Significance
Since the discovery of Orsay, the first virus that naturally infects nematodes, it has been widely expected that Caenorhabditis elegans-Orsay would serve as a highly tractable model for studying viral pathogenesis. Here we report the crystal structure of the Orsay virus. The Orsay capsid contains 180 copies of the capsid protein, each consisting of a jelly-roll β-barrel and a protrusion domain. Although sequence analyses indicate that Orsay is related to nodaviruses, the structure reveals substantial differences compared with the insect-infecting alphanodaviruses. Small plant RNA viruses are the closest homologs for Orsay when their β-barrel domains are compared. Our results have not only shed light on the evolutionary lineage of Orsay but have also provided a framework for further studies of Orsay–host interaction.
Keywords: virology, crystallography, microscopy
Abstract
Orsay, the first virus discovered to naturally infect Caenorhabditis elegans or any nematode, has a bipartite, positive-sense RNA genome. Sequence analyses show that Orsay is related to nodaviruses, but molecular characterizations of Orsay reveal several unique features, such as the expression of a capsid–δ fusion protein and the use of an ATG-independent mechanism for translation initiation. Here we report the crystal structure of an Orsay virus-like particle assembled from recombinant capsid protein (CP). Orsay capsid has a T = 3 icosahedral symmetry with 60 trimeric surface spikes. Each CP can be divided into three regions: an N-terminal arm that forms an extended protein interaction network at the capsid interior, an S domain with a jelly-roll, β-barrel fold forming the continuous capsid, and a P domain that forms surface spike projections. The structure of the Orsay S domain is best aligned to T = 3 plant RNA viruses but exhibits substantial differences compared with the insect-infecting alphanodaviruses, which also lack the P domain in their CPs. The Orsay P domain is remotely related to the P1 domain in calicivirus and hepatitis E virus, suggesting a possible evolutionary relationship. Removing the N-terminal arm produced a slightly expanded capsid with fewer nucleic acids packaged, suggesting that the arm is important for capsid stability and genome packaging. Because C. elegans-Orsay serves as a highly tractable model for studying viral pathogenesis, our results should provide a valuable structural framework for further studies of Orsay replication and infection.
In the past four decades, the nematode Caenorhabditis elegans has become an important model organism for studying various biological processes such as development, metabolism, aging, the cell cycle, and gene regulation (1). Many key biomedical discoveries about common diseases, including Alzheimer’s disease, diabetes type 2, and even depression, were first made in C. elegans (2). It has been estimated that around 70% of human genes have homologs in C. elegans, including the majority of human disease genes and pathways (3).
It was not until very recently that Orsay, the first virus that naturally infects C. elegans in the wild, was discovered (4). In addition, two other nematode viruses, Santeuil and Le Blanc, both of which infect C. briggsae, have also been identified (4, 5). Infections by these newly identified viruses cause abnormal intestinal morphologies without obvious effect on longevity or brood size. Orsay viral particles have been observed intracellularly in both intestinal cells and the somatic gonad (4). Sequence analysis indicated that all three nematode viruses are related to viruses in the family Nodaviridae (4, 5). Nodaviruses are nonenveloped RNA viruses that can be further classified as alphanodaviruses and betanodaviruses, which are known to infect insects and fish, respectively (6).
Orsay virus has a bipartite, positive-sense RNA genome ∼6.3 kb in size (4). The RNA1 segment of the Orsay virus encodes a predicted RNA-dependent RNA polymerase (RdRP) of 982 amino acids. RNA2 has two ORFs: one coding for a putative capsid protein (CP) and the other for a nonstructural protein δ of unknown function. The protein translation of the Orsay CP uses an AUG-independent, noncanonical initiation mechanism that is likely shared among all three nematode viruses (7). In addition to the viral CP, a CP–δ fusion protein, which is likely generated by ribosomal frameshifting, has been detected in infected cells as well as purified viruses (7). The Orsay CP and RdRP share up to 26–30% amino acid identity with those from betanodaviruses in the Nodaviridae, but the δ protein does not have any sequence homologs in GenBank.
The combination of a simple multicellular host organism and a small naturally infecting virus makes C. elegans-Orsay a highly tractable model for studying host–virus interactions (8). To characterize the molecular properties of the newly discovered Orsay virus, we report the crystal structure of an Orsay virus-like particle (VLP), which shows a T = 3 icosahedral symmetry with a total of 60 trimeric spikes. Each CP molecule can be divided into three linear regions, namely the N-terminal arm, the S domain forming the continuous capsid shell, and the P domain, which forms trimeric surface protrusions. The jelly-roll β-barrel in the S domain closely resembles those from small plant RNA viruses. The P domain consists of a distorted β-barrel that is only remotely related to the P1 domain from a norovirus CP (9). The N-terminal arm of the Orsay CP forms an extensive network inside the capsid through a β-annulus structure similar to tomato bushy stunt virus (10). An N-terminally truncated mutant, CP42–391, produces a slightly expanded capsid with less RNA packaged inside, suggesting important roles for the N-terminal arm in capsid stabilization and genome packaging. Our results also demonstrate that the Orsay capsid is structurally distinct from alphanodaviruses. Together with primary sequence-based phylogeny, these results suggest that the nematode-infecting viruses and betanodaviruses have a common evolutionary history.
Results
Orsay VLP Expression and Purification.
