Significance
Symmetric, football-shaped GroEL:GroES2 particles are the species involved in assisted protein folding. Here we show that the two rings of GroEL are simultaneously functional. The GroEL–GroES nanomachine is thus a parallel-processing device and not an alternating engine as previously thought. Crystals of the GroEL:GroES2 particles containing the ATP analog ADP-BeF3 were prepared both with and without an encapsulated molecule of substrate protein (Rubisco) in each of the central chambers. The structures of both of these GroEL:GroES2 particles have been solved to ∼3.7-Å resolution. Structural changes in the equatorial plate suggest a mechanism for directly linking the ATP-binding sites of the two rings, thus accounting for inter-ring negative cooperativity.
Keywords: symmetric, crystal structure, order-of-ligand-addition, encapsulation
Abstract
The GroE chaperonins assist substrate protein (SP) folding by cycling through several conformational states. With each cycle the SP is, in turn, captured, unfolded, briefly encapsulated (t1/2 ∼1 s), and released by the chaperonin complex. The protein-folding functional form is the US-football–shaped GroEL:GroES2 complex. We report structures of two such “football” complexes to ∼3.7-Å resolution; one is empty whereas the other contains encapsulated SP in both chambers. Although encapsulated SP is not visible on the electron density map, using calibrated FRET and order-of-addition experiments we show that owing to SP-catalyzed ADP/ATP exchange both chambers of the football complex encapsulate SP efficiently only if the binding of SP precedes that of ATP. The two rings of GroEL thus behave as a parallel processing machine, rather than functioning alternately. Compared with the bullet-shaped GroEL:GroES1 complex, the GroEL:GroES2 football complex differs conformationally at the GroEL–GroES interface and also at the interface between the two GroEL rings. We propose that the electrostatic interactions between the ε-NH3+ of K105 of helix D in one ring with the negatively charged carboxyl oxygen of A109 at the carboxyl end of helix D of the other ring provide the structural basis for negative inter-ring cooperativity.
The chaperonin proteins GroEL and GroES assist substrate proteins (SPs) to reach their native states, often under conditions when that otherwise spontaneous event does not occur (1–3). In the absence of SP GroEL/GroES operates via an asymmetric cycle in which the dissociation of ADP is the rate-determining step and the predominant species is an asymmetric, bullet-shaped GroEL:GroES1 complex (4, 5). The structure of this “bullet” GroEL:GroES1 complex has long been known (6, 7) and it has been assumed that this is the species that assists protein folding (8, 9). However, there is much evidence for the involvement of symmetric, “football”-shaped GroEL:GroES2 complexes (10–18). The formation of the football-shaped complex is promoted by SP (5, 17). Unfolded SP changes the kinetic mechanism, accelerating the rate of ADP/ATP exchange such that the dissociation of ADP is no longer rate-determining (Fig. 1A) (5). Thus, SP shifts the equilibrium between the footballs and bullets in favor of the former, consequently making them the predominant species (Fig. 1A) (5, 17).
Fig. 1.
The role of the football complex and its overall structure. (A) The symmetric chaperonin cycle (4, 5). In the presence of unfolded SP the chaperonin cycle operates in the symmetric mode. In this model the rate-determining step (0.5 s−1) is the hydrolysis of ATP that occurs in both rings of the symmetric GroEL:GroES2 football complex, which is consequently the predominant species. BeF3 arrests the cycle by indefinitely stabilizing the footballs, permitting all of the encapsulated SP molecules to fold to the native state. (B) Crystal structures of the MT-football complex and the SP-football complex with encapsulated Rubisco. Three domains of GroEL are colored in gray (equatorial domain), blue (intermediate domain), and cyan (apical domain). GroES is colored in lime. (Left) The overall view of the football complex as a ribbon diagram. (Center) The cross-section of the MT-football complex in surface representation (along a plane containing the axis of sevenfold symmetry). (Right) The cross-section of the SP-football complex containing encapsulated Rubisco. Electron densities are shown as gray mesh. All 2Fo-Fc electron density maps in this manuscript are contoured at 1σ.
To elucidate the mechanism of chaperonin-assisted protein folding by the football complex, we investigated the conditions permitting the formation of the football complexes. Using calibrated FRET we show that owing to SP-catalyzed ADP/ATP exchange the football complex efficiently encapsulates SP in both GroEL chambers. Thus, the two rings of GroEL behave as a parallel processing machine, rather than functioning alternately. We also determined structures of two football complexes; one is empty, the other contains encapsulated SP in both chambers. However, encapsulated SP is not visible on the electron density map. Compared with the bullet-shaped complex (1AON), the football complexes differ at the interface between the rings, suggesting a structural basis for negative inter-ring cooperativity.
