Significance
Autophagy is conserved and essential for cellular survival during starvation and stress, and is initiated by the autophagy-related 1 (Atg1) complex. In yeast, the three subunits Atg17, Atg29, and Atg31 assemble first, and their structure is known. There has been a debate over how the Atg1 and Atg13 subunits assemble when autophagy is triggered. Here we use mass spectrometry to show that the C-terminal domain of Atg1 is highly dynamic on its own. Atg1 forms a stable complex when it binds with high affinity to Atg13. The combined Atg1–Atg13 subcomplex then binds with moderate affinity to the preformed Atg17–Atg31–Atg29 scaffold. This highlights the binding of Atg1–Atg13 to Atg17–Atg31–Atg29 as a pivotal step in autophagy initiation.
Keywords: analytical ultracentrifugation, isothermal titration calorimetry, membrane tethering, intrinsically disordered proteins, protein structure
Abstract
The autophagy-related 1 (Atg1) complex of Saccharomyces cerevisiae has a central role in the initiation of autophagy following starvation and TORC1 inactivation. The complex consists of the protein kinase Atg1, the TORC1 substrate Atg13, and the trimeric Atg17–Atg31–Atg29 scaffolding subcomplex. Autophagy is triggered when Atg1 and Atg13 assemble with the trimeric scaffold. Here we show by hydrogen–deuterium exchange coupled to mass spectrometry that the mutually interacting Atg1 early autophagy targeting/tethering domain and the Atg13 central domain are highly dynamic in isolation but together form a stable complex with ∼100-nM affinity. The Atg1–Atg13 complex in turn binds as a unit to the Atg17–Atg31–Atg29 scaffold with ∼10-μM affinity via Atg13. The resulting complex consists primarily of a dimer of pentamers in solution. These results lead to a model for autophagy initiation in which Atg1 and Atg13 are tightly associated with one another and assemble transiently into the pentameric Atg1 complex during starvation.
The engulfment of cytosolic contents by autophagy is an ancient mechanism for cell survival and homeostasis (1, 2). This process is conserved throughout the Eukarya. Autophagy consists of the surrounding of cellular material in a double-membrane structure known as the phagophore (1), which matures into the autophagosome and fuses with the lysosome. The small-molecule metabolites generated by lysosomal degradation replenish energy stores and biosynthetic precursors. Autophagy, or its dysfunction, has roles in neurodegenerative disease, cancer, infection, inflammation, and aging (3). Despite its central importance in human health and disease, current knowledge of autophagosome biogenesis at the structural and molecular mechanistic level is limited (4). Our laboratory and many others have therefore embarked on a protein-by-protein effort to dissect the structures and interactions responsible for the remarkable process of autophagosome biogenesis.
In yeast, autophagosome biogenesis commences at a single locus known as the phagophore assembly site (PAS). The autophagosome is nucleated, at least in part, from a cluster of a small number of vesicles with radii of 15–30 nm that contain the integral membrane protein autophagy-related 9 (Atg9) (5–7). The Atg1 complex, consisting of the subunits Atg1, Atg13, Atg17, Atg29, and Atg31, is thought to have a central role in autophagy initiation at the PAS. Atg1 is a protein kinase, yet the Atg1 complex is thought to have essential roles very early in autophagy that are independent of its kinase activity (8). These probably include organizing the vesicle cluster that goes on to form the phagophore (9). The kinase activity of Atg1 is also essential, in part because it phosphorylates Atg9 (10). In human cells, the Unc51-like kinase 1 (ULK1) and ULK2 complexes are largely conserved and thought to serve similar functions (11). The subunits Atg17, Atg29, and Atg31 appear to be capable of assembling at the PAS constitutively. They thus appear to serve as a preexisting scaffold for the recruitment of Atg1 and Atg13 upon activation.
The crystal structure of the Atg17–Atg31–Atg29 complex showed that it dimerizes into a structurally unique double crescent (9, 12). Dimerization occurs via the C terminus of Atg17, and is required for formation of the PAS and for autophagy (9). Autophagy initiation also requires the recruitment of Atg1 and Atg13 to the PAS downstream of Atg17–Atg31–Atg29 (13). Atg1 consists of an N-terminal protein kinase domain, a predicted flexible linker, and a C-terminal early autophagy targeting/tethering (EAT) domain. Atg13 consists of an N-terminal HORMA domain (14) and a very long predicted unstructured central and C-terminal region. The presence of extensive regions of presumed intrinsic disorder in the Atg1 and Atg13 subunits has slowed progress in understanding the structure and assembly of the complete pentameric Atg1 complex. Given the essential role of the Atg1 complex in autophagy initiation, we set out to probe its dynamics using hydrogen–deuterium exchange (HDX) coupled to mass spectrometry (MS). In HDX-MS, the intrinsic exchange rate of amide protons is used to measure protein dynamics (15–17). Highly ordered regions of proteins exchange protons slowly, whereas dynamic regions exchange them rapidly.
