Abstract
Pasteurella multocida is the causative agent of fowl cholera in birds. In a previous study using signature-tagged mutagenesis, we identified a mutant, AL251, which was attenuated for virulence in mice and in the natural chicken host. Sequence analysis indicated that AL251 had an insertional inactivation of the gene waaQPM, encoding a putative heptosyl transferase, required for the addition of heptose to lipopolysaccharide (LPS) (M. Harper, J. D. Boyce, I. W. Wilkie, and B. Adler, Infect. Immun. 71:5440-5446, 2003). In the present study, using mass spectrometry and nuclear magnetic resonance, we have confirmed the identity of the enzyme encoded by waaQPM as a heptosyl transferase III and demonstrated that the predominant LPS glycoforms isolated from this mutant are severely truncated. Complementation experiments demonstrated that providing a functional waaQPM gene in trans can restore both the LPS to its full length and growth in mice to wild-type levels. Furthermore, we have shown that mutant AL251 is unable to cause fowl cholera in chickens and that the attenuation observed is not due to increased serum sensitivity.
Pasteurella multocida is an encapsulated, gram-negative coccobacillus and is the causative agent of a wide range of animal diseases including avian fowl cholera. Several P. multocida virulence factors have previously been identified, including the capsule in serogroups A and B (4, 6), PMT toxin in strains causing atrophic rhinitis in pigs (18), putative filamentous hemagglutinins PfhB1 and PfhB2 (19), and several iron acquisition proteins such as TonB, ExbD and ExbB (3, 19, 27).
During infections by gram-negative bacteria, the presence of lipopolysaccharide (LPS) stimulates the innate immune system whereby the inflammatory response plays a critical role in helping to clear the bacteria and prevent infection. This initial response to gram-negative bacteria can be elicited by a number of bacterial components, the most potent being lipid A, a component of the core structure of LPS. If the inflammatory response in the host is unable to clear the bacteria and the infection is allowed to proceed, the presence of large amounts of systemic LPS can result in endotoxic shock, in which an overproduction of inflammatory mediators causes damage to tissues, septic shock, organ failure, and death (31).
LPS is considered to play an important role in the pathogenesis of disease due to P. multocida. Recently it has been shown that LPS from P. multocida assists in adhesion to neutrophils and transmigration through endothelial cells (20). However, there are conflicting reports about the endotoxic properties of LPS isolated from P. multocida. LPS isolated from a serotype B:2 strain was shown to be endotoxic, and intravenously administered LPS could reproduce clinical signs of hemorrhagic septicemia in buffalo (24). However, turkey poults were found to be relatively resistant to the lethal effects of LPS isolated from serogroup A strains of P. multocida, although the inflammatory response and microscopic hepatic lesions were similar to those observed in mammalian hosts (29, 34). In contrast, chicken embryos and mice were found to be highly susceptible to the toxic effects of P. multocida LPS (21).
It is clear that the LPS of P. multocida stimulates humoral immunity, and it is considered to be a protective antigen. P. multocida strains are classified into Heddleston serotypes based on the antibody responses to LPS, while antibodies raised against heat-killed P. multocida vaccines are directed primarily against LPS and protect the host against strains within the same serotype (5). Early studies demonstrated that LPS purified using the Westphal method and injected into mice and rabbits resulted in a poor antibody response and no protection against P. multocida infection. In contrast, LPS injected into chickens induced a good antibody response which passively protected recipients against disease (33). Monoclonal antibodies raised against the LPS from a serotype A strain were shown to be bactericidal and to completely protect mice against homologous challenge (43). In addition, an opsonic monoclonal antibody against LPS from a serotype B strain of P. multocida was shown to partially protect mice against P. multocida infection (32).
A modified LPS structure clearly affects the viability of P. multocida in vivo. Recently in our laboratory, we identified three strongly attenuated mutants that each had a single transposon insertion in the pm1294 gene (designated waaQPM) (22). This gene is predicted to encode a heptosyltransferase, based on 72% amino acid similarity to the heptosyltransferase III from Haemophilus ducreyi, a member of the Pasteurellaceae family (17). Furthermore, a P. multocida galE mutant has been constructed previously and was attenuated in mice (16). galE encodes an enzyme required for the epimerization of UDP-glucose to UDP-galactose prior to LPS assembly, and this mutant probably expresses an altered LPS, although no structural analysis of the LPS was reported (16).