The Orsay CP was overexpressed in Escherichia coli to produce recombinant VLPs. Because the translation initiation site of the Orsay CP had not been mapped at the time these studies were initiated, we performed a multisequence alignment with existing betanodavirus CPs to determine a likely coding region, assuming that CP ORFs should be well-conserved. Our CP construct had a total of 392 amino acid residues, with its N terminus ANKNN- starting from 138 residues upstream of the first Met in the originally annotated CP ORF (4). Recombinant CP was purified by chromatography and separated by gel filtration into two size populations (Fig. S1). The first, which was found in the void volume, contained VLPs with an ∼35-nm diameter according to negative-staining electron microscopy (EM). The overall size and morphology of these VLPs were indistinguishable from those of authentic virions purified from infected worms (7). The second peak eluted much later and had an estimated molecular mass of ∼90 kDa, likely corresponding to CP dimers. Orsay VLPs were further separated into two distinct bands when subjected to CsCl gradient ultracentrifugation. Particles from these two bands were similar in size but different in their nucleic acid contents, as samples from the top and bottom bands had an A260nm:280nm ratio of 1.23 and 1.75, respectively.
The Overall Structure of Orsay VLP.
Orsay VLP from the lower band of the gradient was crystallized in the space group I222 with two particles per unit cell. The structure was determined to 3.25-Å resolution by molecular replacement using a 9-Å resolution cryo-EM reconstruction as a phasing model (Fig. 1 and Table S1). Solvent flattening and 15-fold noncrystallographic symmetry averaging were applied during phase extension (11, 12). The final structural model contains three polypeptide chains making up one icosahedral asymmetric unit, which includes residues 30–384 in subunit A, residues 45–389 in subunit B, and residues 44–389 in subunit C.
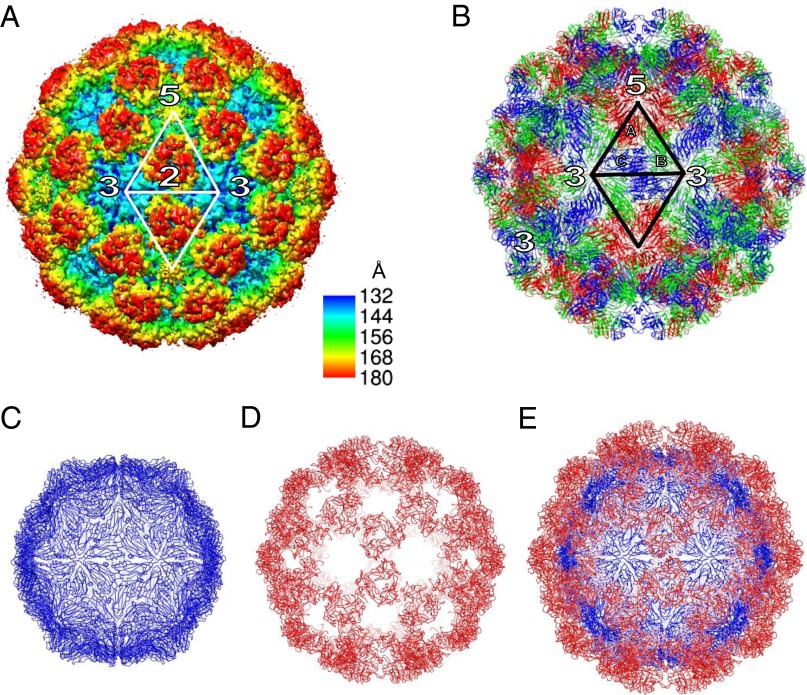
Fig. 1.
Structure of the Orsay virus-like particle. (A) Cryo-EM reconstruction at 6.9-Å resolution. The surface is colored by radial depth cue from red to yellow to cyan. Only a 9-Å map was used for molecular replacement structure determination though. (B) Crystal structure of the Orsay VLP. The three subunits A, B, and C are colored red, green, and blue, respectively. One asymmetric unit is highlighted along with icosahedral symmetry axes. (C) Orsay VLP with only the S domain. (D) Orsay VLP with only the P domain. (E) Orsay VLP with the S and P domains colored in blue and red, respectively.
The crystal structure of the Orsay VLP shows that the particle has an outer diameter of ∼360 Å and an inner diameter of ∼230 Å. Each particle contains 180 copies of the CP that are arranged in a T = 3 icosahedral symmetry (Fig. 1). A total of 60 trimeric spikes are observed on quasi-threefold axes, with each one in direct contact with three surrounding spikes. The capsid shell is ∼30 Å thick, and the spikes are ∼35 Å tall. Small plateaus are observed on fivefold axes, whereas large depressions are formed at the center of CP hexamers on threefold axes.
Structure of the Orsay CP.
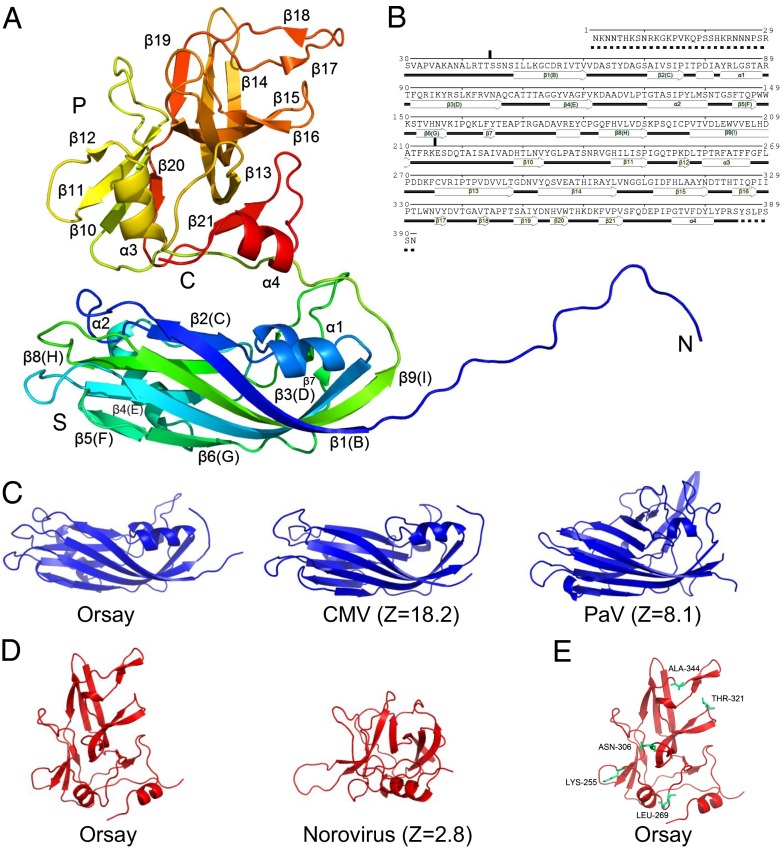
The structure of each Orsay CP can be divided into three modular regions: an N-terminal arm (residues 1–43), a shell (S) domain (residues 44–214), and a protrusion (P) domain (residues 215 to the end) (Fig. 2 A and B). The S domain exhibits a canonical jelly-roll, β-barrel fold that is frequently seen in nonenveloped icosahedral viruses (13). In the jelly-roll β-barrel, eight antiparallel β-strands, which are often named sequentially from B to I, form two twisted β-sheets with four β-strands arranged in the order of BIDG and CHEF, respectively. The S domain by itself forms a continuous capsid shell without any notable holes or cavities (Fig. 1C).
