Phylogenetics, the study of evolutionary relationships, is critical to understanding the history of life on Earth, integrating and synthesizing information collected across diverse organisms, and testing hypotheses about evolutionary patterns and processes. Most phylogenetic studies are now based on detailed analyses of gene sequence evolution. Over the last several decades, these studies have provided a much better understanding of the structure of the Tree of Life. This progress has also highlighted and helped define many tough phylogenetic questions that have not been answered yet with available data and analysis methods. This has inspired the search for new categories of data that could succeed where analyses of gene sequence evolution have not yet provided strong, consistent phylogenetic support. In PNAS, Thomson et al. (1) examine one of these new categories of data (2–5): the presence and absence of microRNA (miRNA) families. They find that there are several reasons why they may not be as useful for estimating phylogenies as has been proposed.
miRNAs are small RNA genes that are involved in posttranscriptional gene regulation (6, 7). They originated independently in plants, animals, and several other organisms (8). miRNAs and the enzymes needed to process them are missing entirely from comb jellies (ctenophores), suggesting that they may have arisen within animals rather than before the most recent common ancestor of animals (9). Different miRNA families are found in different animals, providing a potential source of information on animal relationships. The sequence of each miRNA family is highly conserved, which makes them relatively easy to tell apart.
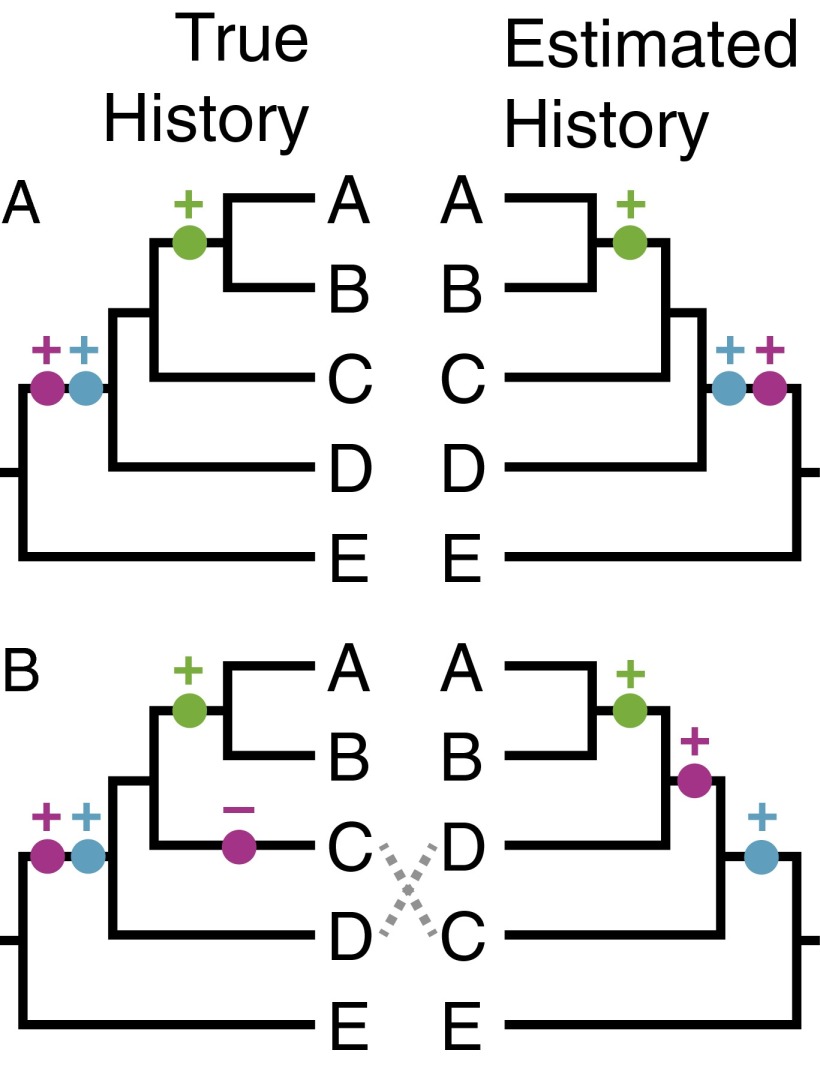
Why was it believed that investigating the presence and absence of miRNAs could succeed where other types of phylogenetic character data have failed? It comes down to homoplasy: any evolutionary pattern of character evolution that deviates from the simplest possible evolutionary history (Fig. 1). Homoplasy can be due to a variety of processes, including convergence and secondary reversal. Homoplasy is common in molecular sequence data, which is one of the main reasons that answering some phylogenetic questions has been so tough. The proponents of miRNA phylogenetic analyses have argued that homoplasy is exceptionally rare in miRNA presence/absence data and that miRNA therefore provides a clear picture of evolutionary relationships (2–5). Because of the way they originate, the convergent acquisition of miRNA families is extremely unlikely, and each is thought to have a single evolutionary origin (3). This means that if the same miRNA family is present in two different species, it was also present in the most recent common ancestor of these species. In addition, however, it has also been claimed that miRNA is rarely lost once it is gained (2, 3, 5). If this is true, then all species that share a particular miRNA are more closely related to each other than to any species that lacks this miRNA. The lack of homoplasy would lead to a ratchet-like evolution of miRNA content through time. miRNA families would accumulate in different lineages without being lost, and their distribution in living species would provide an unambiguous picture of phylogenetic relationships. This has led some to conclude that miRNA “are potentially the near homoplasy-free data set that systematists have long wished for” (3).