The sugar composition of LPS isolated from two serotype A strains of P. multocida was analyzed previously, and a partial structure has been proposed that included the identification of a triheptose unit linked to a 2-keto-3-deoxyoctulosonic acid (KDO) residue (12-14). In this report we present the core structure of LPS from a highly virulent P. multocida serotype A:1 strain, VP161, that causes fowl cholera and compare it with the structure of LPS from a transposon mutant deficient in heptosyltransferase III. We also present virulence data that demonstrate a significant role for LPS in P. multocida disease progression.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown routinely in Luria-Bertani broth. P. multocida strains were grown in either brain heart infusion (BHI) broth or nutrient broth (NB) supplemented with 3% yeast extract (Oxoid, Basingstoke, United Kingdom). Solid media were obtained by the addition of 1.5% agar. When required, the media were supplemented with tetracycline at 2.5 μg/ml. For structural studies, P. multocida strains VP161 and AL251 were grown in a 28-liter fermentor containing 24-liters of BHI broth for 18 h at 37°C with 20% dissolved-oxygen saturation. The cells were killed by addition of phenol to 2%, and 3 h after the phenol addition, 1 g of hyaluronidase (Roche Chemicals) was added; the mixture was stirred for 1 h before cells were harvested by using a Sharples continuous-flow centrifuge.
TABLE 1.
Bacterial strains, plasmids and oligonucleotides used in this study
| Strain, plasmid or oligonucleotide | Relevant description | Source or reference |
|---|---|---|
| Strains | ||
| P. multocida | ||
| VP161 | Serotype A:1, Vietnamese isolate from chickens | 44 |
| AL251 | VP161 Tn916EΔC waaQPM mutant | 22 |
| AL298 | AL251 with plasmid pAL170 | This study |
| AL438 | AL251 with plasmid pAL99 | This study |
| E. coli | ||
| DH5α | deoR endA1 gyrA96 hsdR17(rk− mk+) recA1 relA1 supE44 thi-1 (lacZYA-argFV169) φ80lacZ ΔM15, F− | Bethesda Research Laboratories |
| Plasmids | ||
| pPBA1100 | P. multocida/E. coli shuttle vector | 23 |
| pAL99 | 240-bp EcoRI fragment containing P. multocida tpiA promoter region cloned into pPBA1100 EcoRI site | This study |
| pAL170 | pAL99 containing waaQPM gene | This study |
| Oligonucleotides | ||
| BAP2146 | Forward primer for waaQPM amplification; has BamHI site for cloning; GAGTAGGATCCTGAAACATGTTCCC | This study |
| BAP2147 | Reverse primer for waaQPM amplification; has SalI site for cloning; GGTTGGGTCGACCAAGCCACATTACTG | This study |
Transposon stability studies.
P. multocida AL251 was grown in 10 ml of NB at 37°C with shaking. After approximately 10, 34, and 58 generations, samples of the culture were taken, diluted appropriately, and plated onto NB agar. After overnight incubation, 100 colonies were patched onto NB agar with tetracycline and incubated overnight at 37°C. Transposon loss was expressed as the percentage of tetracycline-sensitive colonies.
SDS-PAGE and silver staining.
LPS was analyzed with a Bio-Rad mini protein gel apparatus, using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (25). LPS was then visualized by silver staining (40).
DNA manipulations.
Restriction digests, ligations, and PCR amplifications were performed as specified by the manufacturers, using enzymes obtained from NEB (Beverley, Mass.) or Roche Diagnostics GmbH (Mannheim, Germany). Plasmid DNA was prepared using alkaline lysis (2) and purified using Qiagen columns (Qiagen GmbH, Hilden, Germany) or by polyethylene glycol precipitation (1). Genomic DNA was prepared using the cetyltrimethylammonium bromide (CTAB) method (1). PCR amplification of DNA was performed using Taq DNA polymerase or the Expand High Fidelity PCR system (Roche Diagnostics), and the DNA was purified using the Qiagen PCR purification kit. The oligonucleotides used in this study are listed in Table 1. The DNA sequence was determined on a model 373A DNA sequencing system (Applied Biosystems) and analyzed with Sequencher version 3.1.1 (GeneCodes, Ann Arbor, Mich.).
In trans complementation of waaQPM.
The complete waaQPM gene was amplified from P. multocida VP161 genomic DNA using oligonucleotides BAP2146 and BAP2147 (Table 1). The amplified 1.1-kb DNA fragment was ligated to SalI- and BamHI-digested vector pAL99 (Table 1), such that transcription was driven by the P. multocida tpiA promoter. E. coli transformants were screened for the presence of the correct plasmid, and one, designated pAL170, was used to transform P. multocida AL251, generating strain AL298. As a control, the pAL99 vector was transformed separately into AL251, generating strain AL438 (Table 1).
Competitive growth assays.
