Significance
Cell surface proteins are regulated by a constant cycle of internalization and recycling from intracellular compartments called endosomes. From these organelles, two protein sorting platforms, sorting nexin 27 (SNX27) and the retromer complex, play a critical role in the retrieval of various proteins responsible for ion transport, glucose metabolism, neurotransmission, and other cell functions. Based on the three-dimensional structure of SNX27 in complex with the retromer subunit VPS26, we define the mechanism by which these proteins cooperate to drive endosomal cargo sorting. Retromer and SNX27 dysfunction is implicated in various disorders, including diabetes, Down syndrome, Parkinson disease, and Alzheimer’s disease, and this work provides important insights into the assembly of this essential endosomal sorting machinery.
Keywords: Alzheimer's disease, Down syndrome, Parkinson disease, endosomal recycling
Abstract
The sorting nexin 27 (SNX27)-retromer complex is a major regulator of endosome-to-plasma membrane recycling of transmembrane cargos that contain a PSD95, Dlg1, zo-1 (PDZ)-binding motif. Here we describe the core interaction in SNX27-retromer assembly and its functional relevance for cargo sorting. Crystal structures and NMR experiments reveal that an exposed β-hairpin in the SNX27 PDZ domain engages a groove in the arrestin-like structure of the vacuolar protein sorting 26A (VPS26A) retromer subunit. The structure establishes how the SNX27 PDZ domain simultaneously binds PDZ-binding motifs and retromer-associated VPS26. Importantly, VPS26A binding increases the affinity of the SNX27 PDZ domain for PDZ- binding motifs by an order of magnitude, revealing cooperativity in cargo selection. With disruption of SNX27 and retromer function linked to synaptic dysfunction and neurodegenerative disease, our work provides the first step, to our knowledge, in the molecular description of this important sorting complex, and more broadly describes a unique interaction between a PDZ domain and an arrestin-like fold.
After endocytosis, transmembrane proteins (here termed cargos) are delivered to the early endosome, where endosome-associated protein complexes direct their sorting to one of two fates (1). Either the cargo is sorted for inclusion into forming intraluminal vesicles, which leads to its ultimate degradation in the lysosome (2), or the cargo is recycled to either the trans-Golgi network (TGN) (3) or the plasma membrane for reuse (4). By regulating the residency time and transport of transmembrane proteins through the endosome network, cargo sorting plays a vital role in maintaining the functionality of the plasma membrane and TGN, and in allowing active remodeling of the protein and lipid composition of these membranes during organism development and in response to cellular demand.
Retromer is an ancient, stable heterotrimer composed of VPS26, VPS29, and VPS35 (5, 6). Although retomer initially was characterized through its role in recycling cargo proteins from endosomes to the TGN (7), the recent identification of the sorting nexin-27 (SNX27)-containing retromer complex as an important regulator of cargo recycling from endosomes to the plasma membrane (8) has established retromer as a master regulator of multiple endosome recycling pathways (9). SNX27 itself serves as an endosome-associated cargo adaptor that interacts with transmembrane cargo proteins containing Asn-Pro-Xaa-Tyr (NPxY) motifs through its carboxy-terminal 4.1/ezrin/radixin/moesin (FERM)-like domain (10), and via its amino-terminal PSD95, Dlg1, zo-1 (PDZ) domain with transmembrane proteins that have class I PDZ-binding motifs (PDZbms) (S/T-x-Φ) (11–13). SNX27-retromer cargos containing PDZbms include the β2-adrenergic receptor (8), ionotrophic glutamate receptors (NMDA and AMPA receptors) (14), potassium channels (15), somatostatin receptor (16), and the glucose transporter 1 (GLUT1) (17). Indeed, our recent proteomics analysis identified more than 80 transmembrane proteins, including ion and amino acid transportors, nutrient transporters, and signaling receptors, that depend on the SNX27-retromer complex for their endosome-to-plasma membrane recycling (17).
Given the importance of SNX27-retromer for the recycling of cargo proteins that perform a wide variety of essential cellular functions, it is not surprising that homozygous knockout animal models are embryonically lethal (18). However, disruption of retromer-mediated endosome sorting, either as a result of reduced expression or through an autosomal recessive mutation, is observed in various neurodegenerative disorders, including Parkinson disease (19, 20), Alzheimer’s disease (21), and, for SNX27, Down syndrome (22). Moreover, the identification of a small molecule that appears to act by stabilizing retromer expression and shows potential for future treatment of Alzheimer’s disease (23) underscores the importance of understanding the assembly, function, and regulation of retromer complexes.
Here we establish the molecular mechanism that governs the core interaction in the assembly of the SNX27-retromer. We identify a β-hairpin insertion in the PDZ domain of SNX27 that mediates binding to a conserved groove between the two lobes of the arrestin-fold structure of VPS26A. Informed by a high-resolution crystal structure, we establish that disruption of the binding interface leads to loss of cargo recycling from endosomes to the plasma membrane. Furthermore, we reveal cooperativity between the sorting complex assembly and the sequence-dependent capture of cargo, with binding of VPS26A to the SNX27 PDZ domain increasing the affinity for PDZbms by an order of magnitude. Our work provides the first step, to our knowledge, in the molecular description of this important sorting complex, and more broadly describes a unique interaction between a PDZ domain and an arrestin-like fold.
Results
A β-Hairpin Insertion of the SNX27 PDZ Domain Mediates VPS26 Binding.
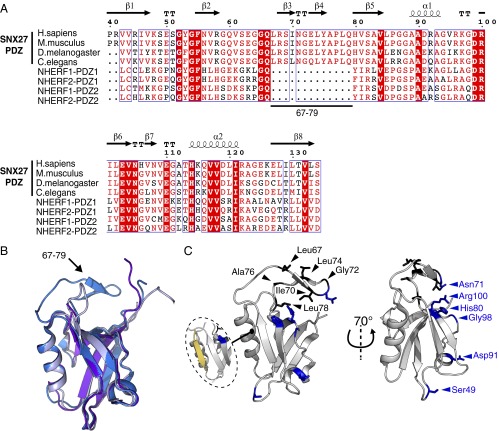
By aligning the amino acid sequence of the SNX27 PDZ domain with the sequences of the Na+/H+ exchanger regulatory factor (NHERF) 1 (24) and NHERF2 PDZ domains (Fig. 1A), which are similar in sequence and structure but do not bind retromer, we identified a conserved stretch of 11 amino acids (Leu67–Pro77) required for binding to VPS26 (17). As demonstrated by the crystal structure of the SNX27 PDZ domain (15), residues Leu67–Gln79 form a β-hairpin loop containing β-strands 3 and 4 of the SNX27 PDZ domain (the β3-β4 loop) that extends from the core of the PDZ domain (Fig. 1B).
Fig. 1.
Identification of the VPS26-binding surface on SNX27 by NMR. (A) Alignment of the amino acid sequences of PDZ domains of human SNX27, C. elegans SNX27, and human NHERF1 and NHERF2. Perfectly conserved residues are highlighted in red. Alignment was generated with ESPript 2.2. (B) Structural comparison of the crystal structures of SNX27 PDZ domain (blue) and the first PDZ domain of NHERF1 (gray) and NHERF2 (purple) (PDB ID codes 3QDO, 1G9O, and 2OCS, respectively). The arrow indicates the conserved loop in SNX27 identified by the sequence alignment. (C) Residues showing significant line broadening and change of chemical shift on formation of the SNX27-VPS26A complex are shown in black and blue, respectively, and displayed as sticks on the SNX27 PDZ domain-Kir3.3 peptide complex crystal structure (PDB ID code 3QDO). Affected residues (see the HSQC spectra in Fig. S1) map to a region close to the Leu67-Pro77 loop, away from the binding site for cargos such as the Kir3.3 potassium channel (in yellow).
