Significance
Our understanding of the molecular basis of our sense of hearing and balance has improved significantly, although some of the key players in sensory hair cells have yet to be identified. Sensory hair cells depend on extracellular filaments known as tip links to transduce mechanical stimuli into electrical signals. We demonstrate that the tip link protein PCDH15 interacts with two integral member proteins, TMC1 and TMC2, which have recently been put forth as candidates for the mechanotransduction channel.
Abstract
The tip link protein protocadherin 15 (PCDH15) is a central component of the mechanotransduction complex in auditory and vestibular hair cells. PCDH15 is hypothesized to relay external forces to the mechanically gated channel located near its cytoplasmic C terminus. How PCDH15 is coupled to the transduction machinery is not clear. Using a membrane-based two-hybrid screen to identify proteins that bind to PCDH15, we detected an interaction between zebrafish Pcdh15a and an N-terminal fragment of transmembrane channel-like 2a (Tmc2a). Tmc2a is an ortholog of mammalian TMC2, which along with TMC1 has been implicated in mechanotransduction in mammalian hair cells. Using the above-mentioned two-hybrid assay, we found that zebrafish Tmc1 and Tmc2a can interact with the CD1 or CD3 cytoplasmic domain isoforms of Pcdh15a, and this interaction depends on the common region shared between the two Pcdh15 isoforms. Moreover, an interaction between mouse PCDH15-CD3 and TMC1 or TMC2 was observed in both yeast two-hybrid assays and coimmunoprecipitation experiments. To determine whether the Pcdh15–Tmc interaction is relevant to mechanotransduction in vivo, we overexpressed N-terminal fragments of Tmc2a in zebrafish hair cells. Overexpression of the Tmc2a N terminus results in mislocalization of Pcdh15a within hair bundles, together with a significant decrease in mechanosensitive responses, suggesting that a Pcdh15a–Tmc complex is critical for mechanotransduction. Together, these results identify an evolutionarily conserved association between the fish and mouse orthologs of PCDH15 and TMC1 and TMC2, supporting the notion that TMCs are key components of the transduction complex in hair cells.
Sensory hair cells are specialized mechanosensory cells that respond to auditory and vestibular stimuli with their apical hair bundles. Each bundle consists of rows of stereocilia that are interconnected by extracellular tip links. Collective evidence suggests that tip link filaments are composed of protocadherin 15 (PCDH15) and cadherin 23 (CDH23) (1), which are each encoded by deafness genes identified in humans, mice, and fish (1–5). Tip links are required for proper gating of mechanotransduction channels (1, 6, 7), and calcium imaging of transduction in rodent auditory hair cells indicates that transduction channels are associated with the lower end of tip links (8), where PCDH15 localizes (6, 9). Thus, integral membrane proteins that interact with PCDH15 are likely to be critical for mechanotransduction and may constitute subunits or accessory proteins of the transduction channel. To date, neither the identity of the transduction channel nor the mechanism of interaction between the tip link and the channel are known.
Recently, TMC1 and TMC2 were put forth as candidates for the pore-forming subunits of the mechanotransduction channel in sensory hair cells (10). Each protein is predicted to have multiple membrane-spanning domains reminiscent of an ion channel structure (11, 12); in addition, the Caenorhabditis elegans family member TMC-1 has been shown to be a putative channel sufficient for detection of high salt (13). Mammalian TMC1 has been implicated in both recessive and dominant forms of nonsyndromic hearing loss in humans and mice (14–16). Moreover, double knockout of mouse Tmc1 and Tmc2 results in severe auditory and vestibular deficits, along with the complete absence of normal mechanotransduction currents in auditory and vestibular hair cells (17). Changes in calcium permeability through the transduction channel of cochlear hair cells were observed for Tmc1 Tmc2 double-mutant mice, as well as in single mutants of either gene (10, 18, 19). In further support of the idea that TMCs are pore-forming subunits of the transduction channel, mouse vestibular hair cells that express only the dominant Beethoven (M412K) allele of Tmc1, in the absence of any wild-type TMC1 or TMC2, display altered single-channel transduction currents (10). Curiously, an 180° phase-shifted transduction current can be evoked in cochlear hair cells expressing only the deafness allele of Tmc1 (Tmc1dn/dn Tmc2−/−) (19). The source of this anomalous current is not clear, but a similar current is also observed in hair cells where tip links have been disrupted by either genetic lesions (Pcdh15av3J, Cdh23v2J) or by 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) treatment (7, 19, 20). Although the anomalous current seen in Tmc1dn/dn Tmc2−/− hair cells calls into question whether TMC1 and TMC2 serve as pore-forming subunits of the mechanotransduction complex, it is nonetheless clear that these proteins are essential for normal transduction and modulate the channel’s properties. However, how the TMCs interact with other components of the mechanotransduction complex has not yet been investigated.
Given that PCDH15 may be located at the site of mechanotransduction, we performed a split-ubiquitin yeast two-hybrid screen with zebrafish Pcdh15a to identify proteins that could serve as subunits of the mechanotransduction complex. Our unbiased screen identified an interaction with the N terminus of zebrafish Tmc2a, an ortholog of mammalian TMC2. Both Tmc1 and Tmc2a from zebrafish interact with the CD1 and/or CD3 isoforms of Pcdh15a, and these interactions require the common region present in both cytoplasmic domains. Likewise, using two different methods of yeast two-hybrid assays and immunoprecipitation experiments in a heterologous expression system, we show that mouse PCDH15-CD3 can interact with either TMC1 or TMC2. Finally, we overexpressed an N-terminal fragment of Tmc2a to disrupt the Tmc–Pcdh15 interaction in zebrafish hair cells and found a reduction in mechanically evoked calcium transients. Thus, our work shows that a Tmc–Pcdh15 interaction is critical for hair cell mechanosensitivity.