Competitive growth assays were performed as described previously (22) and were used to quantify the relative growth rates of the P. multocida LPS mutant AL251 and the complemented mutant AL298. The competitive index (CI) was determined by dividing the percentage of tetracycline-resistant colonies obtained from the output culture (in vitro or in vivo) by the percentage of tetracycline-resistant colonies obtained from the input culture. The relative competitive index (rCI), which measures the difference between growth in vivo and growth in vitro, was determined by dividing the in vivo CI by the in vitro CI. Mutants were identified as attenuated if the rCI value was significantly less than 1.0 as determined by statistical analysis using the one sided z test (P < 0.05).
Virulence trials.
Groups of 10 commercially obtained Leghorn-cross chickens aged 12 weeks were infected with P. multocida VP161 or AL251 at two different doses by injection of 100 μl into the breast muscle. Blood samples were obtained at various time points after infection with AL251, and the birds deemed incapable of survival were euthanized in accordance with animal ethics requirements. Blood samples were diluted twofold in BHI broth containing heparin and plated onto BHI plates. P. multocida colonies isolated from the blood were patched onto NB agar and NB agar with tetracycline.
Serum sensitivity assays.
The sensitivity of P. multocida and E. coli to fresh chicken serum was determined as described previously (6).
Purification of LPS.
P. multocida cells (210 g [wet weight] of VP161; 254 g [wet weight] of AL251) were freeze-dried, yielding 56 g of VP161 and 52 g of AL251. Freeze-dried cells were washed with organic solvents to remove lipids and other lipophilic components in order to enhance the efficiency of the LPS extraction (26). Washed cells (42 g of VP161; 50 g of AL251) were extracted by the hot-phenol-water method (42), and the aqueous phases were combined and dialyzed against running water for 48 h. The retentate was freeze-dried, made up to a 2% solution in water, and treated with DNase and RNase at 37°C for 4 h and then with proteinase K at 37°C for 4 h. Small peptides were removed by dialysis. After a freeze-drying step, the retentate was made up to a 2% solution in water and centrifuged at 8,000 × g for 15 min, and the supernatant was further centrifuged at 100,000 × g for 5 h. The pellet, containing purified LPS, was redissolved and freeze-dried. The core oligosaccharide (OS) was isolated by treating the purified LPS with 1% acetic acid (10 mg/ml) at 100°C for 1.5 h, with subsequent removal of the insoluble lipid A by centrifugation (5,000 × g).
Analytical methods.
Sugars were identified on the basis of their alditol acetate derivatives (35) by gas-liquid chromatography and mass spectrometry (GLC-MS). LPS was hydrolyzed for 4 h using 4 M trifluoroacetic acid at 100°C, reduced overnight with NaBD4 in H2O, and then acetylated with acetic anhydride at 100°C for 2 h using residual sodium acetate as the catalyst. The GLC-MS apparatus was equipped with a 30-m DB-17 capillary column (180 to 260°C at 3.5°C/min), and MS was performed in the electron impact mode on a Varian Saturn II mass spectrometer. Methylation analysis was carried out by the NaOH-dimethyl sulfoxide-methyl iodide procedure (7) and analyzed by GLC-MS as above.
MS analysis.
Capillary electrophoresis electrospray ionization MS (CE-ESI-MS) was performed on a crystal model 310 capillary electrophoresis (CE) instrument (AYI Unicam) coupled to an API 3000 mass spectrometer (Perkin-Elmer/Sciex) via a microIonspray interface (9). A sheath solution (isopropanol-methanol, 2:1 [vol/vol]) was delivered at a flow rate of 1 μl/min to a low-dead-volume tee (250-μm inner diameter [Chromatographic Specialties]). All aqueous solutions were filtered through a 0.45-μm pore-size filter (Millipore) before use.
Nuclear magnetic resonance.
Nuclear magnetic resonance (NMR) spectra were acquired on a Varian Inova 500 MHz spectrometer using a 3-mm triple-resonance (1H, 13C, and 31P) probe. The lyophilized sugar sample was dissolved in 140 μl of 99% D2O. The experiments were performed at 25°C with suppression of the HOD (deuterated H2O) signal at 4.78 ppm. The methyl resonance of acetone was used as an internal or external reference at 2.225 ppm for 1H spectra and 31.07 ppm for 13C spectra. Standard homo- and heteronuclear correlated two-dimensional pulse sequences from Varian, COSY, TOCSY, NOESY, 13C-1H HSQC, 13C-1H HSQC-TOCSY, and 13C-1H HMBC were used for general assignments.
RESULTS
An attenuated P. multocida mutant produces a truncated LPS that is restored to full-length LPS by complementation with a functional waaQPM gene.
