Significance
In mammals, NO produced by NOS acts as a signaling molecule in the nervous and cardiovascular systems and as a cytotoxin in the immune system’s response to infection. Although structures of smaller portions of NOS have been solved, the structures of the entire NOS homodimer have remained elusive. In this study, we use single-particle EM to directly visualize the 3D structures, domain organization, and conformations accessed by all three mammalian NOSs. The observed conformational changes provide insight into the mechanisms through which calmodulin binding results in efficient NO formation.
Keywords: heme, flavin, electron microscopy, conformational heterogeneity
Abstract
NOSs are homodimeric multidomain enzymes responsible for producing NO. In mammals, NO acts as an intercellular messenger in a variety of signaling reactions, as well as a cytotoxin in the innate immune response. Mammals possess three NOS isoforms— inducible, endothelial, and neuronal NOS—that are composed of an N-terminal oxidase domain and a C-terminal reductase domain. Calmodulin (CaM) activates NO synthesis by binding to the helical region connecting these two domains. Although crystal structures of isolated domains have been reported, no structure is available for full-length NOS. We used high-throughput single-particle EM to obtain the structures and higher-order domain organization of all three NOS holoenzymes. The structures of inducible, endothelial, and neuronal NOS with and without CaM bound are similar, consisting of a dimerized oxidase domain flanked by two separated reductase domains. NOS isoforms adopt many conformations enabled by three flexible linkers. These conformations represent snapshots of the continuous electron transfer pathway from the reductase domain to the oxidase domain, which reveal that only a single reductase domain participates in electron transfer at a time, and that CaM activates NOS by constraining rotational motions and by directly binding to the oxidase domain. Direct visualization of these large conformational changes induced during electron transfer provides significant insight into the molecular underpinnings governing NO formation.
NO has emerged as an integral signaling molecule in biology. NOSs are the only enzymes responsible for NO production in mammals. Disruptions in NO signaling are linked to hypertension, erectile dysfunction, neurodegeneration, stroke, and heart disease (1–3). Three NOS isoforms, which vary slightly in size and composition from 260 to 321 kDa for the NOS homodimer, are present in mammals; each is a distinct gene product differing in subcellular localization, tissue distribution, and mode of regulation. The constitutively expressed NOS isoforms are found primarily in endothelial cells [endothelial NOS (eNOS)] and neuronal cells [neuronal NOS (nNOS)], and NO produced by these isoforms initiates diverse signaling processes, including vasodilation, platelet aggregation, myocardial functions, and neurotransmission (4). The activity of the constitutive NOS isoforms is dependent on intracellular Ca2+ concentrations. The third isoform, inducible NOS (iNOS), is transcriptionally controlled and produces cytotoxic concentrations of NO at sites of infection or inflammation. Unlike eNOS and nNOS, the activity of iNOS is independent of the intracellular Ca2+ concentration.
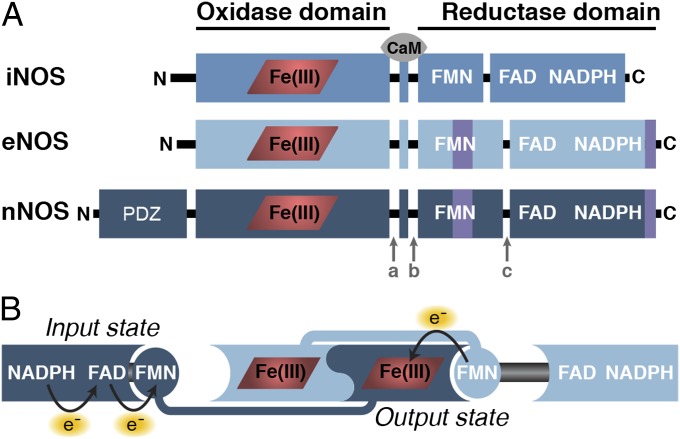
Mammalian NOS isoforms use a complex assembly of domains and cofactors to convert l-arginine to l-citrulline and NO, via the intermediate N-hydroxy-l-arginine. Mammalian NOS isoforms are active as homodimers and composed of two primary domains: the N-terminal oxidase domain and the C-terminal reductase domain (Fig. 1A). The reductase domain can be further divided into the FAD/NADPH-binding and the FMN-binding subdomains. The FMN subdomain is connected to the oxidase domain via a polypeptide linker approximately 30 aa in length, which forms a helical binding site for calmodulin (CaM) (5). During catalysis, the reductase domain passes electrons from the reduced NADPH substrate through the bound FAD and FMN cofactors and ultimately to the heme cofactor in the oxidase domain of the opposite monomer (4, 6). The oxidase domain binds the arginine substrate directly above the heme and tetrahydrobiopterin cofactors. Electron transfer is initiated when NADPH reduces FAD by two electrons. FMN accepts one electron through a direct interaction between the FAD/NADPH and FMN subdomains. This interaction state, where the FMN subdomain is buried within the rest of the reductase domain, is known as the input state (Fig. 1B). To transition into the output state, a large conformational change occurs that allows the FMN subdomain to donate an electron to the oxidase domain. Although progress has been made elucidating the interaction surfaces present in the input and output states (7–12), the trajectory of this conformational change between the input and output is only beginning to be explored (13). Further understanding of this conformational change is particularly important as electron transfer is rate-limiting during NOS catalysis (6).
Fig. 1.
Mammalian NO synthases. (A) NOS domain organization. Each NOS monomer is composed of two domains: the oxidase domain and the reductase domain. The heme cofactor (red) binds within the oxidase domain. The reductase domain is divided further into the FAD/NADPH-binding and the FMN-binding subdomains. CaM binds to a small helical segment that connects the oxidase and reductase domain. Three linker regions connect these domains: linker-a is between the oxidase and the CaM-binding helix; linker-b is between the CaM-binding helix and the FMN subdomain; and linker-c is between the FMN subdomain and the FAD/NADPH subdomain. Although largely similar, each NOS isoform possesses a few unique features. iNOS is the smallest (260 kDa for the homodimer without CaM bound) and lacks the autoinhibitory helix within the FMN subdomain and the C-terminal extension (both shown in purple) present in eNOS and nNOS. eNOS has a molecular weight of 266 kDa for the homodimer, and nNOS is the largest at 321 kDa for the homodimer as a result of an N-terminal extension of ∼250 aa containing a PDZ domain. (B) NO synthase electron transfer pathway. NOS is shown as its native homodimer. The monomer on the left shows the input state, and the monomer on the right shows the output state. In the input state, the FMN subdomain interacts with the FAD/NADPH subdomain allowing electron transfer between the flavin cofactors. In the output state, the FMN subdomain interacts with the heme domain allowing electron transfer between FMN and heme.
For NO formation to occur, NOS requires CaM to bind the linker region between the oxidase and reductase domains. This binding is thought to release the FMN subdomain from the input state (6). Our recent studies investigating iNOS through hydrogen-deuterium exchange MS (HDX-MS) suggested an intermediate state as the enzyme transitions from input to output state (7). During this transition, it is thought that first CaM and then the FMN subdomain bind sequentially and directly to the oxidase domain. Despite these advances in domain placement during various stages of electron transfer, the precise role of CaM during catalysis remains poorly understood.
As the NOS holoenzyme has proven recalcitrant to crystallization, direct high-resolution structural information is limited to the individual NOS domains and subdomains. The oxidase domain structure has been solved for all three NOS isoforms (14, 15). Structures of the nNOS reductase domain (8, 16) and the iNOS FMN subdomain and CaM-binding helix bound to CaM (17) have also been reported. Previous computational docking studies (7, 9, 17, 18) provide an emerging view of the output state in NOS holoenzymes, but these methods provide numerous models, and selecting the catalytically relevant structure or structures without direct experimental evidence is difficult. Our recent HDX-MS studies provided detailed models of the iNOS output state, but, because the FAD/NADPH subdomain was not included in the truncations used in this study, the relative positioning of the entire reductase and oxidase domains could not be determined (7). Based on the crystal structure of the nNOS reductase domain (8), it has been hypothesized that the reductase domains dimerize in the NOS holoenzyme, but this hypothesis remains a point of major uncertainty and contention (6, 19, 20). Furthermore, despite significant homology, it is unknown whether all three NOS isoforms adopt a similar overall structure.
