Significance
In somatic cells of female mammals, one of the two X chromosomes is randomly silenced, a phenomenon called X-chromosome inactivation (XCI). XCI is initiated in cis by a noncoding RNA called Xist, but trans-acting factors that mediate XCI remain largely unknown. In this study, we perform a large-scale RNA interference screen and identify new trans-acting factors that are required for mammalian XCI. Chemical inhibitors of some of these factors can reversibly reactivate the inactive X chromosome. Our results have therapeutic implications for certain human diseases, in particular the neurodevelopmental disorder Rett syndrome, which is caused by loss-of-function mutations in the X-linked MECP2 gene. Reactivation of the silenced wild-type MECP2 allele is a potential strategy for treating the disease.
Keywords: MECP2, RNA FISH, RNA-seq
Abstract
X-chromosome inactivation (XCI), the random transcriptional silencing of one X chromosome in somatic cells of female mammals, is a mechanism that ensures equal expression of X-linked genes in both sexes. XCI is initiated in cis by the noncoding Xist RNA, which coats the inactive X chromosome (Xi) from which it is produced. However, trans-acting factors that mediate XCI remain largely unknown. Here, we perform a large-scale RNA interference screen to identify trans-acting XCI factors (XCIFs) that comprise regulators of cell signaling and transcription, including the DNA methyltransferase, DNMT1. The expression pattern of the XCIFs explains the selective onset of XCI following differentiation. The XCIFs function, at least in part, by promoting expression and/or localization of Xist to the Xi. Surprisingly, we find that DNMT1, which is generally a transcriptional repressor, is an activator of Xist transcription. Small-molecule inhibitors of two of the XCIFs can reversibly reactivate the Xi, which has implications for treatment of Rett syndrome and other dominant X-linked diseases. A homozygous mouse knockout of one of the XCIFs, stanniocalcin 1 (STC1), has an expected XCI defect but surprisingly is phenotypically normal. Remarkably, X-linked genes are not overexpressed in female Stc1−/− mice, revealing the existence of a mechanism(s) that can compensate for a persistent XCI deficiency to regulate X-linked gene expression.
X-chromosome inactivation (XCI), the random transcriptional silencing of one X chromosome in somatic cells of female mammals, is a mechanism that ensures equal expression of X-linked genes in both sexes (1). XCI is initiated by X inactive specific transcript (Xist), a 17-kb noncoding RNA whose expression during early embryogenesis is both necessary and sufficient for silencing (2, 3). Xist represses transcription in cis by coating only the X chromosome from which it is produced. Once Xist has been up-regulated during early development or differentiation, it continues to be expressed from the inactive X (Xi) even in fully differentiated somatic cells. Before the initiation of XCI, TSIX transcript, XIST antisense RNA (Tsix) an antisense repressor of Xist, blocks Xist up-regulation on the future active X chromosome (Xa) (4).
An understanding of the factors and mechanisms involved in XCI is directly relevant to certain human diseases. In particular, loss-of-function mutations in the X-linked methyl-CpG binding protein 2 (MECP2) gene lead to the neurodevelopmental disorder Rett syndrome (RTT) (5–7). Most RTT patients are females who are heterozygous for MECP2 deficiency due to random XCI. Significantly, in a mouse model of RTT, reactivation of wild-type Mecp2 expression can reverse the disease phenotype even in late-stage adult animals (8). Thus, reactivation of the silenced wild-type MECP2 allele is a potential strategy for treating RTT.
We have previously demonstrated how large-scale short hairpin RNA (shRNA) screens can be used to identify factors involved in epigenetic silencing of tumor suppressor genes (9–11). Here, we perform a large-scale RNA interference (RNAi) screen using a genome-wide collection of shRNAs to identify trans-acting factors that are required for mammalian XCI.
Results
Identification of Factors Required for Mammalian XCI.
We used a previously derived female mouse embryonic fibroblast cell line [H4SV (12)] in which genes encoding green fluorescent protein (GFP) and hypoxanthine guanine phosphoribosyltransferase (HPRT) are present only on the Xi. Knockdown (KD) of a factor required for XCI is expected to reactivate expression of the Gfp and Hprt reporter genes (Fig. 1A).
Fig. 1.
Identification of factors required for mammalian XCI. (A) Schematic summary of the shRNA screen. The Xi is designated as such due to deletion of Xist on the Xa. (B) H4SV cells expressing an shRNA against 1 of the 13 candidates or, as a control, a nonsilencing (NS) shRNA were FACS sorted, and GFP-positive cells were isolated. For each KD cell line, the percentage of GFP-positive cells was expressed as the fold increase relative to that obtained with the NS shRNA, which was set to 1. (C) Two-color RNA FISH monitoring expression of G6pdx (red) and Lamp2 (green; Left) and Pgk1 (red) and Mecp2 (green; Right) in each of the 13 XCIF KD BMSL2 cell lines. DAPI staining is shown in blue. The experiment was performed at least twice, and representative images are shown (Upper) and the results quantified (Lower) from one experiment.
A genome-wide mouse shRNA library comprising 62,400 shRNAs (13) was divided into 10 pools, which were packaged into retrovirus particles and used to transduce H4SV cells. GFP-positive cells were selected by fluorescence-activated cell sorting (FACS) and expanded, and the shRNAs were identified by sequence analysis. To validate the candidates, single shRNAs directed against each candidate gene were transduced into H4SV cells, and the number of GFP-positive cells was measured by FACS analysis. The results of these experiments identified 13 candidate genes whose knockdown resulted in an increased percentage of GFP-positive cells relative to that obtained with a control, nonsilencing (NS) shRNA (Fig. 1B). The cell viability assay of Fig. S1A shows that knockdown of each candidate enabled growth in hypoxanthine–aminopterin–thymidine (HAT) medium, indicating that the Xi-linked Hprt gene was reactivated. As expected, the mRNA levels of the 13 candidate genes were decreased in the corresponding KD H4SV cell line (Fig. S1B). To rule out off-target effects, we showed for all 13 candidates that a second, unrelated shRNA also reactivated the Xi-linked Hprt gene (Fig. S1C) and decreased mRNA levels of the targeted gene in the corresponding KD H4SV cell line (Fig. S1D). The 13 X-chromosome inactivation factors (XCIFs) are listed in Table S1 and include proteins that are known, or predicted, to be involved in diverse processes including cell signaling [3-phosphoinositide dependent protein kinase 1 (PDPK1), aurora kinase A (AURKA), LAYN, ACVR1, and NF1], transcription [DNA methyltransferase (cytosine-5) 1 (DNMT1), PYGO1, SOX5, and ZFP426] and ubiquitin-dependent regulation (RNF165 and FBXO8). Significantly, DNMT1 has been previously shown to be involved in XCI (14, 15), validating the screening strategy.
