Abstract
The biosynthesis of methionine in bacteria requires the mobilization of sulfur from Cys by the formation and degradation of cystathionine. Cystathionine β-lyase, encoded by metC in bacteria and STR3 in Schizosaccharomyces pombe, catalyzes the breakdown of cystathionine to homocysteine, the penultimate step in methionine biosynthesis. This enzyme has been suggested to be the target for pyridinamine antimicrobial agents. We have demonstrated, by using purified enzymes from bacteria and yeast, that cystathionine β-lyase is not the likely target of these agents. Nonetheless, an insertional inactivation of metC in Salmonella enterica serovar Typhimurium resulted in the attenuation of virulence in a mouse model of systemic infection. This result confirms a previous chemical validation of the Met biosynthetic pathway as a target for the development of antibacterial agents and demonstrates that cystathionine β-lyase is important for bacterial virulence.
Microorganisms require sulfur for the production of the amino acids Cys and Met, which are essential for protein biosynthesis. Furthermore, Met, in the form of S-adenosylmethionine, is the methyl donor for a number of essential biochemical reactions. The biosynthesis of Met is therefore of vital importance to microbial growth. This has been validated by the fact that several natural products, including 2-amino-5-hydroxy-4-oxopentanoic acid (8), azoxybacilin (1), and rhizocticin (10), target important enzymes for Met biosynthesis and have antimicrobial properties and that the disruption of genes such as STR3, which is important in the fungal sulfur assimilation pathway, results in avirulence in a Cryptococcus neoformans infection model (17).
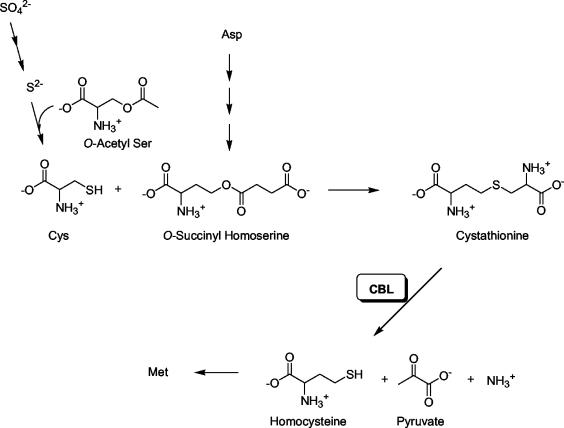
In Escherichia coli and Salmonella, sulfur is assimilated by the sulfate pathway, by which sulfate (SO42−) is reduced to sulfide (S2−) and then incorporated into Cys by O-acetylserine thiolyase (9). Cys then serves as a sulfur donor in the biosynthesis of Met through the formation of cystathionine, followed by its degradation to pyruvate, ammonia, and homocysteine, the penultimate step prior to Met production (Fig. 1). Homocysteine production from cystathionine is catalyzed by the enzyme cystathionine β-lyase (CBL). The three-dimensional structure of E. coli CBL has been determined to a high resolution, confirming the presence of an active-site pyridoxal phosphate cofactor (3).
FIG. 1.
Role of CBL in bacterial Met biosynthesis.
Bacteria can bypass the requirement for cystathionine biosynthesis by directly fixing sulfur with O-succinylhomoserine, generating homocysteine, but this is physiologically irrelevant due to the high concentrations of sulfide required, making cystathionine a critical metabolite (9). In contrast, direct sulfide incorporation into homocysteine is significant in Schizosaccharomyces pombe and other fungi (16). Paradoxically, CBL has been implicated as the target for the pyridinamine class of antifungal agents, such as pyrimethanil, in the fungus Botrytis cinerea, an important plant pathogen (5). Given this reported precedent for the targeting of CBL in a pathogenic microbe and the fact that the cystathionine pathway is prevalent in bacteria, we hypothesized that CBL may be important to bacterial virulence. We have probed the importance of CBL as a potential target for these antimicrobial agents and demonstrate that while the E. coli and yeast CBLs are not a target for pyrimethanil and chemically related pyridinamines, CBL is important for the virulence of Salmonella and is therefore a potential target for antibacterial drug development.
MATERIALS AND METHODS
Overexpression and purification of CBL from E. coli and yeast.