We have previously used signature-tagged mutagenesis (STM) to identify mutants attenuated for growth in mice and chickens (22). During this previous analysis, a mutant was identified (designated AL251) that grew equally well as the wild type in vitro (mean CI = 0.96) but was attenuated in both chickens (mean CI = 0.01) and mice (mean CI = 0.59). Sequence analysis of the mutant revealed a single transposon insertion within the waaQPM gene that is predicted to encode a heptosyltransferase, a glycosyltransferase responsible for the addition of heptose to LPS (17,22).
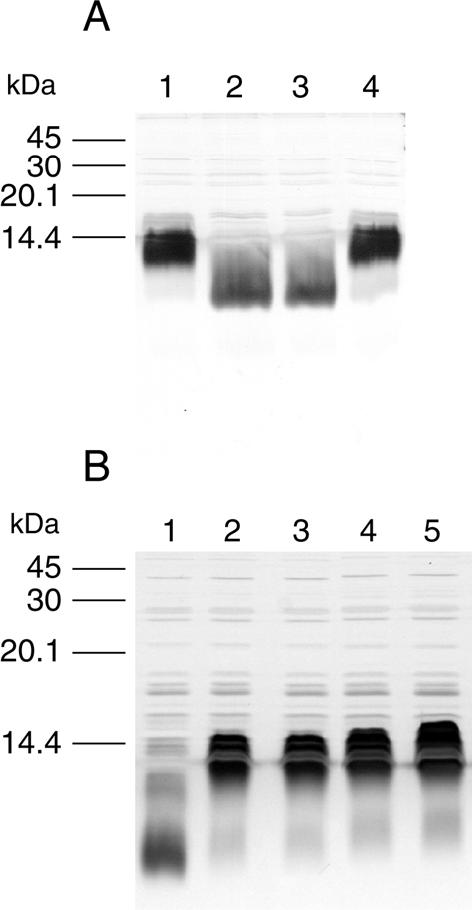
We compared the LPS profile of AL251 with that of its wild-type parent, VP161, and the complemented mutant, AL298, using SDS-PAGE followed by silver staining. The LPS from AL251 migrated further within the gel compared to wild-type LPS, indicating that the LPS produced by the mutant was significantly truncated (Fig. 1A). Furthermore, the LPS profile of the complemented mutant AL298 was identical to that observed for the wild type, indicating that complementation of the AL251 mutant with an intact waaQPM gene was able to restore the synthesis of wild-type LPS (Fig. 1A).
FIG. 1.
Analysis of P. multocida LPS by SDS-PAGE and silver staining of whole-cell lysates. (A) Comparison of P. multocida LPS profiles from wild-type VP161 (lane 1), heptosyltransferase mutant AL251 (lane 2), control strain AL438 (AL251 containing vector plasmid pAL99) (lane 3), and the complemented mutant strain AL298 (lane 4). (B) Comparison of LPS profiles of P. multocida heptosyltransferase mutant AL251 (lane 1), wild-type VP161 (lane 2), and P. multocida wild-type revertants isolated from three different chickens inoculated with AL251 (lanes 3 to 5).
Complementation of AL251 with waaQPM also restores in vivo growth to wild-type levels.
Since complementation of AL251 with waaQPM restored production of wild-type LPS levels, we wanted to determine if complementing the inactivated waaQPM also restored the mutant AL251 to wild-type levels of growth in vivo. Initial studies with the complemented mutant AL298 indicated that there was significant loss of the complementing plasmid pAL170 once antibiotic selection for the plasmid was removed (44% retention after 6 h). For this reason, mice were chosen instead of chickens for the competitive growth assay, since previous studies had demonstrated that the infection time required to harvest bacteria from mice was only 6 h compared with more than 12 h for infections in chickens (22). Three mice were injected with an equal mix of VP161 and the complemented strain AL298. As controls, two mice were injected with an equal mix of wild-type VP161 and the control strain AL438 (AL251 with the pAL99 vector). The complemented mutant AL298 was able to compete equally with wild-type VP161, with an average rCI value of 1.0, while the control strain AL438 had an average rCI value of 0.57 (P = 0.03), similar to the rCI values previously reported for AL251 in mice (22). These results demonstrate that waaQPM is required both for production of full-length LPS and for normal growth during infection.
The P. multocida waaQPM mutant is unable to cause disease in chickens.