Results
Membrane-Based Screen for Components of the Mechanotransduction Complex.
To identify members of the mechanotransduction complex in hair cells, we performed a molecular screen for proteins that bind to the cytoplasmic tail of Pcdh15a. We took advantage of the split-ubiquitin two-hybrid screening method, which is specifically designed to detect membrane protein interactions at the cytoplasmic face of the endoplasmic reticulum of yeast cells (21). This method is ideal for bait proteins that contain single or multiple transmembrane domains, such as cadherins. To generate the bait vectors, we first identified splice variants of pcdh15a in 5-d-old wild-type larvae by using RACE PCR to amplify the 3′ end of pcdh15a. In mice and humans, three major isoforms of PCDH15 have been reported: CD1, CD2, and CD3 (22, 23). As reported in previously (5), we detected the CD1 isoform in zebrafish, along with amplicons corresponding to a CD3 isoform (Fig. S1). We were unable to detect any products corresponding to a CD2 isoform. As in other vertebrates, a common region adjacent to the transmembrane domain is shared by both CD1 and CD3 isoforms of Pcdh15a (Fig. S1).
For the bait vectors, we used truncated versions of Pcdh15a-CD1 or Pcdh15a-CD3 containing the transmembrane domain and the full cytoplasmic domain (Fig. S1). Prey vectors contained cDNA generated from the total RNA of ∼1,500 zebrafish larval ears at 5 d postfertilization (dpf). After performing the screen and validating positive interactions, we focused on an interaction between Pcdh15a-CD1 and the first 117 aa of the N terminus of Tmc2a, an ortholog of mammalian TMC2 (Fig. S2).
There are two zebrafish tmc2 genes in the Ensembl genome database (release 73), and we designated the paralogous gene isolated in the screen as tmc2a, and the gene duplicate as tmc2b. By contrast, searches of the zebrafish Ensembl database did not reveal a gene duplication of tmc1. All three genes are located on chromosome 5, with tmc1 and tmc2b directly adjacent to one another (Fig. S2). The two Tmc2 proteins are more closely related to each other and other vertebrate Tmc2 orthologs than to Tmc1 and its orthologs (Fig. S2).
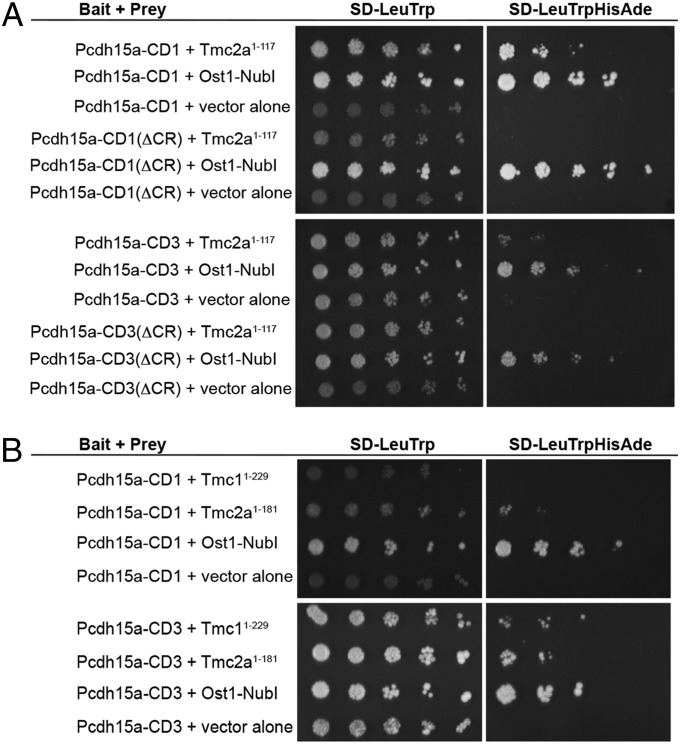
Next, we tested whether the 117-aa fragment of Tmc2a was able to interact with both isoforms of Pcdh15a. Using the split-ubiquitin assay, we observed growth of yeast colonies for both CD1 and CD3 constructs (Fig. 1A). Because this result was unexpected given the divergence of the cytoplasmic tails of CD1 and CD3, we sought to determine whether binding was mediated via the common region shared between both Pcdh15 isoforms. As shown in Fig. 1A, the interaction between Tmc2a1–117 and Pcdh15-CD1 or -CD3 was abolished when the common region is deleted from either isoform. This result highlights the importance of the common region in mediating the interaction between the Pcdh15a cytoplasmic domain and the Tmc2a N terminus.
Fig. 1.
Interaction of zebrafish Pcdh15a with Tmc2a and Tmc1 using a split-ubiquitin yeast two-hybrid assay. (A) The N terminus of zebrafish Tmc2a (1–117 aa) can interact with either CD1 or CD3 isoforms of Pcdh15a, and binding is dependent upon the common region (CR). Transfection efficiency is shown on SD-LeuTrp medium (Left). Protein–protein interactions were detected on SD-LeuTrpHisAde selective media (Right). Shown from left to right on each plate is a fivefold serial dilution series of yeast cells. The Ost1-NubI ubiquitin moiety was used as a positive control, and empty prey vector was used as a negative control. (B) Both zebrafish Tmc2a (1–181 aa) and Tmc1 (1–229 aa) can interact with Pcdh15a isoforms.
