Significance
Poly-ADP-ribosylation (PARylation) is an abundant posttranslational modification in eukaryotes. The responsible enzyme, poly [ADP-ribose] polymerase 1 (PARP-1), binds to chromatin and shapes its architecture. It is activated by DNA damage and other triggers, and catalyzes the addition of long chains of poly-ADP ribose (PAR) mainly to itself. The biological effects of PARylation are unknown. Here, we show that PARylation confers the ability to bind histones and to assemble nucleosomes upon PARP-1, and also affects its interaction with chromatin. The rapid turnover of PAR groups provides a mechanism for switching PARP-1 function from nucleosome binder to nucleosome assembler. This explains the involvement of active PARP-1 in both transcription and DNA damage repair, because histone chaperone activity is required during both processes.
Keywords: posttranslational modification, dissociation constant, HI-FI FRET, linker DNA
Abstract
Poly [ADP-ribose] polymerase 1 (PARP-1) is a highly abundant chromatin-associated enzyme. It catalyzes the NAD+-dependent polymerization of long chains of poly-ADP ribose (PAR) onto itself in response to DNA damage and other cues. More recently, the enzymatic activity of PARP-1 has also been implicated in the regulation of gene expression. The molecular basis for the functional switch from chromatin architectural protein to transcription factor and DNA damage responder, triggered by PARP-1 automodification, is unknown. Here, we show that unmodified PARP-1 engages in at least two high-affinity binding modes with chromatin, one of which does not involve free DNA ends, consistent with its role as a chromatin architectural protein. Automodification reduces PARP-1 affinity for intact chromatin but not for nucleosomes with exposed DNA ends. Automodified (AM) PARP-1 has the ability to sequester histones (both in vitro and in cells) and to assemble nucleosomes efficiently in vitro. This unanticipated nucleosome assembly activity of AM–PARP-1, coupled with the fast turnover of the modification, suggests a model in which DNA damage or transcription events trigger transient histone chaperone activity.
The highly abundant nuclear enzyme and chromatin binding protein poly [ADP-ribose] polymerase 1 (PARP-1) binds nucleosomes and shapes chromatin architecture. At a nuclear concentration of about one PARP-1 molecule per 20 nucleosomes, it has the potential to affect many nuclear processes. Historically, the majority of PARP-1–related studies have addressed its role as a first responder to DNA damage (1, 2) and as a popular target for cancer therapy. More recently, a role of PARP-1 in transcription regulation has been described (3, 4). How PARP-1 functionally switches from a chromatin architectural protein to a transcription regulator and DNA damage responder is largely unknown (3, 5). The enzymatic activity of PARP-1 is required for its function in both transcription regulation and DNA repair.
PARP-1 is a multidomain protein that is functionally divided into two halves: an N-terminal DNA and chromatin binding portion (N-parp) and a C-terminal catalytic domain (C-parp) (6, 7) (Fig. 1A). PARP-1 is activated by a host of factors, most effectively by binding to ssDNA and dsDNA breaks and cruciform DNA. Interactions with a variety of nuclear proteins (including histones and histone variants) and a range of posttranslational modifications may also trigger activation (reviewed in ref. 4). Upon enzymatic activation in the presence of NAD+, PARP-1 polymerizes long chains of poly [ADP-ribose] (PAR) onto itself and a variety of acceptor proteins, including histones (8–10). PARP-1 activity is antagonized by poly [ADP-ribose] glycohydrolases (PARGs), a class of enzymes that hydrolyzes nascent PAR chains (11). Thus, PARylation is a dynamic, heterogeneous, and relatively short-lived posttranslational modification.
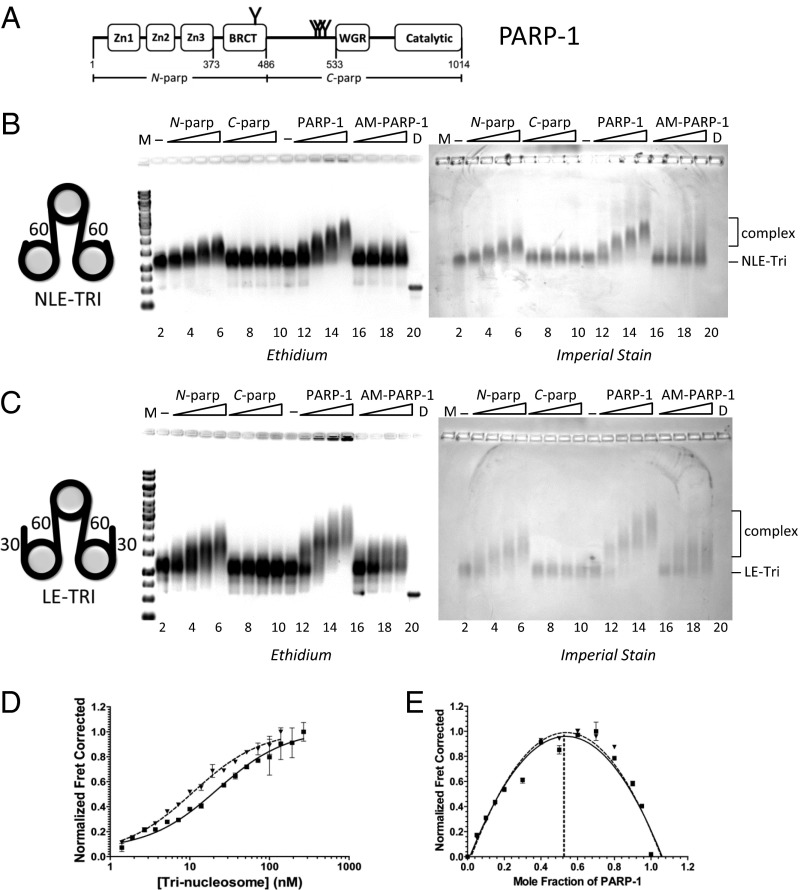
Fig. 1.