To confirm the importance of the β3-β4 loop insertion in binding of SNX27 to VPS26, we investigated perturbations in SNX27 PDZ domain residues on binding of VPS26A using nuclear magnetic resonance (NMR) spectroscopy. 15N/13C-labeled SNX27 PDZ domain was produced, and backbone assignments were obtained to map the chemical shift variations induced on the addition of VPS26A (Fig. S1). Calculated Cα and Cβ chemical shift variations from random coil values for the assigned residues closely matched the secondary structure elements observed in the SNX27 PDZ domain crystal structure. These values correlated very well with the theoretical chemical shifts calculated using the ShiftX program (25), implying that the topology of the soluble protein is highly similar to that of the crystal form (Fig. S2). Affected residues were mapped onto the SNX27 PDZ domain crystal structure to identify the VPS26A-binding surface. Residues corresponding to broadening peaks in the 15N-1H HSQC spectra (displayed in black: Leu67, Ile70, Gly72, Leu74, Ala76, and Leu78) are all found within the β3-β4 loop of SNX27 PDZ domain (Fig. 1C), suggesting involvement in binding of VPS26A. A number of residues with peaks showing altered chemical shifts on VPS26 titration are mapped to distant structural elements (displayed as blue sticks: 49S, 71N, 80H, 91D, 98G, and 100R), and might undergo subtle structural rearrangements induced by engagement with VPS26A.
Crystal Structure of the SNX27 PDZ Domain in Complex with VPS26A.
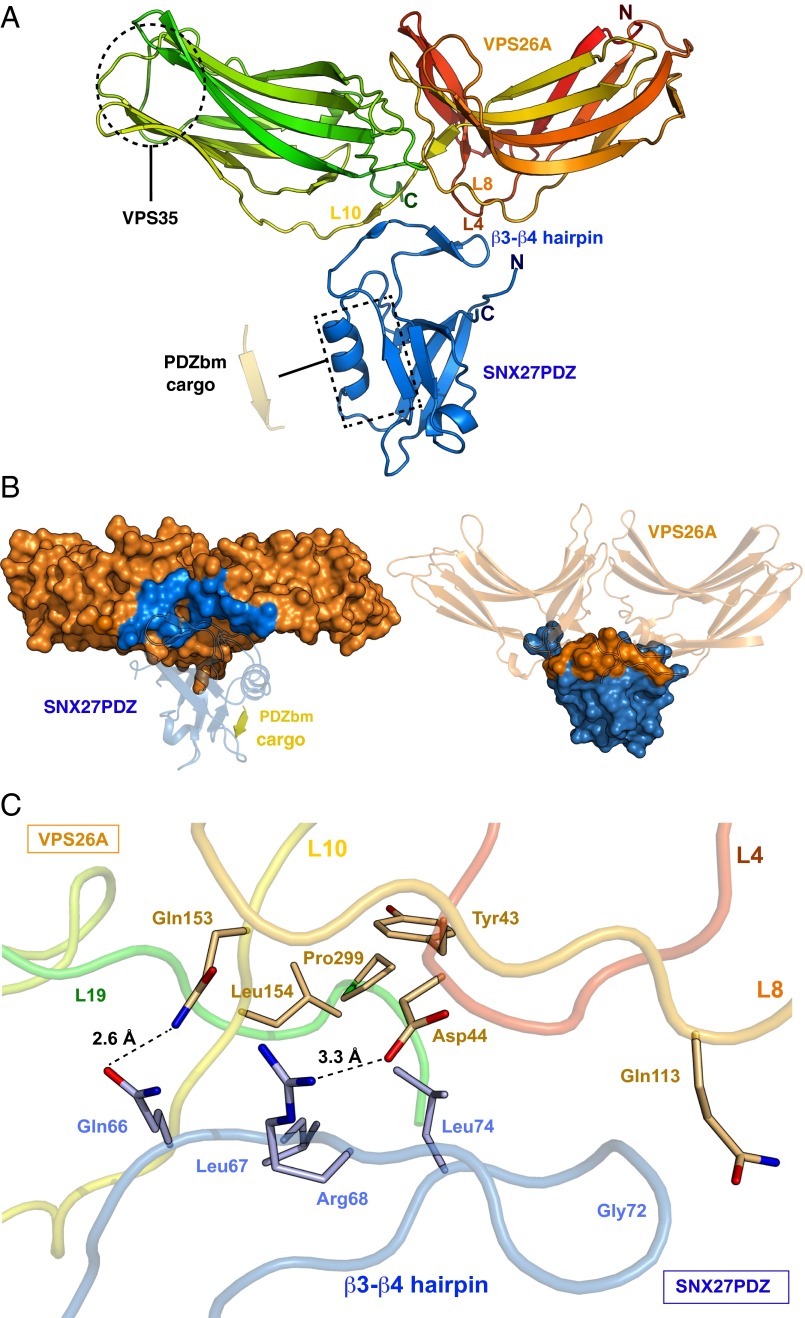
To investigate VPS26 recognition in more depth, we determined the crystal structure of the SNX27 PDZ domain bound to VPS26A at 2.7-Å resolution (Fig. 2A and Table 1). There is a single complex in the asymmetric unit, and the final model contains residues 9–301 for VPS26A and residues 40–135 for the SNX27 PDZ domain. Similar to previously described apo crystal structures, the C-terminal VPS26A residues 302–327 are not visible in the electron density, confirming that this “tail” is structurally flexible (26) and remains so on SNX27 PDZ domain binding. The topology of both proteins is similar to previously published high-resolution structures (PDB ID codes 2FAU and 3QDO) (15, 26). Overall, the structure of SNX27 PDZ domain bound to VPS26A is T-shaped in appearance, with the SNX27 PDZ domain forming the base and VPS26A the upper platform. The interaction involves docking of SNX27 into a highly complementary cleft of the VPS26 protein between the N- and C-terminal subdomains, with a relatively small buried surface area of 536 Å2 (PDBePISA server: www.ebi.ac.uk) (Fig. 2B).
Fig. 2.
Crystal structure of the SNX27 PDZ domain-VPS26A complex. (A) Ribbon representation of the crystal structure of the SNX27 PDZ domain (blue) bound to VPS26A (spectrum red-yellow-green). The dashed black rectangle indicates a putative PDZbm-binding pocket. The dashed black circle indicates the VPS35 interaction surface. Loops 4, 8, and 10 of VPS26A and the β3-β4 hairpin of SNX27PDZ, which make direct contact, are highlighted. (B) Interaction surfaces of VPS26A and SNX27. VPS26A is gold, with its SNX27-binding surface in blue. SNX27 is blue, with its VPS26A-binding surface in gold. The PDZbm peptide of the Kir3.3 cargo protein (yellow) is modeled on the published structure with the SNX27 PDZ domain. (C) Detailed view of the interaction surface of the complex between the SNX27 PDZ domain β3-β4 loop and VPS26A loops (L4–L19). The SNX27 and VPS26A main chains are blue and spectrum red-yellow-green, respectively. Selected residues involved in the interaction are displayed as sticks. Electrostatic and hydrogen bonds, represented by dashed black lines, are measured in angstroms.
Table 1.