The metC gene was amplified from E. coli genomic DNA with primers 5′-GGAATTCCATATGGCGGACAAAAAGCTTGATACTCAACTGG and 5′-GGAATTCGGATCCGTTATACAATTCGCGCAAAACCGGCGTCC (reading frames are indicated in italics and restriction sites are underlined). The DNA fragment was digested with NdeI and BamHI and ligated with plasmid pET28a(+) digested with the same enzymes. The new plasmid, pET28-CBL, was used to transform E. coli BL21 Star (DE3).
An overnight culture of E. coli BL21(DE3)(pET28-CBL) was grown from a single colony in Luria-Bertani (LB) broth supplemented with 50 μg of kanamycin/ml. One liter of LB broth Lennox containing 50 μg of kanamycin/ml was inoculated with 10 ml of the overnight culture and grown at 37°C with shaking at 250 rpm to an optical density at 600 nm of 0.6. Sterile isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. The cells were grown for an additional 4 h at 37°C with shaking at 250 rpm and then harvested by centrifugation at 7,500 × g for 15 min. The cell pellet was washed with cold 0.85% NaCl and frozen at −20°C.
The frozen pellet was resuspended in 7 ml of lysis buffer (50 mM HEPES [pH 7.5], 500 mM NaCl, 20 mM imidazole, 1 mM EDTA, 0.1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). Approximately 1 mg each of RNase and DNase was added, and the cells were lysed by three passages through a French pressure cell. Cell debris was removed by centrifugation at 48,000 × g for 20 min.
All purification steps were performed at 4°C. Approximately 6 ml of Ni-nitrilotriacetic acid-agarose (Qiagen) (approximately 3 ml of resin) was added to the supernatant and stirred for 1 h. The mixture was poured into a column fitting, washed with buffer A (50 mM HEPES [pH 7.5], 500 mM NaCl, 20 mM imidazole), and then eluted with a step-wise gradient of buffer A and buffer B (50 mM HEPES [pH 7.5], 500 mM NaCl, 250 mM imidazole). Fractions containing CBL were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and enzyme activity assays, pooled, and dialyzed in 25 mM HEPES (pH 7.5)-2 mM EDTA.
The protein concentration was determined by the Bradford method. Approximately 65 mg of purified CBL was obtained from 1 liter of culture. The purified enzyme was stored at −20°C in 5% glycerol.
Similarly, the STR3 gene, encoding CBL in the yeast S. pombe, was amplified with primers 5′-GTCTCTAGAGCTAGCATGCCCAGCGATTGTAAATATTCTGTC and 5′-CGCGGATCCAAGCTTCAATTCTGTTTAAAATTTGCTTGAGCA (reading frames are indicated in italics and restriction sites are underlined). The DNA fragment was digested with NheI and HindIII and ligated with plasmid pET28a(+) digested with the same enzymes. The new plasmid, pET28-pomCBL, was used to transform E. coli BL21(DE3), and the enzyme was expressed and purified as described above, with the exception that the induction of expression was performed at 25°C for 10 h.
Enzyme assay.
CBL activity was measured by monitoring the formation of homocysteine at 412 nm, using 5,5′-dithio-bis-(2-nitrobenzoic acid) as a detector of free thiol in a SpectraMax plate reader from Molecular Devices. The assay mixture contained 50 mM TAPS (pH 8.5), 1 mM 5,5′-dithio-bis-(2-nitrobenzoic acid), 25 nM CBL, and 100 μM l-(+)-cystathionine. The total reaction volume was 100 μl. Potential inhibitors dissolved in dimethyl sulfoxide were added to the mixture, to a final concentration of 5% (vol/vol). Reactions were set up to contain all components except cystathionine, incubated for 10 min at room temperature, and initiated by the addition of cystathionine.
Construction of CBL null strain of Salmonella enterica serovar Typhimurium.
The sequence of the metC gene in the S. enterica serovar Typhimurium LT2 genome (12) was identified, and a region flanking roughly 300 bp 5′ and 3′ of the gene was amplified from genomic DNA isolated from S. enterica serovar Typhimurium (ATCC 14028) by using the oligonucleotide primers 5′-CACGACAATATCGGCGTGCTG and 5′-CATCCTGGGCGATCAACGCGCC. This fragment was cloned into pCR4Blunt-TOPO (Invitrogen) and verified by sequencing. The construct was digested with PmeI and NotI to recover the metC fragment, which was inserted into the vector pKO3 (11) digested with SmaI and NotI, generating pKO3(CBL). pKO3 includes a chloramphenicol resistance cassette along with a temperature-sensitive origin of replication and the Bacillus subtilis sacB gene, which is lethal when E. coli is grown on sucrose-containing media (11). The gentamicin resistance cassette aacC1 was excised from plasmid pUCGm (14) by digestion with PstI and was inserted into the metC gene at position 355 of metC, generating pKO3(CBL-Gm).