Fig. 1.
A simplified example of the impact of unrecognized homoplasy on phylogenetic analysis. Evolutionary changes in each miRNA family are indicated by a different color. (A) If miRNA evolution proceeds without homoplasy, there would be a single origin (indicated by a plus) and no losses for each miRNA family. A simple model can recover true relationships. (B) If there is homoplasy, in this case a single loss (indicated by a minus), and it is not recognized with additional data and robust analysis tools, phylogenetic estimation can provide the wrong answer (C and D are switched).
Thomson et al. (1) evaluate three specific issues: the hypothesis that secondary loss of miRNA is rare, the suitability of the analysis methods that have been applied to miRNA evolution, and the ability to correctly detect the presence or absence of miRNA. They find critical problems on each of these fronts. Their analyses indicate that secondary loss of miRNA is widespread. This fundamentally undermines what had been proposed as their unique value, remarkably low homoplasy, for phylogenetic inference. Thomson et al. (1) then apply Bayesian phylogenetic inference methods that can better accommodate secondary loss, revealing considerable phylogenetic uncertainty. Finally, Thomson et al. find that miRNA families often go undetected using standard observation methods. This suggests that, in addition to often being lost, miRNAs are sometimes not detected. Thomson et al. (1) reexamine five previously published phylogenies based on miRNA and find that when these problems are taken into account, the results of all these studies are fundamentally altered. The support for stated claims is found to be very weak in two of the studies, and in three of the studies, a strongly supported contradictory conclusion is reached.
Why were these strong claims about low homoplasy in miRNA evolution made in the first place, and why did it take so long to systematically examine them? The amount of homoplasy that can be observed depends on how well a character is sampled. It is only in the last decade that a concerted effort has been made to collect miRNA data across phylogenetically diverse animals (2). Initial glimpses of miRNA diversity were consistent with low homoplasy (2), but the sampling was not yet sufficient to rigorously test the hypothesis that they had low rates of loss. The original claims that there are no losses have been tempered through the years as more data became available (5), but it was still claimed that they were remarkably low and little was done analytically to accommodate these losses.
There are multiple historical precedents for the problems identified by Thomson et al. (1). Considerations of each type of new phylogenetic character data often initially assert that homoplasy is low, and these claims are later refuted when sufficient data become available to actually test them (10). Early animal phylogenies were based on the assumption that complex morphological traits such as body cavities and segmentation were difficult to gain and lose and therefore exhibited little homoplasy. It is only when other categories of data, particularly molecular sequence data, were obtained that these old assumptions became testable hypotheses. Many have been soundly rejected (11, 12). Segmented animals such as arthropods and annelids, for example, do not form an evolutionary clade to the exclusion of unsegmented animals. Arthropods and annelids are distantly related (13), and segmentation has been repeatedly lost within annelids. Another historical precedent for underestimated homoplasy is the phylogenetic analysis of short and long interspersed elements, mobile genetic elements that replicate in the genome. The insertion of these elements was proposed to be nearly homoplasy free (14), making them an ideal phylogenetic marker that did not even require statistical analysis (15). In a striking parallel to miRNA, later analyses made it clear that there was homoplasy and that observation problems compromised their phylogenetic utility (10). Rather than continue to repeat this history of underestimating homoplasy, future considerations of new categories of phylogenetic data should start with the null hypothesis that homoplasy is higher. Low homoplasy should be invoked only if sufficient data later reject this null hypothesis.
What are the implications of the findings of Thomson et al. (1)? There is bad news and good news. The bad news is that miRNAs are less useful than had been proposed for resolving difficult phylogenetic problems. This does not necessarily mean that miRNAs cannot make useful contributions to estimating phylogenies; they just have the same limitations as other data. They may still prove to be useful when analyzed in combination with other data. Scoring the presence and absence of miRNAs based on full genome sequences rather than tissue-specific transcriptomes will help resolve some of the
Thomson et al. find that miRNA families often go undetected using standard observation methods.
sampling issues described by Thomson et al. (1). The major challenge, however, will be to develop phylogenetic analysis methods that can simultaneously evaluate many different types of data, including sequence evolution, gene gain and loss, and genomic rearrangements, in a single unified framework. This echoes past attempts to integrate morphological and molecular sequence data. As molecular datasets grew much larger than morphological datasets, the question became how to weight the datasets. Developing methods that can simultaneously assess multiple categories of data will be one of the major methodological challenges for the field of phylogenetics in coming years.