The translocation of the Atg1 and Atg13 subunits to the PAS upon TORC1 inactivation is a critical event in early autophagy and has been the topic of intensive investigation and debate. Dephosphorylated Atg13 is thought to act as a bridge between Atg1 and Atg17, triggering the assembly of the subunits into a dimer of pentamers. Mutation of the eight identified Atg13 phosphorylation sites to Ala induces Atg1 complex formation in yeast in the absence of autophagy induction (18). Recently, a constitutive interaction between Atg1 and Atg13 was observed (19), consistent with findings in human (20–22) and Drosophila (23) cells. The newer report suggests that the Atg1–Atg13 complex is constitutive and is regulated primarily at the level of its conformation, rather than its assembly. In this work, we probe the dynamics and stability of the Atg1 EAT domain and find that much of it is mobile in the absence of Atg13. We also find that the affinity of Atg1 and Atg13 for one another is very high, on the order of ∼100 nM. By contrast, the affinity of the preassembled Atg1–Atg13 and Atg17–Atg31–Atg29 complexes for one another is two orders of magnitude weaker. These findings have implications for models of Atg1 recruitment to the PAS and so for the mechanism of autophagy initiation.
Results
How Atg13 Binds to Atg1 and Atg17–Atg31–Atg29.
Atg13 directly binds to both Atg1 and Atg17 (9, 24–27). To map which portion of Atg13 binds to the Atg1 EAT domain (henceforth Atg1EAT), a series of deletion constructs was generated using proteins from Kluveromyces lactis (Kl) and tested in pull-down experiments (Fig. 1A). The systematic truncations showed that the Atg13 fragment that comprises the minimal binding domain for Atg1EAT spans residues 400–475. The same strategy was applied to map the determinants for binding to the Atg17–Atg31–Atg29 scaffold. The boundaries of the minimal fragment capable of binding to the scaffolding subcomplex were residues 375–525 (Fig. 1 B and C). Because the Atg13350–525 construct included both sites, it was used for all of the experiments in this study. The deletion mapping of the Atg1 binding site was corroborated and refined upon by HDX-MS. The entirety of the Atg13350–525 construct evidenced rapid exchange in the absence of Atg1EAT (Fig. 1D), consistent with the expectation that this was an intrinsically disordered region. However, residues 417–426 of Atg13 had sharply reduced exchange in the presence of Atg1EAT, implicating this region as the primary binding site for Atg1. Residues 448–451 showed reduced exchange to a lesser extent, suggesting a secondary role in binding to Atg1EAT.
Fig. 1.
Atg1 binding site of Atg13. (A and B) Atg1EAT (A) and Atg17–Atg31–Atg29 (B) were immobilized on Talon resin and the indicated Atg13 fragments were pulled down. (C) Schematic of the Atg1 and Atg17–Atg31–Atg29 binding sites of Atg13. (D) HDX-MS mapping of the Atg1 binding site on Atg13. Atg1EAT-induced changes are localized within two regions between residues 410 and 460 of Atg13, and therefore only these regions are shown. Relative deuteration is indicated by a color gradient. Peptide coverage is shown above the deuteration map. (E) HDX-MS heat map at 10 s onto the crystal structure of Km Atg13 as bound to Atg1EAT. (F) ITC titration of Atg1EAT into a solution of MBP-tagged Atg13350–525. The thermogram was fit to a single-site binding model. Error bars are shown for each individual injection according to the fitting of the baseline (42). DP, differential power.
While this work was under review, a crystal structure analysis of the K. marxianus (Km) Atg1–Atg13 and Lachancea thermotolerans (Lt; Fig. S1) Atg13–Atg17 interactions was published (28). The structure confirmed the prediction that Atg1EAT consists of six α-helices, and showed that these helices correspond to two microtubule-interacting and transport (MIT) domains. The regions of Kl Atg13 that were protected from HDX by Atg1 correspond to regions termed N-terminal MIT interacting motif (MIM-N) and MIM-C by Fujioka et al. (28). MIM-N was found to be the main driver for the affinity of the interaction (28), consistent with our finding that this region has the highest protection from HDX upon binding to Atg1. Leu424 in this highly protected region of Kl Atg13 corresponds to Leu476 of Saccharomyces cerevisiae Atg13 (Fig. 1E and Fig. S1), which is essential for PAS formation and for autophagy (28). The high degree of protection observed for this residue by HDX thus correlates with its essential role in PAS formation.