Because both TMC1 and TMC2 are implicated in mechanotransduction in mouse hair cells (10, 17), we tested whether zebrafish Tmc1 is also able to bind to the C terminus of Pcdh15a. The corresponding amino acids 1–229 of Tmc1 are predicted by the COILS program to include two coiled-coil domains, which may mediate protein–protein interactions (24). To use a comparable fragment of Tmc2a in our subsequent experiments, we included the single predicted coiled coil within the N terminus (amino acids 1–181). Using the split-ubiquitin assay, we observed that Tmc2a1–181 was capable of binding to both Pcdh15a-CD1 and Pcdh15a-CD3 as seen with the smaller fragment of Tmc2a (Fig. 1B). In contrast, we observed growth of yeast colonies with the N terminus of Tmc1 only in the presence of the CD3 isoform, but not the CD1 isoform (Fig. 1B). Our yeast two-hybrid data suggest that either Tmc1 or Tmc2a can bind to the CD3 isoform of zebrafish Pcdh15a, and in the case of Tmc2a, interaction with either the CD1 or CD3 isoform is mediated through the common region.
Conservation of PCDH15–TMC Interactions.
We next determined whether similar interactions occur between the mammalian orthologs of PCDH15 and TMC1 and TMC2. Expression of the N-terminal amino acids of mouse TMC2 (1–232 aa) with the cytoplasmic tail of either of the CD1 or CD3 isoforms of mouse PCDH15 in yeast resulted in growth on selective media (Fig. S3). In addition, the N terminus of TMC1 (1–179 aa) also interacted with the cytoplasmic tail of each isoform of PCDH15 (Fig. S3).
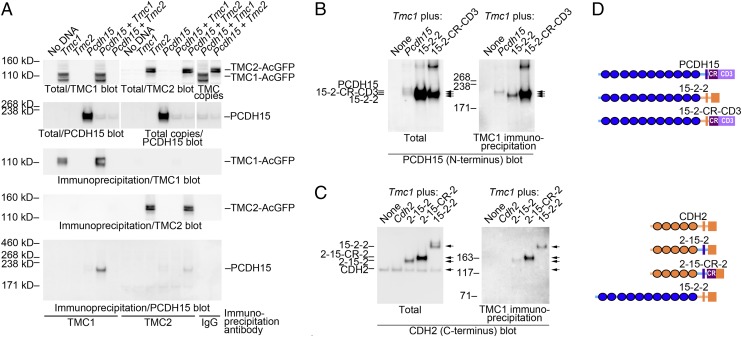
With either mouse or zebrafish proteins, the CD3 variant interacted with the TMC N terminus. Accordingly, we next investigated whether the mouse TMC1 or TMC2 proteins could interact with mouse PCDH15-CD3 in a heterologous expression system. We expressed full-length TMC1 or TMC2, tagged with monomeric AcGFP at the C terminus (17), in the presence or absence of full-length PCDH15-CD3 in HEK 293T cells (Fig. S4). We verified that the bands recognized by antibodies against TMC1 or TMC2 were also recognized by anti-GFP antibodies (Fig. S5). Expression of either TMC1 or TMC2 reduced PCDH15 levels, although the converse was not true. To test for an interaction between TMC and PCDH15, we immunoprecipitated TMCs from detergent-solubilized crude membrane preparations, and then immunoblotted the precipitates with antibodies against PCDH15. Even though PCDH15-CD3 expression was relatively low, pulldown of TMC1 resulted in robust coimmunoprecipitation of PCDH15-CD3 (Fig. 2A, Bottom). Only the faster migrating of the two PCDH15 bands was immunoprecipitated. An interaction between TMC2 and PCDH15-CD3 was also detected (Fig. 2A, Bottom). These results demonstrate that the PCDH15–TMC interaction can occur in mammalian cells and support the results from the initial yeast two-hybrid screen.
Fig. 2.
The interaction between PCDH15 and TMC1 and TMC2 is evolutionarily conserved. (A) Coimmunoprecipitation of mouse PCDH15-CD3 with mouse TMC1 or TMC2 in tissue culture cells. Combinations of full-length Pcdh15-CD3, Tmc1::AcGFP, or TMC2::AcGFP plasmids were expressed in HEK 293T cells as indicated. TMC proteins were precipitated with anti-TMC1, anti-TMC2, or IgG (control) covalently coupled to magnetic beads. TMC1-AcGFP (∼115 kDa) and TMC2-AcGFP (∼130 kDa) were present in total lysates (upper blots) and immunoprecipitates when detected with anti-TMC immunoblotting. PCDH15-CD3 was detected in appropriate lanes in total lysates (10% load relative to immunoprecipitates) and in the TMC1 and TMC2 immunoprecipitates. Total lysate lanes labeled “copy” are digital duplications of corresponding lanes in the left-hand total lysate blots, placed above corresponding lanes for reference. (B–D) Role of the PCDH15 common region and extracellular domain in mediating the interaction between mouse PCDH15 and mouse TMC1. (B) Interaction of mouse CDH2-PCDH15 chimeras with TMC1, detected with N-terminal PCDH15 antibody. (C) Immunoprecipitation of mouse CDH2-PCDH15 chimeras with TMC1, detected with C-terminal CDH2 antibody; this antibody detects endogenous HEK cell CDH2. In B and C, totals are ∼1% load relative to immunoprecipitates. (D) Schematic diagram of PCDH15-CDH2 chimeras.