Data collection, phasing, and refinement statistics
| SNX27PDZ-VPS26A* | |
| Data collection | |
| Beamline | MX-1-AS |
| Space group | P321 |
| Cell dimensions, Å | 113.81, 113.81, 94.75 |
| Cell dimensions, ° | 90.0, 90.0, 120.0 |
| Wavelength | 1.002 |
| Resolution, Å | 68.3–2.70 (2.85–2.70)† |
| Rsym or Rmerge | 0.105 (0.964) |
| I/σI | 14.8 (2.8) |
| Completeness, % | 100 (100) |
| Redundancy | 12.0 (12.3) |
| Refinement | |
| Resolution, Å | 56.9–2.70 (2.76–2.70) |
| No. of reflections | 19,443 |
| Rwork/Rfree | 0.219/0.259 |
| No. of atoms | 3,111 |
| Protein | 3,074 |
| Ligand/ion | 2 |
| Water | 35 |
| B-factors | 82.58 |
| Protein | 82.79 |
| Ligand/ion | 174.6 |
| Water | 64.83 |
| rmsd | |
| Bond length, Å | 0.011 |
| Bond angle, ° | 1.36 |
The SNX27PDZ-VPS26A complex structure was solved from a single crystal.
Values in parentheses are for the highest-resolution shell.
The crystal structure confirms that the conserved β-hairpin insertion, implicated in the binding of VPS26A by our NMR studies, does indeed mediate the interaction with VPS26. The SNX27 β3-β4 loop is well ordered in both free and complexed crystal structures, and its orientation relative to the core SNX27 PDZ domain structure does not change notably on binding to VPS26A (discussed further below). The two antiparallel β-strands, β3 and β4, composing a β-hairpin in the SNX27 PDZ domain, confer rigidity to this section of the protein, shaping it into an interaction platform that associates neatly within the complementary groove between the N- and C-terminal fibronectin type III domains of VPS26A. In detail (Fig. 2C), the interface is governed by hydrophobic contacts between SNX27 and VPS26A, as well as several specific polar and electrostatic interactions. Leu67 and Leu74 within the SNX27 PDZ domain β4-strand lie in a hydrophobic cavity formed by VPS26 residues Tyr43, Leu154, and Pro299 on loop (L) 4, L10, and L19, respectively. Gln66 of SNX27 forms a hydrogen-bonding interaction with Gln153 on the VPS26A L10, and similarly the Arg68 side chain of SNX27 forms an electrostatic bond with the Asp44 carboxyl group of VPS26A on L4. The Ser69-Ile70 amide group present in the β3 strand of the SNX27 PDZ domain forms a hydrogen bond with Asp44 on L4, and the Gly72 backbone amide group interacts similarly with the VPS26 Asn113 carboxyl group on L8. The structure also indicates that part of the binding is governed by main chain contacts that result in formation of a short stretch of intermolecular β-sheet involving residues 155–156 in L10 of VPS26A and residues 65–66 preceding the β3-β4 hairpin in the SNX27 PDZ domain.
The foregoing interactions appear to constitute the core of the SNX27-VPS26A association mechanism, a hypothesis supported by the high degree of conservation of these residues (Fig. S3). Indeed, the association of the SNX27 PDZ domain with VPS26 is conserved in metazoans, as demonstrated by the ability of the SNX27 PDZ domain from Caenorhabditis elegans to directly bind VPS26 from the same organism (Fig. S4). Taken together, these data indicate that the SNX27 PDZ domain β3-β4 loop insertion acts as a conserved and apparently unique feature of the SNX27 PDZ domain to bind the retromer VPS26A subunit within a complementary cleft between N- and C-terminal subdomains.
Disruption of VPS26 Binding by SNX27 Prevents Recycling of Glucose Transporter 1.
To validate the crystal structure of the SNX27 PDZ domain bound to VPS26, we designed mutations in key residues in SNX27 at the interface with VPS26A and tested the ability of these mutant proteins to coimmunoprecipitate VPS26. We removed the β3-β4 loop by deleting residues 67–77 (Δ67–77) and also individually mutated residues Leu67, Ile70, and Leu74 to alanine. GFP-tagged SNX27 mutant constructs expressed in HEK293T cells were immunoprecipitated with a high-affinity GFP antibody (GFP-trap) (Fig. 3A). GFP-SNX27 precipitated both VPS26A and its paralog VPS26B, indicating that a retromer complex with either VPS26 paralog can interact with SNX27. SNX27 Δ67–77, L67A, and L74A mutant constructs did not precipitate either VPS26A or VPS26B, whereas the I70A mutation markedly reduced the association. We also confirmed these results using bacterially expressed and purified SNX27 PDZ domain and VPS26A proteins (Fig. 3B).
Fig. 3.
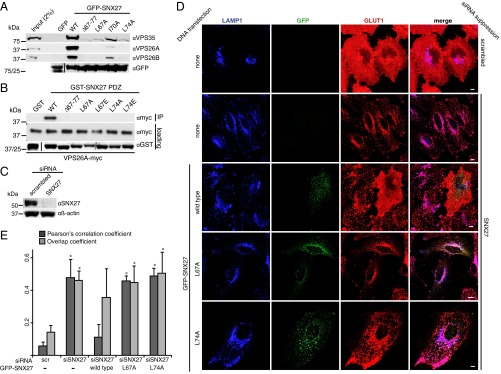
Mutation of the VPS26-binding site of SNX27 prevents endosome-to-plasma membrane recycling of GLUT1. (A) GFP-SNX27 mutant constructs expressed in HEK293T cells were analyzed for binding to the retromer VPS subcomplex by GFP-trap immunoprecipitation and immunoblotting with the indicated antibodies. (B) GST-SNX27 mutant constructs purified from bacteria were tested for binding to purified VPS26A-myc in a direct binding assay. (C) Effective suppression of SNX27 expression using siRNA in HeLa cells. (D) HeLa cells in which SNX27 expression was suppressed using siRNA were transiently transfected with GFP-SNX27 constructs, stained for lysosomal membrane-associated protein 1 (LAMP1) and GLUT1, and imaged on a confocal microscope. (Scale bar: 10 µm.) (E) Pearson correlation coefficient (PCC) and overlap coefficient (Ky) for LAMP1 and GLUT1 in SNX27-suppressed cells expressing the indicated GFP-SNX27 construct. The graph shows the mean of three independent experiments, in each of which at least 30 cells were analyzed. Error bars represent SD. *P < 0.05, two-tailed t test compared with scrambled.
To investigate the effect of abrogating the interaction between SNX27 and VPS26 on endosome-to-plasma membrane recycling, we analyzed the trafficking of a model cargo glucose transporter 1 (GLUT1), a transmembrane protein recycled in a SNX27-retromer–dependent manner (17). When SNX27 expression in HeLa cells is reduced by siRNA (Fig. 3C), the distribution of GLUT1 changes from a predominantly cell surface localization to a late endosome/lysosome localization as the protein fails to undergo endosome-to-plasma membrane recycling and enters the degradative pathway (Fig. 3 D and E). Expression of siRNA-resistant GFP-SNX27 rescued this mistrafficking phenotype; GLUT1 recycled to the plasma membrane was observed in control cells. In contrast, in cells transfected with siRNA-resistant GFP-SNX27 L67A or L74A mutants, GLUT1 remained redistributed in late endosomes/lysosomes.
VPS26A and VPS26B Act Redundantly in SNX27-Retromer–Mediated Recycling.
Similar to what we found in SNX27 mutants, we found that mutating key residues on the VPS26 side of the interface also abrogated the SNX27–VPS26 interaction. We targeted two residues in VPS26A making key contacts with the SNX27 PDZ domain in the cocrystal structure (Fig. 2C). VPS26A Asp44 was mutated to alanine (D44A) to test the requirement for one of the two salt bridges observed in the crystal structure. Leu154 was also targeted (L154A), owing to its involvement in forming a hydrophobic cavity with SNX27 Leu67 and Leu74. In GFP-trap assays, these mutants precipitated only modest levels of SNX27 (Fig. 4A). Disruption of the interaction was specific; these mutations did not noticeably affect precipitation of VPS35. Furthermore, in direct binding assays using recombinant proteins GST-VPS26A L154A failed to precipitate the SNX27 PDZ domain (Fig. 4B).