Here, we use EM to determine structures of the iNOS, eNOS, and nNOS holoenzymes bound to CaM. To gain insight into the mechanism of CaM activation of the constitutive NOS isoforms, we also determined structures of eNOS and nNOS without bound CaM. Fitting the NOS domain crystal structures into the EM reconstructions provides a detailed model for the higher-order architecture and quaternary organization of NOS isoforms that is consistent with all reported structural and biochemical data. For each NOS isoform, numerous structures in distinct conformations were determined that correspond to snapshots along a continuous conformational trajectory. These structures illustrate with exquisite detail a well-defined pathway for electron transfer between the reductase and oxidase domains and a mechanism of CaM activation for efficient NOS catalysis.
Results
iNOS Adopts a Wide Range of Conformations.
To determine the structure and domain organization of a full-length NOS holoenzyme, purified murine iNOS was visualized by negative-stain EM. With this technique, the 260-kDa iNOS homodimer bound to CaM is distinguishable as highly flexible particles (Fig. S1A). Individual particles were chosen and sorted by using iterative reference-free and reference-based techniques (Materials and Methods). Approximately 17,500 particles were clustered into 78 2D class averages, revealing that iNOS is highly heterogeneous and adopts a wide range of conformations (Fig. S1B).
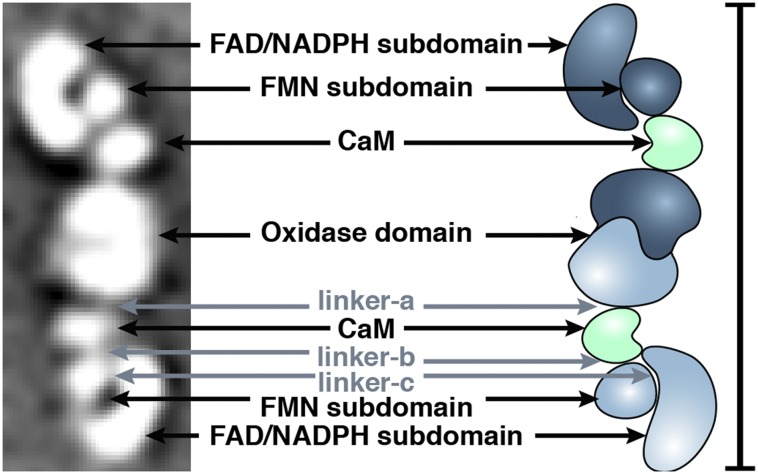
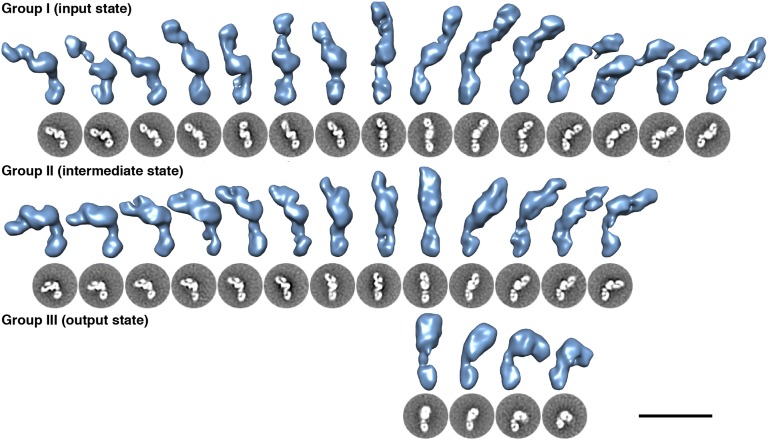
All observed conformations exhibit several common features: an ellipsoid density in the center of the class averages as well as a similarly sized C-shape motif and two circular densities at the peripheries (Fig. 2). The striking detail and recognizable repeating shapes in the 2D averages allowed for immediate assignment of domains based on silhouette and size information provided by known mammalian NOS domain crystal structures (8, 14–16, 21). The central ellipsoid density is the dimerized NOS oxidase domain; the C-shaped density is the FAD/NADPH subdomain; the circular density typically bound within the C-shaped density is the FMN subdomain; the other circular density positioned between the reductase and oxidase domains is CaM bound to the CaM-binding helix of NOS (Fig. 2). In all the 2D class averages, at least four densities are found: the oxidase domain and one reductase domain/CaM feature that projects away from the oxidase domain (the bottom reductase domain in each 2D class average in Fig. 3). The 2D class averages can be divided into three groups based on the densities observed for the other reductase/CaM feature (the top reductase domain in each 2D class average in Fig. 3). All the observed conformations in the 2D class averages lie along a continuous trajectory from the input to the output state. Therefore, these groups are suggested solely to provide a means to discuss critical conformational steps along this continuum, not to define three discrete states. In particular, group II conformations near the transition from group I or to group III are difficult to definitively classify. In group I, which is occupied by ∼50% of the particles, seven densities are clearly delineated in 2D, each corresponding to a known NOS domain or subdomain. Approximately 30% of the particles belong to group II, which differs from group I in that the second FAD/NADPH subdomain is still distinguishable, but the adjacent FMN subdomain and CaM form one continuous density in group II, resulting in six total distinct densities. Group II conformations likely lie along the continuous transition from group I to group III. Group III has the fewest number of particles (∼20%), and the details of the second reductase domain/CaM are not individually defined because the protein is folded compactly on itself; thus, five densities are readily discernable for the group III conformations. To ensure that the acidic pH of the stain was not adversely affecting the observed conformations, iNOS was examined by using a negative stain with a neutral pH (nanoW), and this confirmed that the protein adopts the same range of overall conformations (Fig. S2).
Fig. 2.
Domain assignment. (Left) A 2D class average of iNOS composed of 136 individual particles averaged together shows clearly defined densities for each domain and subdomain. The oxidase is dimerized in the center with a bound CaM on either side. The round FMN subdomain and C-shaped FAD/NADPH subdomain of the reductase domain are easily distinguished. The approximate locations of the three linker regions are also shown: linker-a is between the oxidase and the CaM-binding helix; linker-b is between the CaM-binding helix and the FMN subdomain; and linker-c is between the FMN subdomain and the FAD/NADPH subdomain. (Right) An NOS schematic showing CaM in green and one iNOS monomer in navy blue and the other in light blue. (Scale bar: 28.5 nm, i.e., the same length as the structure.)
Fig. 3.
iNOS 2D class averages (gray scale) with their corresponding 3D reconstructions (blue). Images were collected as detailed in Materials and Methods. iNOS is highly flexible, and the observed iNOS conformations can be organized into three main groups. Thirty-two structures (of 45 total) are shown, selected to illustrate the range of motion accessible in each state. Group I is defined by the presence of two well-defined reductase domains that contain easily distinguishable FAD/NADPH and FMN subdomains. In group II, the overall range of motion remains similar to group I, but one of the FMN subdomains has been released from the FAD/NADPH subdomain, causing the observed density to merge with the adjacent CaM in the 2D class averages. In group III, the second reductase domain has folded over to interact directly with the oxidase domain. Furthermore, the range of motion of the interacting oxidase/reductase cluster is restricted relative to the other reductase domain in group III. (Scale bar: 20 nm in relation to 3D reconstructions.)
iNOS Reconstructions Reveal 3D Flexibility.
To better understand the range of flexibility indicated by the 2D class averages of iNOS, the random conical tilt (22, 23) strategy was used to visualize in 3D the multitude of conformations adopted by iNOS. Because no other validated model of full-length NOS was available, this reference-free ab initio method allowed for 2D classes to be reconstructed in an unbiased manner into 3D density maps, which importantly does not rely on the generation of initial models or masks. From 78 2D classes, 45 3D maps were reconstructed that were fully consistent with their respective 2D class averages (Fig. 3). These models represent snapshots of a continuous movement between the distinct NOS domains. To align the structures in a consistent orientation to facilitate comparison, an iNOS reductase domain homology model was constructed from the crystal structure of the complete rat nNOS reductase domain (8) (Materials and Methods). This homology model was low-pass filtered to 30 Å and used to align to a single reductase domain from each reconstruction. This analysis revealed that the relative orientation of the CaM adjacent to the aligned reductase domain adopted a variety of positions, suggesting a flexible linker (linker-b; Fig. 1A) between the FMN subdomain and CaM-binding helix. The pronounced flexibility of this linker was previously observed in the crystal structure of the human iNOS subdomain and CaM-binding helix bound to CaM (17). Our data further suggest a flexible linker between the CaM-binding helix and the oxidase domain (linker-a; Fig. 1A), as these two components were also positioned at a wide variety of angles relative to one another in the 2D class averages (Fig. 3). The 3D motion in and out of plane of the oxidase domain relative to the reductase domain illustrated by the 3D reconstructions (Fig. 3) suggests that the overall motion may be more complex than a simple 2D hinge. The linkers allow for motions that provide structures in which the complex ranges from a straight/extended conformation to a condensed more compact structure. When in an elongated conformation, the maximum vertical length of the protein is 285 Å, with a width of 20 Å at the narrowest point (at the bound CaM) and as much as 60 Å at the widest point (oxidase domain; Fig. 3). To verify that the primarily extended conformations observed by EM represent conformations also present in solution, iNOS was subjected to analytical ultracentrifugation. Fitting the data to a cylindrical model provided a length of 234 Å and diameter of 52 Å in complete agreement with the primarily elongated conformations observed by using EM.