To assess the binding affinity between Kl Atg1EAT and Atg13350–525, Atg1EAT was titrated into a cell containing Atg13350–525 (Fig. 1F). A dissociation constant (Kd) value of 125 ± 10 nM was obtained with n = 0.78 ± 0.02. Using Km Atg1EAT and Km Atg13460–521, a Kd value of 360 nM was independently obtained by isothermal titration calorimetry (ITC) (28), in excellent agreement with our observations.
Dynamics of Atg1EAT.
To gain insights into the dynamic properties of Atg1EAT in solution, we performed continuous-labeling HDX-MS (29–31). We initially measured deuteron incorporation into isolated Atg1EAT to analyze the overall kinetics of the HDX reaction. Atg1EAT exchanged about 60% of its exchangeable amide hydrogens within 10 s in D2O (Fig. 2 and Figs. S2–S4). The overall exchange characteristics indicate the presence of a large fraction of highly dynamic, solvent-accessible structure.
Fig. 2.
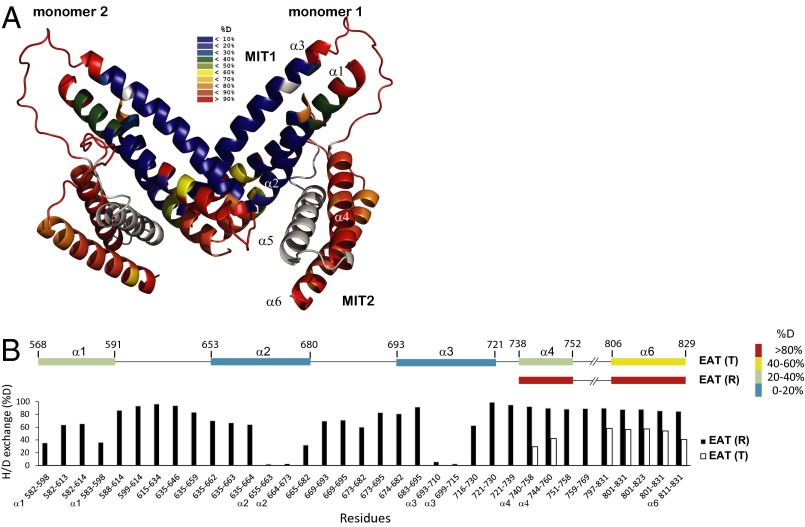
Dynamics of Atg1EAT. (A) HDX-MS heat map (10 s) for the R state onto the crystal structure of Km Atg1EAT. Atg13 is omitted from the structure for clarity. (B) Localization of fast- and slow-exchanging regions of Atg1EAT. Bar graphs showing the changes in % deuteron (D) for the relaxed (R) and tense (T) conformation state of Atg1EAT α-helices after 10 s in D2O. The numbers of the corresponding Atg1 peptides are given at the bottom of the graph. (Upper) Secondary structure representation of Atg1EAT with α-helical segments colored according to the relative deuteron incorporation after 10 s in D2O as indicated. The peptic fragments used to assign the exchange percentage to the various segments are indicated by helix labels preceding the peptide numbers.
To localize fast- and slow-exchanging regions in Atg1EAT, we performed protein digestion under quenched conditions after the HDX reaction (30, 32). The peptides generated by proteolysis were analyzed by electrospray ionization mass spectrometry. The observed fragments covered about 90% of the Atg1EAT sequence. Atg1EAT contains six α-helices (α1–α6) (Fig. 2A). Labeling for 10 s is sufficient to selectively deuterate fully solvated amides (33, 34). The HDX data are consistent with the presence of stable structure in helices α1, α2, and α3 (Fig. 2 A and B). These regions incorporate deuterons to a much lower degree than the interconnecting loop regions. No usable peptides were generated for the region corresponding to helix α5. However, helices α4 and α6 are highly solvent-accessible, with almost complete deuteration within 10 s (Fig. 2 A and B). These data indicate significant flexibility of these helices with transient opening of hydrogen bonds.
The isotopic peaks corresponding to helices α4 and α6 formed a clear bimodal distribution, revealing the presence of two Atg1EAT states with different solvent accessibility. The less accessible state is termed the “T” state, following terminology used to describe the slow-exchanging conformation of Hsp90 (35). The T state rapidly converts into a more accessible “R” state (Fig. S2A). From the isotopic peak area of the two states, we determined a rate constant for the T-to-R conversion of about 0.05 s−1 (Fig. S2B). The two different conformational states were primarily localized to the C-terminal half of Atg1EAT. These data clearly show that the N-terminal subdomain (α1–α3) is well-folded in solution, consistent with its role in forming the dimer interface in the crystal structure (28). The C-terminal subdomain (α4–α6) is highly dynamic and able to alternate between two conformational states.