The ability to express full-length proteins in cell lines led us to test whether the common region and other regions of mouse PCDH15 are important for the interaction with mouse TMC1. We therefore created chimeric proteins with an unrelated cadherin, CDH2 (Fig. 2 B–D). Using the same method of immunoprecipitation of TMC1 from HEK 293T cells, we found that TMC1 failed to associate with CDH2 (Fig. 2C). We then tested whether the transmembrane domain and C terminus of PCDH15-CD3 were necessary for interaction using a chimera that lacked these domains (15-2-2). Although replacing these regions with CDH2 counterparts did not eliminate binding (Fig. 2B), the presence of the cytoplasmic domain greatly enhanced the interaction (15-2-CR-CD3; Fig. 2B). When we swapped the membrane-spanning region of PCDH15 for the corresponding region of CDH2 in a CDH2 chimera (2-15-2), we were able to detect a faint band of the chimeric protein (Fig. 2C); however, a somewhat stronger interaction was observed when we also included the common region of PCDH15 (2-15-CR-2; Fig. 2C). The pulldown of the 15-2-2 chimeric protein with TMC1 antibody suggests that the extracellular domain of PCDH15 also plays a role in interaction with the TMCs. Collectively, the yeast two-hybrid data and coimmunoprecipitation experiments suggest that the interaction between PCDH15-CD3 and TMC1 or TMC2 is conserved among fish and mice. The data also suggest that the common region of PCDH15 is required for robust association of the complex, although several domains of PCDH15 participate in its interaction with the TMCs.
Hair Cell-Specific Pattern of Expression.
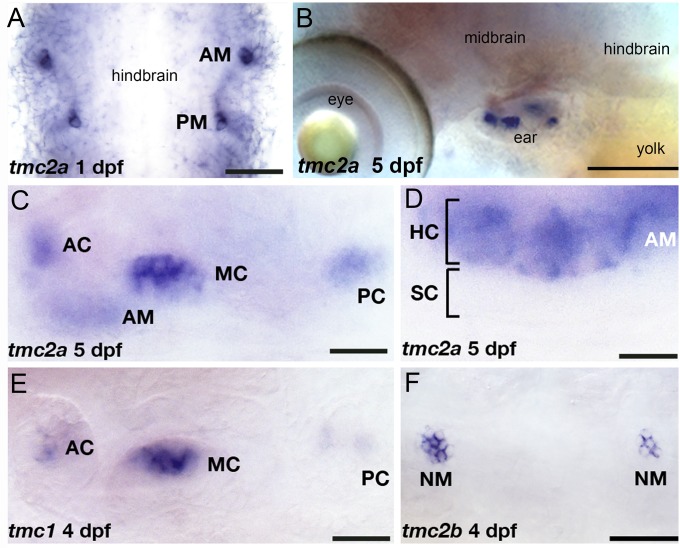
To determine whether the zebrafish tmc1, tmc2a, and tmc2b genes are coexpressed in tissues with pcdh15a, we investigated the pattern of mRNA expression in adult and larval tissues, using RT-PCR and in situ hybridization. With either method, we observed qualitatively that all three transcripts were present at low abundance, requiring 35 cycles of amplification for detectable PCR products or relatively long coloration reactions for in situ hybridization. In contrast to the broad expression pattern of pcdh15a in the larval brain and sensory hair cells (5), expression of tmc1, tmc2a, and tmc2b genes was restricted to hair cells of the inner ear and lateral line organ (Fig. 3). RT-PCR from RNA isolated from various adult tissues revealed that tmc1, tmc2a, and tmc2b transcripts were detectable only in adult ear tissue (Fig. S6). The tmc genes were differentially expressed in sensory organs; tmc2a was detected at higher levels in the inner ear than tmc1 and tmc2b, whereas the opposite pattern was seen with transcripts isolated from lateral-line neuromasts (Fig. S6). In situ hybridization confirmed the tissue-specific expression pattern of the tmc genes (Fig. 3 A–F). We observed that tmc2a is expressed in the nascent hair cells of the developing ear as early as 1 dpf, and expression in the inner ear continues throughout development (Fig. 3 A–C). tmc1 expression was also detectable in the sensory hair cell patches of the ear (Fig. 3E). In the lateral-line neuromasts, only tmc2b was robustly detected by in situ hybridization (Fig. 3F). In either mechanosensory organ, expression of all three paralogs is restricted to hair cells (Fig. 3D). The hair cell-specific expression pattern of tmc1, tmc2a, and tmc2b suggest that these genes play specialized roles in zebrafish hair cells.
Fig. 3.
Embryonic and larval expression pattern of the tmc1/2 genes. In situ hybridization with specific probes for each transcript was performed on stages 1–5 dpf. Expression of tmc2a at 1 dpf (A, flat mount) and 5 dpf (B). Higher magnification view of tmc2a expression in the inner ear (C) and anterior macula at 5 dpf (D). Label is restricted to the upper hair cell layer of the neuroepithelium. (E) Expression of tmc1 at 4 dpf (focal plane: cristae). (F) Expression of tmc2b in neuromasts, 4 dpf. AC, anterior crista; AM, anterior macula; HC, hair cells; MC, medial crista; NM, neuromast; PC, posterior crista; SC, supporting cells. [Scale bars: 50 µm (A); 130 µm (B); 22 µm (C and E); 12 µm (D); 43 µm (F).]
Reduction of Hair Cell Mechanosensitivity by the N Terminus of Tmc2a.
