google Scholar]
- 12.Becker NS, et al. Indirect evidence for the genetic determination of short stature in African Pygmies. Am J Phys Anthropol. 2011;145(3):390–401. doi: 10.1002/ajpa.21512. [DOI] [PubMed] [Google Scholar]
- 13.Price AL, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5(6):e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 2013;93(2):278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lettre G, et al. Diabetes Genetics Initiative; FUSION; KORA; Prostate, Lung Colorectal and Ovarian Cancer Screening Trial; Nurses’ Health Study; SardiNIA Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40(5):584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weedon MN, et al. Diabetes Genetics Initiative; Wellcome Trust Case Control Consortium; Cambridge GEM Consortium Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40(5):575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanna S, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40(2):198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soranzo N, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5(4):e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics. 2008;180(2):977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer MC, Foll M, Excoffier L, Heckel G. Enhanced AFLP genome scans detect local adaptation in high-altitude populations of a small rodent (Microtus arvalis) Mol Ecol. 2011;20(7):1450–1462. doi: 10.1111/j.1365-294X.2011.05015.x. [DOI] [PubMed] [Google Scholar]
- 25.Foll M, Fischer MC, Heckel G, Excoffier L. Estimating population structure from AFLP amplification intensity. Mol Ecol. 2010;19(21):4638–4647. doi: 10.1111/j.1365-294X.2010.04820.x. [DOI] [PubMed] [Google Scholar]
- 26.Wright S. The genetical structure of populations. Ann Eugen. 1951;15(4):323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 27.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller A, et al. GeneTrailExpress: A web-based pipeline for the statistical evaluation of microarray experiments. BMC Bioinformatics. 2008;9:552. doi: 10.1186/1471-2105-9-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lachance J, et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150(3):457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20(4):R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard JK, Di Rienzo A. Adaptation - not by sweeps alone. Nat Rev Genet. 2010;11(10):665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu W, Akey JM. Selection and adaptation in the human genome. Annu Rev Genomics Hum Genet. 2013;14:467–489. doi: 10.1146/annurev-genom-091212-153509. [DOI] [PubMed] [Google Scholar]
- 33.Turchin MC, et al. Genetic Investigation of ANthropometric Traits (GIANT) Consortium Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat Genet. 2012;44(9):1015–1019. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patin E, et al. Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet. 2009;5(4):e1000448. doi: 10.1371/journal.pgen.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veeramah KR, et al. An early divergence of KhoeSan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal resequencing data. Mol Biol Evol. 2012;29(2):617–630. doi: 10.1093/molbev/msr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batini C, et al. Insights into the demographic history of African Pygmies from complete mitochondrial genomes. Mol Biol Evol. 2011;28(2):1099–1110. doi: 10.1093/molbev/msq294. [DOI] [PubMed] [Google Scholar]
- 37.Destro-Bisol G, et al. The analysis of variation of mtDNA hypervariable region 1 suggests that Eastern and Western Pygmies diverged before the Bantu expansion. Am Nat. 2004;163(2):212–226. doi: 10.1086/381405. [DOI] [PubMed] [Google Scholar]
- 38.Bahuchet S. History of the inhabitants of the Central African rainforest: Perspectives from comparative linguistics. In: Hladik CM, et al., editors. Tropical Forests, People and Food. Paris: UNESCO; 1993. pp. 37–54. [Google Scholar]
- 39.N’Diaye A, et al. Identification, replication, and fine-mapping of Loci associated with adult height in individuals of african ancestry. PLoS Genet. 2011;7(10):e1002298. doi: 10.1371/journal.pgen.1002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eggert MKH. The Central African rain forest: Historical speculation and archaeological facts. World Archaeol. 1992;24(1):1–24. [Google Scholar]