High-affinity interactions of PARP-1 with chromatin do not require exposed DNA ends. (A) PARP-1 domain structure. Commonly described automodification sites are indicated by Y. The N-parp and C-parp constructs used in this study are indicated. BRCT, BRCA1 C terminus. (B and C) EMSA of trinucleosomes without free linker DNA (NLE-Tri, B) or with 30-bp linker DNA extensions (LE-Tri, C) bound to various PARP-1 constructs. Trinucleosomes (lanes 2 and 11) were incubated with increasing amounts of N-parp (lanes 3–6), C-parp (lanes 7–10), PARP-1 (lanes 12–15), or AM–PARP-1 (lanes 16–19). Gels were stained as indicated. D, DNA; M, marker (521 bp in B and 621 bp in C). (D) Representative HI-FI FRET curves of PARP-1 with LE-Tri and NLE-Tri. Error bars are from duplicates within a single experiment. The solid line indicates LE-Tri, and the dashed line indicates NLE-Tri. The Kd values from these and similar experiments are listed in Table 1. (E) Job plots for stoichiometry for samples shown in D. LE-Tri (■, solid line) and NLE-Tri (▲, dashed line) are shown.
The preferred target of PARP-1 enzymatic activity is PARP-1 itself (6), yet the functional outcomes of PARP-1 automodification are not well understood. Proposed downstream effects of PARylation include increased redistribution of PARP-1 on activated genes (12), recruitment of the DNA repair machinery (13), and effects on chromatin structure (8, 14, 15), with the latter possibly augmented by PARylation of histones (5, 16–18). Some evidence suggests that upon automodification and/or PARylation of histones, PARP-1 dissociates from chromatin, resulting in chromatin decompaction (19, 20). However, this notion has not been substantiated by quantitative measurements of the interactions of PARP-1 with intact chromatin. The crystal structure of PARP-1 domains in complex with a DNA fragment describes how PARP-1 recognizes a DNA double-strand break through extensive interactions between the PARP-1 zinc finger domains and the terminal base pair (6), but it does not explain how PARP-1 binds and compacts intact chromatin in the absence of an exposed DNA base pair, as demonstrated earlier by visualizing PARP-1 bound to chromatin assembled on circular DNA (8). Compaction was relieved in the presence of NAD+. Kim et al. (8) also showed that PARP-1 binds to, and protects, nucleosomal linker DNA. However, biochemical ramifications of auto-PARylation of PARP-1 have yet to be identified in a quantitative manner.
We have previously reported that unmodified PARP-1 binds to mononucleosomes containing linker DNA with high affinity, and that its activity is efficiently stimulated by such substrates (7). Here, we quantify the affinity of PARP-1 (unmodified and automodified) for chromatin in the presence and absence of free DNA ends. We further describe a previously unidentified histone chaperone and nucleosome assembly function for automodified (AM) PARP-1 and present a model for PARP-1 roles in DNA repair and transcription.
Results
PARP-1 Binds Trinucleosomes with High Affinity.
Because PARP-1 is a chromatin architectural protein that is abundantly bound to undamaged chromatin throughout the genome (21), we wanted to quantify its interactions with nucleosomes in the absence of free DNA ends. We assembled three nucleosomes on a triple repeat of the 601 positioning sequence. This sequence positions the two terminal nucleosomes right at the end of the DNA, with no extending linker DNA (Fig. 1B, cartoon). In these non–linker-ended trinucleosomes (NLE-Tri), the central nucleosome is connected to a flanking nucleosome on either side via 60 bp of linker DNA, representing a minimal model of native chromatin. To mimic double-strand breaks in a nucleosomal context, and to probe the contributions of exposed DNA ends to the interaction of PARP-1 with chromatin, we also prepared linker-ended trinucleosomes (LE-Tri), in which the two terminal nucleosomes each have an additional 30 bp of extranucleosomal DNA (22) (Fig. 1C, cartoon, and Fig. S1 A–C). Unmodified PARP-1 binds to both substrates without releasing free DNA, demonstrating that the nucleosomal arrays remain intact upon PARP-1 binding (Fig. 1 B and C). Trinucleosomes also form complexes with N-parp, which contains only the DNA binding domains, but not with C-parp, the catalytic domain (Fig. 1 A–C). Both types of chromatin substrates are visibly compacted by PARP-1 (Fig. S1 D and E). In the absence of PARP-1, individual nucleosomes can be easily distinguished and have a height profile of ∼1.5–2 nm, as previously reported (23). Addition of PARP-1 results in a much closer spacing of the individual nucleosomes and an increased height profile to ∼3–5 nm (Fig. S1E).
We determined dissociation constants of the various PARP-1–chromatin complexes using high-throughput interactions by fluorescence intensity FRET (HI-FI FRET) (24) (Table 1). Due to the strong distance dependence, FRET can be used as an indicator for the presence or absence of interaction between binding partners. Unexpectedly, full-length PARP-1 binds both trinucleosome substrates with comparably high affinities, despite the absence of freely accessible DNA ends in NLE-Tri (Fig. 1D and Table 1). Stoichiometry measurements show that one PARP-1 is bound per trinucleosome (Fig. 1E and Fig. S2B). We conclude that the central nucleosome with its two internal 60-bp DNA linkers is the likely primary PARP-1 binding site in both trinucleosomes, and that this interaction sterically prevents additional PARP-1 molecules from binding. Our finding that NLE-Tri binds PARP-1 with high affinity is unexpected in light of PARP-1–DNA crystal structures showing substantial interactions arising from the two zinc finger motifs that stack on the terminal bases of linear DNA (6, 25, 26). This mode of interaction is incompatible with nucleosomes lacking extranucleosomal DNA, such as NLE-Tri and Nuc147 (7).
Table 1.