To confirm these results, we analyzed expression of four X-linked genes, glucose-6-phosphate dehydrogenase X-linked (G6pdx), lysosomal-associated membrane protein 2 (Lamp2), phosphoglycerate kinase 1 (Pgk1), and Mecp2, using two-color RNA fluorescence in situ hybridization (FISH) in BMSL2 cells, an unrelated female mouse fibroblast cell line (16). In BMSL2 cells expressing a control NS shRNA, RNA FISH revealed, as expected, a single nuclear signal for G6pdx, Lamp2, Pgk1, and Mecp2, indicative of monoallelic expression (Fig. 1C and Fig. S2A). Knockdown of each of the 13 XCIFs substantially increased the fraction of cells containing two nuclear G6pdx, Lamp2, Pgk1, and Mecp2 signals, indicative of biallelic expression. Reactivation of G6pdx, Pgk1, Mecp2, and Hprt in the 13 XCIF KD BMSL2 cell lines was also demonstrated by a ∼1.5- to 2-fold increase in mRNA levels as monitored by quantitative real-time RT-PCR (qRT-PCR) (Fig. S2B). Reactivation of the Xi-linked Pgk1 gene in representative XCIF KD BMSL2 cell lines was also demonstrated using a single-nucleotide primer extension (SNuPE) assay (Fig. S2C), which could distinguish expression of the Xi- and Xa-linked Pgk1 alleles by virtue of a single-nucleotide polymorphism (16). DNA FISH experiments using an X-chromosome–specific paint probe indicated that the X-chromosome content of the XCIF KD BMSL2 cell lines was similar to that of the control BMSL2 cell line expressing a NS shRNA (Fig. S2D).
The XCIFs Are Required for Initiation of XCI in Mouse Embryonic Stem Cells.
We next asked whether the XCIFs were required to initiate XCI in female mouse embryonic stem (ES) cells. Undifferentiated female mouse PGK12.1 ES cells were transduced with a retrovirus expressing an XCIF shRNA. Cells were then treated with retinoic acid (RA), which induces predominantly, but not exclusively, neuronal differentiation (17). X-linked gene expression was monitored by two-color RNA FISH. Fig. 2A and Fig. S3A show that biallelic expression of the X-linked G6pdx, Lamp2, Pgk1, and Mecp2 genes was substantially increased following knockdown of each XCIF. As above, the X-chromosome content of the XCIF KD ES cells was similar to that of the control ES cell line expressing a NS shRNA (Fig. S3B).
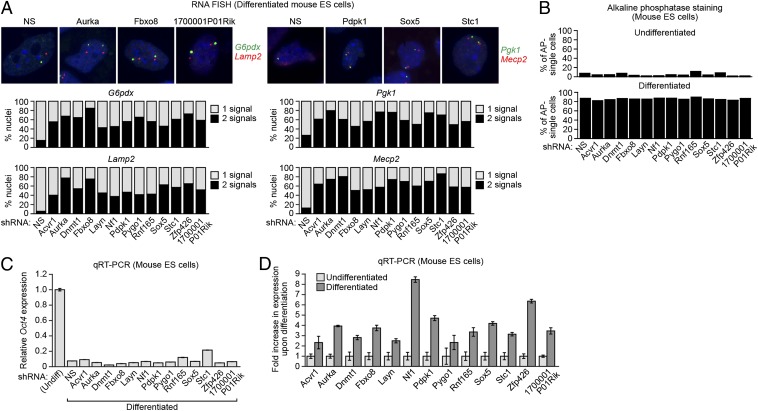
Fig. 2.
The XCIFs are required for initiation of XCI in mouse embryonic stem cells. (A) Two-color RNA FISH monitoring expression of G6pdx (green) and Lamp2 (red; Left) and Pgk1 (green) Mecp2 (red; Right) in the 13 XCIF KD ES cell lines following differentiation. DAPI staining is shown in blue. Representative images are shown (Upper), and the results quantified (Lower). (B) Percentage of alkaline phosphatase-negative single cells in the 13 XCIF KD ES cell lines before (Upper; undifferentiated) and after (Lower; differentiated) treatment with RA. (C) qRT-PCR analysis monitoring expression of Oct4 in the 13 XCIF KD ES cell lines following treatment with RA. As a control, expression of Oct4 in undifferentiated ES cells is shown and was set to 1. Error bars indicate SD. (D) qRT-PCR analysis of XCIFs in undifferentiated and differentiated mouse ES cells. Expression in differentiated ES cells was normalized to that observed in undifferentiated cells, which was set to 1. Error bars indicate SD.
A possible explanation for the failure of 1 or more of the 13 XCIF KD ES cell lines to undergo XCI is that the XCIF is required for differentiation. Following RA treatment, differentiation of the 13 XCIF KD ES cell lines was normal, as evidenced by monitoring two well-established markers of undifferentiated ES cells, alkaline phosphatase activity (Fig. 2B) and Oct4 expression (Fig. 2C). Likewise, several markers of differentiated cells that increase after RA treatment [Eomes (neuronal), Tcf712 (mesoderm), and Cdx2 (epithelial)] were unaffected by XCIF knockdown (Fig. S3C). Finally, the qRT-PCR results of Fig. 2D show that expression of all 13 XCIFs was up-regulated following differentiation, explaining, at least in part, the selective onset of XCI following differentiation.
XCIFs Function by Promoting Xist Expression and/or Localization to the Xi.
We next asked whether the XCIFs were required for Xist expression and/or localization to the Xi. Following knockdown of the 13 XCIFs in mouse ES cells, RA was added to induce differentiation and XCI, and Xist expression was analyzed by qRT-PCR. The results of Fig. 3A show that Xist levels were reduced to varying extents in all XCIF KD ES cell lines. In differentiated female ES cells, Xist is detected by RNA FISH as a large, diffuse nuclear signal referred to as a “cloud” that colocalizes with the Xi (18). Fig. 3B shows that knockdown of each of the XCIFs reduced to varying extents the percentage of cells with the Xist localization pattern characteristic of XCI (Fig. S4A). Taken together, these results indicate that XCIFs promote Xist expression and/or localization of Xist to the Xi.