Dynamics of Atg1EAT–Atg13350–525.
The systematic truncations and the HDX analysis of Atg13 showed that a portion of the intrinsically disordered middle region of Atg13, residues 350–525, was important for binding to Atg1EAT. To characterize the dynamics of Atg1EAT as bound to Atg13, we coexpressed and copurified Atg1EAT with Atg13350–525. To localize Atg13-induced changes in Atg1EAT dynamics, we compared deuteron incorporation into different segments of the Atg1EAT–Atg13 complex with Atg1EAT alone (Fig. 3A). The presence of Atg13350–525 had a prominent stabilizing effect on Atg1EAT. Multiple peptides are more protected in the Atg1EAT–Atg13350–525 complex, including the loop between helices α2 and α3 and the C-terminal regions corresponding to helices α4 and α6 (28). The main Atg13–MIM-N interaction is with the second helical bundle (α4–α6; corresponding to MIT2) of Atg1EAT (28). This is consistent with Atg13 reducing EAT domain dynamics by directly binding to α4 and α6. Atg13 binding had no significant effect on the regions corresponding to helices α1, α2, and α3 at times shorter than ∼5 min. At longer times, the amide protons of these helices can be seen to exchange more rapidly in the absence of Atg13 (Fig. S3). The relative degree of stabilization, however, especially as measured at shorter times, is much more pronounced for α4 and α6.
Fig. 3.
Dynamics of the Atg1EAT–Atg13 subcomplex. (A) Structure of the Atg1EAT–Atg13 complex colored according to the percentage of deuterons incorporated in the presence of Atg13 minus deuterons incorporated in the absence of Atg13 into Atg1EAT residues after 10 s in D2O. Atg13 is in white and regions of Atg1 lacking peptide coverage are in gray. (B) Difference plot of % D incorporated into Atg1EAT α-helices in the presence of Atg13 minus deuterons incorporated in the absence of Atg13 into Atg1EAT segments after 10 s in D2O. The numbers of the corresponding Atg1EAT peptides are given at the bottom of the graph. The Atg1EAT secondary structure drawing (Upper) illustrates the two states that were compared in the HDX-MS analysis. Six predicted α-helices (α1–α6) are colored according to % D exchange. (C) Representative mass spectra (m/z scale) of two selected peptides of Atg1EAT before incubation in D2O (Bottom; monoisotopic mass-to-charge ratio and charge state are given), after 10 s in D2O in the absence (Atg1EAT) and presence of Atg13 (Atg1EAT–Atg13350–525), or after Atg1EAT unfolding with 6 M guanidinium hydrochloride followed by complete deuteration. Arrows above the spectra indicate the bimodal isotope distribution after a 10-s incubation in D2O. The bimodal isotope distribution was attributed to the relaxed and tense states of Atg1EAT. The numbers above the spectra refer to the corresponding backbone amides. (D) Relative amount of R state versus incubation time in D2O. T-to-R transition kinetics for selected peptides of Atg1EAT in isolation or in the context of Atg1EAT–Atg13350–525 and the Atg1EAT–Atg13350–525–Atg17–Atg31–Atg29 complex. A first-order rate equation was fitted to the data (solid lines) to give the T-to-R conversion rate constants of 0.05 s−1 and 0.001 s−1 for Atg1EAT and Atg1EAT–Atg13350–525, respectively. Data are averages of three experiments and error bars represent the SEM.
The specific protection of the C-terminal subdomain of Atg1EAT suggests that it is directly involved in Atg13 binding. In support of this, we found that in the presence of Atg13350–525 the isotope peaks corresponding to the Atg1EAT helices α4 and α6 formed a unimodal distribution after 10 s of labeling (Fig. 3B). The isotope clusters correspond to a single, rigid conformational state with reduced solvent accessibility. The bimodal distribution could be observed only after prolonged incubation in D2O, probably reflecting Atg13 dissociation. The T-to-R conversion constant of the C-terminal subdomain in complex with Atg13 was reduced by more than 50-fold in comparison with free Atg1EAT (Fig. 3C). The T-to-R conversion kinetics of Atg1EAT–Atg13350–525 was not significantly altered by adding the saturating amounts of the Atg17–Atg31–Atg29 complex (Fig. 3C). The findings are consistent with the concept that Atg13 is the dominant modulator of Atg1EAT dynamics. This suggests that once Atg13 binds to and rigidifies Atg1, incorporation with Atg17–Atg31–Atg29 does not restore Atg1 to the R state (Fig. 3C).
Quaternary Structure of Atg1EAT–Atg13350–525.