PARP-1 affinities measured by HI-FI FRET
| N-parp | PARP-1 | AM–PARP-1 | |||||||
| Binding substrate | Approximate Kd, nM | R2 | No. rep. | Approximate Kd, nM | R2 | No. rep. | Approximate Kd, nM | R2 | No. rep. |
| 30Nick | 27.8 ± 5.6 | 0.95 | 5 | 23.4 ± 4.8* | 0.98 | 2 | 33.2 ± 23.5* | 0.94 | 5 |
| Nuc207 | 48.8 ± 21.2 | 0.97 | 2 | 1.0 ± 0.2 | 0.90 | 2 | 13.2 ± 2 | 0.96 | 5 |
| LE-Tri | 20.3 ± 2.6 | 0.97 | 1 | 12.7 ± 6.4 | 0.95 | 2 | 10 ± 2 | 0.95 | 4 |
| NLE-Tri | 22.8 ± 4.8 | 0.95 | 1 | 4.8 ± 2.1 | 0.98 | 3 | 101 ± 23 | 0.94 | 3 |
| H2A-H2B | >500 | 0.94 | 1 | >500 | 0.93 | 4 | 2.3 ± 0.8 | 0.94 | 4 |
| H3-H4 | >500 | 0.98 | 1 | >500 | 0.99 | 3 | 16 ± 3 | 0.96 | 3 |
Amount used is 250 mM NaCl instead of 200 mM NaCl.
Each data point is an average of two to five individual experiments, each performed in duplicate; experiments with N-parp were performed once, in duplicate. Values listed in italics are published (7), but they are shown here for comparison. No. rep., number of independent replicates for each value.
Together, these data demonstrate that although linker DNA is essential, exposed DNA bases are not required for high-affinity PARP-1 interactions with chromatin. Thus, PARP-1 engages in at least two high-affinity binding modes, one driven by interactions with the exposed bases of damaged DNA (6) and one that promotes genome-wide binding of PARP-1 to chromatin in the absence of DNA damage.
PARP-1 Automodification Weakens Its Affinity for Chromatin, but Not for Free DNA.
To test whether automodification of chromatin-bound PARP-1 indeed has an impact on its ability to act as a chromatin architectural protein, as suggested previously (8), we automodified Alexa488-labeled PARP-1 in the presence of 600 μM NAD+ and nicked 30-bp DNA (30Nick). This results in extensive PARP-1 automodification, as demonstrated by smears of PARylated PARP-1 on SDS/PAGE (Fig. S3B).
We tested the ability of AM–PARP-1 to bind free DNA, mononucleosomes, or trinucleosomes. Gel-shift experiments show that AM–PARP-1 (Fig. S3A) loses its ability to shift NLE-Tri but maintains residual interactions with LE-Tri (Fig. 1 B and C); neither array is visibly compacted by AM–PARP-1 (Fig. S1F). HI-FI FRET measurements reveal that automodification of PARP-1 does not significantly weaken its interaction with free DNA or LE-Tri (Fig. 2A and Table 1). Indeed, AM–PARP-1 binds all substrates with free DNA ends [i.e., DNA, Nuc207 (Fig. S2C), LE-Tri] with comparable affinities (Table 1). In contrast, AM–PARP-1 affinity for NLE-Tri is reduced by a factor of 20 (∼100 nM; Fig. 2A). The number of AM–PARP-1 molecules bound to either trinucleosome remains unaffected by automodification (Fig. 2B). Our results suggest that the interaction of PARP-1 with undamaged chromatin is more sensitive toward the effects of automodification than any mode of PARP-1 interaction that engages free DNA ends.
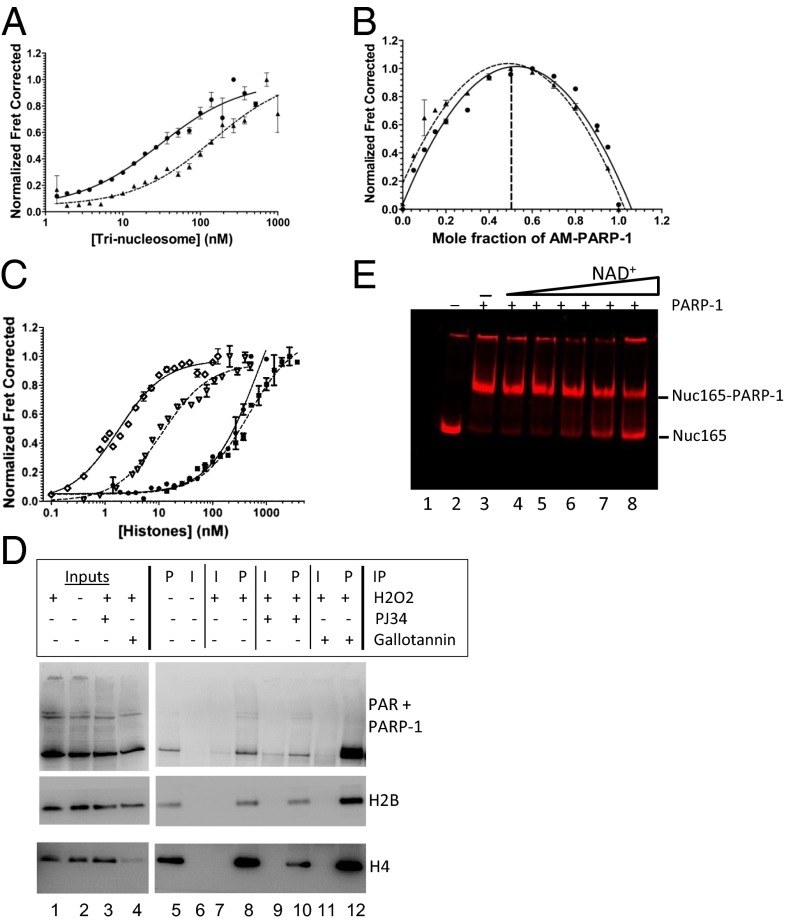
Fig. 2.