This plasmid was introduced into S. enterica serovar Typhimurium (ATCC 14028) by electroporation, and recombinants were selected on LB agar supplemented with 25 μg of gentamicin/ml and 5% sucrose at the nonpermissive temperature of 42°C. Colonies were subsequently screened on LB agar supplemented with 50 μg of gentamicin/ml at 30°C in the presence and absence of chloramphenicol. Chloramphenicol-sensitive, gentamicin-resistant colonies were analyzed by Southern blotting to confirm the insertional inactivation of the metC gene of S. enterica serovar Typhimurium by the aacC1 gene, generating the mutant S. enterica serovar Typhimurium metC::aacC1.
To confirm that the observed phenotype of Met auxotrophy was the result of a disruption of the metC gene, we reintroduced the gene into the S. enterica serovar Typhimurium metC::aacC1 mutant via the same procedure that we used to introduce the aacC1 gene in the first place. We therefore introduced pKO3(CBL) into S. enterica serovar Typhimurium metC::aacC1, selected for plasmid integration at 42°C on LB agar supplemented with chloramphenicol, and then subsequently plated cells on LB agar containing 5% sucrose to select for the loss of plasmid sequences. Confirmation of the reintegration of the metC gene was done by PCR and Southern blot analysis.
We also prepared a complementation plasmid by introducing metC downstream of the arabinose-inducible promoter of pBAD18 (6) and introduced this plasmid into S. enterica serovar Typhimurium metC::aacC1 in trans for complementation of the metC disruption.
Mouse infection with S. enterica serovar Typhimurium.
Disease in C57BL/6J mice after infection with S. enterica serovar Typhimurium and S. enterica serovar Typhimurium metC::aacC1 was evaluated by survival analysis and the monitoring of bacterial loads in reticuloendothelial organs (spleens and livers). Mice of 12 weeks of age were challenged with 103 CFU via intravenous injection in the tail vein or with 105 CFU via the oral route as described previously (2, 15). To determine the growth rates of the bacteria within reticuloendothelial organs, we sacrificed the mice at days 3 and 7 postinoculation. The spleens and livers were aseptically removed and prepared for the determination of infection kinetics. Serial dilutions of each homogenate were plated on Trypticase soy agar to enumerate the numbers of CFU within each organ.
Statistical analysis.
We used a parametric survival analysis to compare the effects of groups (the wild type versus S. enterica serovar Typhimurium metC::aacC1) and the route of administration (intravenous versus oral) on the survival of mice, assuming that the time to death followed a Weibull distribution. We used the statistical package R to perform the analysis (7).
RESULTS AND DISCUSSION
CBL is not inhibited by pyridinamine antimicrobial agents.
The pyridinamine pyrimethanil and the structurally related compounds cyprodinil and mepanipyrim have antifungal and modest antibacterial activities (Table 1). The purification of CBL from E. coli and the yeast S. pombe (34% similar to the E. coli enzyme) permitted a direct evaluation of the suggestion that these antimicrobial agents target this enzyme. A steady-state kinetic analysis of purified recombinant enzymes gave the following parameters: for the E. coli enzyme, Km = 260 μM and kcat = 20 s−1; for the S. pombe enzyme, Km = 380 μM and kcat = 1.5 s−1. Neither enzyme showed sensitivity to pyrimethanil, cyprodinil, or mepanipyrim at the limits of compound solubility in our assay (0.25 mM). This demonstrates that CBL is not a direct target for these antimicrobial agents in these organisms.
TABLE 1.
MIC values of pyridinamine antimicrobial agents for bacterial and fungal species
| Compound | MIC (μg/ml) for indicated species or strain
|
|||||
|---|---|---|---|---|---|---|
| Candida albicans ATTC 90028 | Saccharomyces cerevisiae YC128 | E. coli ATCC 25922 | E. coli MC1061 | Staphylococcus aureus ATCC 29213 | S. enterica | |
| Cyprodinil | 64 | 16 | >128 | 64 | 128 | >128 |
| Mepanipyrim | 128 | 64 | >128 | >128 | >128 | >128 |
| Pyrimethanil | 128 | 128 | >128 | 128 | >128 | >128 |
Characterization of S. enterica serovar Typhimurium metC::aacC1.