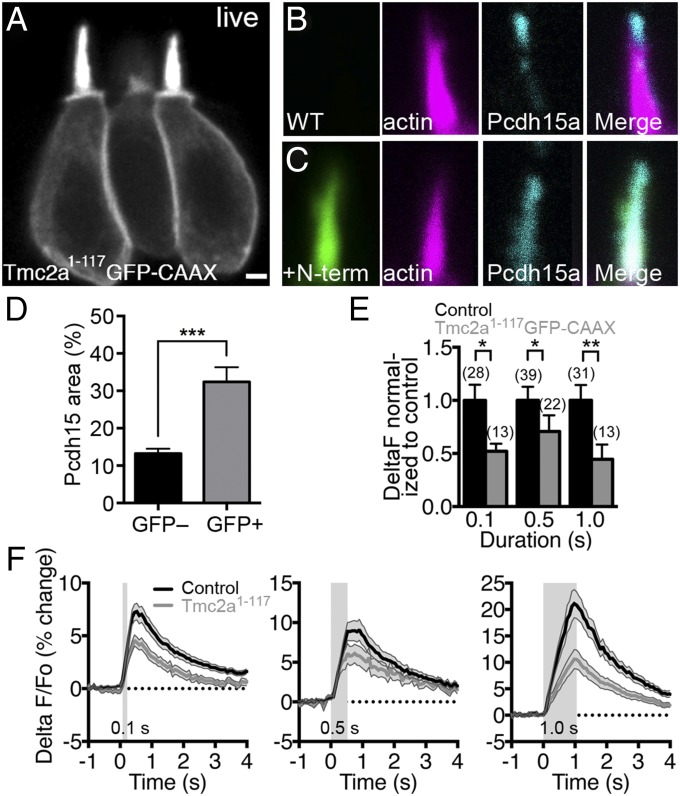
We hypothesized that because Pcdh15 interacts directly with the N terminus of Tmc2a, overexpression of an N-terminal fragment of Tmc2a could disrupt the function of the mechanotransduction complex in a dominant-negative fashion. To test this idea in vivo, we mosaically expressed Tmc2a1–117 in zebrafish hair cells. To enable visualization of the Tmc protein fragment in live cells, we fused GFP with a CAAX prenylation site to the C terminus of the Tmc2a1–117 fragment (Tmc2a1–117-GFP-CAAX). Prenylation of the CAAX sequence targets proteins to membranes. The membrane-associated GFP signal was brightest within mature hair bundles, which are rich in plasma membrane (Fig. 4A). In contrast, tagging of the N-terminal fragments of two distantly related Tmcs, Tmc4 and Tmc5, with GFP-CAAX resulted in accumulation that was almost exclusively in the soma (Fig. S7). We first examined whether expression of exogenous Tmc2a1–117-GFP-CAAX disrupts the morphology of hair bundles. Hair bundles within the zebrafish inner-ear cristae are well suited for live imaging of gross morphology because the stereocilia are long (up to 5 µm) and lateral views are easily obtained in undissected fish. As depicted in the representative example in Fig. 4A, the gross morphology of GFP-positive hair cells and stereociliary bundles was unchanged, suggesting that moderate levels of exogenous Tmc2a1–117-GFP-CAAX do not alter the structure of hair bundles. In contrast, very high expression levels of Tmc2a1–117-GFP-CAAX led to morphological defects of the hair bundles and perturbation of the apical membranes (see example in Fig. S7); these cells therefore were not included in our experiments.
Fig. 4.
Expression of the N-terminal fragment of Tmc2a decreases mechanically evoked responses in zebrafish hair cells. (A) Live confocal image (0.35-μm z sections) of mosaic expression of Tmc2a1–117-GFP-CAAX in ampullary hair cells at 3 dpf. Two mature hair cells are shown in the focal plane. (B–D) Altered distribution of Pcdh15a in hair cells expressing Tmc2a1–117-GFP-CAAX (4 dpf). Control hair bundle (B) and Tmc2a1–117-GFP-CAAX–positive hair bundle (C) were labeled with anti-GFP antibody (green), phalloidin (magenta), and anti-Pcdh15a antibody (cyan). (D) Quantification of the increase in pixel area of Pcdh15a immunolabel within hair bundles (n = 19 hair bundles for each condition; Student t test). [Scale bar: 1.75 µm (A); 1 µm (B and C)]. (E) Response magnitude in Tmc2a1–117-GFP-CAAX–expressing cells normalized to control signals. The number of cells is indicated above the bars. A nonparametric Mann–Whitney U test (unpaired) was used to compare differences between Tmc2a1–117-GFP-CAAX and control hair cell calcium responses. (F) Averaged traces of mechanically evoked calcium responses are plotted as a percentage (ΔF − F0/F0 × 100) and as a function of time in response to 0.1-, 0.5-, and 1.0-s steps. Duration of the step stimulus is indicated by the gray rectangles. The gray shading around traces in F and error bars in E indicates SEM.
Next, we determined whether endogenous Pcdh15a was correctly localized in hair cells expressing Tmc2a1–117-GFP-CAAX. In fixed samples, we found that Pcdh15a immunolabel was distributed more broadly throughout hair bundles in cells expressing Tmc2a1–117-GFP-CAAX (Fig. 4 B–D; see control immunolabel in Fig. S8). This result suggests that exogenous expression of the Tmc2a N-terminal fragment produces a dominant-negative effect on Pcdh15a localization in hair bundles.