PARylation converts PARP-1 from a chromatin architectural protein to a histone chaperone. (A and B) Representative binding curves and stoichiometry measurements of AM–PARP-1 to LE-Tri (●, solid lines) and NLE-Tri (▲, dashed lines). (C) Binding curves for PARP-1 and AM–PARP-1 to histones. H2A-H2B–PARP-1 (●, solid line), H3-H4–PARP-1 (■, dashed line), H2A-H2B–AM–PARP-1 (♢, solid line), and H3-H4–AM–PARP-1 (▿, dashed line) are shown. (D) Endogenous PARylated PARP-1 coimmunoprecipitates soluble histones. U2OS cells were treated with H2O2 in combination with PJ34 or gallotannin as indicated. PAR antibodies (P) or control IgG (I) were used in immunoprecipitation (IP) assays from soluble lysates. Bound proteins were detected by immunoblotting with a mixture of PARP-1 and PAR antibodies (Top) and H2B and H4 antibodies (Middle and Bottom). (E) AM–PARP-1 does not disassemble nucleosomes. Labeled Nuc165 with PARP-1 and 30Nick DNA was incubated with increasing amounts of NAD+. Samples were analyzed by native PAGE and visualized by gel FRET between H2B (cyanine 5 633/670) and H4 (Alexa488 488/520). Lane 2 contains nucleosomes alone; lane 3 contains nucleosomes with PARP-1 and 30Nick DNA; and lanes 4–8 contain nucleosomes with PARP-1, 30Nick DNA, and 0.1, 1, 10, 20, and 40 μM NAD+, respectively. FRET between histones H2B and H4 is observed in all lanes, indicating that nucleosomes remain intact throughout.
AM–PARP-1 Binds Histones with High Affinity.
PAR chains are negatively charged and chemically represent a “third type of nucleic acid.” We therefore quantified whether PARP-1 binds nonnucleosomal histones in a PAR-dependent manner. Unmodified PARP-1 interacts only weakly (>500 nM) with either H2A–H2B or H3–H4 histone complex (Fig. 2C and Table 1). In striking contrast, AM–PARP-1 (Fig. S3B) binds free nonnucleosomal H2A–H2B and H3–H4 complexes with high affinity (∼2–16 nM; Fig. 2C and Table 1). Importantly, the measured affinities of AM–PARP-1 for histones are similar to those obtained for classical histone chaperones under similar conditions [e.g., nucleosome assembly protein 1 (Nap1) (27)].
To verify the relevance of histone binding by AM–PARP-1 in cells, we performed immunoprecipitation with anti-PAR antibodies, utilizing soluble lysates from cells exposed to oxidative stress using hydrogen peroxide (Fig. 2D). PARP-1 is activated by H2O2 treatment, and the PAR modification can be stabilized by a PAR inhibitor, gallotannin (Fig. 2D, lane 4). Basal levels of H2B and H4 are coimmunoprecipitated with anti-PAR antibodies in the absence of H2O2, or when cells were treated with the combination of H2O2 and PJ34 hydrochloride hydrate (PJ34), a potent PARP-1 inhibitor (Fig. 2D, compare lanes 5 and 10). H2O2 treatment alone results in increased coimmunoprecipitation of H2B and H4 (Fig. 2D, lane 8). This effect is magnified when PARG is inhibited (Fig. 2D, lane 12). We do not detect any PARylated histones under these conditions, consistent with results from others (8). We conclude that automodification of PARP-1 enhances its interaction with soluble histones in cells.
To test whether AM–PARP-1 disassembles nucleosomes due to its high affinity for histones, we incubated PARP-1 with mononucleosomes [nucleosome reconstituted onto 601 165-bp DNA (Nuc165)] in the presence of 30Nick DNA and increasing amounts of NAD+. Under these conditions, PARP-1 modifies itself in an NAD+ dose-dependent fashion, without PARylating nucleosomal histones (Fig. S3 C and D). Nucleosomes were fluorescently labeled on histones H4 and H2B; thus, their integrity in the presence of PARP-1 and NAD+ can be assayed by in-gel FRET (Fig. 2E). In agreement with the effects of PARylation on PARP-1 interactions with Nuc207 (Table 1), increasing amounts of NAD+ decrease the ability of PARP-1 to shift nucleosomes (also reported in ref. 28). However, the nucleosome band remains unchanged and exhibits FRET between histones H2B and H4 throughout the entire titration series, indicative of intact nucleosomes; no free DNA is released (Fig. S3E). This demonstrates that AM–PARP-1, when acting alone, does not disassemble salt dialysis-assembled nucleosomes, despite its high affinity for free histones.
AM–PARP-1 Acts as a Nucleosome Assembly Factor.
To test whether histone binding conferred nucleosome assembly activity onto AM–PARP-1, we used the DNA supercoiling assay. Relaxed plasmid DNA was added to histones that were preincubated with unmodified PARP-1, AM–PARP-1, commercially available PAR (cPAR), or Nap1. Nucleosome assembly was detected by monitoring DNA supercoiling (29) (Fig. 3A). Comparable levels of histone-dependent plasmid supercoiling were observed in the presence of AM–PARP-1 (PARP-1 automodification levels are shown in Fig. S3A) and Nap1 (Fig. 3A; compare lanes 7 and 8 and lanes 11 and 12), whereas no supercoiling was observed in the absence of histone chaperone (lane 4). Similarly, neither PARP-1 nor cPAR displays any supercoiling activity (compare lanes 5 and 6 and lanes 9 and 10).