We had shown previously that strain AL251 displayed a profoundly reduced growth rate in chickens (22); therefore, we wished to determine whether the mutant was still capable of causing disease in these hosts. Chickens were challenged with either VP161 or AL251 at two different doses (Table 2). All of the chickens challenged with wild-type VP161 died within 20 h. In contrast, most chickens challenged with AL251 remained well over the first 20 h but within 4 days all of the chickens inoculated with AL251, irrespective of dose, succumbed to fowl cholera infection. P. multocida was isolated from the blood of AL251-infected chickens in the late or terminal stages of the disease, and it was found that all of the isolated P. multocida colonies were tetracycline sensitive, indicating that the transposon was no longer present in the bacteria. Sequence analysis of waaQPM from the recovered colonies indicated that in all cases the transposon had excised, thereby reconstituting a functional waaQPM gene. Interestingly, for all but one isolate, the sequence analysis also revealed the presence of nucleotide substitutions at the point of transposon excision, resulting in two amino acid changes within waaQPM (amino acids 88 and 89; Ser to Leu and Asp to Cys, respectively). These amino acid changes did not affect the function of waaQPM, since the LPS profiles of the P. multocida isolates recovered from the chickens challenged with AL251 were all identical to those for the wild type (Fig. 1B). Taken together, these results indicate that the later onset of fowl cholera observed in the chickens inoculated with AL251 was due entirely to wild-type revertants of AL251 and that strains with an inactivated waaQPM gene are therefore incapable of causing disease.
TABLE 2.
Virulence of VP161 and AL251 in groups of 10 chickens
| Strain | Dose (CFU) | Mean time to death (range) (h) |
|---|---|---|
| VP161 | 1.5 × 102 | <20 (NDa) |
| 1.5 × 103 | <20 (ND) | |
| AL251 | 70 | 65 (33-120) |
| 7 × 102 | 30 (23-42) |
ND, not determined.
Attenuation of the waaQPM mutant in chickens is not due to increased sensitivity to chicken serum.
To determine the relative sensitivity of the wild-type P. multocida strain VP161 and the LPS mutant AL251 to complement-mediated killing, mid-log-phase cells of each strain and a control strain, E. coli DH5α, were incubated in either normal or heat-treated chicken serum for 3 h. Wild-type P. multocida strain VP161 was able to grow at the same rate in both the heated and the unheated serum, indicating that it is fully serum resistant as previously reported for other P. multocida serotype A strains (11). The bactericidal activity of the chicken serum was confirmed using the E. coli control strain, which multiplied 19-fold in heat-treated serum whereas its viability was reduced approximately 9-fold in untreated serum. Interestingly, for the AL251 mutant, no difference in growth was observed between the heat-treated and normal serum, with approximately a 160-fold increase in growth in the heat-treated serum and an identical growth rate in the normal serum. These results indicate that the attenuation observed for the LPS mutant in chickens was not due to increased sensitivity to complement.
Structural analysis of the LPS from P. multocida VP161 and AL251.
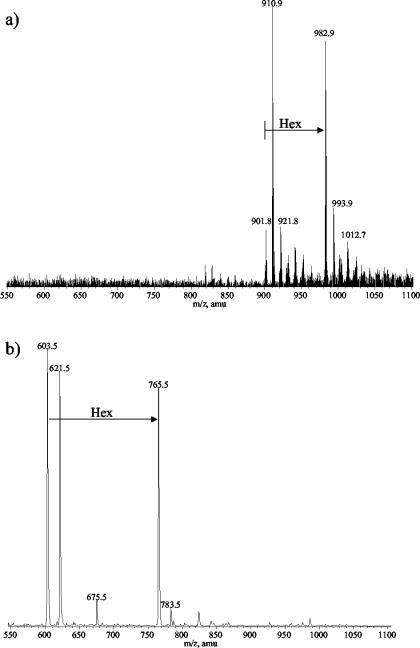
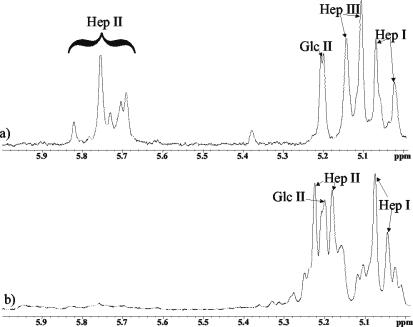
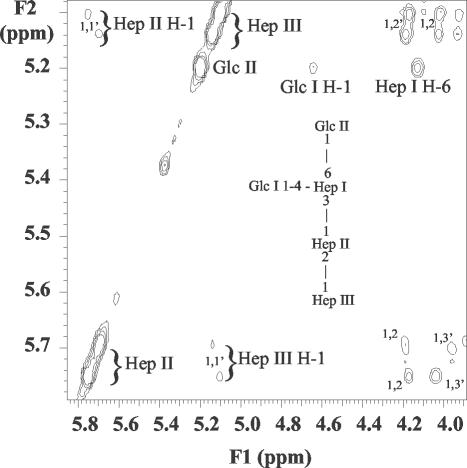