We then examined the consequences of overexpression of Tmc2a1–117-GFP-CAAX on hair cell function. To do so, we measured mechanically evoked calcium transients in a transgenic line that stably expresses the calcium indicator R-GECO1 specifically in hair cells. Derived from GCaMP3 and mApple, R-GECO1 is a red-shifted single fluorescent protein calcium indicator that allows visualization of calcium responses in the presence of spectrally distinct fluorescent proteins such as GFP (25). We selected hair cells that were morphologically mature for our imaging experiments, which ensured that all hair cells tested had similar expression levels of the R-GECO1 indicator and were of comparable developmental stages. To assess whether overexpression of Tmc2a1–117-GFP-CAAX disrupted the mechanosensitivity of hair cells, we focused on lateral-line hair cells (Fig. S9), whose readily accessible hair bundles at the surface of the skin enable the use of a more precise stimulus in an intact preparation (26). To stimulate hair cells, we displaced the hair bundles of lateral-line hair cells with a nonsaturating fluid-jet step stimulus. Fig. 4F shows averaged traces of the calcium transients during stimuli of increasing duration. In response to the step stimulus, wild-type control cells with no detectable GFP signal showed robust increases in R-GECO1 fluorescence above the baseline signal (Fig. 4F, black traces; 0.1 s: 8.6 ± 1.3, 0.5 s: 9.0 ± 1.2, 1.0 s: 22.9 ± 3.3 ΔF/F0). By contrast, mechanosensitivity was reduced for all stimulus durations in neighboring cells expressing Tmc2a1–117-GFP-CAAX (Fig. 4F, gray traces; 0.1 s: 4.47 ± 0.61, 0.5 s: 6.4 ± 1.4, 1.0 s: 10.2 ± 3.2 ΔF/F0). When normalized to the calcium transients of control cells, the reduction of responses was similar for each stimulus tested (Fig. 4E). A similar decrease in activity was obtained with a line stably expressing a fragment of Tmc2a containing the predicted full-length N terminus (260 aa) fused to GFP-CAAX (Fig. S10). However, this fragment did not have a significant impact on calcium transients using the shorter 0.1-s stimulus, possibly due to lower levels of this larger fragment within the bundles or other factors that require further investigation. In a separate set of control experiments, we tested hair cells expressing GFP-CAAX alone and did not detect significant differences in mechanically evoked calcium transients between GFP-positive and -negative cells (stimulus: 5.0 Hz, 2 s; wild-type control, 22.0 ± 1.9 ΔF/F0, n = 82; GFP-CAAX alone, 21.3 ± 2.1 ΔF/F0, n = 30; Tmc2a1–260-GFP-CAAX, 14.0 ± 3.9 ΔF/F0, n = 46; P value compared with control 0.41 and 0.002, respectively). Overall, these in vivo experiments suggest that overexpression of the N terminus of Tmc2a both disrupts the endogenous Pcdh15a–Tmc complex in hair bundles and decreases hair cell mechanosensitivity, which is consistent with a requirement of the Pcdh15a–Tmc complex for mechanosensitivity.
Discussion
Although several studies show that the tip links, composed of PCDH15 and CDH23, are required for normal mechanotransduction, it is unclear how they are coupled to the transduction machinery. Likewise, it has been demonstrated that the transmembrane channel-like proteins TMC1 and TMC2 are required for mechanosensitive responses in hair cells, but how they interact with other components of the mechanotransduction complex is not known. Here, we show that TMC1 and TMC2 can interact with PCDH15, thereby establishing a critical connection between the tip link and these putative components of the mechanotransduction channel in hair cells. Our conclusions are based on three independent sets of results, including (i) protein–protein interactions in yeast two-hybrid experiments, (ii) coimmunoprecipitation experiments in HEK 293T cells, and (iii) mislocalization of Pcdh15a and subsequent reduction of mechanically evoked calcium transients in zebrafish hair cells expressing N-terminal fragments of Tmc2a. Indicative of the importance of this interaction, this association appears to be an early development in evolution, as the interaction occurs among both fish and mouse orthologs. Together, these data support a model in which hair cell mechanosensitivity requires a direct interaction between PCDH15 and TMC proteins, thereby clarifying our understanding of the molecular composition of the mechanotransduction complex in vertebrate sensory hair cells.
Previous work has shown that the cytoplasmic domains of PCDH15 and CDH23 can interact with several proteins, including myosin VIIA, harmonin, and whirlin; mutations in these proteins are associated with hearing loss and all three proteins localize to stereocilia (27). To date, tetraspan membrane protein of hair cell stereocilia (TMHS; official protein name LHFPL5) is the only known example of an integral membrane protein required for hearing that interacts with PCDH15 (28). Although both mechanotransduction channel conductance and latency are changed in Tmhs−/− outer hair cells, TMHS is not thought to form a channel by itself, but instead acts as an accessory subunit that couples PCDH15 to the transduction channel (28). How the TMHS protein is integrated and arranged into the PCDH15–TMC complex remains to be determined.
In our membrane-based two-hybrid assays, N-terminal fragments of zebrafish Tmc2a interacted with both CD1 and CD3 isoforms of Pcdh15a, demonstrating that binding is not restricted to a particular Pcdh15a isoform. The C termini of all vertebrate isoforms of PCDH15 are highly divergent with the exception of a common region next to the transmembrane domain (23). Deletion of the common region from either Pcdh15a isoform abolished interaction with Tmc2a, suggesting that this shared region mediates the association with the Tmc2a. In contrast to Tmc2a, zebrafish Tmc1 specifically interacted with the Pcdh15a-CD3 isoform. Mouse TMC1 also interacted robustly with the CD3 isoform in coimmunoprecipitation experiments. Similar to the zebrafish Pcdh15a–Tmc2a interaction, the association of mouse TMC1 is enhanced by the common region of PCDH15. Nevertheless, the extracellular domain of PCDH15 and, to a more limited extent, the transmembrane domain of PCDH15 also contribute to the association.