Results and Discussion
SP Promotes the Formation of GroEL:GroES2 Footballs.
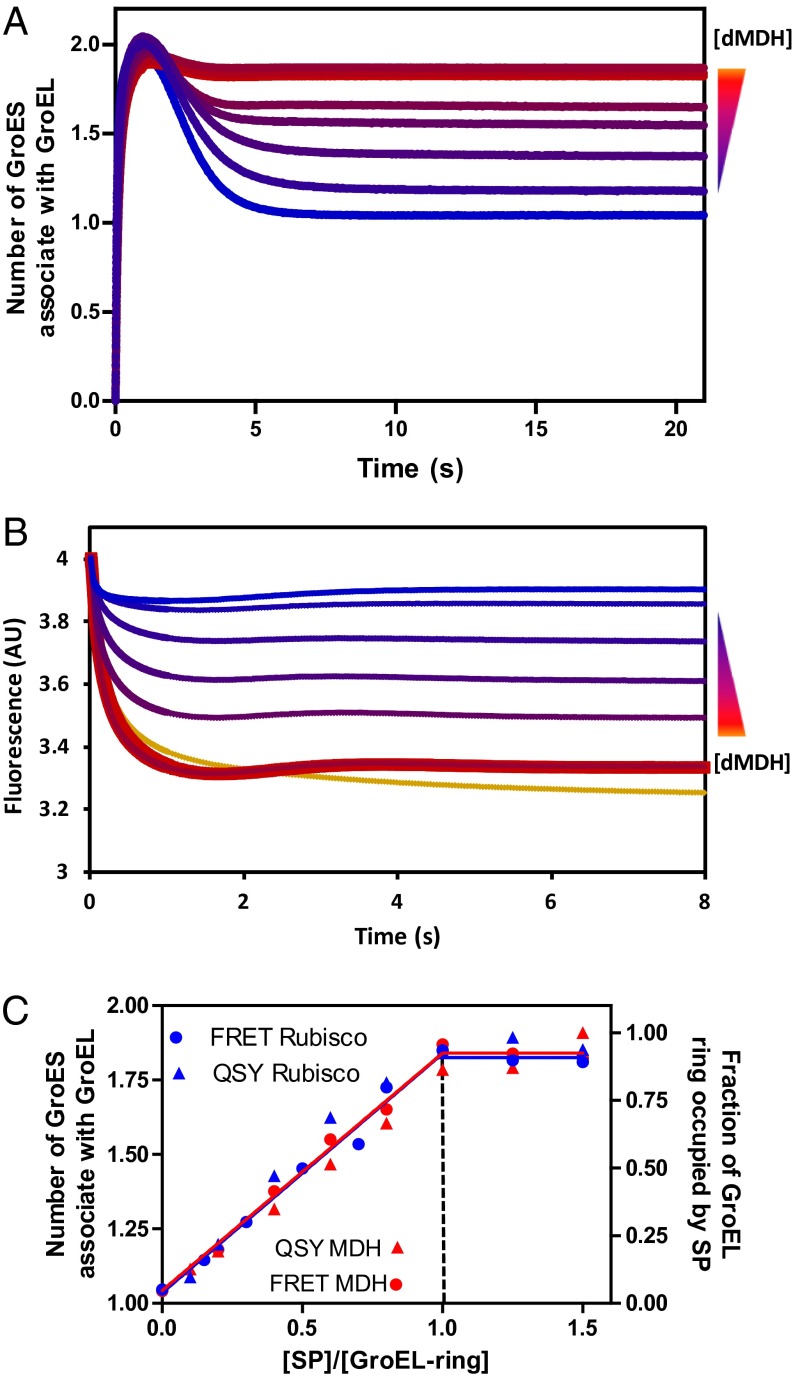
A dynamic, SP-dependent equilibrium exists between GroEL:GroES2 footballs and GroEL:GroES1 bullets that can be quantified by calibrated FRET (17). If the two rings of GroELIAEDANS are first equilibrated with stoichiometric quantities of two tightly bound SPs, malate dehydrogenase (MDH) or ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco), subsequent addition of ATP and GroESF5M leads to the stoichiometric formation of GroEL:GroES2 footballs (Fig. 2 A and C). On a much longer time scale, the footballs revert to GroEL:GroES1 bullets as the respective SPs fold to their native states (17).
Fig. 2.