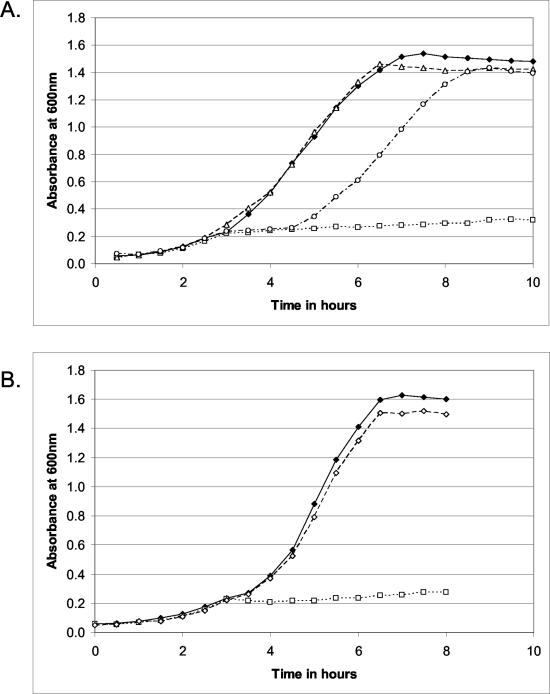
Despite the biochemical evidence that CBL is not the target of the pyridinamine antimicrobial agents, the importance of CBL in sulfur-containing amino acid metabolism and the low concentrations of these amino acids in serum (23 μM for Met and 74 μM for Cys [4]) suggested to us that CBL may play an important role in bacterial virulence by maintaining adequate concentrations of sulfur-containing amino acids, which are necessary for protein synthesis and the formation of S-adenosylmethionine. We therefore insertionally inactivated the metC gene of S. enterica serovar Typhimurium (94% similar to the E. coli enzyme) with the aacC1 gentamicin resistance cassette. As expected, the metC::aacC1 mutant was a Met auxotroph, as it was unable to grow in minimal medium without supplemental Met (Fig. 2A). The reintroduction of the metC gene into this mutant, followed by selection for reintegration of the gene and the associated gentamicin sensitivity, rescued the wild-type ability to grow in minimal medium (Fig. 2B). Furthermore, complementation of the disruption by the introduction of metC in plasmid pBAD18 also reversed the Met auxotrophy (not shown). These results support the hypothesis that the phenotype was solely the result of metC inactivation.
FIG. 2.
Insertional inactivation of S. enterica serovar Typhimurium metC results in Met auxotrophy. Studies were conducted in M9 minimal liquid medium. (A) Results of insertional inactivation of metC with aacC1. ♦, wild-type S. enterica serovar Typhimurium; □, metC::aacC1 mutant; ▵, metC::aacC1 mutant plus 0.1 mM Met; ○, metC::aacC1 mutant plus 0.1 mM Met for 4.5 h. (B) Reintroduction of metC into the metC::aacC1 mutant background rescues wild-type phenotype. ♦, wild-type S. enterica serovar Typhimurium; □, metC::aacC1 mutant; ⋄, reintegration of metC into the metC::aacC1 mutant.
Experimental infection with S. enterica serovar Typhimurium.