The PCDH15–TMC interactions shown here suggest that the mechanotransduction complex can vary in its molecular content. Such variability has also been supported by previous studies in mice. In cochlear hair cells, the CD3 isoform of PCDH15 is concentrated in stereociliary tips, whereas the CD1 isoform is localized throughout the hair bundle (22). However, neither isoform is essential, as single knockouts of CD1 or CD3 each have no phenotype (29). Like the CD1 and CD3 isoforms, the TMC1 and TMC2 genes also show redundancy in that expression of either gene alone can rescue mechanotransduction in double knockouts (17). Overall, there appears to be some flexibility in combining different PCDH15 isoforms with TMC1 and TMC2. Our data suggest that multiple interactions are permitted, albeit with preference for the PCDH15-CD3 isoform that is normally present near the site of cation influx in hair bundles (22).
As members of the transduction complex, TMC1 and TMC2 appear to play specialized roles in hair cells. The highly specific expression patterns of the tmc1 and tmc2 genes in zebrafish hair cells coincide with the restricted expression patterns of Tmc1 and Tmc2 in mouse hair cells. Differential expression also appears to be a common theme; although both Tmc1 and Tmc2 genes are expressed in all hair cells in the mouse, Tmc1 is more highly expressed in the auditory hair cells (17). In the fish ear, all three tmc1/2 paralogs are detected in both macular and ampullary hair cells by in situ hybridization. However, semiquantitative PCR with larval tissues suggests that tmc2a is expressed predominantly in the inner ear, whereas tmc1 and tmc2b appear to be more abundant in lateral-line hair cells. The reason for differential patterns of tmc expression is not clear, but may reflect the needs of the particular end organ.
Our finding that both fish and mouse orthologs of PCDH15 interact with TMC1 and TMC2 lends support to the idea that the TMC proteins are conserved components of the mechanotransduction complex in vertebrate hair cells. Consistent with a role for the TMCs in mechanotransduction, we identified a dominant-negative effect upon overexpression of N-terminal fragments of Tmc2a. The resulting decrease in mechanosensitive responses is likely due to a disruption of the interaction between Pcdh15a and endogenous Tmcs. Although our results do not prove that a Pcdh15–Tmc interaction is required for mechanosensitivity, the parallel finding that Pcdh15a takes on a more widespread distribution within hair bundles following Tmc2a N-terminal fragment expression is consistent with this interpretation. It is possible, however, that expression of the N-terminal fragments of Tmc2a may also disrupt interactions of the Tmcs with other proteins as well. Nevertheless, collective evidence strongly suggests that TMC1 and TMC2 are part of the transduction complex, and a link to PCDH15 provides compelling evidence for this notion. Whether the TMCs serve as channels or accessory proteins in mechanotransduction awaits further investigation.
Materials and Methods
Two-Hybrid Interaction Assays.
Yeast expressing bait and prey vectors were grown at 30 °C on SD-LeuTrp selective plates to assess transformation efficiency and SD-LeuTrpHisAde selective plates to detect interactions for 3–6 d. Background growth with the CD3 constructs necessitated the addition of 7.5–15 mM 3-amino-1,2,4-triazole (3-AT) to the SD-LeuTrpHisAde plates. Each experiment was repeated in three independent trials.
Coimmunoprecipitation Experiments.
HEK 293T cells were transfected with PCDH15 and TMC plasmids; membranes were extracted with detergent, and anti-TMC antibodies (SI Materials and Methods) were used to immunoprecipitate TMC–PCDH15 complexes. Anti-PCDH15 antibody PB811 (6) was used to detect complexes. Chimeras were constructed using the following regions: 15-2-2: PCDH15 (1–1,265), CDH2 (718–906); 15-2-CR-CD3: PCDH15 (1–1,265), CDH2 (718–755), PCDH15 (1,413–1,682); 2-15-2: CDH2 (1–717), PCDH15 (1,266–1,412), CDH2 (756–906); 2-15-CR-2: CDH2 (1–716), PCDH15 (1,266–1,460), CDH2 (753–906). Complexes with chimeras containing C-terminal CDH2 were detected with Abcam antibody ab6529. See SI Materials and Methods for details.
In Situ Hybridization and Immunohistochemistry.
In situ hybridizations and immunohistochemistry were performed essentially as previously described (30, 31). See SI Materials and Methods for details.
Calcium and Confocal Imaging.
Tg(myo6b:R-GECO1) fish were mated with wild-type fish (TLF × Tuebingen hybrid), and the embryos were microinjected at the one-cell stage with Gateway plasmids (myo6b:Tmc2a1–117-GFPCAAX or myo6b:GFPCAAX as a control). R-GECO1 was visualized on a Zeiss Axio Examiner microscope using an Orca ER CCD camera (Hamamatsu), a 63×, 1.0 N.A. Plan-Apochromat Zeiss water-immersion lens and the following filter set: excitation, 535/50 565LP, and emission, 620/60 (Chroma). Larvae were sedated, mounted, and stimulated using a fluid jet to deliver a square step stimulus, and data were acquired and processed as previously described (26). Care was taken to avoid using any recordings with movement artifacts. Even in strongly expressing Tmc2a-GFP-CAAX cells, for the settings and filter sets selected, no bleed-through was observed.