Fig. 4.
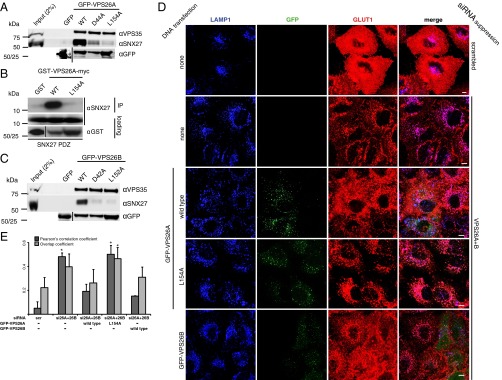
VPS26A and VPS26B function redundantly in SNX27-retromer mediated endosome-to-plasma membrane recycling. (A) GFP-VPS26A mutant constructs expressed in HEK293T cells were analyzed for binding to SNX27 by GFP-trap immunoprecipitation and immunoblotting with the indicated antibodies. (B) GST-VPS26A-myc constructs purified from bacteria were tested for binding to the purified SNX27 PDZ domain in a direct binding assay. (C) GFP-VPS26B mutant constructs expressed in HEK293T cells were analyzed for binding to SNX27 by GFP-trap immunoprecipitation and immunoblotting with the indicated antibodies. (D) HeLa cells, in which VPS26A and VPS26B expression was suppressed using siRNA (knockdown efficiency shown in Fig. S5), were transiently transfected with GFP-VPS26 constructs, stained for LAMP1 and GLUT1, and imaged on a confocal microscope. (Scale bar: 10 µm.) (E) PCC and overlap coefficient (Ky) for LAMP1 and GLUT1 in VPS26-suppressed cells expressing the indicated GFP-VPS26 construct. The graph shows the mean of three independent experiments, in each of which at least 30 cells were analyzed. Error bars represent SD. *P < 0.05, two-tailed t test compared with scrambled.
Because VPS26A is very similar in structure to its paralog VPS26B (27), we investigated whether the equivalent amino acids, Asp42 and Leu152, in VPS26B are required for binding to SNX27. WT GFP-VPS26B precipitated SNX27 in a GFP-trap assay, whereas GFP-VPS26B D42A and L152A constructs did not (Fig. 4C), indicating that the mechanism of interaction with SNX27 is conserved between the paralogs. We next investigated the effect of loss of VPS26 on GLUT1 trafficking. Using siRNA to suppress VPS26A or VPS26B individually had little effect on GLUT1 recycling, with GLUT1 remaining primarily at the plasma membrane as in control cells (Fig. S5). In contrast, suppression of the unique core retromer subunit VPS35 resulted in a similar degree of GLUT1 rerouting to late endosomes/lysosomes as SNX27 suppression. These observations suggest redundancy between VPS26A and VPS26B proteins in endosome-to-plasma membrane recycling, in contrast to the previous finding that VPS26A-retromer can mediate endosome-to-Golgi trafficking of the mannose-6-phosphate receptor, but VPS26B retromer cannot (28). Confirming this redundancy, when both VPS26A and VPS26B were suppressed, GLUT1 mislocalized to late endosomes/lysosomes (Fig. 4D and Fig. S5).
We next tested whether reexpression of siRNA-resistant GFP-VPS26 constructs is able to rescue the knockdown phenotype (Fig. 4 D and E). Expression of either GFP-VPS26A or GFP-VPS26B restored GLUT1 cell surface localization to normal levels; however, the GFP-VPS26A L154A SNX27-binding mutant did not, as expected. These data confirm the requirement for direct engagement of VPS26 retromer subunits by SNX27 for endosome-to-plasma membrane recycling of PDZbm-containing cargo, and demonstrate that either VPS26A- or VPS26B-containing retromer complexes are active in this pathway.
Affinity of SNX27 for PDZ-Binding Motifs Is Enhanced by VPS26 Interaction.
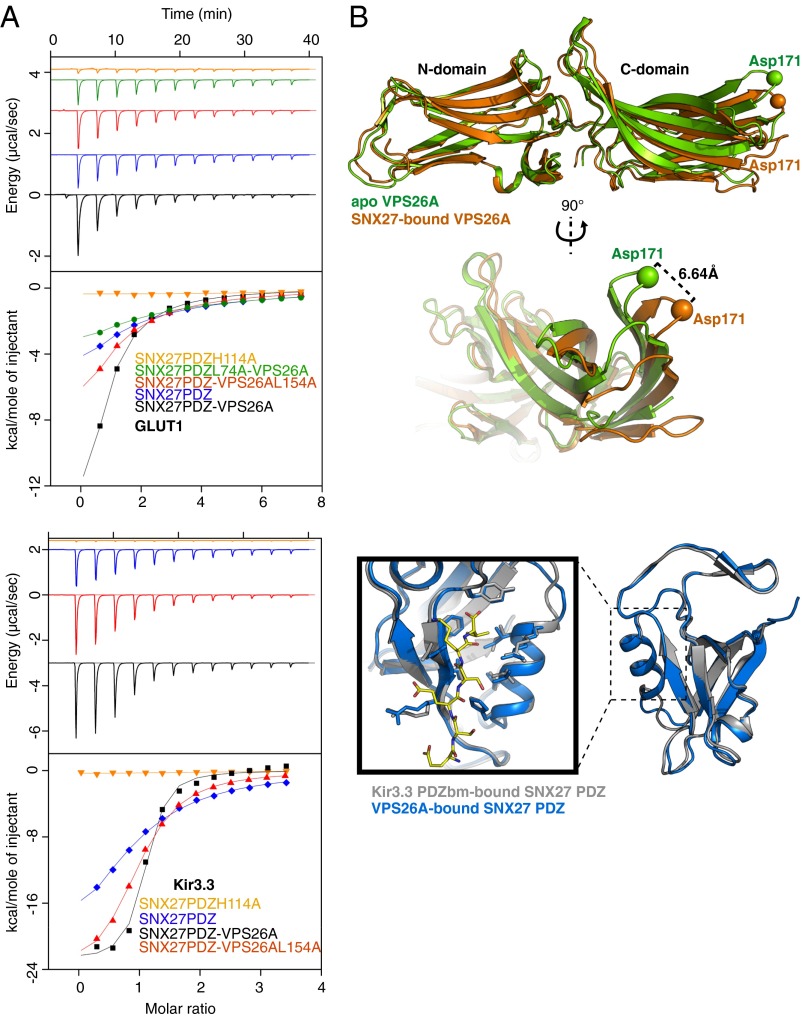
The present structure indicates the VPS26-binding site on the SNX27 PDZ domain is distinct from the site of interaction with PDZbm-containing cargo (15) (Fig. 2 A and B); nonetheless, retromer association may still influence the affinity of SNX27 for cargo. To investigate this possibikity, we used isothermal titration calorimetry (ITC) to measure the strength of the interaction between the GLUT1 PDZbm and the SNX27 PDZ domain in both the presence and absence of VPS26A (Fig. 5A and Table S1). In isolation, an eight-residue synthetic peptide including the C-terminal GLUT1 type I PDZbm (DSQV) bound to the SNX27 PDZ domain with a low to moderate affinity (Kd ∼154 μM), and mutation of H114A in the PDZbm-binding pocket of the SNX27 PDZ domain (13) abolished the interaction, as expected. In the presence of VPS26A, GLUT1 affinity was increased by an order of magnitude (Kd ∼15 μM). Confirming the importance of direct VPS26A binding for this affinity increase, replacing WT proteins with binding-defective mutants abrogated this cooperative effect; SNX27 PDZ domain L74A almost completely reversed the effect, and VPS26A L154A reduced it. When assayed for binding to the Kir3.3 PDZ-ligand in the same experimental setup, an even more striking increase in affinity was observed in the presence of VPS26A (from Kd 17 μM to 1.1 μM) (Fig. 5A and Table S1). Thus, this allosteric bivalent recognition effect is not limited to GLUT1, but is likely a general feature of PDZbm binding by SNX27-retromer.