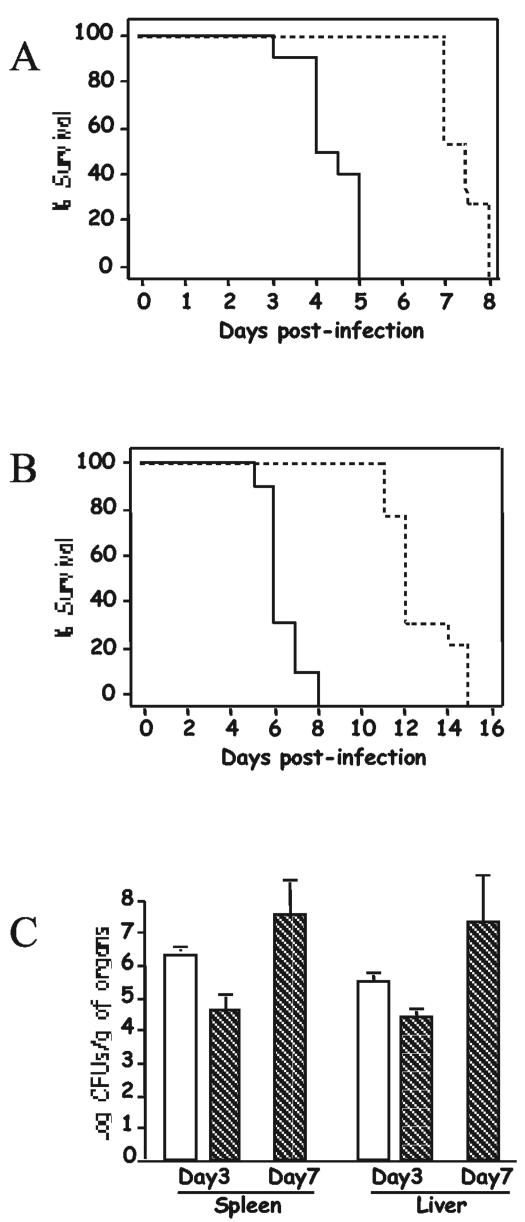
Initially, we examined the survival of C57BL/6J mice after infection with the virulent S. enterica serovar Typhimurium strain ATCC 14028 (wild type) or the isogenic metC::aacC1 mutant by using two routes of infection (103 CFU given intravenously or 105 CFU given orally). Postinfection survival was monitored for a 2-week period. Mice infected with the wild-type Salmonella strain died earlier than mice infected with the mutant strain (Fig. 3A and B). This observed difference in survival was statistically significant for both routes of infection: χ2 = 42.5 with 1 degree of freedom (P = 7.2 × 10−11) for intravenous inoculation and χ2 = 27.5 with 1 degree of freedom (P = 1.6 × 10−7) for oral administration. As reported previously, the route of administration of the bacteria had little impact on disease development in mice, with the exception of a delay in the expression of the phenotype, as was seen for the oral administration of Salmonella (Fig. 3). To determine if the increase survival times of mice infected with the S. enterica serovar Typhimurium metC::aacC1 mutant were due to reduced microbial burdens or delayed growth rates, we measured the bacterial load within the spleens and livers of mice infected with wild-type Salmonella (day 3) and with the isogenic metC::aacC1 mutant (day 3 or 7) (Fig. 3C). Mice infected with wild-type Salmonella had 50 times more bacteria in their spleens than mice infected with the metC::aacC1 mutant at day 3 (P = 0.00029). At day 7, the bacterial loads in the spleens of mice infected with the Salmonella mutant reached values (7.6 ± 1.1 log10 CFU/g of spleen) equivalent to those observed for mice infected with wild-type Salmonella the day prior to death (2). Similar results were observed for the liver (Fig. 3C). These results suggest that the disruption of metC in Salmonella has an impact on the virulence of the bacteria, as seen by the delayed onset of disease in the susceptible host. This may reflect diminished bacterial growth of the metC mutant or this mutant may be more effectively killed by the host.
FIG. 3.
Survival analysis and bacterial loads of mice infected with wild-type S. enterica serovar Typhimurium and an isogenic metC::aacC1 mutant. C57BL/6J mice were infected with wild-type S. enterica serovar Typhimurium intravenously (103 CFU) (A) or orally (105 CFU) (B) (five males and five females for intravenous infections and five males and four females for oral infections) or with the metC::aacC1 mutant strain (five males and six females for intravenous infections and four males and five females for oral infections). Survival curves are representative of mice infected with the wild type (solid line) and the metC::aacC1 mutant (dashed line). The survival analysis was repeated two times, with similar results. (C) C57BL/6J mice were infected with 103 CFU of wild-type S. enterica serovar Typhimurium (white bars) or the metC::aacC1 mutant strain (hatched bars). Each bar represents the data for groups of four mice (two males and two females). Results are expressed as means + 1 standard deviation.
Conclusions.
The proposal that CBL was the target of the pyridinamine antifungal agents prompted our investigation of the importance of CBL as a potential antibacterial target. While we demonstrated biochemically that these agents do not inhibit bacterial or fungal CBL directly, we suspected that an attenuation of the ability to biosynthesize sulfur-containing amino acids would impact an organism's ability to live in an environment such as serum, where concentrations of these key amino acids are relatively low. This would be consistent with our knowledge that the blockage of early steps in Met biosynthesis, such as aspartate semialdehyde dehydrogenase, results in a loss of virulence in Salmonella (13). We therefore constructed an S. enterica serovar Typhimurium metC::aacC1 mutant and demonstrated that this organism was less virulent than the wild-type bacterium in a mouse model of bacteremia. Therefore, the biosynthesis of sulfur-containing amino acids represents a potential new target for antibacterial agents, supporting previous chemical and genetic evidence for targeting this pathway in fungi.