Fig. 2.
Structure of the Orsay capsid protein. (A) One CP molecule (subunit A). The polypeptide is rainbow-colored, with the N terminus in blue and the C terminus in red. (B) Secondary structure assignment. Helices are indicated by tubes, β-strands by arrows, loops by lines, and disordered regions by dashed lines. (C) The CP S domain of Orsay, CMV, and PaV. (D) The CP P domain of Orsay and the P1 domain of norovirus. (E) The Orsay P domain showing five residues (in cyan) where Orsay differs from Le Blanc and Santeuil.
The P domain, which forms the trimeric protrusions on the capsid surface, is also rich in β-strands. The P domain can be further divided into an apical and a cradle subdomain that occupies the upper and bottom half of the capsid protrusion, respectively. The apical subdomain, which is made from a single polypeptide excursion from the cradle subdomain, is folded into a distorted β-barrel made of seven antiparallel β-strands (residues 275–253, including β13, β14, β15, β16, β17, β18, and β19) (Fig. 2A). In the cradle subdomain, two noncontiguous polypeptide fragments (residues 228–274 and 354–384) intertwine with each other, forming a cradle-shaped platform that holds the apical subdomain on top. Whereas the cradle subdomain mainly interacts with the underlying capsid shell, the apical subdomain mediates trimeric spike formation by interacting with symmetry-related molecules around quasi-threefold symmetry axes (Fig. 1).
Structural Comparison with Other Viruses.
When Dali (14) was used to search for structural homologs, the S domain of the Orsay CP was best aligned to the coat proteins of T = 3 plant RNA viruses, which include, in descending order, carnation mottle virus (CMV; Z score = 18.2, with greater than 2.0 being significant) (15), melon necrotic spot virus (MNSV; Z = 17.9) (16), and tobacco necrosis virus (Z = 17.2) (17) from Tombusviridae, and ryegrass mottle virus (Z = 17.2) (18), rice yellow mottle virus (RyMV; Z = 16.8) (19), and sesbania mosaic virus (SeMV; Z = 16.7) (20) from Sobemoviridae (Fig. 2C). The top-ranking animal viruses were rabbit hemorrhagic disease virus (Z = 15.2) (21) in Caliciviridae, Triatoma virus (Z = 14.5) (22) and equine rhinitis A virus (Z = 14.2) (23) in Picornaviridae, and hepatitis E virus (HEV; Z = 13.7) (24, 25) in Hepeviridae. Surprisingly, the four available alphanodavirus structures, including Pariacoto virus (PaV; Z = 8.1) (26), Flock House virus (FHV; Z = 7.4) (27), nodamura virus (Z = 7.3) (28), and the black beetle virus (Z = 7.2) (29), were ranked much lower on the list (Fig. 2C).
The Orsay virus P domain exhibits significant variations in overall structure compared with its structural homologs (Fig. 2D). According to Dali, its closest structural homolog is the P1 domain (9) of a Norovirus CP in Caliciviridae, with a rather modest Z score of 2.8 (30).
Pairwise sequence alignment demonstrated that the viral CP is more highly conserved among the three nematode viruses (i.e., Orsay, which infects C. elegans, and Santeuil and Le Blanc, which infect C. briggsae) than the other two viral proteins (i.e., the δ protein and the RdRP), with amino acid sequence identities ranging from 52 to 68% (Table S2). It is important to note that C. elegans lacks adaptive immunity and relies solely on innate immunity to fight off microbial attacks (1). Consequently, the CPs of these nematode viruses do not experience the same selective pressure as those from vertebrate viruses, possibly explaining the high level of sequence conservation. Consistent with our speculation, the same trend is also seen in the insect-infecting alphanodaviruses but not in the fish-infecting betanodaviruses. Among the three Orsay CP polypeptide regions the S domain is the most conserved, whereas the P domain and the N-terminal arm show greater sequence variation (Table S1).
The three nematode viruses have high host specificity: Orsay only infects C. elegans, and Le Blanc and Santeuil only infect C. briggsae. Because cell receptor recognition is important for host restriction, it is possible that the Orsay virus uses a receptor that is not found in C. briggsae, thus limiting its infection to C. elegans. In an effort to map host specificity determinants, we performed a multisequence alignment for all three nematode viral CPs (Fig. S2). The results show a total of 13 common residues between Santeuil and Le Blanc but not in Orsay. Among these 13 residues, 5 are exposed on the outer surface of the Orsay capsid spike: K255, L269, N306, T321, and A344 (Fig. 2E). In Santeuil and Le Blanc, the equivalent sequence locations are assumed by Arg, Ala, Asp, Val, and Thr, respectively (Fig. S2). Whether these residues are indeed involved in host specificity may be addressed once a reverse genetics system becomes available.