Higher-Order Domain Architecture of iNOS.
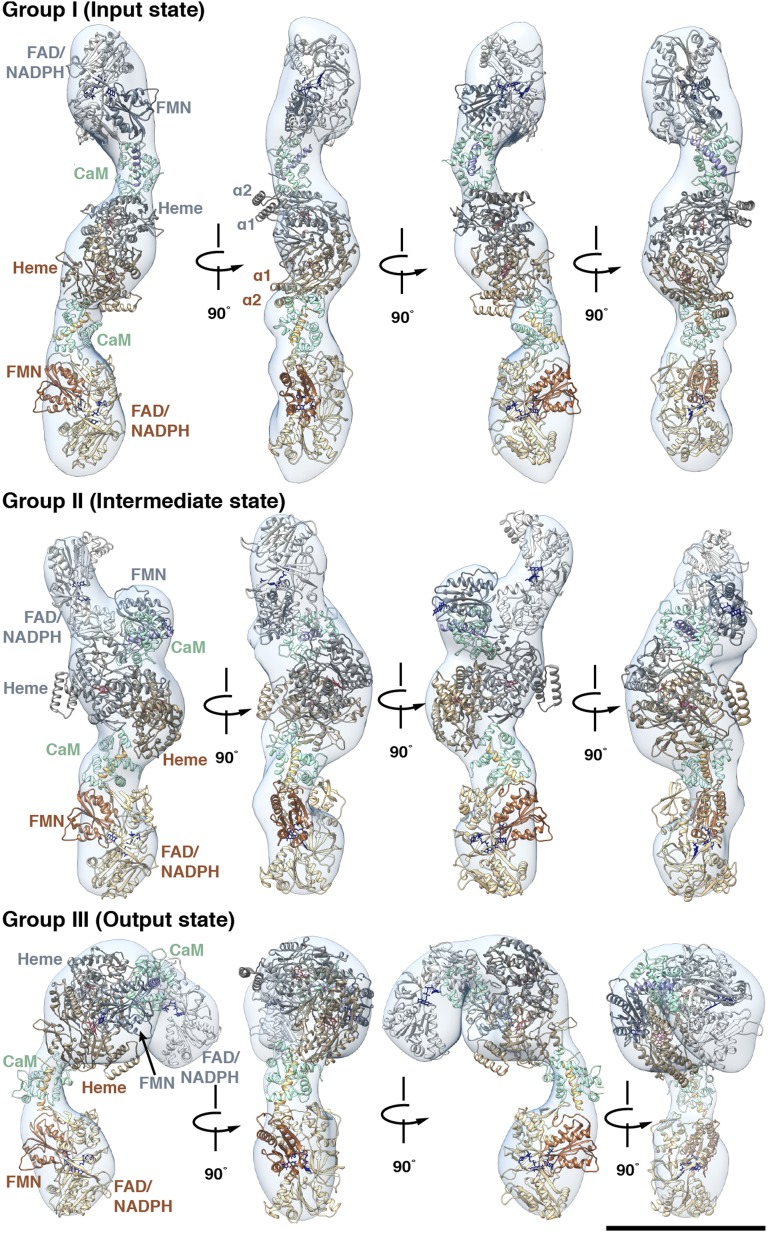
To provide a more detailed understanding of the domain orientations and interactions in the iNOS holoenzyme complex, homology models and previously solved crystal structures of the murine iNOS domains and CaM (8, 17) were fit the EM maps (Fig. 4 and Movies S1–S3). A representative structure for each group is shown, but the crystal structures were fit and are well accommodated by all EM maps shown in Fig. 3, indicating that these EM maps represent full-length NOS protein and not partially proteolyzed NOS truncations accessed during purification. To further verify the consistency of these fittings, the 2D class averages were compared with forward projections of the proposed models (Fig. S3). For group I, two copies of the reductase homology model were docked manually into the corresponding terminal densities and further refined using the “fit in map” function of UCSF Chimera (24). Crystal structures of CaM bound to the CaM-binding helix of iNOS (17) and the murine iNOS oxidase domain (25) were fit into the remaining densities by the same method. The best fits placed the C-termini of the oxidase domain dimer and the N-termini of the CaM-binding helices ∼27 Å apart—compatible with the expected length of a ∼20-aa linker region (26). Likewise, the C-termini of the CaM-binding helices and the N-termini of the FMN subdomains are located within the predicted 27-Å, ∼20-aa-length linker between these domains. In the group I conformations, the oxidase domain does not interact with the reductase domain or a specific binding surface on CaM, suggesting that the group I conformations are representative of the input state (Fig. 4 and Movie S1).
Fig. 4.
The higher-order domain architecture of iNOS. Crystal structures and homology models of the individual domains and subdomains are shown as ribbon structures and were fit into a representative EM map from the input (group I), the intermediate (group II), and the output (group III) state. The ribbon structures shown in shades of brown belong to one monomer, and those in shades of gray belong to the second monomer. CaM is shown in green, the heme cofactors are shown in red, and FAD and FMN are shown in navy blue. The progression of electron transfer can be observed by tracing the movement of the FMN subdomain in the gray monomer. In group I, the FMN subdomain is bound within the FAD/NADPH subdomain. In group II, CaM has docked to the oxidase domain, and the FMN subdomain has begun to pull away from the rest of the reductase domain. By group III, the FMN subdomain has also docked to the oxidase domain allowing electron transfer from FMN to the heme cofactor in the oxidase domain of the opposite (brown) monomer. The 2D class average used to create the 3D reconstruction, and the corresponding forward projection of the proposed models for each of the structures are shown in Fig. S3. (Scale bar: 10 nm.)
For the group II structures, the iNOS domains and CaM were fit in the same manner as described earlier (Fig. 4 and Movie S2). However, unlike group I, in which the reductase domains both adopt the input state conformation present in the crystal structure of the nNOS reductase domain (8), in group II, one of the FMN subdomains (of the top reductase domain in Fig. 3) is released from its interaction with the FAD/NADPH subdomain. This FMN subdomain and the nearby CaM are closely positioned and appeared merged in group II and therefore could not be independently docked. In addition, the density of this CaM is positioned near the oxidase domain dimer interface, suggesting a direct interaction between CaM and the oxidase domain. The location of this direct interaction between CaM and the oxidase domain matched precisely with the interaction proposed in our recent iNOS HDX-MS and modeling study (7). Therefore, our NOS models of CaM docking on the oxidase domain from our recent HDX-MS study (7) were used to place CaM in the group II densities. Because, in these conformations, CaM is interacting with the oxidase domain and the FMN subdomain is released from the FAD/NADPH domain but not yet interacting with the oxidase domain, the group II conformations belong to an intermediate, transitional CaM-docked state the existence of which we previously hypothesized (7).
Group III structures show conformations whereby one of the reductase domains and CaM have folded over to directly interact with the oxidase domain (Fig. 4 and Movie S3). In these conformations, CaM remains in the same position as in group II, but the FMN subdomain is extended further to interact with the oxidase domain of the opposite monomer in the NOS homodimer. This interdimer interaction is necessary for the interdimer electron transfer between the FMN and heme cofactors known to occur during NOS catalysis (4, 6). We recently developed computational models of the output state complex between CaM, the FMN subdomain, and the oxidase domain that are entirely consistent with all available HDX-MS and mutagenesis data (7). Importantly, these output state complex models (7) were fit without any alteration into the EM density, and the FAD/NADPH subdomain fits snugly into the remaining density (Fig. 4 and Movie S3). The combination of the previous HDX-MS data (7) and the present EM data allowed unambiguous fitting of the similarly sized FMN subdomain and CaM into the group III 3D reconstructions. Furthermore, the relative positioning of CaM, the FMN subdomain, and the oxidase domain in group III is in agreement the proposed positioning in a recent cryo-EM study of eNOS with and without bound CaM (27). Thus, all available structural and biochemical data support the conclusion that group III represents the output state of iNOS.