Fig. 5.
SNX27 binding to cargo motifs is allosterically enhanced by VPS26. (A) (Upper) The GLUT1 PDZbm peptide binds the SNX27 PDZ domain in vitro and has enhanced affinity for the SNX27 PDZ domain-VPS26A complex. Mutation in the SNX27 PDZbm pocket (H114A) abolishes the peptide interaction. Mutations in the VPS26A- and SNX27-binding interface reverse the allosteric effect. (Lower) This effect was observed for the Kir3.3 potassium channel peptide as well. Raw data are at the top; integrated normalized data, at the bottom. Colors indicate the proteins used in the ITC cell. (B) (Upper) Overlay of free (green) and complexed (orange) forms of VPS26A, aligned based on their N-terminal subdomains to highlight the conformational flexibility between the N- and C-terminal subdomains in the free and bound structures. Asp171 Cα, represented as a sphere, is used as an arbitrary reference point to quantify C-terminal domain flexibility. (Lower) Overlay of the SNX27 PDZ domain bound to the Kir3.3 PDZbm (gray) or VPS26A (blue) (PDB ID code 3QGL and this study, respectively). The SNX27 PDZ domain protein superposition highlights the rigidity of the β3-β4 β-hairpin and the canonical PDZ-binding pocket with the Kir3.3 peptide in yellow.
Comparing the structures of VPS26A and the SNX27 PDZ domain within the complex to their free forms does not provide an obvious explanation for this allosteric effect (Fig. 5B). VPS26A has an arrestin-like fold, characteristic of the arrestin family of proteins (26), composed of two curved β-sandwich domains with a short linker that confers some flexibility (27). When bound to the SNX27 PDZ domain, the VPS26A N- and C-terminal domains undergo moderate movement in relation to each other compared with the apo VPS26A structure (26). In the case of SNX27, comparing the structures of the SNX27 PDZ domain bound to either VPS26A or to the Kir3.3 PDZbm (15) shows only small-scale variations in the PDZ domain, with no major structural changes in the PDZbm-binding site that readily explain the allosteric effect. Thus, we speculate that either (i) the increased affinity is a result of long-range stabilization of the SNX27 PDZ domain by VPS26, an effect similar to that observed in the context of the Par6PDZ/Cdc42 interaction (29, 30), or (ii) additional interactions occur between peptide residues upstream of the PDZbm and a secondary site on the SNX27-VPS26 complex. The increased affinity of PDZbms for the SNX27 PDZ domain when bound to retromer may promote the efficient capture of cargo on entering retromer-decorated endosomal tubules.
Discussion
In this study, we present the crystal structure of the PDZ domain of SNX27 bound to the retromer subunit VPS26A. Our structure shows a previously unidentified binding mechanism between the PDZ and the arrestin protein families, and establishes precisely how VPS26 can engage PDZbm-bound SNX27 while simultaneously binding with high affinity to the retromer subunit VPS35. Our detailed biochemical analysis has revealed cooperativity between retromer binding and cargo recognition through the SNX27 PDZ domain, and our phenotypic rescue experiments have clearly defined the critical importance of the SNX27 retromer association for endosome-to-plasma membrane recycling of PDZbm-containing cargos. Overall, these data provide, to our knowledge, the first detailed structural insight into the assembly of the evolutionary conserved SNX27-retromer coat complex.
The PDZ domain of SNX27 binds to retromer through its evolutionarily conserved β3-β4 hairpin loop. Although extensions of the classic PDZ fold exist in other proteins and often contribute to the assembly of signaling protein complexes, the exact sequence of this β3-β4 hairpin insertion is found only in the PDZ domain of SNX27. Many structures of PDZ domain complexes have established the molecular basis for their homodimerization (31) and heterodimerization (32) via structural embellishments. Outside of canonical PDZbm interactions, the only other known structure of a PDZ domain bound to a different protein family is that of Cdc42 bound to the CRIB motif present in the Par6 PDZ domain (33). Thus, our structure presents a unique example of a PDZ domain insertion mediating the interaction with a distinct protein family. In addition, our structure adds to recent insights into arrestin family ligand interactions that have revealed how different members of the arrestin-like protein family provide distinct surfaces for binding their respective partners by apparently diverse mechanisms (34, 35). As shown in Fig. S6, the α-arrestin family member thrioredoxin-interacting protein (TXNIP) and the β-arrestin1 proteins use entirely distinct interfaces to bind their related known interactors.
Within the β3-β4 loop of the SNX27 PDZ domain, Leu67 and Leu74 contact the VPS26A surface via insertion into a hydrophobic pocket, and the interaction appears to be stabilized by two salt bonds involving Asp44 and Gln153 on each side of the cavity. The matching residue for Gln153 in VPS26B is Thr151, which likely is unable to establish a salt bridge with the SNX27 residue Arg68. This is the only notable structural difference between VPS26A and VPS26B at the interaction surface, although it remains to be seen whether this difference has any biological relevance by distinguishing the functions of the two paralogs. Indeed, we found that VPS26A and VPS26B seemingly function redundantly in SNX27-retromer–mediated endosome-to-plasma membrane recycling in a model cell system, because suppression of each protein individually had no effect on trafficking of the SNX27-retromer transmembrane cargo protein GLUT1 (Fig. S5). Different tissue distributions of VPS26A and VPS26B in vivo (28) may indicate cell-specific roles for each paralog at the organism level, however.
To validate the biological relevance of the VPS26A-SNX27 complex crystal structure, we generated mutant constructs that specifically abrogate the interaction of SNX27 and VPS26. In rescue experiments, these mutants were unable to revert the missorting of GLUT1 to lysosomes, thereby establishing the central role of the SNX27–VPS26 interaction for efficient endosome-to-plasma membrane recycling. These point mutants will be useful tools for examining the recycling of other SNX27-retromer cargos, including those involved in diverse cellular processes such as neurologic function, metabolism, and metal ion transport, among others (13, 15, 17, 18). Thus, the involvement of SNX27-retromer in so many housekeeping functions makes the described interaction between SNX27 and VPS26 of critical biological significance. This is reinforced by the evolutionary conservation of the binding mechanism in C. elegans.
Recent studies have shown that PDZ-binding specificity determinants are not limited to the final three amino acids of the canonical PDZbm, but follow a more complex code optimized across the whole proteome to minimize cross-reactivity (36). Here we have described that the binding of VPS26 to SNX27, although not dramatically affecting the architecture of the PDZ domain, can greatly enhance binding of the PDZ domain to PDZbm in the cytoplasmic tails of GLUT1 and Kir3.3; we anticipate that this phenomenon will extend to other cargos as well. These data are suggestive of a model in which the precise sequence and structural context of the PDZbms of cargo proteins determine not only their straightforward affinity for the SNX27 PDZ domain, but also the increase in this affinity induced by association of SNX27 with VPS26. In the competition between PDZ domain-containing proteins for binding to the PDZbm of cargo proteins, the higher affinity observed on assembly of the endosome-associated SNX27-retromer would act to outcompete those with lower affinity (e.g., proteins that engage the PDZbm at the cell surface), thereby allowing retrieval of the cargo from lysosomal degradation and its ultimate recycling back to the plasma membrane. Whether our description of the binding interface between the SNX27 PDZ domain and VPS26 points to a mode of regulation in controlling the decision between cargo degradation and recycling remains to be addressed.