- 41.Cornelissen E. Human responses to changing environments in Central Africa between 40,000 and 12,000 B.P. J World Prehist. 2002;16(3):197–235. [Google Scholar]
- 42.Bahuchet S, McKey D, Garine I. Wild yams revisited: Is independence from agriculture possible for rain forest hunter-gatherers? Hum Ecol. 1991;19(2):213–243. [Google Scholar]
- 43.Brosius JP. Foraging in tropical rain forests: The case of the Penan of Sarawak, East Malaysia (Borneo) Hum Ecol. 1991;19(2):123–150. [Google Scholar]
- 44.Colinvaux PA, Bush MB. The rain-forest ecosystem as a resource for hunting and gathering. Am Anthropol. 1991;93(1):153–160. [Google Scholar]
- 45.Dwyer PD, Minnegal M. Hunting in a lowland, tropical rain forest: Towards a model of non-agricultural subsistence. Hum Ecol. 1991;19(2):187–212. [Google Scholar]
- 46.Endicott K, Bellwood P. The possibility of independent foraging in the rain forest of peninsular Malaysia. Hum Ecol. 1991;19(2):151–185. [Google Scholar]
- 47.Stearman AM. Making a living in the tropical forest: Yuqui foragers in the Bolivian Amazon. Hum Ecol. 1991;19(2):245–260. [Google Scholar]
- 48.Mercader J. Forest people: The role of African rainforests in human evolution and dispersal. Evol Anthropol. 2002;11(3):117–124. [Google Scholar]
- 49.Yasuoka H. Long-term foraging expeditions (molongo) among the Baka hunter-gatherers in the Northwestern Congo Basin, with special relevance to the “Wild yam question”. Hum Ecol. 2006;34(2):275–296. [Google Scholar]
- 50.Hart TB, Hart JA. The ecological basis of hunter-gatherer subsistence in African rain forests: The Mbuti of Eastern Zaire. Hum Ecol. 1986;14(1):29–55. [Google Scholar]
- 51.Headland TN. The wild yam question: How well could independent hunter-gatherers live in a tropical rain forest ecosystem? Hum Ecol. 1987;15(4):463–491. [Google Scholar]
- 52.Bailey RC, et al. Hunting and gathering in tropical rain forest: Is it possible? Am Anthropol. 1989;91(1):59–82. [Google Scholar]
- 53.Headland TN, Bailey RC. Introduction: Have hunter-gatherers ever lived in tropical rain forest independently of agriculture? Hum Ecol. 1991;19(2):115–122. [Google Scholar]
- 54.Mercader J, Garralda MD, Pearson OM, Bailey RC. Eight hundred-year-old human remains from the Ituri tropical forest, Democratic Republic of Congo: The rock shelter site of Matangai Turu Northwest. Am J Phys Anthropol. 2001;115(1):24–37. doi: 10.1002/ajpa.1053. [DOI] [PubMed] [Google Scholar]
- 55.Phillipson DW. African Archaeology. Cambridge, UK: Cambridge Univ Press; 2005. [Google Scholar]
- 56.Meachen JA, Samuels JX. Evolution in coyotes (Canis latrans) in response to the megafaunal extinctions. Proc Natl Acad Sci USA. 2012;109(11):4191–4196. doi: 10.1073/pnas.1113788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans AR, et al. The maximum rate of mammal evolution. Proc Natl Acad Sci USA. 2012;109(11):4187–4190. doi: 10.1073/pnas.1120774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319(5866):1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 59.Astle W, Balding DJ. Population structure and cryptic relatedness in genetic association studies. Stat Sci. 2009;24(4):451–471. [Google Scholar]
- 60.Zhou X, Carbonetto P, Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9(2):e1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 62.Günther T, Schmid KJ. Improved haplotype-based detection of ongoing selective sweeps towards an application in Arabidopsis thaliana. BMC Res Notes. 2011;4:232. doi: 10.1186/1756-0500-4-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: Genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18(11):1814–1828. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paten B, et al. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008;18(11):1829–1843. doi: 10.1101/gr.076521.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19(5):826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Stat. 2003;31(6):2013–2035. [Google Scholar]
- 67.Hansen MC, et al. High-resolution global maps of 21st-century forest cover change. Science. 2013;342(6160):850–853. doi: 10.1126/science.1244693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.