The football complex encapsulates two misfolded proteins simultaneously. (A) Pre-steady-state formation of GroEL:GroES2 monitored by FRET. Concentrations of dMDH are indicated as follows (from top to bottom): 0.43, 0.357, 0.286, 0.229, 0.171, 0.114, 0.057, and 0 μM; 2 μM GroELIAEDANS subunit (0.3-μM rings), 4 μM GroESF5M subunit, and 0.5 mM ATP were used for all of the measurements. The reactions were initiated by introducing ATP+GroESF5M into solution containing GroELIAEDANS preincubated with dMDH. The summary plot of this set of experiment is shown in C as a red circle. (B) Encapsulation of dMDH by GroEL:GroES2 shown by titrating GroEL ring with dMDHQSY7 reported by the quenching of F5M labeled GroES. The experiments were performed under the same condition as those in A. The bottom yellow trace was generated by also including 1 mM BeCl2 and 10 mM NaF to restrict the measurement to single turnover condition and was used as 100% SP occupation level. The summary plot of this set of experiment is shown in C as red triangles. (C) Determination of SP (dMDH in red and dRubisco in blue) encapsulation stoichiometry by the GroEL:GroES2 complex. Two different ways are used: (i) the FRET signal between GroELIAEDANS and GroESF5M using unlabeled SPs (circles) (primary data of dMDH shown in A and that of dRubisco shown in Fig.S1B) and (ii) quenching of GroESF5M by SPQSY7 upon SP encapsulation and formation of the GroEL:SP2:GroES2 complex (triangles) (primary data of dMDH shown in B and that of dRubisco in Fig. S1C).
SPs Can Be Transiently and Simultaneously Trapped in Both Rings of GroEL:GroES2 Footballs.
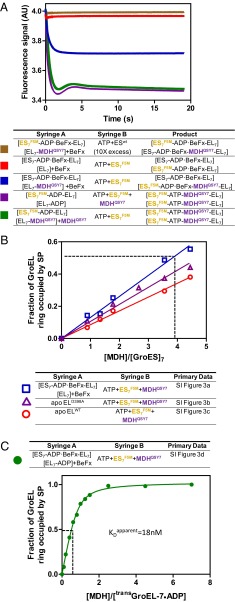
To quantify the number of SPs encapsulated per football, we labeled unfolded SP with QSY7, which quenches the FRET donor (fluorescein: R0 = 6.1 nm) but does not fluoresce itself. Changes in GroESF5M emission can then be assigned to the close proximity of SPQSY7 to GroESF5M by association with the same GroEL7 ring (Fig. 3A, red and brown, and Fig. S1A). Using SPQSY7/GroESF5M we measured the number of SPs encapsulated by GroEL in two distinct ways. First, we replaced the GroELIAEDANS:GroESF5M FRET system with GroEL:SPQSY7 and GroESF5M and followed the quenching of F5M by QSY7 (Fig. 2B). The resulting stoichiometric binding curves also break at one SP per GroEL7 (Fig. 2C), indicating that both rings of the football are occupied by SPQSY7. Second, in the absence of BeF3 multiple turnovers are permitted and the GroEL rings eventually bind both SPQSY7 and GroESF5M (Fig. 3A, green and purple) regardless of the order of addition of the reagents. In the presence of BeF3, the cis ring of the acceptor complex is “locked” by ADP-BeF3 and cisGroELwt/GroESwt remains spectroscopically silent. Then, only events on the transGroEL ring are reported. Half as much F5M quenching was seen in the presence of BeF3 as was seen in its absence (Fig. 3A, blue), indicating that both rings can be simultaneously occupied by SPQSY7.
Fig. 3.
SP catalyzed ADP/ATP exchange guarantees efficient SP encapsulation. (A) Calibration of SP encapsulation with the MDHQSY7/GroESF5M quench pair. The experimental setup is listed in the table beneath the graph and the color code is to the left of the table. The square brackets define the ligand content of each GroEL ring. Both the red and the purple traces were offset by a slight fluorescence signal value (0.025) to reveal the brown and green traces, respectively. (B) The consequences of permitting ATP to bind to GroEL before GroESF5M and SP (MDHQSY7). The experimental setup is listed in the table beneath the graph and the color code is to the left of the table. The inset (primary data in Fig. S3 and also for a description of how the plot is generated from the primary data) plots the mole fraction of encapsulated MDH against [MDH]/[GroES]7. The slope of the regression line is used to calculate the [MDH]/[GroES7] ratio required to achieve half occupancy of GroEL rings by SP, which is marked by the dashed line in the plot. (C) The green trace was generated by introducing varying amounts of MDHQSY7 to the asymmetric resting state [cisGroEL7-(ADP-BeF3)7-GroES7]–[transGroEL7-ADP7] complex in the presence of BeF3. Both MDHQSY7 and ATP (0.5 mM) were introduced simultaneously. The presence of ADP on the trans ring ensures that SP binds before ATP and GroESF5M. This SP-catalyzed ADP/ATP exchange permits efficient SP encapsulation, which is evident from the much lower [MDH]/[GroES7] required to achieve 50% occupancy of the GroEL ring. Assuming that an equilibrium between the unfolded protein and GroEL is established before encapsulation, an apparent dissociation constant of 18 nM was obtained by fitting the green dataset with the quadratic binding equation: .