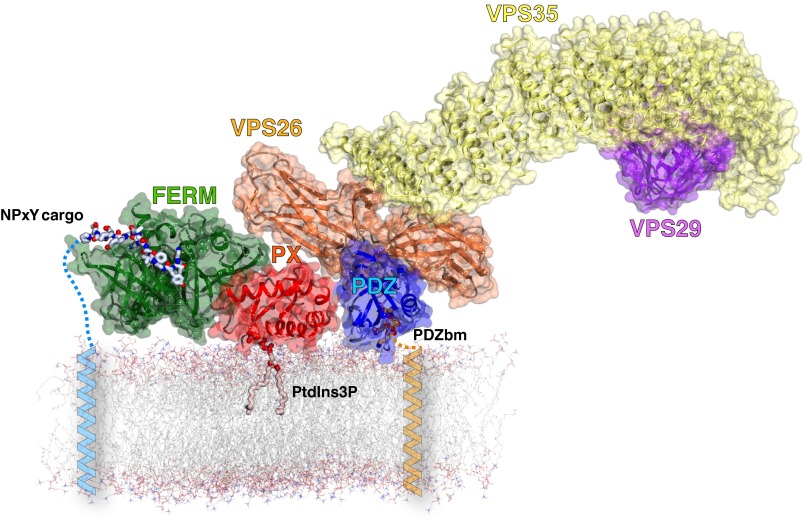
The structure of the VPS26A-SNX27 PDZ domain complex also reveals how VPS26 engages SNX27 while simultaneously binding with high affinity to the retromer subunit VPS35. Although no high-resolution structure exists for this complex, mutagenesis, electron microscopy, and small angle X-ray scattering experiments define a loop region of the VPS26 C-terminal subdomain as the VPS35-binding site, which is remote from the SNX27-binding site in the SNX27 PDZ domain-VPS26 structure (6, 26, 27, 37). Moreover, in presenting the first crystal structure, to our knowledge, of a retromer subunit associated with another protein other than another subunit of the core trimer, we explain precisely how SNX27 functions as a cargo adaptor that is able to bind retromer and PDZbms simultaneously. We propose a speculative model to describe how the SNX27-retromer complex may assemble on the endosome membrane (Fig. 6). This is based on the structure described herein, as well as on previously published crystal structures of retromer subunits (5, 6, 26); the solution structure of retromer (37); crystal structures of the PX and FERM-like domains of SNX17 (10, 38), a related member of the PX-FERM family to which SNX27 belongs; and the previously published solution structure of full-length SNX27 (10). The model shows how SNX27 engages both NPxY and PDZbm sorting signals in the cytoplasmic tails of transmembrane cargo proteins while associated with phosphatidylinositol-3-phosphate (PtdIns3P) in the endosome membrane. VPS26 provides the interface between SNX27 and retromer, whereas the elongated structure of VPS35 bound to the opposite face of VPS26 extends out from the membrane-tethered complex. Such an extended architecture will allow space for association of the various other known retromer interactors, including the WASH complex (39), SNX3 (40), RME-8 (41), and TBC1D5 (42). A detailed structural description of this network will provide much-needed molecular information to describe the workings of this biologically critical sorting machine. In this regard, the data presented here not only identify a mode of interaction between a PDZ domain and an arrestin-fold protein, but also represent a significant step toward this ultimate goal.
Fig. 6.
Model of the SNX27-retromer complex. Low-resolution retromer trimeric structure (VPS26-VPS35-VPS29) and full length SNX27 structure were obtained from previous SAXS experiments. The overall complex was derived from overlaying these complexes with the VPS26A-SNX27 PDZ domain crystal structure. The PDZ domain (blue) and the FERM-like domain (green) of SNX27 bind PDZbm- and NPxY/NxxY-containing transmembrane cargo, respectively, whereas the PX domain of SNX27 (red) associates with the endosomal lipid phosphatidylinositol 3-phosphate (PtdIns3P). VPS26 (orange) binds the PDZ domain of SNX27, VPS35 (yellow) binds VPS26, and VPS29 (purple) in turn binds VPS35.
Materials and Methods
Antibodies and Peptides.
The following antibodies were used in this study: mouse monoclonal β-actin (Sigma-Aldrich; clone AC-15 A1978, 1:5,000 dilution), mouse monoclonal GFP (clones 7.1/13.1; Roche; 11814460001, 1:5,000), rabbit polyclonal GLUT1 (Abcam; 15309, 1:100), rabbit polyclonal KIDINS220 (Proteintech; 21856-1-AP, 1:1,000), sheep polyclonal myc-tag (made in-house for University of Bristol; 1:1,000), mouse monoclonal SNX27 antibody (clone 1C6; Abcam; AB77799, 1:500), and rabbit polyclonal VPS26A (Epitomics; S1181, 1:500), VPS26B (Proteintech; 15915-1-AP, 1:400), and VPS35 (Abcam; 97545, 1:2,000). The GLUT1 and Kir3.3 synthetic peptides used for ITC were obtained from Genscript.
Western Blot Analysis.
Western blot analysis was performed as described previously (17).
Immunoprecipitation.
HEK293T cells were transfected with respective GFP-tagged constructs using polyethylene imine. The cells were incubated for 48 h, followed by lysis in Tris-based immunoprecipitation buffer (50 mM Tris⋅HCl pH 7.4, 0.5% Nonidet P-40, and Roche protease inhibitor mixture) and then precipitation with GFP-trap beads (Chromotek).
Protein Expression, Purification, and Crystallization.
cDNAs of residues 9–327 of VPS26A (mouse) and 40–135 of SNX27 (rat) were separately cloned in a pMW172KanH6 plasmid downstream of a hexahistidine tag, and a pGEX4-T2 yielding an N-terminally GST-tagged protein, respectively. The Stratagene QuikChange II Kit was used for site-directed mutagenesis. Proteins were expressed in Escherichia coli BL21 (DE3) strain overnight at 20 °C and then purified using affinity chromatography followed by gel filtration. VPS26 proteins were purified on a nickel-NTA gravity column and eluted with 300 mM imidazole in 200 mM NaCl and 20 mM Hepes (pH 7). The SNX27 PDZ domain was purified on a glutathione Sepharose gravity column and eluted after 3 h of thrombin cleavage in 200 mM NaCl and 25 mM Tris (pH 8). Proteins were then gel-filtered in 200 mM NaCl and 25 mM Tris (pH 8) for SNX27PDZ, and in 200 mM NaCl and 25 mM Hepes (pH 7) for VPS26A using a Sepharose S200 16/60 column attached to an AKTA system (GE Healthcare). Fractions were pooled and concentrated separately, then directly mixed together to a 1.5:1 molar ratio of SNX27 PDZ domain to VPS26A, with a final VPS26A concentration of 20 mg/mL. Eight 96-well crystallization hanging-drop screens were set up using a Mosquito Liquid Handling robot (TTP LabTech) at 20 °C. Optimized diffraction-quality crystals were obtained using the sitting drop vapor diffusion method and a buffer containing 100 mM sodium citrate (pH 6) and 4 M NaCl.