The known crystal structures of the domains are well accommodated into the EM density maps in each of the three states with one exception. When fitting iNOS oxidase domain crystal structures, the α1 and α2 helices of each monomer of the oxidase domain are consistently and symmetrically positioned outside the oxidase domain EM density. This minor discrepancy between the iNOS oxidase domain crystal structures and EM density maps suggests either that the position of these helices in the oxidase domain crystal structures does not reflect the positioning in the iNOS holoenzyme or that these helices are dynamic and adopt a variety of conformations that are lost as a result of averaging during data processing. Despite this single discrepancy, the consistency of the fittings, suggest that the domains (or subdomains in the case of the reductase portion) remain relatively rigid, intervened by flexible linkers.
Destabilization of the iNOS Input State Results in Greater Occupancy of the Output State.
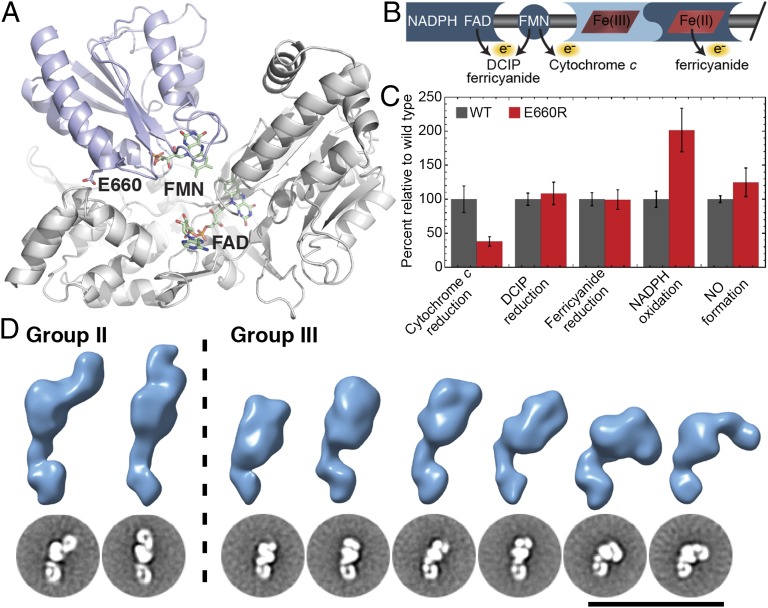
To further confirm that group I represents the input state and group III represents the output state (Fig. 3), we perturbed the equilibrium toward the output state by construction of a point mutant (iNOS E660R) that, based on our recent HDX-MS studies (7), was predicted to selectively disrupt the interaction of the FMN subdomain with the FAD/NADPH subdomain but not the oxidase domain (Fig. 5A). To verify that the iNOS E660R mutant behaved as expected, the steady-state rate of cytochrome c, 2,6-dichlorophenol-indophenol (DCIP), and ferricyanide reduction as well as NADPH oxidation and NO formation were determined as detailed in Materials and Methods (Table S1). In NOS isoforms, cytochrome c is reduced exclusively by the FMN cofactor; DCIP is reduced by FAD or FMN; ferricyanide is reduced by FAD, FMN, or heme (Fig. 5B) (6, 10). A mutant that selectively perturbs the interface between the FMN subdomain and the FAD/NADPH subdomain should result in a decreased efficiency of electron transfer from FAD to FMN. This decreased efficiency would be more pronounced for cytochrome c reduction compared with DCIP or ferricyanide reduction. As predicted, iNOS E660R displayed a significantly decreased rate of cytochrome c reduction (0.57 ± 0.1 s−1) compared with iNOS WT (1.50 ± 0.29 s−1) whereas the rates of DCIP and ferricyanide reduction were not significantly changed from the rates determined for iNOS WT (Fig. 5C and Table S1). The E660R mutation may also disrupt the interaction of cytochrome c with the FMN subdomain (28). In addition, a decreased efficiency of electron transfer from FAD to FMN was predicted to uncouple NADPH oxidation from NO formation. Indeed, the rate of NADPH oxidation for iNOS E660R (2.08 ± 0.33 s−1) was nearly double that of iNOS WT, but the rate of NO formation for iNOS E660R (0.60 ± 0.10 s−1) corresponded to the rate for iNOS WT (0.48 ± 0.02 s−1) within error (Fig. 5C and Table S1), indicative of significant uncoupling in iNOS E660R. The increased rate of NADPH oxidation for iNOS E660R is likely a result of increased solvent exposure of the FAD and FMN cofactors and resulting increased rates of electron transfer to alternative electron acceptors (e.g., oxygen) in addition to the heme reduction required for NO formation. The observed unchanged rate of NO formation suggests that precise tuning of the NOS interdomain interfaces is more important to prevent uncoupling than to optimize the rate of the electron transfer to the oxidase domain for NO formation. Taken together, the aforementioned results support the hypothesis that the E660R mutant selectively disrupts the interaction of the FMN subdomain with the FAD/NADPH subdomain. EM data were then acquired and processed for the iNOS E660R mutant by using the same methods used for the WT protein. As predicted, the majority of the conformations observed for the iNOS E660R mutant corresponded to group II and group III, and no 2D class averages were found corresponding to group I of iNOS WT (Fig. 5D). This observation provides strong evidence that the group I conformations observed for iNOS WT but not present in the iNOS E660R mutant represent the input state and the group III conformations enriched for the iNOS E660R mutant represent the output state.
Fig. 5.
Kinetic and EM analysis of iNOS E660R. (A) Homology model of the murine iNOS reductase domain based on the structure of the nNOS reductase domain (8). E660, FMN, and FAD are shown as sticks. The FMN subdomain is colored light blue and the FAD/NAPDH subdomain is colored light gray. (B) NOS cofactor specificity for the alternative electron acceptors used in this study. DCIP can accept electrons from FAD or FMN. Cytochrome c selectively accepts electrons from FMN. Ferricyanide can accept electrons from FAD, FMN, or ferrous heme. (C) Bar graphs of steady-state rates of cytochrome c reduction, DCIP reduction, ferricyanide reduction, NADPH oxidation, and NO formation for iNOS WT (dark gray) and E660R (red). Rates were normalized to the rates for iNOS WT. Assays were performed as described in Materials and Methods. Error bars represent SD from the mean for at least five independent replicates. (D) The constricted range of motion of iNOS E660R visualized through 2D class averages and 3D reconstructions. Approximately 25% of iNOS E660R particles occupy group II conformations, whereas an overwhelming 75% of particles belong to group III. No group I conformations characterized by two defined reductase domains locked in the input state were observed. The abundance of group III particles with iNOS E660R revealed several new group III conformations that were not defined with iNOS WT because of the lower abundance of group III particles in the iNOS WT dataset. Steady-state kinetics are shown in Table S1.
All Three NOS Isoforms Adopt Similar Conformations and a Similar Range of Motion.
The iNOS EM data described thus far provides a detailed conformational landscape for the progression between the input and output states. However, it was unclear if the constitutive NOS isoforms adopt similar overall structures and ranges of conformations. In addition, as iNOS activity does not respond to changes in intracellular Ca2+ concentration as a result of formation of a tight complex with CaM, this precluded determination of the mechanisms through which CaM binding and dissociation regulate NOS activity. We thus determined the structures of full-length human eNOS and rat nNOS in the presence and absence of bound CaM. Although closely related in sequence to iNOS, the nNOS and eNOS isoforms respond to the intracellular Ca2+ concentration for CaM binding and subsequent NO formation; thus, both eNOS and nNOS could be examined under two physiologically relevant conditions: with and without CaM bound. EM sample preparation, data collection, and data processing methods closely followed those described for the iNOS datasets detailed earlier (Materials and Methods).