Sugar analysis of the LPS from the parent strain VP161 revealed glucose (Glc), galactose (Gal), N-acetylglucosamine (GlcNAc), and l-glycero-d-manno-heptose (ld-Hep) in the ratio of 2:1:1:3. In contrast, the LPS from mutant strain AL251 revealed only Glc and ld-Hep in the ratio of 1:3 with traces of Gal and GlcNAc. It is possible that GlcNAc could be contributed by residual amounts of the hyaluronic acid capsule known for this serogroup in addition to the anticipated two residues in the lipid A region of each LPS molecule. CE-ESI-MS analyses of the core OS sample derived from the parent VP161 LPS revealed a simple mass spectrum (Fig. 2a) corresponding to two glycoforms with masses of 1,805 and 1,967 Da, the smaller glycoform being consistent with a composition of 2PCho + 3Hex + 4Hep + KDO, where KDO is the unique LPS sugar 2-keto-3-deoxyoctulosonic acid and PCho is the phosphate moiety phosphocholine. The larger glycoform contains an additional hexose species (Table 3; Fig. 2a). Consistent with SDS-PAGE analysis of the mutant AL251 LPS, CE-ESI-MS analyses of the core OS sample derived from the mutant AL251 LPS revealed a simple mass spectrum of a highly truncated molecule with a mass of 605 Da (Fig. 2b). This mass corresponds to a composition of 2Hep + KDO. A larger glycoform was observed, consistent with the presence of an additional hexose residue (767 Da). Trace amounts of a more extended glycoform of 1,448 Da (PCho + 3Hep + 3Hex + KDO) were observed, but no glycoforms were observed that contained the full parental complement of four Hep residues (Table 3). To characterize completely the nature of the truncation in the core OS of the mutant strain, 1H-NMR experiments were performed on the parent- and mutant-derived core OS. Since MS data had suggested that a Hep residue was missing in the mutant OS and amino acid homology comparisons had suggested that WaaQPM was a heptosyltransferase, close attention was paid to the resonances from the heptose residues of the molecule. Examination of the 1H-NMR spectra revealed some heterogeneity for the heptose residues closest to KDO, as is often observed for core oligosaccharides due to rearrangement of the KDO residue under the acidic hydrolysis conditions used to obtain the core OS (Fig. 3). For the core OS derived from the parent strain, chemical shifts for the Hep II anomeric proton were identified at ∼5.7 ppm, consistent with 2-substitution of this residue (Fig. 3a) (9). However, in the AL251 mutant OS 1H-NMR spectrum (Fig. 3b), the anomeric resonance for the Hep II residue had shifted up-field about 0.5 ppm, consistent with this residue no longer being 2-substituted. The Hep II residue is now a terminal moiety, as we have observed previously for a Haemophilus somnus strain, and the chemical shifts to the H-4 resonance were consistent with this assignment (8). Definitive evidence for the structural nature of the mutation was obtained from two-dimensional NOESY experiments (Fig. 4). Characteristic nuclear Overhauser effects (NOEs) were observed for VP161 core OS between the Hep III and Hep II anomeric protons that are diagnostic for the α-1-2 linkage between the Hep III and Hep II residues (Fig. 4) (9). The NOESY spectrum of the AL251 mutant core OS confirmed the lack of 2-substitution of Hep II as the characteristic NOEs described above were absent, confirming that the Hep III residue is no longer present in the mutant OS. Chemical shift and NOE data for the Hep II and Hep III residues for the parent OS and Hep II residue for the mutant OS are summarized in Table 4. Structural techniques have therefore demonstrated that the effect on the LPS structure of mutating gene waaQPM is to preclude the addition of Hep III to Hep II (Fig. 5), and this function is consistent with strong similarity of the encoded protein to known heptosyltransferases.
FIG. 2.
Negative-ion CE-ESI-MS of P. multocida core OS. (a) Doubly charged region of core OS from parent strain VP161; (b) singly charged region of core OS from mutant strain AL251.
TABLE 3.