Fig. 3.
XCIFs function by promoting Xist expression and/or localization, and DNMT1 is a transcriptional activator of Xist on the Xi. (A) qRT-PCR analysis monitoring Xist expression in the 13 XCIF KD ES cell lines following differentiation. Expression in differentiated ES cells was normalized to that obtained with the NS shRNA, which was set to 1. Error bars indicate SE. (B) RNA FISH monitoring localization of Xist in the 13 XCIF KD ES cell lines following differentiation. Cells were categorized as having either a typical Xist cloud or “other” pattern, which includes either the lack of a detectable Xist signal or presence of two small Xist signals, as in undifferentiated ES cells. (C) RNA FISH monitoring expression of Xist (Upper) and Mecp2 (Lower) in BMSL2 cells treated with an Xist locked nucleic acid antisense oligonucleotide (LNA ASO) or a control LNA ASO. (D) ChIP analysis monitoring binding of DNMT1 and POL2 to the Xist promoter and exon 2 in BMSL2 cells expressing a NS or Dnmt1 shRNA. Error bars indicate SD. (E) Nuclear run-on assay monitoring transcription of Xist, Hprt, and Tbp in BMSL2 cells expressing a NS or DNMT1 shRNA. (F) qRT-PCR analysis monitoring Xist levels in BMSL2 cells expressing a NS or Dnmt1 shRNA following treatment with actinomycin D. Actin mRNA was used as a normalization control. Error bars indicate SD. (G) qRT-PCR analysis monitoring Xist expression in MEFs isolated from female Dnmt1+/+ and Dnmt1−/− embryos. Four different litters were analyzed (n = 4 mice total per genotype), and the results were averaged. Expression was normalized to that observed in Dnmt1+/+ MEFs, which was set to 1. Error bars indicate SD. *P < 0.001 (Student t test). (H) qRT-PCR monitoring levels of Xist and Tsix in H4SV cells expressing a NS or DNMT1 shRNA. Expression was normalized to that obtained with the NS shRNA, which was set to 1. Error bars indicate SD. (I) qRT-PCR analysis monitoring Hprt and Xist expression in BMSL2 cells treated in the absence or presence of 5-azacytidine (5-AZA). Expression was normalized to that observed in the absence of 5-AZA, which was set to 1. Error bars indicate SD.
Several previous studies have suggested that Xist is required for the initiation but not maintenance of XCI (19–21). However, the results of Fig. 3 A and B implied that Xist was also necessary for maintenance of XCI. To provide independent evidence for this model, we abrogated Xist function in mouse BMSL2 fibroblasts using an Xist antisense locked nucleic acid (LNA) oligonucleotide (22). The results of Fig. 3C show, consistent with previous results (22), that the Xist antisense LNA oligonucleotide perturbed the normal pattern of Xist expression/localization. Most importantly, the Xist antisense LNA oligonucleotide substantially increased biallelic expression of X-linked Mecp2. Thus, Xist is required for both the initiation and maintenance of XCI.
DNMT1 Is a Transcriptional Activator of Xist on the Xi.
We were surprised that DNMT1, which typically functions as a transcriptional repressor (23, 24), was required for Xist expression and/or localization to the Xi. To further investigate this finding, we performed chromatin immunoprecipitation (ChIP) experiments in BMSL2 cells in which the Xa harbors a deletion encompassing the Xist promoter and several genes including Hprt (16, 25). Fig. 3D shows that DNMT1 and, as expected, RNA polymerase II (POL2) were bound near the Xist transcription start site on the Xi. The fact that DNMT1 was required for Xist transcription and bound to the Xist promoter suggested that DNMT1 might function as a direct transcriptional activator of Xist. Consistent with this idea, following knockdown of DNMT1 the level of POL2 bound to the Xist promoter substantially decreased (Fig. 3D). Moreover, in a nuclear run-on assay DNMT1 knockdown reduced Xist transcription but increased Xi-linked Hprt transcription, as expected (Fig. 3E). As a control, transcription of the TATA-box-binding protein (Tbp) gene, which is not X-linked and expressed constitutively, was unaffected by DNMT1 knockdown. In addition, knockdown of DNMT1 did not affect the half-life of Xist RNA (Fig. 3F), indicating the decreased levels of Xist RNA following DNMT1 depletion were predominantly transcriptional. Finally, the level of Xist transcripts was significantly lower in Dnmt1−/− compared with Dnmt1+/+ mouse embryonic fibroblasts (MEFs) (Fig. 3G). Collectively, these results indicate that DNMT1 is a transcriptional activator of Xist on the Xi.
We considered the possibility that DNMT1 indirectly activated Xist transcription by repressing expression of Tsix, which negatively regulates Xist (4). However, knockdown of DNMT1 in fibroblasts (Fig. 3H and Fig. S4B) or murine ES (Fig. S4C) cells substantially decreased Xist expression but did not affect Tsix levels. An alternative mechanism we considered was that DNMT1-mediated methylation at the Xist promoter could block the binding of a transcriptional repressor. Consistent with this possibility, following addition of 5-azacytidine, which inhibits DNMT1 enzymatic activity resulting in DNA demethylation, Xist levels were markedly reduced, whereas expression of the Xi-linked Hprt gene increased, as expected (Fig. 3I). Collectively, these results suggest that DNMT1 promotes Xist transcription by antagonizing a repressor.
Reactivation of the Xi-Linked Mecp2 Gene by Small-Molecule XCIF Inhibitors.