Acknowledgments
We thank Rosalie Wilkinson for technical assistance.
This work was supported by the Canadian Bacterial Disease Network Centre of Excellence. D.M. is supported by a salary award of the Canadian Institutes of Health Research (Scientist) and by a Dawson McGill Professorship. G.D.W. is supported by a Canada Research Chair in Antibiotic Biochemistry.
Editor: A. D. O'Brien
REFERENCES
- 1.Aoki, Y., M. Yamamoto, S. M. Hosseini-Mazinani, N. Koshikawa, K. Sugimoto, and M. Arisawa. 1996. Antifungal azoxybacilin exhibits activity by inhibiting gene expression of sulfite reductase. Antimicrob. Agents Chemother. 40:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bihl, F., L. Salez, M. Beaubier, D. Torres, L. Larivière, L. Laroche, A. Benedetto, D. Martel, J.-M. Lapointe, B. Ryffel, and D. Malo. 2003. Over-expression of Toll-like receptor 4 (Tlr4) amplifies the host response to LPS and provides a survival advantage in transgenic mice. J. Immunol. 170:6141-6150. [DOI] [PubMed] [Google Scholar]
- 3.Clausen, T., R. Huber, B. Laber, H. D. Pohlenz, and A. Messerschmidt. 1996. Crystal structure of the pyridoxal-5′-phosphate dependent cystathionine beta-lyase from Escherichia coli at 1.83 Å. J. Mol. Biol. 262:202-224. [DOI] [PubMed] [Google Scholar]
- 4.Fasman, G. D. 1976. Handbook of biochemistry and molecular biology, 3rd ed. CRC Press, Cleveland, Ohio.
- 5.Fritz, R., C. Lanen, V. Colas, and P. Leroux. 1997. Inhibition of methionine biosynthesis in Botrytis cinerea by the anilinopyrimidine fungicide pyrimethanil. Pestic. Sci. 49:40-46. [Google Scholar]
- 6.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics. J. Comput. Graphic. Stat. 5:299-314. [Google Scholar]
- 8.Jacques, S. L., A. Mirza, L. J. Ejim, K. Koteva, D. W. Hughes, K. Green, R. Kinach, J. F. Honek, H. K. Lai, A. M. Berghuis, and G. D. Wright. 2003. Enzyme assisted suicide: molecular basis for the antifungal activity of 5-hydroxy-4-oxonorvaline by potent inhibition of homoserine dehydrogenase. Chem. Biol. 10:989-995. [DOI] [PubMed] [Google Scholar]
- 9.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, E. B. Low, B. Magasanik, W. S. Reznikoff, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 10.Kugler, M., W. Loeffler, C. Rapp, A. Kern, and G. Jung. 1990. Rhizocticin A, an antifungal phosphono-oligopeptide of Bacillus subtilis ATCC 6633: biological properties. Arch. Microbiol. 153:276-281. [DOI] [PubMed] [Google Scholar]
- 11.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama, K., S. Kelly, and R. Curtiss. 1988. Construction of an Asd+ expression-cloning vector: stable maintenance and high expression of cloned genes in a Salmonella vaccine strain. Bio/Technology 6:693-697. [Google Scholar]
- 14.Schweizer, H. D. 1993. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 15.Sebastiani, G., L. Olien, S. Gauthier, E. Skamene, K. Morgan, P. Gros, and D. Malo. 1998. Mapping of genetic modulators of natural resistance to infection with Salmonella typhimurium in wild-derived mice. Genomics 47:180. [DOI] [PubMed] [Google Scholar]
- 16.Yamagata, S. 1989. Roles of O-acetyl-l-homoserine sulfhydrylases in micro-organisms. Biochimie 71:1125-1143. [DOI] [PubMed] [Google Scholar]
- 17.Yang, Z., R. C. Pascon, A. Alspaugh, G. M. Cox, and J. H. McCusker. 2002. Molecular and genetic analysis of the Cryptococcus neoformans MET3 gene and a met3 mutant. Microbiology 148:2617-2625. [DOI] [PubMed] [Google Scholar]