Considering the basic silhouette of the structures (Fig. 6), the range of conformations observed for all isoforms, with and without CaM bound, are similar, ranging from a straight and elongated morphology to a bent and folded-over form. When CaM is bound, the detailed features of the domains of eNOS and nNOS appear similar to those recognized in iNOS: a central dimerized oxidase domain and two flexible reductase domains with CaM bound in between. In many of the 2D class averages each representing an individual conformation, the FMN subdomain and CaM are readily recognizable, but, in general, the class averages of the constitutive NOS isoforms with CaM bound are not as clearly defined as the corresponding iNOS class averages. This might be a consequence of incomplete occupancy of CaM on the CaM-binding helices of eNOS and nNOS.
Fig. 6.
NOS isoforms share the same basic structure. Two-dimensional class averages of iNOS, eNOS, and nNOS arranged by column (i–viii) to emphasize the similar set of conformations adopted by each NOS isoform. CaM binding enhances the detail in the individual NOS domains and subdomains. iNOS is the most highly resolved isoform; the CaM-binding helix is fully occupied in this NOS isoform. The loss of definition in the unbound forms of eNOS and nNOS (labeled “–CaM”) is probably the result of the increased rotational flexibility in the linking regions in the absence of CaM. Similarly, the loss of details in the domains of CaM-bound eNOS and nNOS (labeled “+CaM”) compared with iNOS may arise from incomplete CaM-binding. (Scale bar: 20 nm.)
In eNOS and nNOS samples without CaM bound, there is a striking lack of well-defined details of the individual domains: neither the characteristic shape of the FAD/NADPH nor the FMN subdomain can be identified in any single 2D class average or 3D reconstructed density map (Fig. 6 and Fig. S4). This blurring of the higher resolution information may arise as a result of an increased range of rotational motion in linker-a and linker-b in the absence of CaM. This increased heterogeneity means that very few particles occupy the exact same conformation, and thus well-defined features are lost because of overlay of many different conformers during averaging.
Although the three NOS isoforms possess several notable differences in their amino acid sequence, these differences are not observed in the EM 2D class averages and 3D maps at the resolution possible for these highly flexible conformers. iNOS lacks two components found in eNOS and nNOS that are necessary for Ca2+ regulation: an autoinhibitory helix in the FMN subdomain and the C-terminal extension, but the ∼42-aa single helix and ∼30-aa extension are not large enough to alter the perceived structure at the resolution attainable for negatively stained NOS isoforms. nNOS incorporates an additional N-terminal extension of ∼250 aa containing a PDZ domain that targets the enzyme to the cell membrane via protein–protein interactions (29). Although extensions of this size are large enough to observe by EM, a distinct extra density in nNOS was not observed compared with eNOS and iNOS. One possible explanation is that, because the size of the oxidase domain dimer is larger in nNOS compared with eNOS and iNOS (as detailed later), the PDZ domain may be closely associated with the nNOS oxidase domain. Alternatively, the PDZ domain may be flexibly positioned relative to the oxidase domain and therefore the density of the PDZ cannot be recovered by averaging.
Measurements of lengths of the 3D structures in their most elongated form (Fig. 6, i, and Fig. S4, i) show that iNOS and eNOS occupy nearly identical dimensions to each other when CaM is bound: a vertical length of ∼285 Å and a width of ∼25 Å at the thinnest point (where CaM is bound) and ∼60 Å at the widest point in the oxidase dimer. CaM-unbound eNOS occupies identical dimensions, but, because the densities of the oxidase and reductase domain are discontinuous, there is no observable thin width. nNOS, both CaM-bound and unbound, has a vertical length of ∼320 Å. The widths of CaM-bound range from ∼25 to ∼70 Å, whereas, when CaM is not bound to nNOS, again there is no minimum width, but the maximum width in the oxidase domain is ∼75 Å. Analytical ultracentrifugation measurements were in agreement the overall shape and size of the unbound and CaM-bound of eNOS and nNOS.
Discussion
The lack of information on higher-order domain architecture and quaternary structure of NOS isoforms was a vital missing piece of NOS enzymology and hampered a full appreciation of the mechanisms controlling NO production. In the absence of structural information, two critical pieces of the NOS mechanism have remained largely unknown: (i) the pathway of the large conformational change that the FMN subdomain undergoes to transition from the input state to the output state; and (ii) the mechanism through which CaM activates this conformational change. Furthermore, it was unclear whether the three mammalian NOS isoforms display similar or distinctive overall structures and mechanisms of CaM activation. By using EM, we captured and compared 3D reconstructions of all three full-length NOS holoenzymes in multiple conformations, in CaM-bound and unbound states in the case of eNOS and nNOS. Exploring this series of structures allowed us to define the higher-order domain architecture, compare the NOS isoforms with each other, verify flexible linker regions, and visualize critical interactions. This additional information also allows in depth visualization of the sequence of conformational changes necessary for efficient electron transfer during NOS catalysis.
By using automated high-throughput single-particle EM (30–33), 512 individual 3D reconstructions of full-length murine iNOS, human eNOS, and rat nNOS were generated. The overall domain structure and range of conformations adopted by the three NOS isoforms are largely similar, with the central oxidase domain dimerized as a single feature and two flexible reductase domains of varying conformations protruding at each end of the oxidase domain (Fig. 6). The known crystal structures of CaM and the individual NOS domains and subdomains are well accommodated in the 3D reconstructions, suggesting that the oxidase domain, FMN subdomain, FAD/NADPH subdomain, and CaM remain relatively rigid, connected by flexible linkers. Three primary flexible linkers were observed: linker-a between the oxidase domain and the CaM-binding helix, linker-b between the CaM-binding helix and the FMN subdomain, and linker-c between the FMN subdomain and the FAD/NADPH subdomain (Fig. 1A). Each of these linkers could adopt a variety of orientations, providing significant conformational flexibility between the reductase and oxidase domains. The observed conformational flexibility was consistent with the 2D class averages of a recent nNOS EM study (13). This conformational flexibility allowed us to examine snapshots of the NOS holoenzyme in the input and the output state as well as many intermediate states. The snapshots were organized sequentially to visualize a transition pathway for the FMN subdomain and CaM between the input and output states (Fig. 7). Understanding of this large conformational change is particularly critical, as this step is rate-limiting during catalysis (6, 14, 25).
Fig. 7.
NOS electron transfer states. In the input state, the two NOS reductase domains are highly flexible with respect to the oxidase domain and adopt a wide range of conformations (dashed gray arrows). When electron transfer is initiated (yellow arrows), a single monomer (dark blue) undergoes a series of conformational changes: CaM docks to the oxidase dimer, which pulls away the FMN subdomain from the NADPH/FAD subdomain (solid gray arrows). This locks the monomer into an intermediate, CaM-docked state. NOS then continues to the output state, and the FMN subdomain undergoes a second transition (gray arrow) to interact with the oxidase domain of the opposite monomer and transfer an electron (yellow arrow) to the heme cofactor. Throughout this electron transfer process, the second monomer (light blue) remains in the input state and adopts a variety of conformations (dashed gray arrows) that are unaffected by the large conformational changes occurring in the opposite monomer.
Direct visualization of the intermediates along the transition from the input state to the output state provided several insights into the mechanisms of CaM activation of NOS catalysis. CaM activates NO synthesis via two proposed mechanisms (4, 6). The first mechanism is unique to the constitutive NOS isoforms. In this mechanism, CaM binding releases the FMN subdomain from its interaction with the FAD/NADPH subdomain, allowing the FMN subdomain to transfer electrons to the oxidase domain (or alternative acceptors such as cytochrome c; Fig. 1B) (34–39). Indeed, in CaM-bound nNOS, many 2D class averages were observed whereby the FMN subdomain is visualized as a blurred cloud of density (Fig. 6), consistent with the FMN subdomain adopting a variety of conformations upon release from the FAD/NAPDH subdomain. This blurring also means that the FMN subdomain is absent in the corresponding 3D reconstructions (Fig. S4).
The second function of CaM in the constitutive NOS isoforms is to increase the efficiency of electron transfer from the FMN subdomain to the oxidase domain (7, 27, 40–42). CaM activates this electron transfer by binding to the CaM-binding helix between linker-a and linker-b, which constrains the rotational movements of the reductase domain relative to the oxidase domain. The increased rotational constraint is manifested as a striking loss of resolution in the unbound compared with the CaM-bound samples of eNOS and nNOS (Fig. 6). Consistent with this interpretation, iNOS binds CaM with the highest affinity [Kd ≤ 0.3 nM (43–45)], and the domains of iNOS are the most clearly resolved. eNOS [Kd = 1.6–4 nM (44, 46, 47)] and nNOS [Kd = 1.8–5.6 nM (44, 48, 49)] display weaker CaM binding, and therefore CaM may partially dissociate at the low concentrations (3.2 nM for eNOS and 4 nM for nNOS) required for EM data collection, which results in increased heterogeneity compared with iNOS. Alternatively, even with CaM fully bound eNOS and nNOS may be intrinsically more flexible than iNOS.