The good news is that we are now in a better position to learn about miRNA function. Homoplasy creates challenges for estimating phylogenies, but it is tremendously helpful for testing mechanistic hypotheses about character evolution. This is because homoplasy provides replication. If a trait has a single evolutionary origin and no losses, it is hard to know if that one change is associated with evolutionary changes in other characters. If the trait has multiple changes, then it is possible to test if other organism characters repeatedly change each time the trait changes. This is particularly helpful in the case of understanding miRNA evolution, as some very bold hypotheses have been proposed for their functional evolution. It has been claimed that increased miRNA diversity has enabled lineages to become more complex than other lineages with fewer miRNA families (2, 16, 17). In this scenario, a ratchet-like accumulation of miRNA families is seen as the cause of a ratchet-like increase in complexity (18). In this way, miRNA has been invoked as an explanation for the “taxonomic hierarchy of animal relationships” (2). However, the relationships of living animals are not hierarchical but are tree-like. Organisms cannot be arrayed on a single spectrum from simple to complex, akin to Aristotle’s scala naturae, with a parallel spectrum of miRNA diversity. Organisms have many traits that vary independently, and each living species, which is at the tip of a branch of the phylogenetic tree, can have a mix of complex and simple traits that are gained and lost independently. With greater homoplasy in miRNA evolution, it will be easier to learn if any of these changes in specific complex traits are associated with gains and losses of particular miRNA families. Homoplasy is not always the enemy; it can sometimes be your friend.
Supplementary Material
Footnotes
The author declares no conflict of interest.
See companion article on page E3659.
References
- 1.Thomson RC, Plachetzki DC, Mahler DL, Moore BR. A critical appraisal of the use of microRNA data in phylogenetics. Proc Natl Acad Sci USA. 2014;111:E3659–E3668. doi: 10.1073/pnas.1407207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sempere LF, Cole CN, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: Insights into evolutionary complexity and constraint. J Exp Zoolog B Mol Dev Evol. 2006;306(6):575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- 3.Sperling EA, Peterson KJ. In: Animal Evolution: Genomes, Fossils, and Trees. Telford MJ, Littlewood DTJ, editors. Oxford, UK: Oxford Univ Press; 2009. pp. 157–170. [Google Scholar]
- 4.Dolgin E. Phylogeny: Rewriting evolution. Nature. 2012;486(7404):460–462. doi: 10.1038/486460a. [DOI] [PubMed] [Google Scholar]
- 5.Tarver JE, et al. miRNAs: Small genes with big potential in metazoan phylogenetics. Mol Biol Evol. 2013;30(11):2369–2382. doi: 10.1093/molbev/mst133. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Tarver JE, Donoghue PCJ, Peterson KJ. Do miRNAs have a deep evolutionary history? BioEssays. 2012;34(10):857–866. doi: 10.1002/bies.201200055. [DOI] [PubMed] [Google Scholar]
- 9.Maxwell EK, Ryan JF, Schnitzler CE, Browne WE, Baxevanis AD. MicroRNAs and essential components of the microRNA processing machinery are not encoded in the genome of the ctenophore Mnemiopsis leidyi. BMC Genomics. 2012;13:714. doi: 10.1186/1471-2164-13-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillis DM. SINEs of the perfect character. Proc Natl Acad Sci USA. 1999;96(18):9979–9981. doi: 10.1073/pnas.96.18.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgecombe GD, et al. Higher-level metazoan relationships: Recent progress and remaining questions. Org Divers Evol. 2011;11(2):151–172. [Google Scholar]
- 12.Wagele JW. In: Deep Metazoan Phylogeny: The Backbone of the Tree of Life. Bartholomaeus T, editor. Berlin: Walter De Gruyter Inc; 2014. [Google Scholar]
- 13.Aguinaldo AM, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387(6632):489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 14.Nikaido M, Rooney AP, Okada N. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interpersed elements: Hippopotamuses are the closest extant relatives of whales. Proc Natl Acad Sci USA. 1999;96(18):10261–10266. doi: 10.1073/pnas.96.18.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murata S, Takasaki N, Saitoh M, Tachida H, Okada N. Details of retropositional genome dynamics that provide a rationale for a generic division: The distinct branching of all the pacific salmon and trout (Oncorhynchus) from the Atlantic salmon and trout (Salmo) Genetics. 1996;142(3):915–926. doi: 10.1093/genetics/142.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heimberg AM, Sempere LF, Moy VN, Donoghue PCJ, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci USA. 2008;105(8):2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12(12):846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler BM, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11(1):50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]