Supplementary Material
Acknowledgments
We thank Andrew Griffith for the TMC1-AcGFP and TMC2-AcGFP clones, and Ulrich Müller for the PCDH15-CD3 clone and PB811 antibody. We are also grateful for the psi7 allele of pcdh15a from Tatjana Piotrowski. This study was supported by the Howard Hughes Medical Institute (T.N.); the National Institute on Deafness and Other Communication Disorders [Grants R01 DC 0013531 (to T.N.) and R01 DC002368 and P30 DC005983 (to P.G.B.-G.)]; and the Uehara Memorial Foundation, the Naito Foundation, and the Japan Society for the Promotion of Science (R.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402152111/-/DCSupplemental.
References
- 1.Sakaguchi H, Tokita J, Müller U, Kachar B. Tip links in hair cells: Molecular composition and role in hearing loss. Curr Opin Otolaryngol Head Neck Surg. 2009;17(5):388–393. doi: 10.1097/MOO.0b013e3283303472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed ZM, et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69(1):25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alagramam KN, et al. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet. 2001;10(16):1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- 4.Alagramam KN, et al. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001;27(1):99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- 5.Seiler C, et al. Duplicated genes with split functions: Independent roles of protocadherin15 orthologues in zebrafish hearing and vision. Development. 2005;132(3):615–623. doi: 10.1242/dev.01591. [DOI] [PubMed] [Google Scholar]
- 6.Kazmierczak P, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449(7158):87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 7.Alagramam KN, et al. Mutations in protocadherin 15 and cadherin 23 affect tip links and mechanotransduction in mammalian sensory hair cells. PLoS One. 2011;6(4):e19183. doi: 10.1371/journal.pone.0019183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beurg M, Fettiplace R, Nam J-H, Ricci AJ. Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nat Neurosci. 2009;12(5):553–558. doi: 10.1038/nn.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodyear RJ, Forge A, Legan PK, Richardson GP. Asymmetric distribution of cadherin 23 and protocadherin 15 in the kinocilial links of avian sensory hair cells. J Comp Neurol. 2010;518(21):4288–4297. doi: 10.1002/cne.22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan B, et al. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron. 2013;79(3):504–515. doi: 10.1016/j.neuron.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keresztes G, Mutai H, Heller S. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics. 2003;4(1):24. doi: 10.1186/1471-2164-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labay V, Weichert RM, Makishima T, Griffith AJ. Topology of transmembrane channel-like gene 1 protein. Biochemistry. 2010;49(39):8592–8598. doi: 10.1021/bi1004377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatzigeorgiou M, Bang S, Hwang SW, Schafer WR. tmc-1 encodes a sodium-sensitive channel required for salt chemosensation in C. elegans. Nature. 2013;494(7435):95–99. doi: 10.1038/nature11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurima K, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30(3):277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- 15.Vreugde S, et al. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet. 2002;30(3):257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- 16.Duman D, Tekin M. Autosomal recessive nonsyndromic deafness genes: A review. Front Biosci (Landmark Ed) 2012;17:2213–2236. doi: 10.2741/4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawashima Y, et al. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J Clin Invest. 2011;121(12):4796–4809. doi: 10.1172/JCI60405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim KX, Fettiplace R. Developmental changes in the cochlear hair cell mechanotransducer channel and their regulation by transmembrane channel-like proteins. J Gen Physiol. 2013;141(1):141–148. doi: 10.1085/jgp.201210913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim KX, et al. The role of transmembrane channel-like proteins in the operation of hair cell mechanotransducer channels. J Gen Physiol. 2013;142(5):493–505. doi: 10.1085/jgp.201311068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcotti W, et al. Transduction without tip links in cochlear hair cells is mediated by ion channels with permeation properties distinct from those of the mechano-electrical transducer channel. J Neurosci. 2014;34(16):5505–5514. doi: 10.1523/JNEUROSCI.4086-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snider J, et al. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat Protoc. 2010;5(7):1281–1293. doi: 10.1038/nprot.2010.83. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed ZM, et al. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J Neurosci. 2006;26(26):7022–7034. doi: 10.1523/JNEUROSCI.1163-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alagramam KN, et al. Promoter, alternative splice forms, and genomic structure of protocadherin 15. Genomics. 2007;90(4):482–492. doi: 10.1016/j.ygeno.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 25.Tewson P, et al. Simultaneous detection of Ca2+ and diacylglycerol signaling in living cells. PLoS One. 2012;7(8):e42791. doi: 10.1371/journal.pone.0042791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kindt KS, Finch G, Nicolson T. Kinocilia mediate mechanosensitivity in developing zebrafish hair cells. Dev Cell. 2012;23(2):329–341. doi: 10.1016/j.devcel.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown SDM, Hardisty-Hughes RE, Mburu P. Quiet as a mouse: Dissecting the molecular and genetic basis of hearing. Nat Rev Genet. 2008;9(4):277–290. doi: 10.1038/nrg2309. [DOI] [PubMed] [Google Scholar]
- 28.Xiong W, et al. TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell. 2012;151(6):1283–1295. doi: 10.1016/j.cell.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webb SW, et al. Regulation of PCDH15 function in mechanosensory hair cells by alternative splicing of the cytoplasmic domain. Development. 2011;138(8):1607–1617. doi: 10.1242/dev.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 31.Obholzer N, et al. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J Neurosci. 2008;28(9):2110–2118. doi: 10.1523/JNEUROSCI.5230-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.