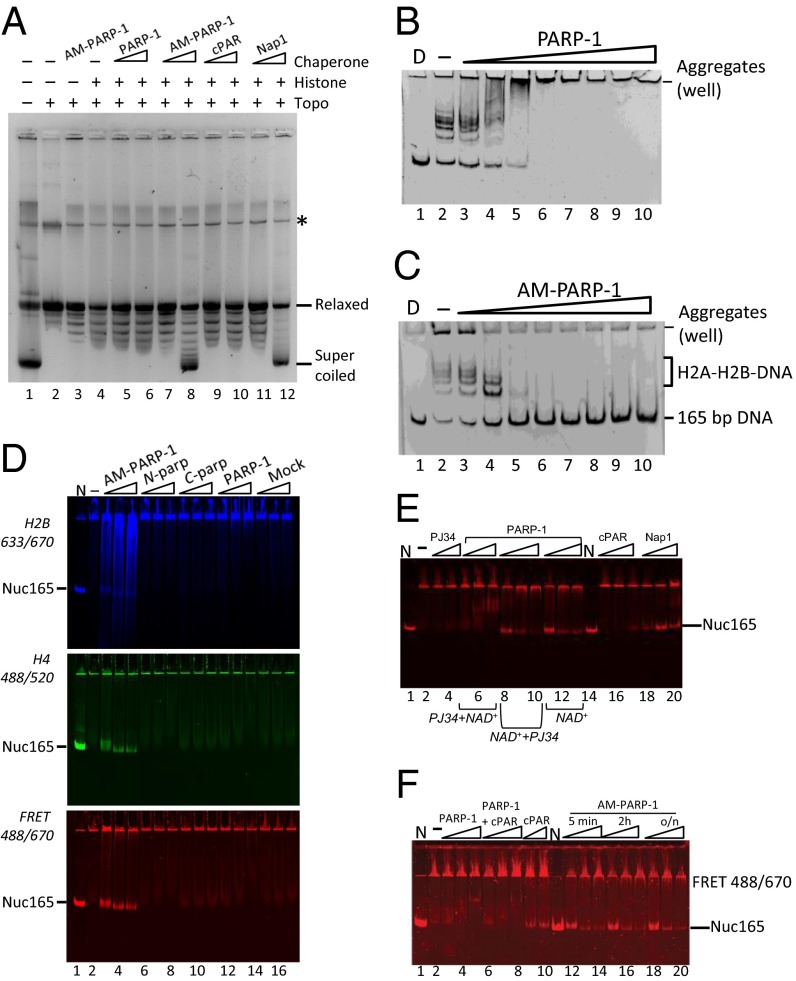
Fig. 3.
AM–PARP-1 is a nucleosome assembly factor. (A) AM–PARP-1 mediates plasmid supercoiling. Lane 1 contains pGEM-3Z plasmid before relaxation. Lane 2 contains relaxed plasmid. Lane 3 contains 200 nM AM–PARP-1 and no histone octamer. Lanes 4–12 contain 40 nM histone octamer. Lanes 5 and 6 contain 40 and 200 nM unmodified PARP-1. Lanes 7 and 8 contain 40 and 200 nM AM–PARP-1. Lanes 9 and 10 contain 40 and 200 nM cPAR. Lanes 11 and 12 contain 40 and 400 nM Nap1. The asterisk (*) indicates a contaminant present in the DNA. Topo, topoisomerase. (B and C) AM–PARP-1, but not unmodified PARP-1, removes H2A-H2B from DNA. Lane 1 contains D only, lane 2 contains 165-bp DNA with a 7 M excess of H2A-H2B. Lanes 3–10 contain 10, 50, 120, 240, 320, 500, 750, and 1,000 nM unmodified PARP-1 or AM–PARP-1. (D) RAC assay, in which 10 nM 165-bp DNA was incubated with 10 nM (H3–H4)2 tetramer (donor) and 120 nM H2A–H2B dimer (acceptor). An increasing amount of PARP-1 was added, and samples were analyzed by 5% native PAGE, followed by visualization of H2B fluorescence (Top), H4 fluorescence (Middle), and FRET between H2B and H4 (Bottom). Lane 1 contains fluorescently labeled nucleosome (N). Lane 2 contains no PARP-1. Lanes 3–5 contain 10, 50, and 120 nM AM–PARP-1 (0.083, 0.42, and 1 M ratio of AM–PARP-1 to H2A–H2B dimer). The same amounts of N-parp, C-parp, unmodified PARP-1, and a mock reaction without PARP-1 were added as indicated (lanes 15–17). (E) Nucleosome assembly activity of AM–PARP-1 cannot be recapitulated by PAR chains and does not require AM–PARP-1 to be enzymatically active. The assay was done as in D; only the FRET scan is shown here. Lanes 1 and 14 contain N. Lane 2 contains DNA, (H3–H4)2 tetramer, and excess H2A–H2B (as in A, lane 2). Lanes 3 and 4 contain 50 and 120 μM PJ34 inhibitor. Lanes 5–7 contain 10, 50, and 120 nM PARP-1 preincubated with a final concentration of 1 mM PJ34 before addition of NAD+ (PJ34 + NAD+). Lanes 8–10 contain 10, 50, and 120 nM PARP-1 preincubated with NAD+ before addition of PJ34 (NAD+ + PJ34). Lanes 11–13 contain 0, 50, and 120 nM PARP-1 together with NAD+ (no inhibitor). Lanes 15–17 contain 10, 50, and 120 nM cPAR. Lanes 18–20 contain 10, 50, and 120 nM Nap1. (F) Length of automodification reaction does not affect assembly activity. The assay was done as in D; only the FRET channel is shown. Lanes 1 and 11 contain N. Lane 2 is DNA, (H3–H4)2 tetramer, and excess H2A–H2B as in D (lane 2). To this sample, 10, 50, and 120 nM PARP-1 (lanes 3–5); PARP-1 together with PAR (lanes 6–8); PAR alone (lanes 9–10); and AM–PARP-1 modified for 5 min (lanes 12–14), 2 h (lanes 15–17), or overnight (lanes 18–20) were added (automodification is shown in Fig. S3A, lanes 15–18).
Nap1 promotes nucleosome assembly by resolving nonproductive interactions of H2A–H2B complex with DNA (30, 31). To test whether AM–PARP-1 has acquired this activity, we incubated DNA with an excess of H2A–H2B dimer and with increasing amounts of unmodified or AM–PARP-1 (Fig. 3 B and C). PARP-1 forms large aggregates with DNA–histone complexes that do not enter the gel (Fig. 3B), whereas AM–PARP-1 efficiently removes histones and releases free DNA (Fig. 3C). This demonstrates that AM–PARP-1 directly competes with DNA for histones, as predicted from our affinity measurements and as observed for other histone chaperones (22, 30).