Capsomere Interactions in the Orsay VLP.
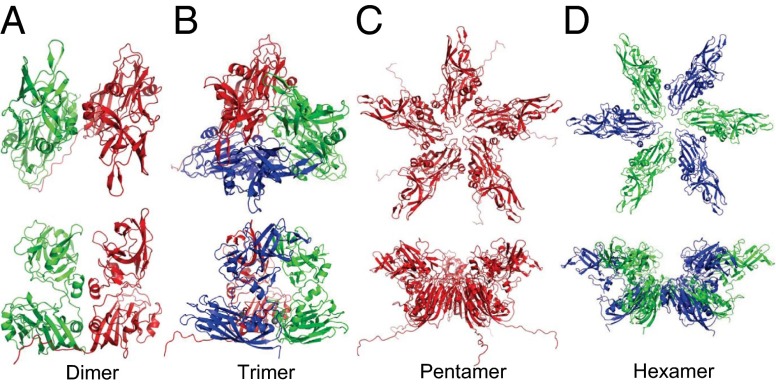
The structure of the Orsay VLP shows that the CP trimer buries the largest interface between adjacent subunits. The three CPs from each asymmetric unit form a triangle-shaped trimer with a notable spike (Fig. 3). Approximately 3,000 Å2 of surface area is buried between adjacent subunits, with the S domain and P domain contributing ∼1,800 and ∼1,300 Å2 of buried surface area, respectively (Table S3). A calcium ion is also found at each of the three CP interfaces, coordinated by amino acid side chains of Q92 and D122 from one molecule together with D125 and E166 from another molecule (Fig. S3). Furthermore, there is an intrasubunit disulfide bond (Cys107–Cys195) found in each CP near the vertices of the triangular-shaped trimer.
Fig. 3.
Orsay capsomeres. (A) A–B dimer. (B) Trimer. (C) Pentamer. (D) Hexamer. (Upper) Views along the symmetry axes from the outside of the VLP. (Lower) Views from the side. The three subunits A, B, and C are shown in red, green, and blue, respectively.
Similar calcium binding sites have been previously observed in several tombusviruses (15–17, 31). Tomato bushy stunt virus (TBSV) was shown to swell by ∼10% in the presence of a metal ion chelator (32). Cowpea chlorotic mottle virus, another small RNA plant virus in Bromoviridae, showed similar expansion, indicating a loosened capsid in the absence of calcium (33). For FHV, which is an alphanodavirus in the Nodaviridae family, removing calcium did not affect the overall architecture of the capsid, but mutations at the calcium binding sites reduced FHV stability and virus infectivity, and it was suggested that calcium removal may be involved in the endosomal entry of the virus (34). To test the effect of calcium on Orsay, the VLPs were treated with 10 mM EDTA overnight before ultracentrifugation. EDTA-treated particles migrated to the same position as untreated VLPs in the CsCl gradient, indicating that the Orsay VLP was able to maintain its integrity in the absence of calcium. The effect of calcium on authentic Orsay virus infectivity, however, remains to be tested.
Orsay CP dimers are found on both icosahedral twofold and quasi-twofold symmetry axes (i.e., C–C dimer vs. A–B dimer) (Fig. 3 and Fig. S4). The two subunits in a dimer bury a total interface of ∼3,100 Å2 for the A–B dimer or ∼1,800 Å2 for the C–C dimer (Table S3). The differences between these two types of dimers are mainly due to two reasons. First, the A subunit has an extended N terminus that is structurally ordered, providing extra contact with the adjacent B subunit. Second, the different bending angle between the two subunits in an A–B dimer results in a more extensive interface between their S domains.
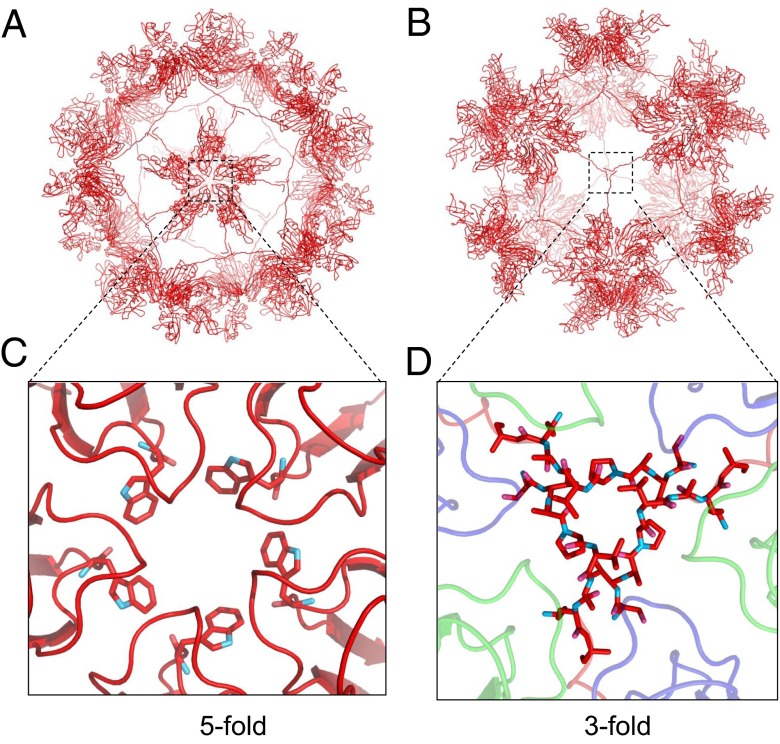
The Orsay capsid shell around fivefold icosahedral axes is slightly elevated. The total buried surface area between adjacent subunits is ∼1,600 Å2 in CP pentamers, compared with ∼1,100–1,400 Å2 in CP hexamers (Table S3). Two structured loops F/G (147PWWKS151) and D/E (109TTT111) from the S domain line up the fivefold symmetry axes (Fig. 4). In particular, a cluster of five tryptophans (W149) is observed around the fivefold axes, in a similar configuration as previously reported in the plant-infecting sobemoviruses, in which a single point mutation of this tryptophan disrupted capsid assembly and produced only dimers in solution (35). The F/G loop, which faces the capsid interior, has also been implicated in genome packaging in sobemoviruses (35).