The derivative crystal was produced by soaking for 3 h in the crystallization solution supplemented with 10 mM HgCl2. Crystals were transferred into a cryoprotectant solution of mother liquor supplemented with 0.34 M sodium malonate (pH 6) and then cooled to 100 K for transport to the synchrotron.
Data Collection and Structure Determination.
Data were collected at 100 K at beamline MX1 at the Australian Synchrotron (Table 1) and integrated and scaled with MOSFLM and SCALA. The structure was determined by combining a molecular replacement and a single-wavelength anomalous dispersion analysis (MR-SAD) using a mercury-soaked crystal as a derivative. A first molecular replacement step was carried out using Phaser-MR (43) with the SNX27 PDZ domain and VPS26A high-resolution structures as input ensembles (PDB ID codes 3QDO and 2FAU, respectively). A partial model was generated and refined against experimental data by phasing using the peak wavelength data of mercury with AUTOSOL in the PHENIX suite (44). A model was built using COOT (45), followed by repeated refinement and model building with PHENIX.REFINE and COOT. Most residues in the final model were built in accordance with Ramachandran statistics (favored, 96.8%; allowed, 2.4%; outliers, 0.8%). The final structure was solved at 2.7-Å resolution and revealed electron density corresponding to mercury atoms attached to the two cysteine residues of VPS26. All structural figures were generated using PyMOL (www.pymol.org).
Direct Recombinant Interaction Studies.
SNX27 PDZ domain and VPS26A-myc (and mutant forms of these proteins) were cloned into pGEX6P-3 and GST fusion proteins purified from BL-21 E. coli using standard procedures. The GST tag was cleaved using precision protease (GE Healthcare) or left as GST fusion proteins bound to beads. To test for interactions, either SNX27 PDZ domain or VPS26A-myc in 50 mM Tris⋅HCl pH 7.4, 200 mM NaCl, 1% (vol/vol) Triton X-100, and 0.1% (wt/vol) BSA were incubated with the bead-coupled GST-VPS26-myc or GST-SNX27 PDZ domain, respectively. After washing, bound proteins were eluted by boiling in SDS sample buffer and detected by Western blot analysis.
ITC.
ITC experiments were performed on a MicroCal iTC200 instrument (GE Healthcare) in 50 mM Tris pH 8 and 100 mM NaCl. The GLUT1 (PLGADSQV) and Kir3.3 (PPESESKV) peptides at a concentration of 1.75 mM were titrated into 50 μM and 40 μM SNX27 PDZ domain (supplemented with 50 μM or 40 μM VPS26A when required) protein solutions at 25 °C. Data were processed using ORIGIN to extract the thermodynamic parameters ΔH, Ka (1/Kd) and the stoichiometry n. ΔG and ΔS were derived from the relationships ΔG = −RTlnKa and ΔG = ΔH – TΔS.
NMR Spectroscopy.
The SNX27 PDZ domain was 15N/13C-labeled by expression in Minimal Media supplemented with 15NH4Cl and 13C-glucose, and then purified into 20 mM Hepes (pH 7) and 200 mM NaCl. All spectra were collected at 298 K on a Bruker 900-MHz spectrometer equipped with a cryoprobe and z-axis gradients. For sequential backbone assignments of the SNX27 PDZ domain, the sample contained 0.8 mM 15N/13C-labeled SNX27 PDZ domain in NMR buffer with 10% (vol/vol) D2O. Sequential backbone assignments were obtained by recording 3D HNCACB, CBCA(CO)NH, HNCO, and HN(CA)CO spectra. All experiments were analyzed using nonuniform sampling (46). For the NMR chemical shift titration studies of the SNX27 PDZ domain with VPS26, unlabeled VPS26 was added to a sample containing 100 μM 15N-labeled SNX27 PDZ domain in NMR buffer with 10% D2O. Two-dimensional 1H-15N HSQC spectra of 15N-labeled SNX27 PDZ domain were collected with increasing amounts of unlabeled VPS26 (0.25, 0.5, and 2 molar ratios). Spectra were processed using NMRPipe or the maximum entropy method and processed using the Rowland NMR toolkit with automatically generated processing scripts (46, 47). Spectra were analyzed with the CCPNMR program.
siRNA-Mediated Suppression of Gene Expression.
For SNX27 suppression, a single oligonucleotide with the published sequence 5′-CCAGGUAAUUGCAUUUGAA(dTdT)-3′ was used. VPS26A suppression was done using either oligonucleotide 1 [sequence 5′-GCUAGAACACCAAGGAAUU(dTdT)-3′] or oligonucleotide 4 [sequence 5′-CCACCUAUCCUGAUGUUAA(dTdT)-3′] of the ON-TARGETplus human SMARTpool (Dharmacon). For VPS26B suppression was done using either oligonucleotide 1 [sequence 5′-CCAUGAGAAUGACACGAUA(dTdT)-3′] or oligonucleotide 2 [sequence 5′-GAAGUUCUCUGUGCGCUAU(dTdT)-3′] of the ON-TARGETplus human SMARTpool (Dharmacon). HeLa cells were reverse-transfected using DharmaFECT (Dharmacon), then transfected again 12 h later using Hiperfect (Qiagen).
Mutagenesis.
SiRNA-resistant SNX27 was generated by introducing three silent base mismatches (a1414c;t1417a;a1420t) into the ORF conferring resistance to SNX27 siRNA described above. SiRNA-resistant VPS26A was generated by introducing six silent base mismatches (c187t;a189g;a192g;c195t;a198g;a201g) into the ORF, conferring resistance to VPS26A siRNA oligonucleotide 1. siRNA-resistant VPS26B was generated by introducing seven silent base mismatches (g795a;c798t;t799a;c800g;g804a;c805a;c807g) into the ORF, conferring resistance to VPS26B siRNA oligonucleotide 2.
Rescue of GLUT1 Lysosomal Missorting with SNX27 or VPS26 Constructs.
Expression of endogenous SNX27 or VPS26A and VPS26B was suppressed in HeLa cells by transfecting SNX27 siRNA or a mixture of VPS26A oligonucleotide 1 and VPS26B oligonucleotide 2 siRNA. At 24 h after transfection, cells were trypsinized, seeded onto fibronectin (Sigma-Aldrich)-coated coverslips, and transfected with the respective GFP-tagged, siRNA-resistant SNX27, VPS26A, or VPS26B constructs using Fugene 6 (Roche). At 72 h after siRNA transfection, cells were fixed and stained for LAMP1 and GLUT1.
Immunofluorescence.
Staining of cells for immunofluorescence was performed as described previously (17).
Image Acquisition and Quantification of Colocalization.
Images were acquired and quantified as described previously (17).
Supplementary Material
Acknowledgments
We acknowledge support from the University of Queensland Remote Operation Crystallization and X-ray facility and the Australian Synchrotron. We thank Suzanne Norwood for assistance with protein expression and discussion of the manuscript, Tom Caradoc-Davies for assistance with X-ray diffraction data collection, and Mehdi Mobli for assistance with NMR spectroscopy. This work is supported by grants from the Wellcome Trust (083474), Australian Research Council (DP120103930), and National Health and Medical Research Council (APP1058734). M.G. is supported by a Wellcome Trust 4-Year PhD Studentship awarded through the Dynamic Cell Biology program (Award 083474). C.M. was supported by a University of Queensland Postdoctoral Fellowship. R.D.T. is supported by a National Health and Medical Research Council Senior Research Fellowship (Award APP1041929). B.M.C. is supported by an Australian Research Council Future Fellowship (Award FT100100027). P.J.C. is supported by the Wellcome Trust (Grants 089928 and 085743), the Medical Research Council (Grant MR/K018299/1), and the Royal Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4P2A).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410552111/-/DCSupplemental.