We previously found that Atg1EAT is a dimer (9). The studies above suggested to us that the Atg1–Atg13 subcomplex is the biologically functional form in which Atg1 is presented to the Atg17–Atg31–Atg29 scaffold. Here we used sedimentation velocity analytical ultracentrifugation (SV-AUC) to probe the quaternary structure of both Atg1EAT alone and the Atg1EAT–Atg13350–525 subcomplex. Atg1EAT exists primarily as a dimer in solution at a sedimentation coefficient at 20 °C in water (s20,w) of 4.2 S (Fig. 4A, Fig. S5, and Table S1), consistent with the previous analysis. Atg1EAT–Atg13350–525 also exists primarily as a dimer in solution at an s20,w value of 4.7 S (Fig. 4A and Table S1). Size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS) was used to independently obtain a molecular mass of 98 kDa for the Atg1EAT–Atg13350–525 complex (Fig. S6A). This is in good agreement with the estimate of 88 kDa from SV-AUC and an expected value of 106.6 kDa computed from the sequences. These data show that Atg1EAT is a dimer irrespective of the presence or absence of Atg13.
Fig. 4.
Assembly of the Atg1EAT–Atg13–Atg17–Atg31–Atg29 complex. (A) Overlay of the c(s) plots of Atg1EAT and Atg1EAT–Atg13350–525. Both Atg1EAT and Atg1EAT–Atg13350–525 primarily exist as dimeric species at an s20,w of 4.2 S and 4.7 S, respectively. The concentration of protein used for both Atg1EAT and Atg1EAT–Atg13350–525 was 20 µM. The c(s) distributions were normalized to the peak area of Atg1EAT. (B) Representative ITC thermogram of Atg1EAT–Atg13350–525 binding to Atg17–Atg31–Atg29, yielding a Kd of 11 ± 2 μM. Atg17–Atg31–Atg29 complex (15 µM) was loaded into the ITC cell and EAT-13 (450 µM) was loaded into the syringe. The thermogram was fit to a single-site binding model. Error bars are shown for each individual injection according to the fitting of the baseline (42). (C) Overlay of the c(s) plots of Atg17–Atg31–Atg29 and the pentamer. Atg17–Atg31–Atg29 exists primarily as an elongated, dimeric species at an s20,w value of 5.5 S with some formation of a tetramer at 8.1 S. To assess the oligomeric state of the pentamer, an 8-M excess of Atg1EAT–Atg13350–525 was mixed with Atg17–Atg31–Atg29 (2 µM). The c(s) distributions show that the pentamer exists primarily as a dimer at an s20,w value of 7.9 S. The excess, unbound EAT-13 sediments at 4.9 S. The c(s) distributions were normalized to the peak area of the Atg17–Atg31–Atg29 dimer.
Assembly of the Atg1–Atg13–Atg17–Atg31–Atg29 Complex.
We used ITC to determine the dissociation constant between Atg17 and Atg13. The Kd for binding of Atg1EAT–Atg13350–525 to Atg17–Atg31–Atg29 was found to be 11 ± 2 μM with n = 0.86 ± 0.17 (Fig. 4B). The figure for the Kl proteins studied here is significantly greater than the figure of 1.2 μM for the binding of the 424–436 peptide from Lt Atg13 to Lt Atg17 (28), which might reflect differences in the species studied or in the size of the constructs. Next, we wanted to understand the oligomeric state of the full pentameric complex, which includes the full-length Atg17–Atg31–Atg29 complex and the Atg1EAT–Atg13350–525 subcomplex. Using SV-AUC, we found that for Atg17–Atg31–Atg29, the majority species is a highly elongated dimer at an s20,w of 5.5 S, with some presence of a tetramer at 8.2 S (Fig. 4C and Fig. S7). These data are generally consistent with a previous sedimentation equilibrium (SE) AUC study of the Atg17–Atg31–Atg29 complex (36), although the tetrameric form was not noted in the SE study. Even though the Atg17 dimer is ∼60 kDa larger than the Atg1EAT–Atg13350–525 dimer, their sedimentation coefficient distributions [c(s)] partially overlap. We attribute this to the highly elongated nature of Atg17–Atg31–Atg29, which slows its sedimentation.
To characterize the oligomeric state of Atg1EAT–Atg13350–525–Atg17–Atg31–Atg29, we kept the concentration of Atg17–Atg31–Atg29 constant and titrated it with a four- or eightfold molar excess of Atg1EAT–Atg13350–525. Atg1EAT–Atg13350–525–Atg17–Atg31–Atg29 exists primarily as a dimer at an s20,w of 7.9 S, with some tetramer formation at 11.7 S (Fig. 4C). The excess unbound Atg1EAT–Atg13350–525 sediments at an s20,w of 4.9 S. On the basis of the SEC-MALS, a molecular mass of 234 kDa was independently obtained (Fig. S6B and Table S1), consistent with a predominant population of Atg1EAT–Atg13350–525–Atg17–Atg31–Atg29 dimers. Taken together with the ITC and HDX-MS data, these results led us to a model for the pathway of Atg1 complex assembly (Fig. 5).