High-Efficiency SP Encapsulation Requires That SP Binding Precede ATP Binding.
A mechanism for SP-catalyzed nucleotide exchange (Fig. 1A) has been proposed (17, 19), which ensures that (i) the machine only embarks on a new round of ATP hydrolysis when SP is present, (ii) the machine avoids the production of the biologically unproductive “empty-football” complexes, and (iii) that encapsulation proceeds with high efficiency. A contrary view in which ATP binding precedes the binding of SP (Fig. S2) has been proposed (20). We tested this proposal by introducing ATP+GroESF5M+SP(MDHQSY7) simultaneously to four different initiation species (summarized in Fig. 3 B and C) using otherwise identical conditions. A control in which GroEL was preincubated with a molar equivalent of MDHQSY7 (to GroEL rings) before mixing with ATP, BeF3, and GroESF5M established the maximum encapsulation. In three out of four cases where SP and ATP were introduced simultaneously to an empty GroEL ring, at least fourfold excess of [MDH] over [GroES7] is required to occupy 50% of GroEL rings by SP (Fig. 3B, dashed line). This result confirms that the mechanism in which the binding of ATP precedes that of SP (Fig. S2) leads to a biologically unproductive event, the encapsulation of nothing. Premature ATP binding and inefficient SP encapsulation can be prevented by the slow dissociation of ADP remaining from the previous cycle. When ATP and MDHQSY7 are simultaneously presented to the resting-state complex with ADP occupying the trans ring, efficient encapsulation of the MDHQSY7 is restored as evident from the much lower [MDH]/[GroES7] ratio needed to achieve half occupancy of GroEL rings by SP (Fig. 3C, dashed line), although there is a slight drop of MDH affinity compared with that of the acceptor state complex (Fig. 3C).
In normal circumstances the football complexes formed in the presence of SP are dynamic, with a lifetime of ∼1 s, after which time only a tiny fraction (<1%) of the transiently encapsulated SP will have folded to the native state (17). Under these cycling conditions it is unclear exactly where the SP folds. Nevertheless, the chaperonins function as parallel processing devices and not as alternating machines. Regardless of SP, the football complexes become indefinitely stable in the presence of ADP+BeF3, enabling the formation of diffraction-quality crystals.
Overall Structures of the Football Complexes, With or Without Encapsulated SP.
We determined the crystal structures of two GroEL:(ADP-BeF3)14:GroES2 football complexes, one devoid of SP (MT football) (Fig. 1B) and the SP football containing encapsulated Rubisco (Fig. 1B and Table S1) The crystal packing of both football complexes is almost identical. One layer of footballs pack with their sevenfold axis parallel to one another and the footballs in the next layer pack with their sevenfold axis orthogonal to the footballs in the first layer (Fig. S4). Both football complexes consist of two heptameric GroEL rings, capped by two heptameric GroES “lids.” All 14 nucleotide-binding sites on GroEL are saturated with the ATP analog ADP-BeF3.
Our biochemical data indicates that the SP football contains Rubisco in both cavities. Given the time for crystal formation it is likely that the Rubisco monomer in the cavity will have assumed a native-like, folded state. However, the encapsulated Rubisco is not visible on the electron density map and does not significantly distort the GroEL/GroES structure overall (rmsd = 0.74; Fig. S5A). Here we are dealing with an asymmetric object (Rubisco) in a container of near-sevenfold symmetry (GroEL). So the conformation and orientation of encapsulated Rubisco varies between different unit cells in the crystal. For the same reason, an SP-containing bullet complex is also devoid of electron density attributable to encapsulated SP (7).
Structural Plasticity of the Football Complexes.