Fig. 4.
Orsay CP subunit A and its extended N-terminal arm. (A) VLP containing only subunit A viewed along the fivefold axis. (B) VLP containing only subunit A viewed along the threefold axis. (C) Fivefold interface. The five tryptophan residues (W149) are shown as sticks. (D) Threefold interface. The β-annulus is shown as sticks. Subunits B (green) and C (blue) are added for completeness. The areas shown in C and D correspond to the dotted boxes highlighted in A and B, respectively.
Orsay CP N-Terminal Arm.
One of the most notable features of the Orsay VLP is the extended structural network mediated by the CP N-terminal peptide (residues 1–43) (Fig. 4). Inside the capsid, the N-terminal arm of the A subunit, which is structurally ordered starting from residue 30, radiates outward from the fivefold axis, running at the B–C subunit interface, and, upon reaching the threefold symmetry axes, forms a tightly knitted annulus structure that is stabilized by hydrogen bonds between three prolines and three valines (Fig. 4). The N-terminal arm becomes disordered after the threefold annulus and terminates inside the viral particle.
Extended but structurally ordered CP N termini have been observed in several other small RNA viruses, and based on structural conformation they can be divided into three categories. (i) In TBSV, MNSV, and SeMV, the N-terminal arm from the C subunit folds back under itself and interacts with the other two CP N termini at the nearby threefold axis (Fig. S5B). The N terminus of SeMV has been found to facilitate particle assembly and regulate capsid size by inserting into the CP dimer interface, thus creating flat C–C CP dimers around the icosahedral twofold symmetry axes (36). (ii) In RyMV, the N terminus of the C subunit passes under the B subunit toward the neighboring threefold axis (Fig. S5C). The resulting β-annulus structure was shown to enhance capsid stability (19). (iii) In PaV, which is an alphanodavirus that infects southern armyworms, the N terminus of subunit A runs at the B–C interface, forming a hydrophobic cluster containing Ala, Leu, and Met around the threefold axis before it extends internally to interact with the viral genome (Fig. S5D) (26). It was believed that this N-terminal arm, together with RNA duplex, could function as a molecular switch to ensure proper capsid assembly (37).
Structure of an N-Terminally Truncated CP Mutant.
In an attempt to characterize the functional role of the Orsay CP N-terminal arm, we overexpressed a CP mutant, CP42–391, in which the first 41 amino acid residues had been removed. EM images showed that the recombinant CP42–391 is capable of self-assembly into T = 3 capsids, like the full-length CP. The mutant VLPs also separated into two bands in CsCl gradient ultracentrifugation. The lower band had an A260nm:280nm ratio of 1.42, which is lower than that of the full-length capsid, indicating a reduced number of nucleic acids.
The structure of the mutant VLP was determined to 3.75-Å resolution by molecular replacement (Fig. 5 and Table S1). The resulting map confirmed that the N-terminal annulus was missing, with the first structurally ordered residue being 44 for subunit A, and 45 for subunits B and C. Except for the absence of the N-terminal arm, the subunit structure of CP42–391 was nearly identical to that of the full-length CP, with an rmsd in distance of 0.2 Å for 341 common Cα atoms. However, superposition of the two VLPs showed that in the mutant, each CP is shifted outward by ∼0.8 Å, implying a diameter expansion of 1.6 Å or ∼0.5%, consistent with the overall increase in crystal unit-cell dimensions of the mutant (Table S1).
Fig. 5.
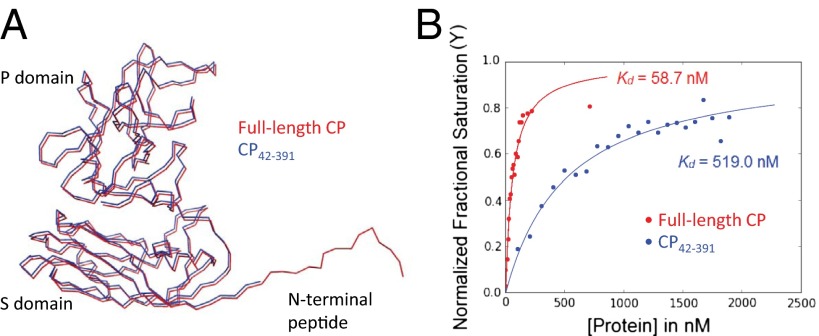
Orsay N-terminal truncated CP mutant. (A) Superimposition of the full-length (red) and N-terminal truncated CP (blue). (B) RNA binding by both full-length (red) and mutant CP (blue). A 28-nt RNA was used for fluorescence anisotropy measurements.
At the capsid level, the removal of the N-terminal arm had several major consequences: (i) the loss of the pentamer tethering network at the interior of the capsid; (ii) significant reduction of the buried surface areas at the A–C and A–B interfaces; and (iii) a slight decrease in buried surfaces for all types of CP interactions by ∼100 Å2, indicating a relaxed capsid at the global level (Table S3). Therefore, although the Orsay N-terminal arm was not essential for capsid assembly under in vitro conditions, the internal polypeptide network mediated by the N-terminal arm is likely to affect capsid stability and the accuracy of assembly, and may also be able to promote capsid assembly at lower protein concentrations.