Fig. 5.
Model for Atg1 complex assembly. Atg1EAT–Atg13 is deliberately shown as a monomer to emphasize interactions with a single copy of Atg17. The binding site for Atg13 is positioned near the tip of the scaffold (28).
Discussion
Atg1EAT was previously shown to tether high-curvature liposomes in vitro (9). It was proposed that this property was dependent on the dimerization of Atg1EAT. We found that Atg1EAT is a dimer in solution by SV-AUC, and that in the absence of other members of the complex, the first three helices (MIT1) of Atg1EAT are much more rigid than the last three (MIT2). The crystal structure of Atg1EAT (28) showed that the asymmetric unit consists of a dimer, and that the dimer interface is formed by the burial of ∼800 Å2 of surface area from MIT1. The formation of this interface explains why α1–α3 are so much less flexible than α4 and α6. The complementary crystallographic and solution data present a consistent picture that Atg1EAT dimerizes through an extensive interface involving MIT1 (Fig. 2A).
Because the recruitment of Atg1 to the PAS upon starvation is a central trigger for autophagy, considerable effort has been spent in understanding how this is regulated. In particular, the mechanism whereby the TORC1 complex phosphoregulates the Atg1 complex has been a major question. In an early model, it was observed that coimmunoprecipitation of Atg1 and Atg13 is inhibited in rich medium under conditions when TORC1 is active (18, 24). More recent investigation found that Atg1 and Atg13 form a constitutive complex in yeast (19). Experimental differences in the mode of cell lysis and immunoprecipitation thus seem to significantly affect the conclusions of these studies. We sought to complement these cellular-level analyses with in vitro biochemistry. We found that Atg1 and Atg13 fragments bind to each other with near–100-nM affinity. These fragments were generated in Escherichia coli and so represent the hypophosphorylated states of these proteins that are thought to exist during starvation in yeast. This figure approaches the concentrations at which they are likely to be present in cells (37), although these levels have not been established rigorously. Subject to the usual limitations of interpreting in vitro studies with respect to cellular mechanism, these results seem potentially consistent with the model that Atg1–Atg13 is a stable complex in yeast (19). It also seems probable that Atg1 exists in cells under some conditions separate from its complex with Atg13. In this event, the exceptional dynamics of the EAT domain will undoubtedly affect the properties of Atg1.
We next sought to understand how Atg1–Atg13 interacts with the preexisting and constitutive Atg17–Atg31–Atg29 scaffold. The preassembled complex of Atg1–Atg13–interacting regions binds to Atg17–Atg31–Atg29 with an affinity of near 10 μM. Thus, the interaction between the subcomplexes is nearly two orders of magnitude weaker than the interaction between Atg1 and Atg13 within the Atg1–Atg13 subcomplex. The Atg17–Atg31–Atg29 assembly is so tight that the separate components cannot be produced, and thus it has not been possible to measure its affinity. The Kd for the dissociation of the two subcomplexes from one another is much greater than the plausible cellular concentrations of these proteins. Therefore, additional factors must participate to drive this interaction. Atg9 and the Rab GTPase Ypt1 (38) are candidates for such factors. Overall, the data are consistent with a model in which the assembly of Atg1–Atg13 with Atg17–Atg31–Atg29 is the most energetically costly step in the Atg1 assembly pathway at cellular concentrations. This suggests that it is likely to be a rate-limiting step as well, and therefore a natural target for regulation.
The analytical ultracentrifugation data add perspective to the larger organization of the complex. The Atg1–Atg13 subcomplex acts, structurally, as a nearly independent module within the larger complex, in the sense that neither the dimerization nor the dynamics of the complex seems to be perturbed by binding to Atg17–Atg31–Atg29. The HDX-MS data also add insight into the role of putative intrinsically disordered regions within the Atg1 complex. The extensive flexibility in the central region of Atg13 allows it to bind to Atg1 and so become ordered within the context of the Atg1–Atg13 complex. Thus, in their functional form, some regions of Atg proteins that appear to be disordered based on prediction may in fact be folded together with their partners. Other regions may genuinely require flexibility. In the current model for the assembly of the Atg1 complex, the Atg1–Atg13 subcomplex functions as an essentially independent module, tethered to the tip of the Atg17–Atg31–Atg29 scaffold by a flexible region within Atg13.