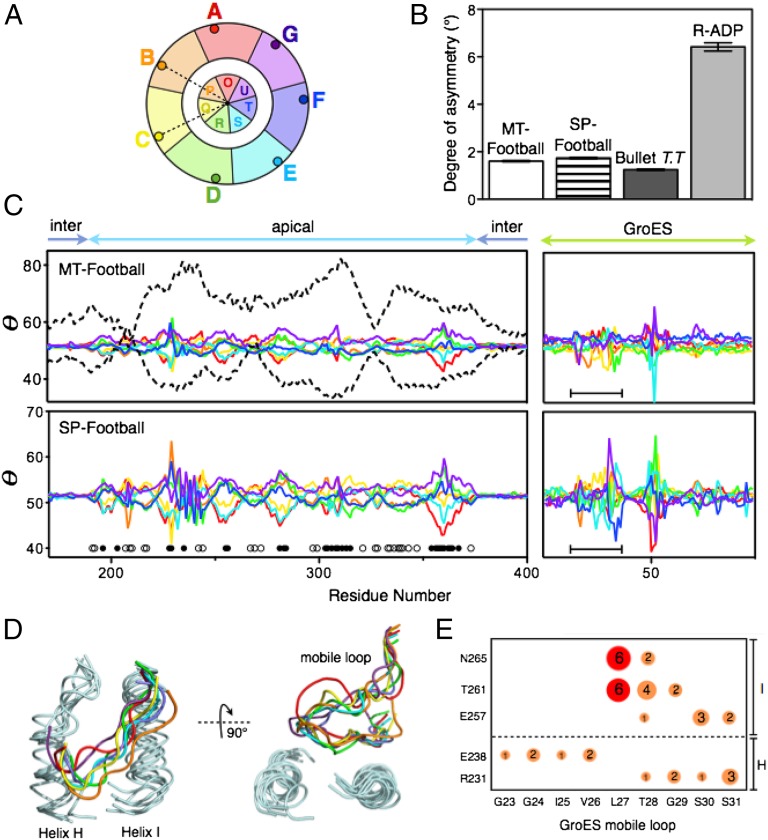
Although both MT footballs and SP footballs seem symmetric, closer inspection reveals that the apical domains of GroEL and the GroES are not truly sevenfold symmetric, but rather pseudosymmetric. The θ plots (19) (Fig. 4 A–C) show that the GroEL apical domains and GroES in both footballs deviate from perfect sevenfold symmetry by up to 10°. We attribute this asymmetry in these football complexes to their intrinsic flexibility, similar to but of a smaller magnitude than the asymmetry in the R state GroEL-ADP14 (19).
Fig. 4.
Asymmetry in the football complexes. (A) Definition of asymmetry probe θ. θ is the angle between two vectors: one from the Cα of a residue i to the sevenfold axis and another from the Cα of residue i to the sevenfold axis in the neighboring GroEL subunit. (B) A histogram showing the average deviation from perfect sevenfold symmetry (θ = 360°/7 = 51.4°) for SP footballs, MT footballs (averaged over all 14 subunits), the cis ring of Thermus thermophilus GroEL:GroES1 (averaged over seven cis subunits) (7), and the R-ADP structure of a GroELD83A/R197A mutant (averaged over seven subunits) devoid of the salt bridges that break during the T-to-R allosteric transition in the normal chaperonin cycle (19). (C) Quantitative θ plots showing that the apical domains and GroES mobile loops (black bars) of both the MT football and the SP football deviate from sevenfold symmetry. Subunit colors are as the same as in A. Also shown (dashed lines) are θ plots of the two most asymmetric subunits of the R-ADP structure. In both football complexes, greater deviations from symmetry occur in solvent-exposed residues of the central cavity (black circles) than in exterior solvent-exposed residues (white circles) (Fig. S6). (D) GroEL–GroES interfaces in the football complexes, showing the GroEL–GroES interactions are heterogeneous. (E) Hydrogen bonds that stabilize GroEL–GroES interfaces are represented by circles. Area of circles represents the relative occurrence of hydrogen bonds. Two hydrogen bonds present in the bullet complex are colored in red.
However, the asymmetry in the football complexes is not identical to the asymmetry we observed in the R state. First, the overall degree of asymmetry decreases by 70% in the football complexes compared with the R state (Fig. 4 B and C). Second, rather than being distributed throughout the entire apical domain, the asymmetry in the football complexes is restricted to several regions of the apical domain. This suggests that when GroES binds to the R state GroEL it gathers GroEL’s flexible apical domains together, making GroEL more rigid. Finally, greater asymmetry occurs at the solvent-exposed residues facing the inside of the cavity, indicating that the encapsulated SP is surrounded with a plastic chamber rather than a rigid cage (Fig. 4C and Fig. S6). Such plasticity might allow GroEL to closely interact with SPs of different sizes and shapes during encapsulation (21). In this regard we note that the average b-factor of apical domains in the SP football is lower than that of the MT football (147 Å2 vs. 225 Å2), which could be caused by SP–GroEL interactions (Fig. S5B).
None of the apical domains in the football complexes have identical conformations and the interaction at the GroEL/GroES interfaces was heterogeneous. Indeed, no two GroES “mobile loops” have the same conformation, and each GroEL/GroES interface is maintained by a unique set of hydrogen bonds (Fig. 4 D and E). This is quite different from the GroEL/GroES interface in the asymmetric bullet complex (PDB ID code 1AON), where, owing to imposed symmetry, all seven GroEL/GroES interfaces consist of the same hydrogen bonds between residues from helix I and the mobile loop of GroES (Fig. 4E, red).