CaM also forms a direct complex with the oxidase domain in all three NOS isoforms (Fig. 6 and Fig. S4). Formation of this complex further restricts the movements of the FMN subdomain to conformations near the oxidase domain, which activates electron transfer from the FMN subdomain to the oxidase domain. Importantly, the location of the CaM-docking site on the NOS oxidase domain is entirely consistent with the location predicted by our recent HDX-MS and modeling studies of iNOS (7), as well as a recent cryo-EM study of eNOS (27). Furthermore, based on our recent HDX-MS studies (7), we hypothesized that the CaM-docked state precedes the output state, and the CaM-docked state is required for efficient alignment of the FMN subdomain and oxidase domain for subsequent electron transfer in the output state. In support of this hypothesis, we observe several conformations whereby CaM has directly bound the oxidase domain, but the FMN subdomain remains unbound (Fig. 7).
CaM activation of iNOS is unique among the three NOS isoforms in that CaM copurifies with iNOS (50) and iNOS activity is not controlled by changes in intracellular Ca2+ concentration. In addition, previous studies have shown that CaM binding to the isolated iNOS reductase domain does not increase the rate of cytochrome c reduction (41, 42, 51), suggesting that CaM binding to the CaM-binding helix by itself does not release the FMN subdomain when the oxidase domain is absent. Therefore, CaM activates iNOS solely by stabilizing the interaction of the FMN subdomain with the oxidase domain (41, 42, 51). Consistent with this hypothesis, conformations in which the FMN subdomain is released from the rest of the reductase domain are not found in the iNOS 2D class averages and 3D reconstructions unless CaM has already docked to the oxidase domain (Fig. 3).
Another interesting feature observed in almost all the iNOS, eNOS, and nNOS reconstructions is that the FMN subdomain of one of the monomers is locked in the input state in the same conformation as the crystallized nNOS reductase domain (8). However, in contrast to the dimeric crystal structure of the nNOS reductase domain (8), the reductase domain was never observed as a dimerized entity in the hundreds of EM structures determined for all three NOS isoforms. Taken together, these observations suggest that only one reductase domain at a time contributes to NOS catalysis, while the other reductase domain remains in the input state. In contrast, a recent negatively stained EM structure of a chemically cross-linked nNOS–CaM complex showed both reductase domains in the output state (13). Because of the high flexibility observed here for NOS isoforms, it is possible that cross-linking the particles may have trapped a minor NOS conformation that is not populated under normal conditions. Furthermore, the enforcement of C2 symmetry during the reconstruction steps may bias the analysis. It is notable that the 2D class averages obtained by Yokom et al. (13) for the nNOS–CaM complex in the absence of chemical cross-linking are in complete agreement with those reported here.
These structures of all three NOS isoforms, consistent with available biochemical and structural data, give substantial insight into the NOS conformational changes required for efficient electron transfer between the reductase and oxidase domains as well as the mechanisms through which CaM restricts and regulates these conformational changes. Each NOS holoenzyme adopts many conformations comprised of a dimerized oxidase domain flexibly attached to two monomeric reductase domains. The observed flexibility of these reductase domains also likely explains why NOS holoenzymes have proven to be recalcitrant to crystallization. The flexibility may be necessary to minimize unproductive electron transfer from the reductase domain to the oxidase domain when CaM is not bound and NO synthesis is not desired, while still allowing the FMN subdomain the necessary flexibility to shuttle electrons from the reductase domain to the oxidase domain when CaM is bound.
Materials and Methods
Mutagenesis, Expression, and Purification of NOS Isoforms.
The murine iNOS E660R mutant was generated in the pCWori vector by using the QuikChange method (Stratagene) according to the manufacturer’s instructions except that the annealing temperature was 50 °C and the extension time was 21 min. iNOS WT and E660R were coexpressed with CaM and purified as previously described (52, 53). Human eNOS in the pCWori vector was gift of Paul R. Ortiz de Montellano (University of California, San Francisco, CA). eNOS was coexpressed with CaM and purified as previously described (54). Rat nNOS in the pCWori vector was a gift of Richard B. Silverman (Northwestern University, Evanston, IL). nNOS was expressed and purified as previously described (55, 56). Protein purity was assessed by SDS/PAGE and concentrations were determined by using the method of Bradford (57) with BSA as the standard.
iNOS Steady-State Activity Assays.
Steady-state ferricyanide reduction, DCIP reduction, cytochrome c reduction, NADPH oxidation, and NO formation kinetic assays with iNOS WT and E660R were performed as previously described (7).
Analytical Ultracentrifugation.
Sedimentation velocity experiments were performed in a ProteomeLab XL-I (BeckmanCoulter) analytical ultracentrifuge. The samples contained 2.4 µM nNOS, eNOS, or iNOS, 100 µM arginine, 150 mM NaCl, 1 mM β-mercaptoethanol, and 100 mM Hepes, pH 7.5. To some samples, 3.6 µM CaM and 3 mM CaCl2 were added. Protein samples were loaded in two-channel cells and spun in an An-50 Ti eight-place rotor at 30,000 rpm at 20 °C for 15 h. Absorbance at 280 nm was used for detection. Sedimentation velocity data were analyzed by using Sedfit software (P. Schuck, National Institutes of Health/National Institute of Biomedical Imaging and Bioengineering). Shape models were generated by using Sednterp software (D. B. Hayes, Magdalen College, Oxford, United Kingdom; T. Laue, University of New Hampshire, Durham, New Hampshire; and J. Philo, Alliance Protein Laboratories).
Negative-Stain EM.
iNOS was diluted to a final concentration of 4.6 × 10−9 M in 50 mM triethanolamine, pH 7.5, 150 mM NaCl, and 5 mM DTT. iNOS E660R was diluted to a final concentration of 6.7 × 10−9 M in 50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM DTT, 100 µM arginine, 50 µM tetrahydrobiopterin, and 200 µM NADP+ (buffer A). eNOS and nNOS were diluted in buffer A to 3.2 × 10−9 M and 4.0 × 10−9 M concentrations, respectively. For the CaM-bound experiments, 1.5 equivalents of CaM relative to the NOS monomer concentration were added to buffer A. As eNOS was coexpressed and purified with CaM, to obtain non–CaM-bound eNOS, 2 mM of EGTA was added to buffer A. For all experiments, 3 µL of the sample was applied to a glow-discharged C-flat grid with 2-µm holes overlaid by a thin (∼1.5 nm) layer of continuous carbon. The specimen was stained as previously described (58) with a solution containing 2% (wt/vol) uranyl formate or nanoW.
Data were acquired by using a Tecnai F20 Twin transmission electron microscope operating at 200 kV using a dose of ∼35 e−/Å2 and a nominal range of 1.0–2.5 μm under focus. Tilt pair micrographs at 0° and −55° at a nominal magnification of 62,000× (pixel size, 0.136 nm) were collected by using the automated software Leginon (31). All images were recorded on a 4k × 4k Tietz F416 CMOS detector by using Leginon software. For iNOS, 2,226 micrographs (1,113 micrograph pairs) were collected; for nNOS, 1,602 micrographs were collected; for CaM-bound nNOS, 1,684 micrographs were collected; for eNOS with EGTA, 1,918 micrographs were collected; and for CaM-bound eNOS, 2,306 micrographs were collected.
Image Processing and Reconstruction.