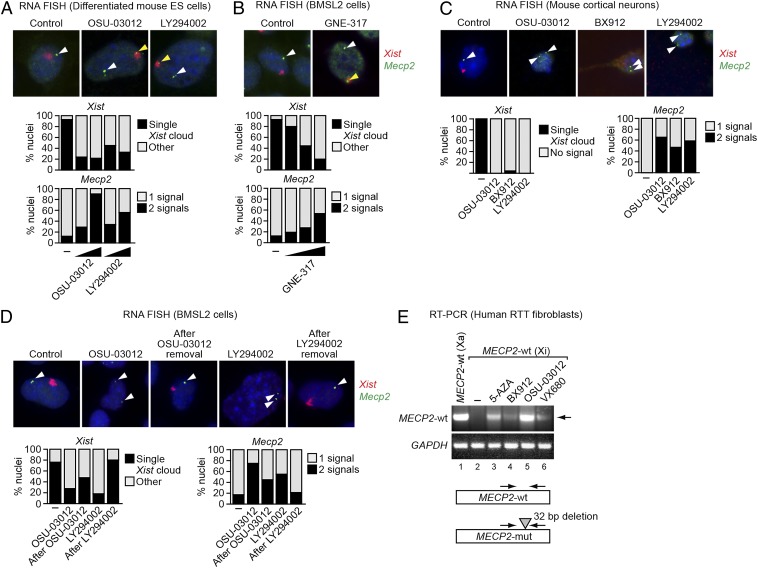
We next asked whether small-molecule XCIF inhibitors could reactivate Xi-linked genes. One of the XCIFs is PDPK1, a serine-threonine kinase that regulates phosphatidylinositol 3-kinase (PI3K)/AKT signaling (26). Fig. 4A and Fig. S5A show that following treatment of differentiated female mouse ES cells with a chemical inhibitor of either PDPK1 (OSU-03012) or PI3K (LY294002), there was a dose-dependent loss of the Xist cloud and increased biallelic expression of Mecp2. Similar results were obtained in BMSL2 cells using GNE-317 (Fig. 4B and Fig. S5B), a PI3K inhibitor specifically designed to cross the blood–brain barrier (27). As expected, with all three inhibitors the majority of cells contained two Mecp2 RNA FISH signals and lacked a detectable Xist cloud. Notably, however, in some cells one of the two Mecp2 RNA FISH signals colocalized with a Xist cloud, which marked the Xi. Similar results were obtained with postmitotic mouse cortical neurons using the PDPK1 inhibitors OSU-03012 and BX912 or the PI3K inhibitor LY294002 (Fig. 4C).
Fig. 4.
Reactivation of the Xi-linked Mecp2 gene by small-molecule XCIF inhibitors. (A and B) Two-color RNA FISH monitoring expression of Xist (red) and Mecp2 (green) in differentiated mouse ES cells treated with DMSO (control or –), OSU-03012, or LY294002 (A), and in BMSL2 cells treated with DMSO or GNE-317 (B). Representative images are shown (Upper) using the higher concentrations of the inhibitors, and the results quantified (Lower). The yellow arrowheads indicate colocalizing Xist and Mecp2 signals; the white arrowheads indicate Mecp2 signals not colocalizing with Xist. (C) Two-color RNA FISH monitoring Xist (red) and Mecp2 (green) expression in mouse cortical neurons treated with DMSO (control or –), OSU-03012, BX912, or LY294002. Representative images are shown (Upper), and the results quantified (Lower). The arrowheads indicate Mecp2 signals. (D) Two-color RNA FISH monitoring Xist (red) and Mecp2 (green) expression in BMSL2 cells treated with DMSO (control or –), LY294002, or OSU-03012, and at least 6 d following removal of the inhibitor. Representative images are shown (Upper), and the results quantified (Lower). The arrowheads indicate Mecp2 signals. (E) qRT-PCR monitoring Xi-linked wild-type MECP2 expression in human RTT fibroblasts treated with DMSO (–), 5-azacytidine (5-AZA), BX912, OSU-03012, or VX680. As a control, Xa-linked wild-type MECP2 expression was monitored in another clonal fibroblast cell line derived from the same RTT patient (lane 1). The arrowhead indicates the wild-type MECP2 qRT-PCR product. GAPDH was monitored as a loading control. (Lower) Schematic of the MECP2 wild-type (wt) and mutant (mut) alleles.
To ask whether reactivation of the Xi is reversible, BMSL2 cells were treated with OSU-03012 or LY294002, resulting, as expected, in biallelic expression of the Xi-linked Mecp2 gene (Fig. 4D and Fig. S5C). Following removal of the drug for at least 6 d, normal Xist expression and localization, and monoallelic expression of Mecp2, was largely restored, indicating that small-molecule–mediated reactivation of Xi-linked genes is reversible.
Finally, we tested whether small-molecule XCIF inhibitors could reactivate an Xi-linked wild-type MECP2 allele in a clonal fibroblast cell line from an RTT patient (28). In this cell line, the Xa-linked mutant MECP2 allele contains a 32-bp deletion, enabling selective detection of Xi-linked wild-type MECP2 mRNA in an RT-PCR assay using a primer within the deleted region (28). Another clonal fibroblast cell line derived from the same RTT patient in which the wild-type MECP2 allele is on the Xa provided a control for the correct RT-PCR product (Fig. 4E, lane 1). The results show, as expected, that the Xi-linked wild-type MECP2 allele was not expressed (lane 2) but could be reactivated by addition of the DNA methyltransferase inhibitor 5-azacytidine (lane 3). Significantly, addition of the PDPK1 inhibitors BX912 and OSU-03012 (lanes 4 and 5), or VX680 (lane 6), an inhibitor of AURKA, another XCIF (Table S1), reactivated the Xi-linked wild-type MECP2 allele. Thus, XCIF chemical inhibitors can reactivate the Xi-linked Mecp2/MECP2 gene in murine fibroblasts, ES cells, and cortical neurons, as well as human RTT fibroblasts.
Defective XCI in Female Stc1−/− Mice.
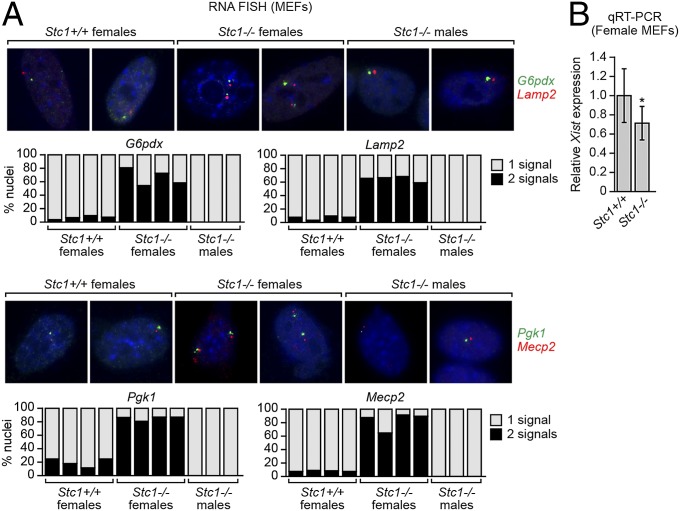
One of the XCIFs, stanniocalcin 1 (STC1), is a glycoprotein found in both the cytoplasm and nucleus and whose function is poorly understood (29). Stc1−/− mice have no obvious phenotype and litters have the expected Mendelian and male/female ratios (30). To determine whether STC1 is required for XCI in the mouse, we intercrossed Stc1+/− mice and analyzed MEFs from the resultant progeny by two-color RNA FISH for expression of G6pdx, Lamp2, Pgk1, and Mecp2. As expected, female Stc1+/+ MEFs, and as a control male Stc1−/− MEFs, displayed monoallelic expression of G6pdx, Lamp2, Pgk1, and Mecp2 (Fig. 5A). By contrast, female Stc1−/− MEFs predominantly displayed biallelic expression of the four genes, indicative of an XCI defect. qRT-PCR analysis revealed reduced Xist levels in female Stc1−/− MEFs compared with female Stc1+/+ MEFs (Fig. 5B). Notably, the X-chromosome content of female Stc1−/− and Stc1+/+ MEFs was comparable (Fig. S6A).