The nonspecific interactions between GroEL and GroES mobile loops could be important for GroES binding. Before the binding of GroES, both the GroEL apical domains, including helices H and I, and the mobile loops of GroES are extremely flexible (19, 22, 23). Having a large number of nonspecific interactions in the GroEL/GroES interface would permit the engagement of these flexible partners with fewer entropic costs that a highly specific interaction would entail. For example, T28 on the mobile loop interacts through hydrogen bonds with up to four residues in helices H and I (Fig. 4E), which increases the probability of GroES capture.
Conformational Changes at the Inter-Ring Interface.
Although the football complex encapsulates two SP molecules simultaneously, the two GroEL/GroES cavities do not act independently. Biochemical evidence has shown that the communication between two GroEL rings is crucial for GroES release (24). When such inter-ring communication is disrupted, as it is in the single-ringed version SR1, GroEL fails to release GroES, SP, and ADP, resulting in a “dead-end” complex (25).
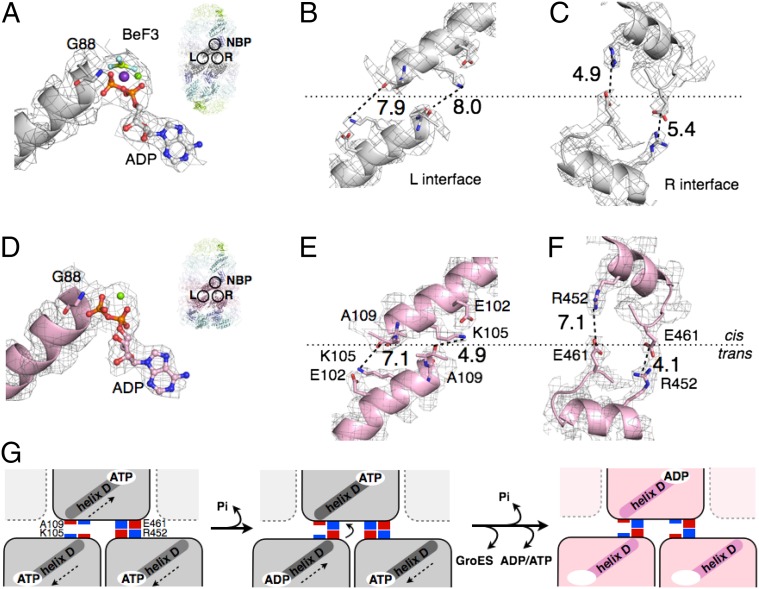
The crystal structure of the football complex at 3.7-Å resolution allows us to analyze the inter-ring communication during chaperonin’s natural catalytic cycle. The two GroEL rings in the football complex communicate through the same two inter-ring interfacial sites, L and R, as previously reported (Fig. 5A, Inset) (6).
Fig. 5.
Switching of electrostatic interactions at the inter-ring interface. (A) Interaction between ADP-BeF3 and the N terminus of helix D in the football complex. The relative position of inter-ring interfaces L, R, and the nucleotide-binding pocket (NBP) is shown in the inset. (B and C) Close views of the inter-ring interfaces L and R in the football complex. The twofold axis of symmetry is shown as a long dashed line. (D) Interaction between ADP and the N termini of helix D in the bullet complex. The relative position of inter-ring interfaces L, R, and the NBP is shown in the inset. (E and F) The same as B and C, except showing the inter-ring interface L and R in the bullet complex. (G) A structure-based mechanism for sensing ATP hydrolysis. The dashed arrows indicate movement of helix D in response to ATP binding and hydrolysis.
The L interface involves interactions between helix D of one subunit with the same helix in the opposite ring. The axes of the two helices D are antiparallel to one another. The C termini of each helix (A109) are slightly offset from one another, across the twofold axis of symmetry (Fig. 5 B and E). The R interface involves interactions between helix P of one subunit and the same helix in the opposite ring. The axes of the two helices P are nearly antiparallel to one another and also to the twofold axis of symmetry (Fig. 5 C and F).
When GroEL/GroES switches from the football complex to the bullet complex, the total contact surface area of L and R interfaces increases only slightly (from 2,232 Å2 to 2,464 Å2). However, close inspection shows the relative strength of the two interfaces has changed. The L interface expands by ∼50% (from 764 Å2 to 1,132 Å2), whereas the R interface shrinks by ∼25% (from 1,472 Å2 to 1,132 Å2). This change in the inter-ring interface is caused by a reduction in the radius (up to 6 Å) of the equatorial plate of the trans ring (Fig. S7A) plus a slight rotation (up to 8°) of the trans GroEL ring relative to the cis GroEL ring (Fig. S7B).