The ability of a histone chaperone to prevent noncanonical interactions of histones with DNA and nucleosomes might be important during transcription and DNA repair, where ATP-dependent remodeling factors release histones (particularly H2A–H2B dimers) from nucleosomes. We therefore tested whether AM–PARP-1 promotes nucleosome assembly under conditions of histone imbalance. We developed an assay in which an overabundance of histone H2A–H2B (or H3–H4) complex over DNA precludes nucleosome assembly. In the absence of a histone chaperone, no nucleosomes are formed and the entire sample remains in the well (Fig. 3D, lane 2). Bona fide histone chaperones (e.g., Nap1), are able to rescue aggregated chromatin in this assay [rescue of aggregated chromatin (RAC) assay], as observed by the appearance of nucleosomes with fluorescent markers on H4 and H2B (Fig. 3E, lanes 18–20). This assay allows for a more direct readout of the appearance of canonical nucleosomes than the supercoiling assay.
PARP-1, N-parp, C-parp, and a mock sample (no PARP-1) were subjected to automodification. As expected, neither N-parp nor C-parp was automodified under these conditions, whereas full-length PARP-1 undergoes extensive automodification (Fig. S3A). Addition of AM–PARP-1 to the RAC assay results in the formation of nucleosomes (Fig. 3D, lanes 3–5), as evident by FRET, between H2B and H4 in a band that comigrates with salt-assembled nucleosomes (Fig. 3D, lane 1). The N-parp, C-parp, PARP-1, and mock sample do not form nucleosomes, suggesting that automodification confers nucleosome assembly activity onto full-length PARP-1. To verify further that the assembled particles are bona fide nucleosomes, we performed micrococcal nuclease digestion of Nuc165 assembled by salt dialysis, yeast Nap1 (yNap1), or AM–PARP-1 (the latter two assembled via RAC assay). In all instances, we observe protection at 147 bp, as expected for canonical nucleosomes (Fig. S4A). Further, the nucleosome bands were excised from the native gel (similar to that shown in Fig. 3D), electroeluted, and analyzed by SDS/PAGE (Fig. S4B). The histone content for salt-assembled or RAC-assembled nucleosomes (by AM–PARP-1 or yNap1) was found to be very similar.
To dissect whether AM–PARP-1 chaperone function is dependent upon PARP-1 activity during assembly itself, we tested the effect of PJ34 on the ability of PARP-1 to assemble chromatin. PARP-1 was incubated with NAD+ and DNA to initiate automodification. PJ34 was added either before or after 12 h of incubation with NAD+. When PJ34 was introduced before NAD+, auto-PARylation was completely inhibited (Fig. S3A, lane 12) and nucleosome assembly activity was abolished (Fig. 3E, lanes 5–7); inhibitor alone has no activity in the RAC assay (Fig. 3E, lanes 3 and 4). In contrast, the addition of PJ34 after incubation with NAD+ did not inhibit nucleosome assembly activity (lanes 8–10). We conclude that catalytically inactive but modified PARP-1 assembles nucleosomes as efficiently as catalytically active AM–PARP-1 (Fig. 3E, lanes 11–13). This demonstrates that PARylation of PARP-1, but not of histones, is required for its histone chaperone and nucleosome assembly activity.
To test whether PAR chains alone can function in nucleosome assembly, we tested cPAR in the RAC assay. We found that cPAR alone yields only trace amounts of ill-defined nucleosomes at the highest concentrations (120 nM) (Fig. 3E, lanes 15–17). The same result was obtained by mixing cPAR with unmodified PARP-1 (Fig. 3F, lanes 6–8, and Fig. S3F, lanes 6–8). This suggests that PAR needs to be covalently attached to PARP-1 for it to assume nucleosome assembly function.
Because cPAR chains are heterogeneous with respect to chain length and degree of branching, we cleaved PAR from overnight-modified PARP-1 and compared its nucleosome assembly activity with that of the same batch of AM–PARP-1 before cleavage (Fig. 3 E, lanes 16–18, and F, lanes 8–10). We found that the lowest concentration of AM–PARP-1 (10 nM) is as efficient in the RAC assay as the highest concentration of PAR (120 nM) tested (Fig. S3F, compare lanes 14–16 with lanes 17–19).
The experiments described above were all performed with PARP-1 that had been incubated overnight with DNA and NAD+ to ensure a maximum degree of automodification. To exclude possible artifacts due to the long incubation times, we performed the RAC assay with PARP-1 that has been automodified for much shorter times (5 min, 2 h, and overnight, respectively; Fig. 3F and Fig. S3A). Consistent with the notion that PARP-1 is a highly active enzyme, we did not observe any significant differences in the degree of automodification at each of the three time points (Fig. S3A, Right). The three AM–PARP-1 preparations were equally active in the RAC assay (Fig. 3F; compare lanes 12–14, lanes 15–17, and lanes 18–20).
We reproducibly observed that AM–PARP-1 works best at substoichiometric amounts compared with histone dimer (∼0.1 AM–PARP-1 per H2A–H2B dimer) in the RAC assay. It is possible that at higher concentrations, AM–PARP-1 binds to the nucleosomes it assembles (Fig. 1C, lanes 16–19), resulting in an apparent loss of nucleosomes on native PAGE. Alternatively, excess AM–PARP-1 may disassemble partially assembled nucleosomes, even though it has no effect when presented with fully assembled nucleosomes (Fig. 2E).
Discussion
PARP-1 affects many nuclear processes through binding and compacting native chromatin (32). We find that the compaction of chromatin arrays by PARP-1 observed previously (20) can be reproduced with trinucleosomes. We show that one PARP-1 binds one trinucleosome and likely compacts trinucleosomes by reorienting the linker DNA, as suggested previously (8). Auto-PARylation of PARP-1 triggered by a variety of stimuli associated with transcription and DNA repair regulates its ability to compact chromatin. Under our conditions, automodification has little effect on PARP-1 interactions with DNA or with chromatin containing exposed free DNA ends (LE-Tri), although significantly reducing the binding affinity for “native” chromatin (NLE-Tri). This suggests chromatin binding activity that is tunable by PARylation, with important implications for transcription and DNA repair.