Negative-ion CE-MS data and proposed compositions for core OS from P. multocida strains VP161 (parent) and AL251 (mutant)a
| Strain | [M − H]− | [M − H]2− | Observed mass of molecular ion | Calculated mass of molecular Ion | Relative intensity | Proposed composition |
|---|---|---|---|---|---|---|
| VP161 | 1,804.4 | 901.8 | 1,805.6 | 1,805.4 | 0.2 | 2PCho + 4Hep + 3Hex + aKDOb |
| 1,822.4 | 910.9 | 1,823.1 | 1,823.4 | 1.0 | 2PCho + 4Hep + 3Hex + KDO | |
| 921.8 | 1,845.1 | 1,845.4 | 0.3 | 2PCho + 4Hep + 3Hex + KDO + Na | ||
| 1,966.3 | 982.9 | 1,967.8 | 1,967.6 | 0.9 | 2PCho + 4Hep + 4Hex + aKDO | |
| 1,988.4 | 993.9 | 1,989.1 | 1,989.6 | 0.3 | 2PCho + 4Hep + 4Hex + aKDO + Na | |
| AL251 | 603.5 | 604.5 | 604.5 | 1.0 | 2Hep + aKDO | |
| 621.5 | 622.5 | 622.5 | 0.9 | 2Hep + KDO | ||
| 765.5 | 766.5 | 766.7 | 0.8 | Hex + 2Hep + aKDO | ||
| 1,447.5 | 1,448.5 | 1,448.2 | 0.1 | PCho + 3Hep + 3Hex + aKDO |
Average mass units were used for calculation of molecular weight based on proposed composition as follows: Hex, 162.15; Hep, 192.17; Kdo, 220.18; PCho, 165.05. Relative intensity is expressed as relative height of either doubly or singly charged ions.
aKDO, anhydro-KDO derivative.
FIG. 3.
Region of the 1H-NMR spectrum of the core OS derived from the LPS of P. multocida parent strain VP161 (a) and P. multocida mutant strain AL251 (b). The spectra were recorded at 25°C and referenced against internal acetone at 2.225 ppm.
FIG. 4.
Region of the NOESY spectrum of P. multocida VP161 core OS. NOE connectivities are as indicated. (Inset), Structure of the inner-core OS from VP161. The spectrum was recorded at 25°C and referenced against internal acetone at 2.225 ppm.
TABLE 4.
1H-NMR chemical shifts for the Hep II and Hep III residues from the core OS derived from strains of P. multocida VP161 (parent) and AL251 (mutant)a
| Strain and Hep | Chemical shift
|
NOEsc
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| H-1 | H-2 | H-3 | H-4 | H-5 | H-6 | H-7 | Inter | Intra | |
| VP161 | |||||||||
| Hep II | 5.76 | 4.17 | 3.85 | 3.83 | 3.60 | 4.05 | 3.76 | 5.11 Hep III H-1 | 4.17 H-2 |
| 3.65 | 4.04 Hep I H-3 | ||||||||
| Hep II | 5.70 | 4.19 | 3.86 | 3.84 | 3.55 | 4.05 | 3.76 | 5.14 Hep III H-1 | 4.19 H-2 |
| 3.65 | 3.96 Hep I H-3 | ||||||||
| Hep III | 5.14 | 4.02 | 3.87 | 3.83 | 3.78 | 4.05 | 3.77 | 5.70 Hep II H-1 | 4.02 H-2 |
| 3.65 | 4.19 Hep II H-2 | ||||||||
| Hep III | 5.11 | 4.01 | 3.87 | 3.83 | 3.78 | 4.05 | 3.77 | 5.76 Hep II H-1 | 4.01 H-2 |
| 3.65 | 4.17 Hep II H-2 | ||||||||
| AL251 | |||||||||
| Hep II | 5.22 | 4.07 | 3.89 | 3.72 | NDb | ND | ND | 4.03 Hep I H-3 | |
| Hep II | 5.17 | 4.06 | 3.88 | 3.65 | ND | ND | ND | ND | |
Recorded at 25°C in D2O. Chemical shifts are referenced to internal acetone at 2.225 ppm. Two resonances were observed for each residue due to heterogeneity of the KDO molecule following core hydrolysis.
ND, not determined.
Inter, between residues; intra, with residues.
FIG. 5.
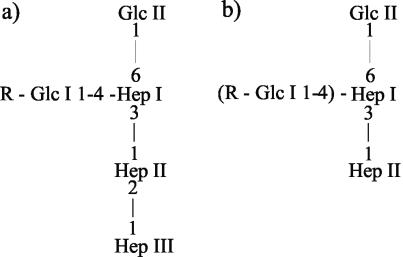
Proposed structures of inner core LPS of P. multocida from parent strain VP161 (a) and mutant strain AL251 (b), where R is the OS chain extension beyond Glc. Based on negative-ion CE-MS data shown in Table 3, extension of the mutant LPS molecule to include the structures shown in parentheses occurs at only low frequency (less than 4%).
DISCUSSION
The P. multocida LPS mutant AL251, first identified using STM in mice and chickens, was shown to have a transposon insertion in a predicted heptosyltransferase gene, waaQPM, and was significantly attenuated in chickens and mice (22). Silver-stained polyacrylamide gels of cell lysates from wild-type VP161 and AL251 showed that the LPS from the mutant was significantly truncated (Fig. 1A), consistent with waaQPM encoding a heptosyltransferase responsible for the addition of a heptose molecule in the core region of the LPS structure.