These results provide a framework for various questions that remain to be explored. Can the effects of TORC1 phosphorylation of Atg13 be accounted for in the context of reduced affinity of Atg1–Atg13 for Atg17–Atg31–Atg29? What additional interactions bolster the moderate 10-μM affinity of the two subcomplexes for one another, such that the cellular thermodynamic equilibrium shifts to the pentameric complex in starvation? Given that both the Atg17–Atg31–Atg29 and Atg1–Atg13 complexes are dimers, how are these different types of dimers arranged into potential higher-order structures at the PAS? How do these interactions regulate the vesicle-tethering and protein kinase activities of the Atg1 complex? Among the Atg proteins, how much of what appears by sequence analysis to be intrinsic disorder is actually disordered functionally? Here we have provided some tools and a conceptual framework to move forward in answering these questions.
Materials and Methods
Analytical Ultracentrifugation.
Sedimentation velocity experiments were conducted at speeds between 98,784 × g and 201,600 × g using a Beckman Coulter XLI analytical ultracentrifuge. The samples were monitored by absorbance at 280 nm and by interference. All proteins were dialyzed in 20 mM Tris⋅HCl (pH 8.0) and 200 mM NaCl, with 1 mM Tris(2-carboxyethyl)phosphine added for Atg1EAT. Atg17–Atg31–Atg29 samples were run at 5, 8, and 10 µM, and Atg1EAT–Atg13350–525 samples were run at 5 and 20 μM. To characterize the formation of higher-order species when assembling the pentamer, Atg17–Atg31–Atg29 (2 μM) was incubated with Atg1EAT–Atg13350–525 at molar ratios of 1:4 and 1:8. The solvent density (1.007 g⋅mL−1), viscosity (0.01026 poise), and partial specific volumes that were used for the analyses, 0.7152 mL⋅g−1 (Atg1EAT), 0.7109 mL⋅g−1 (Atg1EAT–Atg13350–525), and 0.7348 mL⋅g−1 (Atg17–Atg31–Atg29 and Atg1EAT–Atg13350–525–Atg17–Atg31–Atg29), were calculated by SEDNTERP version 20130813 BETA (http://bitcwiki.sr.unh.edu/index.php/Main_Page) (39). The sedimentation coefficients and apparent molecular weights were calculated from c(s) analysis using SEDFIT version 14.3e (40, 41). The figures were prepared using GUSSI (http://biophysics.swmed.edu/MBR/software.html) version 1.0.8d with the sedimentation coefficients standardized to s20,w.
Isothermal Titration Calorimetry.
ITC was conducted with a MicroCal Auto-iTC200 apparatus (GE Healthcare). Proteins were dialyzed in the same buffer used for AUC. The binding affinity between Atg13350–525 and Atg1EAT was determined by using 10 μM maltose-binding protein (MBP)–tagged Atg13350–525 in the cell and 100 μM Atg1EAT in the syringe. As a control, Atg1EAT was injected into 10 μM MBP (Fig. S8). To measure the interaction between the two subcomplexes, the cell contained 15 μM Atg17–Atg31–Atg29 and the syringe contained 450 μM Atg1EAT–Atg13350–525. Experiments were done at 20 °C, with 20 total injections of 2 μL per injection. The data were baseline-corrected with NITPIC (42) and loaded into SEDPHAT (43–47) for global analysis and fitting using a 1:1 model.
HDX-MS Experiments.
Amide HDX-MS was initiated by a 20-fold dilution of stock Atg1EAT, Atg1EAT–Atg13350–525, or MBP-tagged Atg13350–525 (20–40 μM) into D2O buffer containing 20 mM Tris⋅HCl (pD 8.0), 200 mM NaCl, and 1 mM DTT at 30 °C. The pentamer was assembled by incubating Atg1EAT–Atg13350–525 (20 μM) with Atg17–Atg31–Atg29 (354 μM) for a minimum of 1 h at room temperature before 20-fold dilution into D2O. After intervals of 10 s to 1 h, the exchange was quenched at 0 °C with the addition of ice-cold quench buffer (400 mM KH2PO4/H3PO4, pH 2.2). Quenched samples were injected onto an HPLC-MS (Agilent 1100) with in-line peptic digestion and desalting. Desalted peptides were eluted and directly analyzed by an Orbitrap Discovery mass spectrometer (Thermo Scientific). The HPLC system was extensively cleaned between samples. Initial peptide identification was done by running tandem MS/MS experiments. Peptides were identified using a Mascot (48) and PEAKS Studio 7 (www.bioinfor.com) search. Initial mass analysis of the peptide centroids was performed using the software HDExaminer version 1.3 (Sierra Analytics) followed by manual verification of every peptide. The deuteron content of the peptic peptides covering Atg1EAT and Atg13350–525 was determined from the centroid of the molecular ion isotope envelope. The deuteron content was adjusted for deuteron gain/loss during digestion and HPLC. Both nondeuterated and fully deuterated Atg1EAT and Atg13350–525 were analyzed. Fully deuterated samples were prepared by three cycles of drying and resolubilization in D2O and 6 M guanidinium hydrochloride. For the kinetic analysis of the T-to-R transition, the abundance of the two deuterated mass species was calculated by fitting two Gaussian peaks to the bimodal isotope cluster. The isotope cluster area of the R state was divided by the sum of the areas of the bimodal isotope cluster [R/(R+T)] and plotted as a function of time. The data were fitted using a first-order rate equation using GraphPad Prism. Experiments were repeated three times and errors are given as SEM. For full-length protein analysis, samples were trapped on a reversed-phase column and desalted for 3 min before analysis. To analyze deuteron incorporation into full-length proteins, mass spectra of deuterated proteins were analyzed by ProMass Deconvolution software (Thermo Scientific). The deuteron content was adjusted for deuteron gain/loss during HPLC.