Orsay CP Interaction with Nucleic Acids.
The N-terminal region of the Orsay CP polypeptide (residues 1–29) contains a large number of positively charged residues, including four arginines and seven lysines. This region is internally disordered and could potentially interact with genomic RNA. Indeed, the number of nucleic acids packaged in the CP42–391 mutant capsid was significantly reduced compared with the full-length capsid (see section above). In addition to the N-terminal arm, residues from other regions of the Orsay capsid may also participate in genome packaging, as the CP42–391 mutant still retains some of its RNA packaging activity. A number of basic residues (i.e., K36, R41, K51, R83, K95, R97, K100, R102, K121, K150, H154, and H208) and aromatic residues (i.e., F144 and W149) are exposed at the capsid interior, and may potentially interact with nucleic acids through electrostatic as well as base-stacking interactions. We did not observe any ordered density for nucleic acids in the final X-ray electron density maps of the Orsay VLPs, presumably due to a combination of causes, including the inherent structural disorder of the N terminus of the Orsay CP and associated RNA, the lack of specificity in Orsay CP–RNA interaction, and the heterogeneous nature of the RNA packaged in the Orsay VLP. Cryo-EM reconstruction of the VLP does show diffused density at the particle interior that likely comes from the packaged nucleic acids. The nucleic acid density is not connected to the capsid shell, however.
To quantitate the effect of the N-terminal arm on genome packaging, the RNA binding affinity of both full-length and CP42–391 was measured by fluorescence polarization assays using CP dimers (Fig. 5B). The full-length CP dimer showed significantly tighter binding to a 28-nt RNA oligomer (Kd = 58.7 nM) than the CP42–391 dimer (Kd = 519.0 nM). Thus, this result confirms that the N-terminal region of the Orsay CP plays a major role in genome packaging.
When nucleic acids were extracted from full-length and mutant VLPs forming the higher-density band (see previous sections), they both gave rise to a large smeared band spanning from ∼100 to ∼400 bp on an agarose gel (Fig. S6). The nucleic acids were sensitive to RNase V1 and T1 but resistant to DNase I, suggesting that they were heterogeneous RNA molecules containing both single-stranded and double-stranded regions. The extracted RNAs were much smaller in size than those extracted from other RNA viruses (38), possibly due to different packaging specificity associated with the Orsay CP. On the other hand, VLPs from the lower-density band did not produce any obvious band on the agarose gel, maybe because they were empty or contained very few nucleic acids.
Assembly of the Orsay Capsid.
A substantial fraction of overexpressed Orsay CP was purified as a soluble oligomer, suggesting that this oligomer could be the first intermediate in capsid assembly. Our results from analytical ultracentrifugation showed that the molecular mass of the oligomer is 84.3 kDa, consistent with that of a CP dimer, which has a calculated molecular mass of 86.0 kDa (Fig. S1B). In a subsequent cross-linking experiment, we found that the most predominant products were CP dimers and tetramers (Fig. S1C), consistent with the notion that the Orsay CP dimer is the only stable capsomere in solution.
Therefore, despite having trimeric surface spikes, the assembly of the Orsay capsid likely propagates through the orderly addition of CP dimers to nucleating RNA molecules, similar to the assembly process previously proposed for plant RNA viruses (13). Several factors may be able to stimulate the further assembly of CP dimers into capsids, such as the addition of metal ions, the presence of single-stranded RNA, and perhaps the availability of certain cellular factors that have not yet been identified. Once such triggering factors are determined, it may be possible to produce artificial virions in vitro with foreign RNAs packaged inside. Such artificial virions could potentially be used to develop virus-based delivery systems or single–replication-cycle viruses for gene functional analysis.
Other Biological Implications.
The crystal structure of the Orsay VLP reported here represents a major step forward in our understanding of this newly discovered nematode-infecting virus. Among all T = 3 virus structures that are available, Orsay is unique in that it is the only one with trimeric surface spikes made of a single protrusion domain. The crystal structure of the Orsay VLP confirms that the protein translation of the Orsay CP uses an AUG-independent, noncanonical initiation mechanism. Met138, the first and only methionine found in the previously annotated CP ORF (4), is mapped to the middle of the jelly-roll β-barrel domain. Recently, mass spectrometry analyses of authentic purified Orsay virions identified the second residue in our CP construct as the N terminus (7). Because there is no AUG either at this site or upstream, this implies that a noncanonical translation initiation mechanism must be used to express the CP (39). The amino acid numbering of our structural model has been adjusted to match the experimentally mapped coding sequence (7).
Previous phylogenetic analyses indicate that the Orsay virus is related to alpha- and betanodaviruses of the Nodaviridae family (4). Betanodaviruses are an important emerging group of viruses known to infect over 40 marine fish species worldwide, causing viral nervous necrosis or viral encephalopathy and retinopathy (40). At present, the only betanodavirus structure available is a 23-Å resolution cryo-EM reconstruction of the Malabaricus grouper nervous necrosis virus (MGNNV) (41). Although the resolution is rather modest, the reconstruction shows trimeric spikes and capsid morphology that resemble those of the Orsay VLP. Considering an overall amino acid sequence identity of ∼23% between the two viral CPs, we expect MGNNV as well as other betanodaviruses to have capsid structures similar to Orsay.