Fig. 5.
Defective XCI in female Stc1−/− MEFs. (A) Two-color RNA FISH monitoring expression of G6pdx (green) and Lamp2 (red; Upper) and Pgk1 (green) and Mecp2 (red; Lower) in female Stc1+/+ and Stc1−/− MEFs, and as a control male Stc1−/− MEFs. Representative images are shown (Upper), and the results quantified (Lower). (B) qRT-PCR analysis monitoring Xist expression in MEFs isolated from female Stc1+/+ and Stc1−/− embryos. Four different litters were analyzed (n = 4 mice total per genotype), and the results were averaged. Expression was normalized to that of the ribosomal gene RPL4, and Xist expression in Stc1+/+ MEFs was set to 1. Error bars indicate SD. *P < 0.001 (Student t test).
To further validate these findings, we analyzed Xist and Mecp2, or Xist and G6pdx, in cortical neurons from brain sections of Stc1−/− and Stc1+/+ postnatal female mice. In female Stc1−/− mice, biallelic expression of Mecp2 and G6pdx was clearly evident in some cortical neurons (Fig. S6B). Again, in some cells we could observe colocalization of Mecp2 and Xist, or G6pdx and Xist signals, indicative of reactivation of the Xi-linked Mecp2 and G6pdx genes.
Defective XCI in Female Stc1−/− Mice Is Not Accompanied by Increased X-Linked Gene Expression.
We performed transcriptome profiling (RNA-Seq) experiments to determine whether the expression levels of X-encoded genes were elevated in female Stc1−/− MEFs. In these experiments, RNA was prepared from three independent cultures of female Stc1+/+ or Stc1−/− MEFs. RNA samples were processed and amplified followed by deep sequencing (Fig. 6A). The results of Fig. 6B and Dataset S1 shows that total expression levels of the vast majority (98%) of X-linked genes were indistinguishable in Stc1+/+ and Stc1−/− MEFs. The similarity of X-linked gene expression between Stc1+/+ and Stc1−/− MEFs was statistically significant (Fig. 6C and Fig. S7A). Moreover, the vast majority (99%) of autosomal genes were also expressed at statistically comparable levels in female Stc1+/+ and Stc1−/− MEFs (Fig. S7B and Dataset S1).
Fig. 6.
Defective XCI in female Stc1−/− mice is not accompanied by increased X-linked gene expression. (A) Schematic of the RNA-Seq analysis pipeline. (B) Distribution of log2-transformed ratio of X-linked gene expression in MEFs from female Stc1−/− (KO) and Stc1+/+ (WT) embryos (n = 3 per genotype). (C) Box plot of X-linked gene expression (log2-transformed FPKM) in MEFs from female Stc1−/− and Stc1+/+ embryos (n = 3 per genotype). The boxed areas span the first to the third quartile. The whiskers represent 15th and 85th percentiles. (D) qRT-PCR analysis monitoring expression of Mecp2 and Hprt in MEFs from two different litters of female Stc1−/− and Stc1+/+ embryos (n = 2 mice total per genotype). The results were normalized to those obtained in Stc1+/+ MEFs, which was set to 1. Error bars indicate SE. (E) Immunoblot showing MECP2 and STC1 levels in female Stc1+/+ and Stc1−/− MEFs (Left) or brain tissue female Stc1+/+ and Stc1−/− P1 mice (Right) (n = 3 per genotype). α-Tubulin (TUBA) was monitored as a loading control. (F) qRT-PCR analysis of Stc1, Xist, Mecp2, and Hprt expression in BMSL2 cells expressing a NS or STC1 shRNA. The results were normalized to those obtained with the NS shRNA, which was set to 1. Error bars indicate SE. (G) Immunoblot showing MECP2 and STC1 levels in BMSL2 cells expressing a NS or Stc1 shRNA.
To support these RNA-Seq–based results, we also analyzed the levels of X-linked genes Mecp2 and Hprt by qRT-PCR. Fig. 6D shows that Mecp2 and Hprt mRNA levels were equivalent in female Stc1+/+ and Stc1−/− MEFs despite deficient XCI. Furthermore, the immunoblot results of Fig. 6E show that the level of MECP2 protein in Stc1+/+ female MEFs (Left) and brain lysates (Right) was comparable to that in Stc1−/− females.
The experiments described above were performed in Stc1−/− mice in which there was a long-term, stable impairment of XCI. To determine whether X-linked gene expression was increased immediately following abrogation of XCI, we analyzed expression of Mecp2 and Hprt in mouse BMSL2 fibroblasts following shRNA-mediated knockdown of STC1. As expected, in STC1 KD BMSL2 cells there was an approximate twofold increase in Mecp2 and Hprt expression, which was evident at both the mRNA (Fig. 6F and Fig. S2B) and protein (Fig. 6G) level. Collectively, these results suggest the existence of a mechanism(s) that can compensate for a persistent XCI deficiency to regulate X-linked gene expression.
Discussion
In this study, we have performed a large-scale shRNA screen to identify factors, XCIFs, required for mammalian XCI. The XCIFs we have identified are selectively required for silencing of X-linked genes and not for general transcriptional repression. For example, knockdown of the 13 XCIFs did not affect repression of Oct4 following RA-induced differentiation of ES cells (Fig. 2C) or reactivate any of several imprinted genes that we analyzed (Fig. S7C).
For several reasons, we think it is likely that our screen, like other large-scale shRNA screens (31), did not achieve saturation, and thus additional XCIFs remain to be identified. For example, although our screen identified DNMT1, a known XCIF (14, 15), we did not isolate Polycomb subunits, which, consistent with other studies (32–35), we find in directed experiments are required for repression of X-linked genes (Fig. S7D).