We further analyzed the change in electrostatic interactions at both L and R interfaces. Accompanying the release of one GroES, the two interacting D helices at the L interface move closer and twist to established one electrostatic interaction between ε-amino group of K105 of the cis ring and helix dipole-induced charge on the carbonyl oxygen of A109 of the trans ring. (Fig. 5 B and E). At the R interface, the two helices P move apart and twist so one of the two salt bridges between E461 of the trans ring and R452 of the cis ring breaks (Fig. 5 C and F).
Structural Basis for Inter-Ring Communication.
During the chaperonin’s natural catalytic cycle, the dissociation of GroES from the football complex requires the hydrolysis of ATP and the development of nucleotide asymmetry (i.e., the difference in the number of ATPs hydrolyzed between two GroEL rings) (17). We propose that helix D senses and transmits the signal of ATP hydrolysis and ATP asymmetry by exploiting the helix dipole that is positively charged at the N-terminal G88 and negatively charged at the C-terminal A109 (Fig. 5 A and D).
Before the football complex hydrolyzes ATP and releases GroES, the N terminus of helix D (G88) in the two GroEL subunits from opposite rings both form electrostatic interactions with the ATP γ-phosphate (Fig. 5 A and G). The interaction between γ-phosphate and the N terminus of the two helices D draws the two helices apart from one another (arrows in Fig. 5G). Once ATP in one subunit is hydrolyzed and the γ-phosphate is released, helix D in that subunit moves closer to helix D in the other ring and a cross-ring electrostatic interaction forms between A109 and K105 (Fig. 5G). As more ATP is hydrolyzed and a critical number of cross-ring A109–K105 interactions is reached, one or the other GroES departs (16).
The mechanism of inter-ring communication in the football complex is quite different from a previously proposed model that is based on the comparison between the interfaces of apo-GroEL and the bullet complex (8). However, our current understanding of the chaperonin cycle (Fig. 1A) assigns no role whatsoever to apo-GroEL. Exactly how the events at the equatorial plate are transmitted to the apical domains leading to the dissociation of GroES is yet to be determined, however.
Methods
Calibrated FRET-based spectroscopic methods were developed to measure the number of both GroES and SP molecules bound to the symmetric GroEL:GroES2 football complexes (5, 17, 26). SPQSY7 was shown to quench GroESF5M only when both are bound to the same GroEL ring (SI Methods). Stable football particles were obtained by using BeF3 to arrest the natural chaperonin cycle at the football stage, permitting the formation of crystals of MT and SP footballs that contained a molecule of encapsulated Rubisco in each central chamber (SI Methods). The structures of the MT and SP footballs were solved by segmented molecular replacement using AutoMR, Refine, and Coot in PHENIX suites (27), using the cis-GroEL ring (PDB ID code 1AON) as the search model. Hydrogen bonds, salt bridges and interface surface areas are analyzed using PISA (28).
Note Added in Proof.
The results of contemporaneous studies of chaperonin football complexes from E. coli and from human mitochondria have recently been deposited in the PDB under 3WVL and 4PJ1, respectively.
Supplementary Material
Acknowledgments
We thank Drs. Axel Brunger, Wah Chiu, Zbigniew Dauter, and Zhechun Zhang for helpful suggestions. This work is based on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which is supported by National Center for Research Resources Grant 5P41RR015301-10 and National Institute of General Medical Sciences Grant 8 P41 GM103403-10 from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy Office of Science by the Argonne National Laboratory, was supported by US Department of Energy Contract DE-AC02- 06CH11357. X.F. was partly supported by an Anne G. Wylie dissertation fellowship. X.Y. was partly supported by a William Bailey fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4PKO and 4PKN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412922111/-/DCSupplemental.