Meanwhile, the crystal structure of the Orsay VLP reveals several major differences compared with the four known alphanodavirus structures (26–29). First, the CP of the Orsay virus has two linear domains that are connected by a flexible linker, with the S domain forming a continuous capsid shell. In contrast, alphanodaviruses only have a single jelly-roll β-barrel domain in their CPs. Second, the P domain in Orsay forms prominent trimeric protrusions on quasi-threefold symmetry axes. However, the small protrusions on alphanodavirus capsids are made of a β-hairpin insertion from the jelly-roll β-barrel domain. Last, in insect-infecting alphanodaviruses, a C-terminal lipophilic peptide located in the particle interior mediates membrane penetration during host cell entry. However, the C terminus of the Orsay CP is located at the outside of the capsid.
Having an external C terminus implies that the Orsay CP–δ fusion protein, if incorporated into infectious virions, would place the δ protein at the outside of the capsid. Unlike the B2 proteins encoded by alpha- and betanodaviruses, Orsay δ protein does not appear to have any RNAi suppression activity (42). The exact function of δ is not known, but its location at the capsid exterior suggests potential roles in capsid stabilization and/or interaction with the host.
The substantial differences in Orsay and alphanodavirus capsid structures indicate a complex phylogenetic relationship between alpha- and betanodaviruses. For RNA1, which encodes the RdRP, these viruses clearly share common descent according to their RdRP sequence homology. It is possible that an ancestral gene reassortment event introduced a new RNA2 segment, thus generating a new lineage from which either the alpha- or betanodaviruses descended. Indeed, comparative analysis of both gene segments provides evidence for frequent gene reassortment between different betanodaviruses (43). Alternatively, a polymerase-mediated template switching or gene recombination event may also allow alpha- or betanodaviruses to acquire a new CP while keeping the rest of the RNA2 sequence unchanged.
Based on structural homology with other T = 3 RNA viruses, we speculate that the Orsay virus CP may have evolved from an ancestor that closely resembles the present-day plant T = 3 RNA virus capsids. The protrusion P domain of the Orsay CP is remotely related to the P1 domain of the HEV and the caliciviruses, both of which have a second P2 protrusion domain that is missing in Orsay. Hence, HEV and caliciviruses may have acquired their CPs from an Orsay-like ancestor species during evolution.
In summary, the atomic structure of the Orsay capsid provides a structural framework for further studies of cell receptor interaction, host cell entry, RNA encapsidation, and the novel function of the δ protein as a potential capsid–δ fusion protein. Furthermore, comparing the capsid structure of Orsay with other small RNA viruses gives valuable insight into the evolutionary relationship of these viral CPs. This information should be useful for making the appropriate taxonomic assignment for Orsay, as it is currently an unclassified agent. We expect that studies of C. elegans-Orsay will play an important role in advancing our understanding of antiviral responses in nematodes and virus–host interactions in general.
Materials and Methods
Details of experimental procedures are available in SI Materials and Methods. Recombinant Orsay VLPs were obtained by overexpressing Orsay CP in E. coli. The crystal structure of Orsay VLP was solved by molecular replacement using a 9 Å resolution cryo-EM reconstruction as the initial model. RNA binding activity of the Orsay CP was determined using fluorescence polarization assays with CP dimer samples.
Supplementary Material
Acknowledgments
We thank Yunyun Jiang and Ranjana Kishore for helpful discussions. This work was supported by the Welch Foundation (C-1565 to Y.J.T.; Q1242 to W.C.), the National Institutes of Health (NIH) (AI077785 to Y.J.T.; R00HG004724 to W.Z.; P41GM103832 to W.C.), and the Kresge Science Initiative Endowment Fund at Rice University. D.W. holds an Investigator in the Pathogenesis of Infectious Diseases Award from the Burroughs Wellcome Fund. C.F.H. was supported by an NIH training grant from T15LM007093 through the Gulf Coast Consortia.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors for the full-length capsid protein (CP) and CP42–391 reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4NWV and 4NWW, respectively). The EM map has been deposited in the EMDataBank (accession no. EMD-5952).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407122111/-/DCSupplemental.
References
- 1.Riddle DL, Blumenthal T, Meyer BJ, Priess JR. 1997. Introduction to C. elegans. C. elegans II, Cold Spring Harbor Monograph Series, eds Riddle DL, Blumenthal T, Meyer BJ, Priess JR (Cold Spring Harbor Lab Press, Cold Spring Harbor, NY), 2nd Ed, pp 1–12.
- 2.Kaletta T, Hengartner MO. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov. 2006;5(5):387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 3.Kuwabara PE, O’Neil N. The use of functional genomics in C. elegans for studying human development and disease. J Inherit Metab Dis. 2001;24(2):127–138. doi: 10.1023/a:1010306731764. [DOI] [PubMed] [Google Scholar]
- 4.Félix MA, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9(1):e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franz CJ, Zhao G, Félix MA, Wang D. Complete genome sequence of Le Blanc virus, a third Caenorhabditis nematode-infecting virus. J Virol. 2012;86(21):11940. doi: 10.1128/JVI.02025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiery R, et al. Family Nodaviridae. In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic; 2012. pp. 1061–1067. [Google Scholar]
- 7.Jiang H, et al. Orsay virus utilizes ribosomal frameshifting to express a novel protein that is incorporated into virions. Virology. 2014;450-451:213–221. doi: 10.1016/j.virol.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balla KM, Troemel ER. Caenorhabditis elegans as a model for intracellular pathogen infection. Cell Microbiol. 2013;15(8):1313–1322. doi: 10.1111/cmi.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad BV, et al. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286(5438):287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 10.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. Tomato bushy stunt virus at 2.9 Å resolution. Nature. 1978;276(5686):368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 11.Wang BC. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- 12.Bricogne G. Geometric sources of redundancy in intensity data and their use for phase determination. Acta Crystallogr A. 1974;30(Pt 3):395–405. [Google Scholar]
- 13.Harrison SC. 2007. Principles of virus structure. Fields Virology, eds Knipe DM, Howley PM (Lippincott Williams & Wilkins, Philadelphia), 5th Ed, pp 59–98.