In this regard, an RNAi screen similar in design to ours reported a number of factors required for XCI that are distinct from the XCIFs we identified (36). Although we do not exclude the possibility that some of candidates identified by Chan et al. (36) are involved in XCI, in this study RNA FISH experiments were not performed and there were no experiments demonstrating that depletion of these factors resulted in biallelic expression of endogenous X-linked genes. Rather, all that was shown was that short-term knockdown of the candidate genes modestly increased expression of several X-linked genes analyzed, which could be explained by increased expression of the Xa-linked gene rather than reactivation of the Xi-linked gene.
The Xi has multiple inhibitory histone modifications that can be epigenetically inherited and are thought to “lock in” a repressive chromatin structure. This and other considerations have led to the proposal that, once established, XCI may be irreversible (see, for example, ref. 21). However, we have shown that RNAi knockdown and small-molecule XCIF inhibition can reverse XCI, resulting in the reactivation of silenced X-linked genes. Analogous to our results, work from Philpot and colleagues (37) have shown that small molecules can reactivate an epigenetically silenced UBE3A gene.
The ability of small-molecule XCIF inhibitors to reactivate the Xi has important therapeutic implications for the treatment of RTT and perhaps other dominant X-linked diseases. Because reactivation of the Xi is reversible, for therapeutic applications the drug would need to be administered continually. A potential concern of a therapy based upon reactivation of the Xi is that the resultant elevated levels of X-linked gene expression may be deleterious. However, a homozygous mouse knockout of one of the XCIFs, STC1, has an XCI defect but surprisingly is phenotypically normal. Remarkably, despite the XCI deficiency, X-linked genes are not overexpressed in Stc1−/− mice. We interpret these results to mean that there is another mechanism(s) that can compensate for a persistent XCI deficiency in regulating X-linked gene expression. Notably, other mechanisms that regulate the expression of X-linked genes have been proposed (38, 39). These results suggest that pharmacological or genetic reactivation of the Xi-linked wild-type MECP2 allele in an RTT patient would not increase total X-linked gene expression. Moreover, the results of a recent study suggest that the wild-type MECP2 would be functional in a cell also expressing an MECP2 mutant (40).
Consistent with our results, a previous study reported that, in a subtype of breast cancers (sporadic basal-like breast cancers), ∼50% of cases have two Xas and lack an Xi (41). Expression profiling of these cell lines revealed that there was not a general increase in X-linked gene expression. In addition, a recent study reported that long-term conditional depletion of Xist in mouse hematopoietic cells led to increased or decreased expression of specific X-linked genes (42). However, analysis of available datasets from this study revealed that, in Xist-depleted hematopoietic cells, only ∼2–12% of X-linked genes were up-regulated greater than 1.5-fold (Fig. S7E).
Xist−/− mice have an embryonic lethal phenotype (43), which has led to the general belief that XCI is required for normal development and viability. In addition, conditional depletion of Xist in mouse hematopoietic cells resulted in the induction of leukemia accompanied by large-scale changes in expression of autosomal genes (42). By contrast, our results indicate that female Stc1−/− mice have an XCI defect but are phenotypically normal. The normal development and viability of female Stc1−/− mice can be explained by our finding that X-linked genes are not overexpressed.
Collectively, these observations indicate that the phenotypes resulting from complete loss of Xist are more severe than those resulting from the XCI defect resulting from loss of STC1, in which Xist levels are reduced but not completely absent (Figs. 5B and 6F). An intriguing possibility that would explain these findings is that Xist has a function(s) in addition to its role in XCI, which is lost in the complete absence of Xist but retained when Xist levels are merely reduced to a level that impairs XCI.
Materials and Methods
Cell Culture.
H4SV cells (12), BMSL2 (HOBMSL2) cells (16), and human RTT fibroblasts (28) were cultured as recommended by the supplier. PGK12.1 cells were cultured as previously described (44) and differentiated by replating, on gelatinized plastic dishes, in the presence of 100 nM α-retinoic acid (Sigma) and absence of leukemia inhibitory factor for at least 1 wk.
Isolation of MEFs, Brain Tissue, and Cortical Neurons.
MEFs were isolated from embryonic day 8.5 (E8.5) (Dnmt1 mice; The Jackson Laboratory) or E14.5 (Stc1 mice; provided by D. Sheikh-Hamad, Baylor College of Medicine, Houston) embryos as described previously (45), and were PCR genotyped using gene-specific and SRY primers (Table S2). Stc1+/+ and Stc1−/− postnatal day 1 (P1) pup heads were embedded in O.C.T. compound (Tissue-Tek) and frozen in liquid nitrogen. Brain tissue cryosections (5 µm thick) were mounted, fixed, and hybridized with FISH probes as described (46, 47). Neurons were isolated from the cerebral cortexes of E19.5 C57BL/6 embryos and cultured as described (48).
Large-Scale shRNA Screen and Validation.
The mouse shRNAmir library (release 2.16; Open Biosystems/Thermo Scientific) was obtained through the University of Massachusetts Medical School RNAi Core Facility. H4SV cells (1.1 × 106) were transduced at a multiplicity of infection of 0.2 with the retroviral pools, generated as previously described (9), and selected for resistance to puromycin for 7 d. Cells were FACS sorted, and GFP-positive cells were selected. Candidate shRNAs were identified as described previously (9). To validate the candidates, 3 × 105 H4SV or BMSL2 cells were transduced with single shRNAs (Table S3) and puromycin selected for 4 d. For HAT selection, 3 × 105 cells were plated in six-well plates and selected in medium containing 1× HAT (Gibco) for 1 wk, followed by live-cell imaging using a Zeiss Axiovert 200 microscope.
RNA FISH.
RNA FISH experiments were performed as described (49) (see Table S4 for cDNA template sources for probes). Cells were visualized on a Leica DM IRE2 confocal microscope. For quantification, 100–500 cells in total from at least 10 different fields were counted and scored; only cells with a detectable RNA FISH signal were included in the analysis, with the exception of the experiment in Fig. 3B. Images were adjusted consistently for contrast and brightness using AxioVision Software (Zeiss). All RNA FISH experiments were performed at least twice, and representative images and quantification are shown from one experiment.
Alkaline Phosphatase Assay.
ES cells were treated in the presence or absence of retinoic acid (see above) and analyzed using an Alkaline Phosphatase Staining Kit (Stemgent).
qRT-PCR.