Supplementary Material
Acknowledgments
We thank J. Doudna for ITC instrument use, J. Kuriyan for SEC-MALS instrument use, and the MacroLab for analytical ultracentrifuge use. This work was supported by National Institutes of Health Grant GM111730 (to J.H.H.), Ruth Kirschstein National Research Service Award Fellowships GM112301 (to C.W.D.) and GM099319 (to M.J.R.), a Damon Runyon Cancer Research Fellowship (to R.E.S.), and a L’Oreal USA Women in Science Fellowship (to R.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407214111/-/DCSupplemental.
References
- 1.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22(1):R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 2.Reggiori F, Klionsky DJ. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics. 2013;194(2):341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurley JH, Schulman BA. Atomistic autophagy: The structures of cellular self-digestion. Cell. 2014;157(2):300–311. doi: 10.1016/j.cell.2014.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mari M, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190(6):1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto H, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198(2):219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge L, Baskaran S, Schekman R, Hurley JH. The protein-vesicle network of autophagy. Curr Opin Cell Biol. 2014;29C:18–24. doi: 10.1016/j.ceb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Abeliovich H, Zhang C, Dunn WA, Jr, Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14(2):477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151(7):1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papinski D, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53(3):471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22(2):132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Chew LH, Setiaputra D, Klionsky DJ, Yip CK. Structural characterization of the Saccharomyces cerevisiae autophagy regulatory complex Atg17-Atg31-Atg29. Autophagy. 2013;9(10):1467–1474. doi: 10.4161/auto.25687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12(2):209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 14.Jao CC, Ragusa MJ, Stanley RE, Hurley JH. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc Natl Acad Sci USA. 2013;110(14):5486–5491. doi: 10.1073/pnas.1220306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17(11):1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem. 2009;81(19):7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konermann L, Pan J, Liu Y-H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40(3):1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 18.Kamada Y, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30(4):1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraft C, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31(18):3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganley IG, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20(7):2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott SV, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275(33):25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 26.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6(1):79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 27.Kabeya Y, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16(5):2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujioka Y, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21(6):513–521. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- 29.Engen JR. Analysis of protein complexes with hydrogen exchange and mass spectrometry. Analyst (Lond) 2003;128(6):623–628. doi: 10.1039/b212800b. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z, Post CB, Smith DL. Amide hydrogen exchange determined by mass spectrometry: Application to rabbit muscle aldolase. Biochemistry. 1996;35(3):779–791. doi: 10.1021/bi952227q. [DOI] [PubMed] [Google Scholar]
- 31.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25(1):158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 32.Walters BT, Ricciuti A, Mayne L, Englander SW. Minimizing back exchange in the hydrogen exchange-mass spectrometry experiment. J Am Soc Mass Spectrom. 2012;23(12):2132–2139. doi: 10.1007/s13361-012-0476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11(2):150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 34.Pantazatos D, et al. Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc Natl Acad Sci USA. 2004;101(3):751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 2009;28(5):602–613. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabeya Y, et al. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389(4):612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 37.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, et al. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci USA. 2013;110(24):9800–9805. doi: 10.1073/pnas.1302337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. 1992. Computer-aided interpretation of analytical sedimentation data for proteins. Analytical Ultracentrifugation in Biochemistry and Polymer Science, eds Harding SE, Rowe AJ, Horton JC (R Soc Chem, Cambridge, UK), pp 90–125.
- 40.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90(12):4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keller S, et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem. 2012;84(11):5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balbo A, et al. Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation. Proc Natl Acad Sci USA. 2005;102(1):81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vistica J, et al. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326(2):234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 45.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys J. 2005;89(1):619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houtman JC, et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: Application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16(1):30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320(1):104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 48.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.