References
- 1.Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu Rev Biophys Biomol Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 2.Horovitz A, Willison KR. Allosteric regulation of chaperonins. Curr Opin Struct Biol. 2005;15(6):646–651. doi: 10.1016/j.sbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Lin Z, Rye HS. GroEL-mediated protein folding: Making the impossible, possible. Crit Rev Biochem Mol Biol. 2006;41(4):211–239. doi: 10.1080/10409230600760382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sameshima T, Iizuka R, Ueno T, Funatsu T. Denatured proteins facilitate the formation of the football-shaped GroEL-(GroES)2 complex. Biochem J. 2010;427(2):247–254. doi: 10.1042/BJ20091845. [DOI] [PubMed] [Google Scholar]
- 5.Ye X, Lorimer GH. Substrate protein switches GroE chaperonins from asymmetric to symmetric cycling by catalyzing nucleotide exchange. Proc Natl Acad Sci USA. 2013;110(46):E4289–E4297. doi: 10.1073/pnas.1317702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388(6644):741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 7.Shimamura T, et al. Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity. Structure. 2004;12(8):1471–1480. doi: 10.1016/j.str.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Horwich AL. Protein folding in the cell: An inside story. Nat Med. 2011;17(10):1211–1216. doi: 10.1038/nm.2468. [DOI] [PubMed] [Google Scholar]
- 9.Hayer-Hartl MK, Ewalt KL, Hartl FU. On the role of symmetrical and asymmetrical chaperonin complexes in assisted protein folding. Biol Chem. 1999;380(5):531–540. doi: 10.1515/BC.1999.068. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt M, et al. Symmetric complexes of GroE chaperonins as part of the functional cycle. Science. 1994;265(5172):656–659. doi: 10.1126/science.7913554. [DOI] [PubMed] [Google Scholar]
- 11.Azem A, Diamant S, Kessel M, Weiss C, Goloubinoff P. The protein-folding activity of chaperonins correlates with the symmetric GroEL14(GroES7)2 heterooligomer. Proc Natl Acad Sci USA. 1995;92(26):12021–12025. doi: 10.1073/pnas.92.26.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llorca O, Marco S, Carrascosa JL, Valpuesta JM. Symmetric GroEL-GroES complexes can contain substrate simultaneously in both GroEL rings. FEBS Lett. 1997;405(2):195–199. doi: 10.1016/s0014-5793(97)00186-5. [DOI] [PubMed] [Google Scholar]
- 13.Sparrer H, Rutkat K, Buchner J. Catalysis of protein folding by symmetric chaperone complexes. Proc Natl Acad Sci USA. 1997;94(4):1096–1100. doi: 10.1073/pnas.94.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taguchi H, Tsukuda K, Motojima F, Koike-Takeshita A, Yoshida M. BeF(x) stops the chaperonin cycle of GroEL-GroES and generates a complex with double folding chambers. J Biol Chem. 2004;279(44):45737–45743. doi: 10.1074/jbc.M406795200. [DOI] [PubMed] [Google Scholar]
- 15.Sameshima T, et al. Football- and bullet-shaped GroEL-GroES complexes coexist during the reaction cycle. J Biol Chem. 2008;283(35):23765–23773. doi: 10.1074/jbc.M802541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takei Y, Iizuka R, Ueno T, Funatsu T. Single-molecule observation of protein folding in symmetric GroEL-(GroES)2 complexes. J Biol Chem. 2012;287(49):41118–41125. doi: 10.1074/jbc.M112.398628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang D, Ye X, Lorimer GH. Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc Natl Acad Sci USA. 2013;110(46):E4298–E4305. doi: 10.1073/pnas.1318862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corrales FJ, Fersht AR. Kinetic significance of GroEL14.(GroES7)2 complexes in molecular chaperone activity. Fold Des. 1996;1(4):265–273. doi: 10.1016/s1359-0278(96)00040-5. [DOI] [PubMed] [Google Scholar]
- 19.Fei X, Yang D, LaRonde-LeBlanc N, Lorimer GH. Crystal structure of a GroEL-ADP complex in the relaxed allosteric state at 2.7 Å resolution. Proc Natl Acad Sci USA. 2013;110(32):E2958–E2966. doi: 10.1073/pnas.1311996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi NK, Fenton WA, Horwich AL. GroEL/GroES cycling: ATP binds to an open ring before substrate protein favoring protein binding and production of the native state. Proc Natl Acad Sci USA. 2009;106(48):20264–20269. doi: 10.1073/pnas.0911556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A. Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat Struct Mol Biol. 2005;12(3):233–237. doi: 10.1038/nsmb901. [DOI] [PubMed] [Google Scholar]
- 22.Nojima T, Ikegami T, Taguchi H, Yoshida M. Flexibility of GroES mobile loop is required for efficient chaperonin function. J Mol Biol. 2012;422(2):291–299. doi: 10.1016/j.jmb.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Hunt JF, Weaver AJ, Landry SJ, Gierasch L, Deisenhofer J. The crystal structure of the GroES co-chaperonin at 2.8 A resolution. Nature. 1996;379(6560):37–45. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- 24.Todd MJ, Viitanen PV, Lorimer GH. Dynamics of the chaperonin ATPase cycle: Implications for facilitated protein folding. Science. 1994;265(5172):659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 25.Horwich AL, Burston SG, Rye HS, Weissman JS, Fenton WA. Construction of single-ring and two-ring hybrid versions of bacterial chaperonin GroEL. Methods Enzymol. 1998;290:141–146. doi: 10.1016/s0076-6879(98)90013-1. [DOI] [PubMed] [Google Scholar]
- 26.Grason JP, Gresham JS, Widjaja L, Wehri SC, Lorimer GH. Setting the chaperonin timer: The effects of K+ and substrate protein on ATP hydrolysis. Proc Natl Acad Sci USA. 2008;105(45):17334–17338. doi: 10.1073/pnas.0807429105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.