Total RNA was isolated and reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen). qRT-PCR was performed as described previously (9) using primers listed in Table S2. For the experiments shown in Fig. 3 F and H and Fig. S4 B and C, strand-specific cDNA synthesis of Xist and/or Tsix RNAs was performed as described previously (50), and expression of Xist and Tsix were normalized to that of Gapdh.
LNA Nucleofection.
Cy3-labeled Xist and control (scrambled) LNAs (22) were added to 104 BMSL2 cells at a final concentration of 1 µM in OptiMem using Lipofectamine (Invitrogen) every 6–8 h for 48 h.
ChIP Assay.
ChIP assays were performed as described previously (9) using extracts prepared 7 d post-retroviral transduction and puromycin selection, and antibodies against DNMT1 or POL2 (Abcam). Primer sequences used for amplifying ChIP products are listed in Table S2.
Nuclear Run-On Assay.
Assays were performed in the presence of [32P]UTP, and radioactive RNA was isolated using TRIzol reagent. Samples were hybridized to a nylon membrane immobilized with cDNA probes to Xist [prepared from a plasmid containing Xist exons 1 and 6 (51)], Hprt (prepared from a plasmid containing the Hprt coding region PCR-amplified using forward 5′-TCCGCCTCCTCCTCTGCT-3′ and reverse 5′-GGGAATTTATTGATTTGCAT-3′ primers) and Tbp (prepared from a cloned Tbp cDNA; Open Biosystems). After washing the membranes, filters were exposed to a PhosphorImager screen and the signal was quantified on a Fujifilm FLA-7000 imaging system using Image Gauge, version 4.22, software.
Xist RNA Stability Assay.
The assay was performed as described previously (52). After treatment with DNase (Ambion), strand-specific Xist RNA levels, and as a control Actin, were quantified by qRT-PCR (see Table S2 for primer sequences).
Chemical Inhibitor Treatment.
Differentiated mouse ES or BMSL2 cells were treated with dimethyl sulfoxide (DMSO), LY294002 (Cayman Chemicals; 4 or 10 µM), OSU-03012 (Selleck Chemicals; 2.5 or 4 µM), or GNE-317 (Genentech; 1.25, 2.5, or 5 µM) for 3 d before RNA FISH analysis. For XCI reversibility experiments, BMSL2 cells were treated with 8 µM LY294002 or 2.5 µM OSU-03012 for 3 d, washed twice with PBS, and then the media was replaced with fresh media every day for at least 5 d before RNA FISH analysis.
Mouse cortical neurons, isolated as described above, were treated with DMSO, 5 µM BX912 (Axon Medchem), 0.4 µM LY294002, or 2.5 µM OSU-03012 for 4 d before RNA FISH analysis.
RTT fibroblasts were treated with either DMSO, 5-azacytidine (Calbiochem; 10 µM for 3 d), BX912 (10 µM for 3 d), OSU-03012 (10 µM for 2 d followed by 5 µM for 1 d), or VX680 (ChemieTek; 10 µM for 2 d followed by 3 µM for 1 d). The wild-type MECP2 levels were analyzed as previously described (28) using primers listed in Table S2.
RNA Sequencing and Data Analysis.
Total RNA was isolated from MEFs from Stc1+/+ and Stc1−/− embryos (n = 3 for each genotype) using the RNeasy Plus Mini Kit (Qiagen) and treated with RNase-free DNase I (Qiagen). mRNA libraries were generated as described in the TruSeq RNA sample preparation guide (Illumina).
Libraries were sequenced as 50-bp paired ends using an Illumina HiSEq 2000 by the University of Massachusetts Sequencing Core. Raw reads (ranging from 47 to 92 million reads per sample) were trimmed by removing adaptor sequences and demultiplexed with bar codes. Reads with ambiguous nucleotides and Phred quality scores <46 were removed before assembly. Paired-end sequencing reads were aligned using TopHat (version 2.0.6) (53) against mouse genome assembly NCBI38/mm10 (downloaded from prebuilt indexes at http://bowtie-bio.sourceforge.net/) by default parameters, with the exception of expecting an inner distance between mate pairs of 75 bp instead of the default value of 50 bp. The reads aligned by TopHat were processed by Cufflinks (version 2.0.1) (54) to assemble transcripts and to measure their relative abundances in fragments per kilobase of exon per million fragments mapped (FPKM) units. Assembled transcripts from control and knockout samples were compared with the transcriptome downloaded from Ensembl.org and tested for differential expression using the Cuffcompare and Cuffdiff utilities in the Cufflinks package. Cuffdiff was run with classic-FPKM normalization and a false-discovery rate threshold of 0.05. Genes with more than a twofold change in expression between Stc1+/+ and Stc1−/− samples and P < 0.05 (calculated using Cufflinks) were considered significant.
The gene expression results measured by Cufflinks were annotated based on a GTF file downloaded from Ensembl.org using Bioconductor package ChIPpeakAnno (55). All figures were plotted using R/Bioconductor (version 2.15.2) software. The RNA-Seq data have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (56) and are accessible to reviewers through Gene Expression Omnibus Series accession no. GSE47395 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47395).
Immunoblotting.
Cell extracts were prepared and immunoblots proved using antibodies against HPRT (Abcam), MECP2 (Abcam), STC1 (Santa Cruz Biotechnology), and α-tubulin.
Supplementary Material
Acknowledgments
We thank Ingolf Bach, Marisa Bartolomei, Neil Brockdorff, David Corey, Rudolf Jaenisch, Jeannie Lawrence, David Sheikh-Hamad, Art Riggs, and Judith Singer-Sam for providing reagents; Genentech for providing GNE-317; the University of Massachusetts Medical School (UMMS) RNAi Core Facility; Amy Virbasius for virus preparation; the UMMS Diabetes Endocrinology Research Center facility for cryosectioning; Joel Richter, Mike Perkins, and Karen Sargent for neuronal isolation; the UMMS Deep Sequencing Core Facility; Ellie Kittler of the Center for AIDS Research Molecular Biology Core for assistance with the SNuPE assay; and Sara Deibler for editorial assistance. N.W. is a translational scholar of The Sidney Kimmel Foundation for Cancer Research. This work was supported by grants from the National Institutes of Health (R01GM033977), a Pilot Project Program award from the UMass Center for Clinical and Translational Science, and The Rett Syndrome Research Trust (to M.R.G.). M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA-Seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE47395).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413620111/-/DCSupplemental.