- 14.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgunova EYu, et al. The atomic structure of carnation mottle virus capsid protein. FEBS Lett. 1994;338(3):267–271. doi: 10.1016/0014-5793(94)80281-5. [DOI] [PubMed] [Google Scholar]
- 16.Wada Y, et al. The structure of melon necrotic spot virus determined at 2.8 Å resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64(Pt 1):8–13. doi: 10.1107/S1744309107066481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda Y, et al. Crystal structure of tobacco necrosis virus at 2.25 Å resolution. J Mol Biol. 2000;300(1):153–169. doi: 10.1006/jmbi.2000.3831. [DOI] [PubMed] [Google Scholar]
- 18.Plevka P, et al. The three-dimensional structure of ryegrass mottle virus at 2.9 Å resolution. Virology. 2007;369(2):364–374. doi: 10.1016/j.virol.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 19.Qu C, et al. 3D domain swapping modulates the stability of members of an icosahedral virus group. Structure. 2000;8(10):1095–1103. doi: 10.1016/s0969-2126(00)00508-6. [DOI] [PubMed] [Google Scholar]
- 20.Bhuvaneshwari M, et al. Structure of sesbania mosaic virus at 3 Å resolution. Structure. 1995;3(10):1021–1030. doi: 10.1016/s0969-2126(01)00238-6. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, et al. Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS Pathog. 2013;9(1):e1003132. doi: 10.1371/journal.ppat.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Squires G, et al. Structure of the Triatoma virus capsid. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 6):1026–1037. doi: 10.1107/S0907444913004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuthill TJ, et al. Equine rhinitis A virus and its low pH empty particle: Clues towards an aphthovirus entry mechanism? PLoS Pathog. 2009;5(10):e1000620. doi: 10.1371/journal.ppat.1000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guu TS, et al. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc Natl Acad Sci USA. 2009;106(31):12992–12997. doi: 10.1073/pnas.0904848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita T, et al. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Natl Acad Sci USA. 2009;106(31):12986–12991. doi: 10.1073/pnas.0903699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang L, et al. The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat Struct Biol. 2001;8(1):77–83. doi: 10.1038/83089. [DOI] [PubMed] [Google Scholar]
- 27.Fisher AJ, Johnson JE. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature. 1993;361(6408):176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 28.Zlotnick A, Natarajan P, Munshi S, Johnson JE. Resolution of space-group ambiguity and structure determination of nodamura virus to 3.3 Å resolution from pseudo-R32 (monoclinic) crystals. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 6):738–746. doi: 10.1107/S0907444997007427. [DOI] [PubMed] [Google Scholar]
- 29.Wery JP, Reddy VS, Hosur MV, Johnson JE. The refined three-dimensional structure of an insect virus at 2.8 Å resolution. J Mol Biol. 1994;235(2):565–586. doi: 10.1006/jmbi.1994.1014. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, et al. Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens. PLoS Pathog. 2011;7(7):e1002152. doi: 10.1371/journal.ppat.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopper P, Harrison SC, Sauer RT. Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. J Mol Biol. 1984;177(4):701–713. doi: 10.1016/0022-2836(84)90045-7. [DOI] [PubMed] [Google Scholar]
- 32.Robinson IK, Harrison S. Structure of the expanded state of tomato bushy stunt virus. Nature. 1982;297(5867):563–568. [Google Scholar]
- 33.Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3(1):63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee M, et al. Structure and function of a genetically engineered mimic of a nonenveloped virus entry intermediate. J Virol. 2010;84(9):4737–4746. doi: 10.1128/JVI.02670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pappachan A, Chinnathambi S, Satheshkumar PS, Savithri HS, Murthy MR. A single point mutation disrupts the capsid assembly in sesbania mosaic virus resulting in a stable isolated dimer. Virology. 2009;392(2):215–221. doi: 10.1016/j.virol.2009.06.047. [DOI] [PubMed] [Google Scholar]
- 36.Lokesh GL, Gowri TD, Satheshkumar PS, Murthy MR, Savithri HS. A molecular switch in the capsid protein controls the particle polymorphism in an icosahedral virus. Virology. 2002;292(2):211–223. doi: 10.1006/viro.2001.1242. [DOI] [PubMed] [Google Scholar]
- 37.Dong XF, Natarajan P, Tihova M, Johnson JE, Schneemann A. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J Virol. 1998;72(7):6024–6033. doi: 10.1128/jvi.72.7.6024-6033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Routh A, Domitrovic T, Johnson JE. Host RNAs, including transposons, are encapsidated by a eukaryotic single-stranded RNA virus. Proc Natl Acad Sci USA. 2012;109(6):1907–1912. doi: 10.1073/pnas.1116168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93(Pt 7):1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crane M, Hyatt A. Viruses of fish: An overview of significant pathogens. Viruses. 2011;3(11):2025–2046. doi: 10.3390/v3112025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang L, et al. Virus-like particles of a fish nodavirus display a capsid subunit domain organization different from that of insect nodaviruses. J Virol. 2002;76(12):6370–6375. doi: 10.1128/JVI.76.12.6370-6375.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X, Lu R. Characterization of virus-encoded RNA interference suppressors in Caenorhabditis elegans. J Virol. 2013;87(10):5414–5423. doi: 10.1128/JVI.00148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olveira JG, et al. Comparative analysis of both genomic segments of betanodaviruses isolated from epizootic outbreaks in farmed fish species provides evidence for genetic reassortment. J Gen Virol. 2009;90(Pt 12):2940–2951. doi: 10.1099/vir.0.013912-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.