References
- 1.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30(17):3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat Rev Mol Cell Biol. 2010;11(8):556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johannes L, Popoff V. Tracing the retrograde route in protein trafficking. Cell. 2008;135(7):1175–1187. doi: 10.1016/j.cell.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Hsu VW, Bai M, Li J. Getting active: Protein sorting in endocytic recycling. Nat Rev Mol Cell Biol. 2012;13(5):323–328. doi: 10.1038/nrm3332. [DOI] [PubMed] [Google Scholar]
- 5.Collins BM, Skinner CF, Watson PJ, Seaman MN, Owen DJ. Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat Struct Mol Biol. 2005;12(7):594–602. doi: 10.1038/nsmb954. [DOI] [PubMed] [Google Scholar]
- 6.Hierro A, et al. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449(7165):1063–1067. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165(1):111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Temkin P, et al. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13(6):715–721. doi: 10.1038/ncb2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burd C, Cullen PJ. Retromer: A master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6(2):a016774. doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghai R, et al. Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins. Proc Natl Acad Sci USA. 2013;110(8):E643–E652. doi: 10.1073/pnas.1216229110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joubert L, et al. New sorting nexin (SNX27) and NHERF specifically interact with the 5-HT4a receptor splice variant: Roles in receptor targeting. J Cell Sci. 2004;117(Pt 22):5367–5379. doi: 10.1242/jcs.01379. [DOI] [PubMed] [Google Scholar]
- 12.Lunn ML, et al. A unique sorting nexin regulates trafficking of potassium channels via a PDZ domain interaction. Nat Neurosci. 2007;10(10):1249–1259. doi: 10.1038/nn1953. [DOI] [PubMed] [Google Scholar]
- 13.Lauffer BE, et al. SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J Cell Biol. 2010;190(4):565–574. doi: 10.1083/jcb.201004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loo LS, Tang N, Al-Haddawi M, Dawe GS, Hong W. A role for sorting nexin 27 in AMPA receptor trafficking. Nat Commun. 2014;5:3176. doi: 10.1038/ncomms4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balana B, et al. Mechanism underlying selective regulation of G protein-gated inwardly rectifying potassium channels by the psychostimulant-sensitive sorting nexin 27. Proc Natl Acad Sci USA. 2011;108(14):5831–5836. doi: 10.1073/pnas.1018645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauch C, Koliwer J, Buck F, Hönck HH, Kreienkamp HJ. Subcellular sorting of the G-protein coupled mouse somatostatin receptor 5 by a network of PDZ domain-containing proteins. PLoS ONE. 2014;9(2):e88529. doi: 10.1371/journal.pone.0088529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinberg F, et al. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat Cell Biol. 2013;15(5):461–471. doi: 10.1038/ncb2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai L, Loo LS, Atlashkin V, Hanson BJ, Hong W. Deficiency of sorting nexin 27 (SNX27) leads to growth retardation and elevated levels of N-methyl-D-aspartate receptor 2C (NR2C) Mol Cell Biol. 2011;31(8):1734–1747. doi: 10.1128/MCB.01044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilariño-Güell C, et al. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89(1):162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimprich A, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011;89(1):168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muhammad A, et al. Retromer deficiency observed in Alzheimer’s disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc Natl Acad Sci USA. 2008;105(20):7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, et al. Loss of sorting nexin 27 contributes to excitatory synaptic dysfunction by modulating glutamate receptor recycling in Down’s syndrome. Nat Med. 2013;19(4):473–480. doi: 10.1038/nm.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mecozzi VJ, et al. Pharmacological chaperones stabilize retromer to limit APP processing. Nat Chem Biol. 2014;10(6):443–449. doi: 10.1038/nchembio.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karthikeyan S, Leung T, Birrane G, Webster G, Ladias JA. Crystal structure of the PDZ1 domain of human Na(+)/H(+) exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains. J Mol Biol. 2001;308(5):963–973. doi: 10.1006/jmbi.2001.4634. [DOI] [PubMed] [Google Scholar]
- 25.Neal S, Nip AM, Zhang H, Wishart DS. Rapid and accurate calculation of protein 1H, 13C, and 15N chemical shifts. J Biomol NMR. 2003;26(3):215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 26.Shi H, Rojas R, Bonifacino JS, Hurley JH. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nat Struct Mol Biol. 2006;13(6):540–548. doi: 10.1038/nsmb1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins BM, et al. Structure of Vps26B and mapping of its interaction with the retromer protein complex. Traffic. 2008;9(3):366–379. doi: 10.1111/j.1600-0854.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 28.Bugarcic A, et al. Vps26A and Vps26B subunits define distinct retromer complexes. Traffic. 2011;12(12):1759–1773. doi: 10.1111/j.1600-0854.2011.01284.x. [DOI] [PubMed] [Google Scholar]
- 29.Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol Cell. 2004;13(5):665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- 30.Whitney DS, Peterson FC, Volkman BF. A conformational switch in the CRIB-PDZ module of Par-6. Structure. 2011;19(11):1711–1722. doi: 10.1016/j.str.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Im YJ, et al. Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization. J Biol Chem. 2003;278(48):48099–48104. doi: 10.1074/jbc.M306919200. [DOI] [PubMed] [Google Scholar]
- 32.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284(5415):812–815. [PubMed] [Google Scholar]
- 33.Garrard SM, et al. Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. EMBO J. 2003;22(5):1125–1133. doi: 10.1093/emboj/cdg110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shukla AK, et al. Structure of active β-arrestin-1 bound to a G-protein—coupled receptor phosphopeptide. Nature. 2013;497(7447):137–141. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang J, et al. The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein. Nat Commun. 2014;5:2958. doi: 10.1038/ncomms3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stiffler MA, et al. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317(5836):364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norwood SJ, et al. Assembly and solution structure of the core retromer protein complex. Traffic. 2011;12(1):56–71. doi: 10.1111/j.1600-0854.2010.01124.x. [DOI] [PubMed] [Google Scholar]
- 38.Ghai R, et al. Phox homology band 4.1/ezrin/radixin/moesin-like proteins function as molecular scaffolds that interact with cargo receptors and Ras GTPases. Proc Natl Acad Sci USA. 2011;108(19):7763–7768. doi: 10.1073/pnas.1017110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helfer E, et al. Endosomal recruitment of the WASH complex: Active sequences and mutations impairing interaction with the retromer. Biol Cell. 2013;105(5):191–207. doi: 10.1111/boc.201200038. [DOI] [PubMed] [Google Scholar]
- 40.Harterink M, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13(8):914–923. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freeman CL, Hesketh G, Seaman MN. RME-8 coordinates the activity of the WASH complex with the function of the retromer SNX dimer to control endosomal tubulation. J Cell Sci. 2014;127(Pt 9):2053–2070. doi: 10.1242/jcs.144659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seaman MN, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci. 2009;122(Pt 14):2371–2382. doi: 10.1242/jcs.048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mobli M, Maciejewski MW, Gryk MR, Hoch JC. An automated tool for maximum entropy reconstruction of biomolecular NMR spectra. Nat Methods. 2007;4(6):467–468. doi: 10.1038/nmeth0607-467. [DOI] [PubMed] [Google Scholar]
- 47.Mobli M, Maciejewski MW, Gryk MR, Hoch JC. Automatic maximum entropy spectral reconstruction in NMR. J Biomol NMR. 2007;39(2):133–139. doi: 10.1007/s10858-007-9180-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.