The affinity data suggest that PARP-1 interacts differently with native chromatin vs. chromatin at DNA damage sites. Both modes of interactions are of high affinity, but they are fundamentally different in two ways. First, the interaction of PARP-1 with chromatin containing exposed DNA ends (e.g., Nuc207, LE-Tri) does not involve contributions from the C-terminal half of PARP-1, as seen by a similar approximate Kd of full-length PARP-1 and N-parp for these substrates. These interactions are likely dominated by the contacts of the N-terminal domain with the exposed DNA base pair, as seen in the crystal structure of the PARP-1–DNA complex (6). In contrast, the interaction with nucleosomes in absence of free DNA ends (NLE-Tri) involves the C-terminal half of PARP-1 in addition to the N-terminal DNA binding domains. We surmise that the zinc finger domains are no longer able to interact with the terminal DNA base pair, and that this might be compensated for by additional interactions between the tryptophan-glycine-arginine–rich and catalytic domain and nucleosomal components. Second, the interaction of PARP-1 with such native chromatin arrays is reduced 20-fold upon automodification, consistent with the main location of PAR chains on PARP-1 between the N- and C-terminal halves of the protein (Fig. 1A). In contrast, the interaction mode that involves free DNA ends is much less sensitive to automodification.
PARP-1 has been described as a “chromatin-associated factor without any known remodeling or chaperoning activity” (3). However, previous studies have shown qualitative evidence that PARP-1 binds histones, particularly histone H4 (19). Using highly purified PARP-1 and defined histone complexes, we find that unmodified PARP-1 exhibits very weak histone binding activity. It is possible that previous accounts in the literature describing histone binding stem from contamination of histones with nucleic acids or that small amounts of AM–PARP-1 were present in these preparations (19, 33). Automodification dramatically increases the affinity of PARP-1 for histones, firmly placing AM–PARP-1 in the league of bona fide histone chaperones (27). Histone chaperones are defined as nuclear proteins that bind free histones with high affinity, prevent and resolve nonproductive histone–DNA interactions within the cell, and facilitate nucleosome assembly (34). Our data show that AM–PARP-1 fits all of these criteria.
A PARylation-dependent “histone shuttling” mechanism was proposed previously (35). In this model, AM–PARP-1 removes histones from DNA and reassembles nucleosomes in a cyclical manner with contributions from PARP-1 and PARG. In this work, the start and end products of the reactions were not defined; nevertheless, the model is in agreement with our finding that AM–PARP-1 resolves noncanonical histone–DNA interactions, thereby assembling nucleosomes. Additionally, AM–PARP-1 could function by retaining histones during HSP70 activation in Drosophila (12).
To date, none of the histone chaperones studied are able to disassemble properly assembled nucleosomes in the absence of ATP-dependent remodeling factors or transcription, and PARP-1 is no exception. Our finding that excess AM–PARP-1 results in less nucleosome product is consistent with the notion that excess histone chaperone will compete with DNA for histones during the assembly process.
Under our conditions, we did not observe any histone PARylation during chromatin assembly by enzymatically active PARP-1; in fact, inactive AM–PARP-1 assembles nucleosomes with equal efficiency as active AM–PARP-1. The nucleosome assembly activity can only be partially recapitulated with free PAR chains. This suggests that the location of PAR on PARP-1, or perhaps the proper scaffolding of PAR polymers by PARP-1, is required for nucleosome assembly.
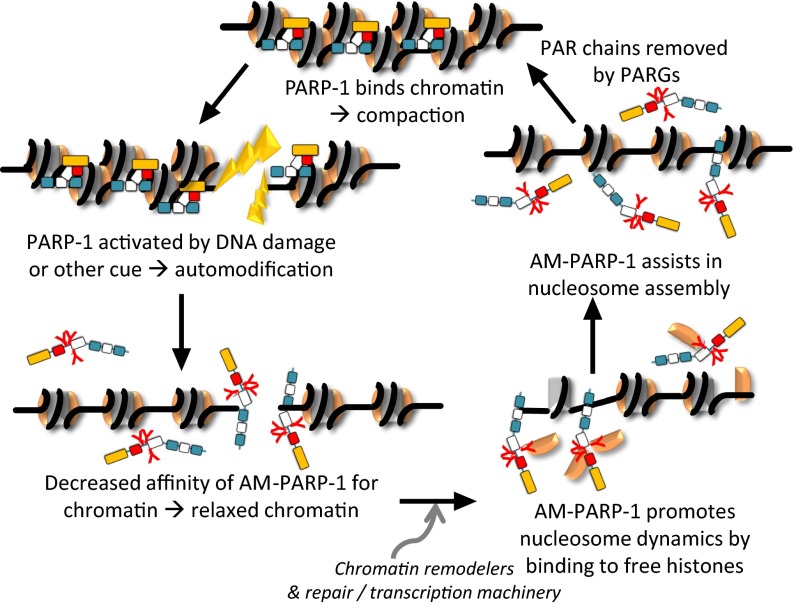
Together, our data demonstrate that enzymatic activation of PARP-1 switches PARP-1 function from a chromatin architectural protein to a histone chaperone and nucleosome assembly factor. In Fig. 4, we propose a tentative model in which PARP-1 binds nucleosomes with high affinity throughout the genome, thereby condensing chromatin (8). Enzymatic activation leads to localized automodification of PARP-1 (8, 12, 19, 28, 36–38). This decreases PARP-1 affinity for nucleosomes and relaxes local chromatin structure. The recruitment of components of the DNA repair or transcription machinery, including chromatin remodeling factors (39–43), results in the destabilization of nucleosomes. The released histones are chaperoned and then reassembled into nucleosomes by AM–PARP-1 in the wake of DNA repair or transcription. Subsequently, PAR chains are removed by PARG, and PARP-1 resumes its function as a chromatin architectural protein. Importantly, only AM–PARP-1 functions as a histone binding protein and a nucleosome assembly factor, making PARP-1, to our knowledge, the first histone chaperone that is switched on by a posttranslational modification.