References
- 1.Lyon MF. X-chromosome inactivation as a system of gene dosage compensation to regulate gene expression. Prog Nucleic Acid Res Mol Biol. 1989;36:119–130. doi: 10.1016/s0079-6603(08)60166-x. [DOI] [PubMed] [Google Scholar]
- 2.Brockdorff N. Chromosome silencing mechanisms in X-chromosome inactivation: Unknown unknowns. Development. 2011;138(23):5057–5065. doi: 10.1242/dev.065276. [DOI] [PubMed] [Google Scholar]
- 3.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 4.Leeb M, Steffen PA, Wutz A. X chromosome inactivation sparked by non-coding RNAs. RNA Biol. 2009;6(2):94–99. doi: 10.4161/rna.6.2.7716. [DOI] [PubMed] [Google Scholar]
- 5.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 6.Moretti P, Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Curr Opin Genet Dev. 2006;16(3):276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 8.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315(5815):1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449(7165):1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palakurthy RK, et al. Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Mol Cell. 2009;36(2):219–230. doi: 10.1016/j.molcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serra RW, Fang M, Park SM, Hutchinson L, Green MR. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. eLife. 2014;3:e02313. doi: 10.7554/eLife.02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153(4):773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37(11):1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 14.Basu R, Zhang LF. X chromosome inactivation: A silence that needs to be broken. Genesis. 2011;49(11):821–834. doi: 10.1002/dvg.20792. [DOI] [PubMed] [Google Scholar]
- 15.Sado T, et al. X inactivation in the mouse embryo deficient for Dnmt1: Distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol. 2000;225(2):294–303. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- 16.Komura Ji, Sheardown SA, Brockdorff N, Singer-Sam J, Riggs AD. In vivo ultraviolet and dimethyl sulfate footprinting of the 5′ region of the expressed and silent Xist alleles. J Biol Chem. 1997;272(16):10975–10980. doi: 10.1074/jbc.272.16.10975. [DOI] [PubMed] [Google Scholar]
- 17.Smith AG. Embryo-derived stem cells: Of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 18.Wutz A. Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12(8):542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 19.Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368(6467):154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 20.Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22(4):323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 21.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5(4):695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 22.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci USA. 2010;107(51):22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10(4):631–647. [PubMed] [Google Scholar]
- 24.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122(Pt 16):2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JG, Chapman VM. CpG island promoter region methylation patterns of the inactive-X-chromosome hypoxanthine phosphoribosyltransferase (Hprt) gene. Mol Cell Biol. 1994;14(12):7975–7983. doi: 10.1128/mcb.14.12.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: More than just a road to PKB. Biochem J. 2000;346(Pt 3):561–576. [PMC free article] [PubMed] [Google Scholar]
- 27.Salphati L, et al. Targeting the PI3K pathway in the brain—efficacy of a PI3K inhibitor optimized to cross the blood-brain barrier. Clin Cancer Res. 2012;18(22):6239–6248. doi: 10.1158/1078-0432.CCR-12-0720. [DOI] [PubMed] [Google Scholar]
- 28.Yu D, Sakurai F, Corey DR. Clonal Rett syndrome cell lines to test compounds for activation of wild-type MeCP2 expression. Bioorg Med Chem Lett. 2011;21(18):5202–5205. doi: 10.1016/j.bmcl.2011.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeung BH, Law AY, Wong CK. Evolution and roles of stanniocalcin. Mol Cell Endocrinol. 2012;349(2):272–280. doi: 10.1016/j.mce.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Chang AC, Cha J, Koentgen F, Reddel RR. The murine stanniocalcin 1 gene is not essential for growth and development. Mol Cell Biol. 2005;25(23):10604–10610. doi: 10.1128/MCB.25.23.10604-10610.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullenders J, Bernards R. Loss-of-function genetic screens as a tool to improve the diagnosis and treatment of cancer. Oncogene. 2009;28(50):4409–4420. doi: 10.1038/onc.2009.295. [DOI] [PubMed] [Google Scholar]
- 32.Mak W, et al. Mitotically stable association of polycomb group proteins eed and enx1 with the inactive X chromosome in trophoblast stem cells. Curr Biol. 2002;12(12):1016–1020. doi: 10.1016/s0960-9822(02)00892-8. [DOI] [PubMed] [Google Scholar]
- 33.Schoeftner S, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25(13):3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva J, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4(4):481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 35.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan KM, Zhang H, Malureanu L, van Deursen J, Zhang Z. Diverse factors are involved in maintaining X chromosome inactivation. Proc Natl Acad Sci USA. 2011;108(40):16699–16704. doi: 10.1073/pnas.1107616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang HS, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481(7380):185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38(1):47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 39.Deng X, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 2011;43(12):1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heckman LD, Chahrour MH, Zoghbi HY. Rett-causing mutations reveal two domains critical for MeCP2 function and for toxicity in MECP2 duplication syndrome mice. eLife. 2014;3:e02676. doi: 10.7554/eLife.02676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson AL, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152(4):727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11(2):156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 44.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 45.Samuelson LC, Metzger JM. Isolation and freezing of primary mouse embryonic fibroblasts (MEF) for feeder plates. CSH Protoc. 2006;2006(2):pii: pdb.prot4482. doi: 10.1101/pdb.prot4482. [DOI] [PubMed] [Google Scholar]
- 46.Capodieci P, et al. Gene expression profiling in single cells within tissue. Nat Methods. 2005;2(9):663–665. doi: 10.1038/NMETH786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lionnet T, et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8(2):165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang YS, Richter JD. Analysis of mRNA translation in cultured hippocampal neurons. Methods Enzymol. 2007;431:143–162. doi: 10.1016/S0076-6879(07)31008-2. [DOI] [PubMed] [Google Scholar]
- 49.Chaumeil J, Augui S, Chow JC, Heard E. Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods Mol Biol. 2008;463:297–308. doi: 10.1007/978-1-59745-406-3_18. [DOI] [PubMed] [Google Scholar]
- 50.Hoki Y, et al. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development. 2009;136(1):139–146. doi: 10.1242/dev.026427. [DOI] [PubMed] [Google Scholar]
- 51.Panning B. X inactivation in mouse ES cells: Histone modifications and FISH. Methods Enzymol. 2004;376:419–428. doi: 10.1016/S0076-6879(03)76028-5. [DOI] [PubMed] [Google Scholar]
- 52.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21(5):617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 53.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu LJ, et al. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.