Experimental data were processed by using the Appion software package (30), which interfaces with the Leginon database infrastructure. The contrast transfer function for each micrograph was estimated by using CTFFIND3 (59). An iterative series of reference-free and reference based sorting and alignment methods were used to effectively deconvolute heterogeneity within the sample. Initial particle picking was done on a subset of untilted micrographs by using a reference-free difference of Gaussians approach (60). To generate templates, these particles were sorted, aligned, and clustered by using the reference-free Xmipp Clustering 2D Alignment (61), which generates initial references by separating the images with the largest correntropies to the class averages from the images with the smallest correntropies. A subset of the resulting 2D class averages were used as references to repick particles on untilted and tilted micrographs that, for iNOS, yielded a total of 16,870 particle pairs that were extracted at a box size of 352 pixels. The new untilted particles were aligned and classified in a reference-free manner by using Xmipp Clustering 2D Alignment and subjected to a sorting strategy to remove outlying or junk particles. The resulting 2D class averages were also used to create a new set of templates. These 21 templates were first manually aligned and then used for iterative rounds of multireference alignment by using SPIDER (62) and multivariate statistical analysis using IMAGIC (63). Resulting 2D classes contained between 149 and 373 particles. Random conical tilt reconstructions (22, 23) were carried out by using the initial model pipeline within Appion, which gave a total of 78 independent 3D maps. Forty-five of these maps gave clear densities that were consistent with their respective 2D averages. Theses maps had reported resolutions from 60 to 90 Å based on the 0.5 Fourier shell correlation criteria. The calculated resolutions are artificially low, as a large masking radius to accommodate the elongated shape of iNOS was used during reconstruction. This larger masking radius contributes random background noise during the Fourier shell correlation calculation. Consistent with this hypothesis, low-pass filtering our models to the calculated resolution resulted in 3D maps with significantly less detail than present in our experimentally determined 3D maps.
The data involving iNOS E662R, nNOS, and eNOS (with and without CaM) were processed in a similar manner as iNOS WT. New, separate templates and references were used for each experiment. For iNOS E660R 29,773 particle pairs led to 100 3D maps; for nNOS, 36,904 particle pairs led to 100 3D maps; for CaM-bound nNOS, 19,833 particle pairs led to 78 3D maps; for eNOS, 23,120 particle pairs led to 78 3D maps; and for CaM-bound eNOS, 18,540 particle pairs led to 78 3D maps.
Supplementary Material
Acknowledgments
We thank Dr. Katherine Barglow and Andrew Birnberg for assistance in protein preparation, Dr. Andrey Bobkov for assistance in collecting and analyzing the analytical ultracentrifugation data, Dr. Anchi Cheng and Dr. Sargis Dallakyan for technical support, and members of the Automated Molecular Imaging group for comments on the manuscript. This work was supported by The Scripps Research Institute, National Institutes of Health National Institute of General Medical Sciences Postdoctoral Fellowship 5F32GM095023 (to B.C.S), and American Heart Association Predoctoral Fellowship 14PRE18870036 (to M.G.C.). The work presented here was conducted at the National Resource for Automated Molecular Microscopy, which is supported by National Institute of General Medical Sciences Grant P41 GM103310. This is Manuscript 28014 from The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The structures reported in this paper have been deposited in the Electron Microscopy Data Bank database, http://www.emdatabank.org (accession nos. EMD-2717–EMD-2749).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413763111/-/DCSupplemental.
References
- 1.Condorelli RA, et al. Vascular regenerative therapies for the treatment of erectile dysfunction: current approaches. Andrology. 2013;1(4):533–540. doi: 10.1111/j.2047-2927.2013.00087.x. [DOI] [PubMed] [Google Scholar]
- 2.Serafim RAM, Primi MC, Trossini GHG, Ferreira EI. Nitric oxide: State of the art in drug design. Curr Med Chem. 2012;19(3):386–405. doi: 10.2174/092986712803414321. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava K, Bath PMW, Bayraktutan U. Current therapeutic strategies to mitigate the eNOS dysfunction in ischaemic stroke. Cell Mol Neurobiol. 2012;32(3):319–336. doi: 10.1007/s10571-011-9777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Förstermann U, Sessa WC. Nitric oxide synthases: Regulation and function. Eur Heart J. 2012;33(7):829–837. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: Structure, function and inhibition. Biochem J. 2001;357(pt 3):593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyanagi T, Xia C, Kim J-JP. NADPH-cytochrome P450 oxidoreductase: Prototypic member of the diflavin reductase family. Arch Biochem Biophys. 2012;528(1):72–89. doi: 10.1016/j.abb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith BC, Underbakke ES, Kulp DW, Schief WR, Marletta MA. Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc Natl Acad Sci USA. 2013;110(38):E3577–E3586. doi: 10.1073/pnas.1313331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcin ED, et al. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J Biol Chem. 2004;279(36):37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 9.Tejero J, Hannibal L, Mustovich A, Stuehr DJ. Surface charges and regulation of FMN to heme electron transfer in nitric-oxide synthase. J Biol Chem. 2010;285(35):27232–27240. doi: 10.1074/jbc.M110.138842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adak S, Ghosh S, Abu-Soud HM, Stuehr DJ. Role of reductase domain cluster 1 acidic residues in neuronal nitric-oxide synthase. Characterization of the FMN-FREE enzyme. J Biol Chem. 1999;274(32):22313–22320. doi: 10.1074/jbc.274.32.22313. [DOI] [PubMed] [Google Scholar]
- 11.Panda K, et al. Surface charge interactions of the FMN module govern catalysis by nitric-oxide synthase. J Biol Chem. 2006;281(48):36819–36827. doi: 10.1074/jbc.M606129200. [DOI] [PubMed] [Google Scholar]
- 12.Haque MM, et al. Neutralizing a surface charge on the FMN subdomain increases the activity of neuronal nitric-oxide synthase by enhancing the oxygen reactivity of the enzyme heme-nitric oxide complex. J Biol Chem. 2009;284(29):19237–19247. doi: 10.1074/jbc.M109.013144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokom AL, et al. Architecture of the nitric-oxide synthase holoenzyme reveals large conformational changes and a calmodulin-driven release of the FMN domain. J Biol Chem. 2014;289(24):16855–16865. doi: 10.1074/jbc.M114.564005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grädler U, et al. Novel nanomolar imidazo[4,5-b]pyridines as selective nitric oxide synthase (iNOS) inhibitors: SAR and structural insights. Bioorg Med Chem Lett. 2011;21(14):4228–4232. doi: 10.1016/j.bmcl.2011.05.073. [DOI] [PubMed] [Google Scholar]
- 15.Li H, et al. Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas. J Inorg Biochem. 2000;81(3):133–139. doi: 10.1016/s0162-0134(00)00099-4. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, et al. Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase. Comparisons with NADPH-cytochrome P450 oxidoreductase. J Biol Chem. 2001;276(40):37506–37513. doi: 10.1074/jbc.M105503200. [DOI] [PubMed] [Google Scholar]
- 17.Xia C, Misra I, Iyanagi T, Kim J-JP. Regulation of interdomain interactions by calmodulin in inducible nitric-oxide synthase. J Biol Chem. 2009;284(44):30708–30717. doi: 10.1074/jbc.M109.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Astashkin AV, Elmore BO, Fan W, Guillemette JG, Feng C. Pulsed EPR determination of the distance between heme iron and FMN centers in a human inducible nitric oxide synthase. J Am Chem Soc. 2010;132(34):12059–12067. doi: 10.1021/ja104461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daff S. NO synthase: Structures and mechanisms. Nitric Oxide. 2010;23(1):1–11. doi: 10.1016/j.niox.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Mowat CG, Gazur B, Campbell LP, Chapman SK. Flavin-containing heme enzymes. Arch Biochem Biophys. 2010;493(1):37–52. doi: 10.1016/j.abb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Liu X-D, Mazumdar T, Xu Y, Getzoff ED, Eissa NT. Identification of a flavin mononucleotide module residue critical for activity of inducible nitrite oxide synthase. J Immunol. 2009;183(9):5977–5982. doi: 10.4049/jimmunol.0902274. [DOI] [PubMed] [Google Scholar]