Fig. 4.
Model for a PAR-induced switch in PARP-1 function. The outcome of PARP-1 enzymatic activation is that the affinity of AM–PARP-1 for chromatin is weakened and its affinity for histones increases dramatically, allowing AM–PARP-1 to function temporarily as a histone chaperone and nucleosome assembly factor until PAR chains are removed.
Materials and Methods
Preparation of reagents (nucleosome substrates and Parp1) is described in SI Materials and Methods. A detailed description of developed assays and ex vivo experiments can also be found in SI Materials and Methods. HI-FI FRET and atomic force microscopy imaging was done as previously described (23, 24).
Supplementary Material
Acknowledgments
We thank the W. M. Keck Protein Purification Facility and H. Scherman for histones, A. E. White for DNA, P. N. Dyer for labeled histones, and R. W. Woody for advice on stoichiometry measurements. This study was supported by National Institutes of Health (NIH) Grants GM067777 and GM088409, by the Howard Hughes Medical Institute (K.L.), and by NIH Grant GM098401 (to T.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405005111/-/DCSupplemental.
References
- 1.Sousa FG, et al. PARPs and the DNA damage response. Carcinogenesis. 2012;33(8):1433–1440. doi: 10.1093/carcin/bgs132. [DOI] [PubMed] [Google Scholar]
- 2.Ko HL, Ren EC. Functional aspects of PARP-1 in DNA repair and transcription. Biomolecules. 2012;2(4):524–548. doi: 10.3390/biom2040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petesch SJ, Lis JT. Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 2012;28(6):285–294. doi: 10.1016/j.tig.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraus WL, Hottiger MO. PARP-1 and gene regulation: Progress and puzzles. Mol Aspects Med. 2013;34(6):1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Krishnakumar R, Kraus WL. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39(1):8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336(6082):728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark NJ, Kramer M, Muthurajan UM, Luger K. Alternative modes of binding of poly(ADP-ribose) polymerase 1 to free DNA and nucleosomes. J Biol Chem. 2012;287(39):32430–32439. doi: 10.1074/jbc.M112.397067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MY, Mauro S, Gévry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119(6):803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37(11):3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao Z, Gao P, Liu HW. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J Am Chem Soc. 2009;131(40):14258–14260. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 11.Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: New players in tumour development. Am J Cancer Res. 2011;1(3):328–346. [PMC free article] [PubMed] [Google Scholar]
- 12.Petesch SJ, Lis JT. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell. 2012;45(1):64–74. doi: 10.1016/j.molcel.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011;25(5):409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.d’Erme M, Yang G, Sheagly E, Palitti F, Bustamante C. Effect of poly(ADP-ribosyl)ation and Mg2+ ions on chromatin structure revealed by scanning force microscopy. Biochemistry. 2001;40(37):10947–10955. doi: 10.1021/bi002742a. [DOI] [PubMed] [Google Scholar]
- 15.Wacker DA, Frizzell KM, Zhang T, Kraus WL. Regulation of chromatin structure and chromatin-dependent transcription by poly(ADP-ribose) polymerase-1: Possible targets for drug-based therapies. Subcell Biochem. 2007;41:45–69. doi: 10.1007/1-4020-5466-1_3. [DOI] [PubMed] [Google Scholar]
- 16.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci USA. 1982;79(11):3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathis G, Althaus FR. Release of core DNA from nucleosomal core particles following (ADP-ribose)n-modification in vitro. Biochem Biophys Res Commun. 1987;143(3):1049–1054. doi: 10.1016/0006-291x(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 18.Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell. 2012;45(6):790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J Biol Chem. 2007;282(44):32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 20.Wacker DA, et al. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27(21):7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraus WL. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20(3):294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286(48):41883–41892. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muthurajan UM, McBryant SJ, Lu X, Hansen JC, Luger K. The linker region of macroH2A promotes self-association of nucleosomal arrays. J Biol Chem. 2011;286(27):23852–23864. doi: 10.1074/jbc.M111.244871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hieb AR, D’Arcy S, Kramer MA, White AE, Luger K. Fluorescence strategies for high-throughput quantification of protein interactions. Nucleic Acids Res. 2012;40(5):e33. doi: 10.1093/nar/gkr1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langelier MF, Planck JL, Roy S, Pascal JM. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem. 2011;286(12):10690–10701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ali AA, et al. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol. 2012;19(7):685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews AJ, Downing G, Brown K, Park YJ, Luger K. A thermodynamic model for Nap1-histone interactions. J Biol Chem. 2008;283(47):32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 29.Fujii-Nakata T, Ishimi Y, Okuda A, Kikuchi A. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J Biol Chem. 1992;267(29):20980–20986. [PubMed] [Google Scholar]
- 30.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37(6):834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Arcy S, et al. Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A-H2B in an unconventional tetrameric form. Mol Cell. 2013;51(5):662–677. doi: 10.1016/j.molcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnakumar R, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319(5864):819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 33.Kim MY. Regulation of chromatin structure by PARP-1. Methods Mol Biol. 2011;780:227–236. doi: 10.1007/978-1-61779-270-0_14. [DOI] [PubMed] [Google Scholar]
- 34.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20(1):14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Realini CA, Althaus FR. Histone shuttling by poly(ADP-ribosylation) J Biol Chem. 1992;267(26):18858–18865. [PubMed] [Google Scholar]
- 36.Martinez-Zamudio R, Ha HC. Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol Cell Biol. 2012;32(13):2490–2502. doi: 10.1128/MCB.06667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang JY, et al. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res. 2006;34(8):2398–2407. doi: 10.1093/nar/gkl241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masaoka A, et al. HMGN1 protein regulates poly(ADP-ribose) polymerase-1 (PARP-1) self-PARylation in mouse fibroblasts. J Biol Chem. 2012;287(33):27648–27658. doi: 10.1074/jbc.M112.370759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahel D, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325(5945):1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottschalk AJ, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA. 2009;106(33):13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA. 2010;107(43):18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29(18):3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costelloe T, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489(7417):581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.