- 22.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146(pt 2):113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 23.Voss NR, et al. A toolbox for ab initio 3-D reconstructions in single-particle electron microscopy. J Struct Biol. 2010;169(3):389–398. doi: 10.1016/j.jsb.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 25.Crane BR, et al. Structures of the N(omega)-hydroxy-L-arginine complex of inducible nitric oxide synthase oxygenase dimer with active and inactive pterins. Biochemistry. 2000;39(16):4608–4621. doi: 10.1021/bi992409a. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran GN, Kolaskar AS, Ramakrishnan C, Sasisekharan V. The mean geometry of the peptide unit from crystal structure data. Biochim Biophys Acta. 1974;359(2):298–302. doi: 10.1016/0005-2795(74)90228-1. [DOI] [PubMed] [Google Scholar]
- 27.Persechini A, Tran Q-K, Black DJ, Gogol EP. Calmodulin-induced structural changes in endothelial nitric oxide synthase. FEBS Lett. 2013;587(3):297–301. doi: 10.1016/j.febslet.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen AL, Kasper CB. Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J Biol Chem. 1995;270(46):27475–27480. doi: 10.1074/jbc.270.46.27475. [DOI] [PubMed] [Google Scholar]
- 29.Tochio H, Zhang Q, Mandal P, Li M, Zhang M. Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide. Nat Struct Biol. 1999;6(5):417–421. doi: 10.1038/8216. [DOI] [PubMed] [Google Scholar]
- 30.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166(1):95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Campbell MG, Underbakke ES, Potter CS, Carragher B, Marletta MA. Single-particle EM reveals the higher-order domain architecture of soluble guanylate cyclase. Proc Natl Acad Sci USA. 2014;111(8):2960–2965. doi: 10.1073/pnas.1400711111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyumkis D, et al. Single-particle EM reveals extensive conformational variability of the Ltn1 E3 ligase. Proc Natl Acad Sci USA. 2013;110(5):1702–1707. doi: 10.1073/pnas.1210041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adak S, Aulak KS, Stuehr DJ. Chimeras of nitric-oxide synthase types I and III establish fundamental correlates between heme reduction, heme-NO complex formation, and catalytic activity. J Biol Chem. 2001;276(26):23246–23252. doi: 10.1074/jbc.M102509200. [DOI] [PubMed] [Google Scholar]
- 35.Ilagan RP, et al. Differences in a conformational equilibrium distinguish catalysis by the endothelial and neuronal nitric-oxide synthase flavoproteins. J Biol Chem. 2008;283(28):19603–19615. doi: 10.1074/jbc.M802914200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman E, et al. Differential activation of nitric-oxide synthase isozymes by calmodulin-troponin C chimeras. J Biol Chem. 2004;279(32):33547–33557. doi: 10.1074/jbc.M403892200. [DOI] [PubMed] [Google Scholar]
- 37.Nishida CR, Ortiz de Montellano PR. Electron transfer and catalytic activity of nitric oxide synthases. Chimeric constructs of the neuronal, inducible, and endothelial isoforms. J Biol Chem. 1998;273(10):5566–5571. doi: 10.1074/jbc.273.10.5566. [DOI] [PubMed] [Google Scholar]
- 38.Richards MK, Clague MJ, Marletta MA. Characterization of C415 mutants of neuronal nitric oxide synthase. Biochemistry. 1996;35(24):7772–7780. doi: 10.1021/bi952582g. [DOI] [PubMed] [Google Scholar]
- 39.Wolthers KR, Schimerlik MI. Reaction of neuronal nitric-oxide synthase with 2,6-dichloroindolphenol and cytochrome c3+: Influence of the electron acceptor and binding of Ca2+-activated calmodulin on the kinetic mechanism. Biochemistry. 2001;40(15):4722–4737. doi: 10.1021/bi0023495. [DOI] [PubMed] [Google Scholar]
- 40.Gachhui R, et al. Characterization of the reductase domain of rat neuronal nitric oxide synthase generated in the methylotrophic yeast Pichia pastoris. Calmodulin response is complete within the reductase domain itself. J Biol Chem. 1996;271(34):20594–20602. doi: 10.1074/jbc.271.34.20594. [DOI] [PubMed] [Google Scholar]
- 41.Montgomery HJ, Romanov V, Guillemette JG. Removal of a putative inhibitory element reduces the calcium-dependent calmodulin activation of neuronal nitric-oxide synthase. J Biol Chem. 2000;275(7):5052–5058. doi: 10.1074/jbc.275.7.5052. [DOI] [PubMed] [Google Scholar]
- 42.Newton DC, Montgomery HJ, Guillemette JG. The reductase domain of the human inducible nitric oxide synthase is fully active in the absence of bound calmodulin. Arch Biochem Biophys. 1998;359(2):249–257. doi: 10.1006/abbi.1998.0917. [DOI] [PubMed] [Google Scholar]
- 43.Anagli J, Hofmann F, Quadroni M, Vorherr T, Carafoli E. The calmodulin-binding domain of the inducible (macrophage) nitric oxide synthase. Eur J Biochem. 1995;233(3):701–708. doi: 10.1111/j.1432-1033.1995.701_3.x. [DOI] [PubMed] [Google Scholar]
- 44.Wu G, Berka V, Tsai A-LA. Binding kinetics of calmodulin with target peptides of three nitric oxide synthase isozymes. J Inorg Biochem. 2011;105(9):1226–1237. doi: 10.1016/j.jinorgbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan T, Vogel HJ, Sutherland C, Walsh MP. Characterization of the Ca2+ -dependent and -independent interactions between calmodulin and its binding domain of inducible nitric oxide synthase. FEBS Lett. 1998;431(2):210–214. doi: 10.1016/s0014-5793(98)00750-9. [DOI] [PubMed] [Google Scholar]
- 46.Golser R, Gorren ACFA, Mayer B, Schmidt K. Functional characterization of Glu298Asp mutant human endothelial nitric oxide synthase purified from a yeast expression system. Nitric Oxide. 2003;8(1):7–14. doi: 10.1016/s1089-8603(02)00131-3. [DOI] [PubMed] [Google Scholar]
- 47.Venema RC, Sayegh HS, Kent JD, Harrison DG. Identification, characterization, and comparison of the calmodulin-binding domains of the endothelial and inducible nitric oxide synthases. J Biol Chem. 1996;271(11):6435–6440. doi: 10.1074/jbc.271.11.6435. [DOI] [PubMed] [Google Scholar]
- 48.Vorherr T, et al. The calmodulin binding domain of nitric oxide synthase and adenylyl cyclase. Biochemistry. 1993;32(23):6081–6088. doi: 10.1021/bi00074a020. [DOI] [PubMed] [Google Scholar]
- 49.Weissman BA, Jones CL, Liu Q, Gross SS. Activation and inactivation of neuronal nitric oxide synthase: Characterization of Ca(2+)-dependent [125I]Calmodulin binding. Eur J Pharmacol. 2002;435(1):9–18. doi: 10.1016/s0014-2999(01)01560-6. [DOI] [PubMed] [Google Scholar]
- 50.Stevens-Truss R, Marletta MA. Interaction of calmodulin with the inducible murine macrophage nitric oxide synthase. Biochemistry. 1995;34(48):15638–15645. doi: 10.1021/bi00048a006. [DOI] [PubMed] [Google Scholar]
- 51.Rafferty S, Malech HL. High reductase activity of recombinant NOS2 flavoprotein domain lacking the calmodulin binding regulatory sequence. Biochem Biophys Res Commun. 1996;220(3):1002–1007. doi: 10.1006/bbrc.1996.0522. [DOI] [PubMed] [Google Scholar]
- 52.Mitchell DA, Erwin PA, Michel T, Marletta MA. S-nitrosation and regulation of inducible nitric oxide synthase. Biochemistry. 2005;44(12):4636–4647. doi: 10.1021/bi0474463. [DOI] [PubMed] [Google Scholar]
- 53.Rusche KM, Spiering MM, Marletta MA. Reactions catalyzed by tetrahydrobiopterin-free nitric oxide synthase. Biochemistry. 1998;37(44):15503–15512. doi: 10.1021/bi9813936. [DOI] [PubMed] [Google Scholar]
- 54.Rodríguez-Crespo I, Ortiz de Montellano PR. Human endothelial nitric oxide synthase: Expression in Escherichia coli, coexpression with calmodulin, and characterization. Arch Biochem Biophys. 1996;336(1):151–156. doi: 10.1006/abbi.1996.0543. [DOI] [PubMed] [Google Scholar]
- 55.Gerber NC, Ortiz de Montellano PR. Neuronal nitric oxide synthase. Expression in Escherichia coli, irreversible inhibition by phenyldiazene, and active site topology. J Biol Chem. 1995;270(30):17791–17796. doi: 10.1074/jbc.270.30.17791. [DOI] [PubMed] [Google Scholar]
- 56.Roman LJL, et al. High-level expression of functional rat neuronal nitric oxide synthase in Escherichia coli. Proc Natl Acad Sci USA. 1995;92(18):8428–8432. doi: 10.1073/pnas.92.18.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradford MM. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 58.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 60.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166(2):205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sorzano COS, et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol. 2010;171(2):197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 63.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116(1):17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.