Analysis of the P. multocida Pm70 genome revealed that the waaQPM gene was probably transcribed independently and therefore the truncated LPS structure and reduced growth in vivo in chickens and mice were due directly to the inactivation of waaQPM and not to polar effects on downstream genes (27). This was confirmed by complementation, since the introduction of a wild-type waaQPM gene in trans restored both the LPS structure (Fig. 1A) and wild-type levels of growth in mice.
Virulence trials using the LPS mutant AL251 in chickens resulted in a delayed onset of fowl cholera symptoms (Table 2), and P. multocida strains isolated from chickens with disease symptoms were tetracycline sensitive, indicating that they no longer carried the transposon. Nucleotide sequence data obtained from P. multocida DNA isolated from infected birds confirmed that the waaQPM gene was intact and, in most cases, had nucleotide base changes at the point where the transposon had previously been inserted, resulting in two amino acid changes. The LPS from the P. multocida isolates recovered from the chickens challenged with AL251 showed that they produced wild-type LPS, confirming that the waaQPM gene was functional despite the amino acid changes (Fig. 1B). Moreover, these results suggest that serine and aspartate, at amino acid positions 88 and 89, respectively, in WaaQPM, are not essential for full enzyme activity since nonconservative changes at these positions did not prevent the formation of full-length LPS. Taken together, these data indicate that the infection observed in chickens inoculated with AL251 was due only to revertant strains that had lost the transposon insertion. The wild-type P. multocida strain VP161 is a highly virulent organism, with fewer than 50 bacteria causing fowl cholera in chickens (44). Our observations measured the transposon excision rate from AL251 cells at 1% after the first 10 generations (overnight in vitro growth), rising to 4% after 58 generations (data not shown). This rate of excision would result in the generation and selection of wild-type revertants in chickens at a rate sufficient to cause lethal fowl cholera infection during the course of the trial. We therefore conclude that a stable inactivation of waaQPM would result in a P. multocida strain incapable of causing fowl cholera. However, a reliable method of constructing defined mutants in P. multocida has not yet been established.
Analysis of the P. multocida VP161 wild-type LPS indicated a “rough” LPS, similar to the LPS or lipooligosaccharide isolated from gram-negative mucosal pathogens such as Haemophilus influenzae, H. ducreyi, Neisseria meningitidis, and N. gonorrhoeae, with only a short nonrepeating polysaccharide unit attached to the lipid A (15). The inner-core structure of P. multocida LPS is similar to that described for H. influenzae, Mannheimia haemolytica, and H. ducreyi, with a triheptose unit linked via a KDO residue to lipid A (Fig. 4) (8, 28, 30). In the AL251 mutant, inactivation of waaQPM resulted in the expression of a highly truncated LPS that lacked the third heptose molecule (Hep III) in the inner-core region (Fig. 2b). The most abundant glycoforms of LPS in the mutant also lacked all sugars distal to the first heptose, suggesting that the inactivation of waaQPM prevented further sugar additions (Table 3). It is therefore probable that conformational changes in the LPS intermediates due to the lack of the third heptose largely prevented the action of subsequent transferases.
The loss of a full-length LPS molecule clearly affects the ability of the mutant AL251 to grow in mice and to cause disease in chickens. The specific reasons for this attenuation are not clear. However, in wild-type P. multocida VP161 LPS, two PCho groups were identified, while the AL251 mutant contained only a single PCho group in a very minor glycoform (Table 3). The presence of more than one PCho residue in the VP161 LPS is unusual; bacteria with PCho-decorated LPS usually have only a single residue attached, although the position of attachment onto the LPS structure varies. Only one other bacterium, a nontypeable H. influenzae strain, is known to have two PCho residues attached to the LPS (M. K. Landerholm et al., unpublished observations). Interestingly, there are no PCho groups on the LPS of the serotype A turkey isolate, PM70 (A. D. Cox, unpublished observations), and this strain is not virulent for chickens (I. W. Wilkie, unpublished observations).
PCho groups are frequently attached to various bacterial structures on the surface of mucosal pathogens such as H. influenzae, Actinobacillus actinomycetemcomitans, Streptococcus pneumoniae, and Neisseria spp and play a key role in adhesion to and invasion of epithelial and endothelial host cells by binding to the platelet-activating receptor (10, 36-38). Nontypeable H. influenzae that has PCho-positive glycoforms of LPS can attach to and invade human bronchial epithelial cells via a series of signaling events (38, 39). However, in both H. influenzae and Neisseria spp., although expression of PCho on LPS was required for adhesion to and invasion of human epithelial cells, its presence reduced survival in some host niches, since strains expressing PCho were more serum sensitive, mediated by binding of PCho to C-reactive protein and subsequent activation of the complement system (37, 41). Interestingly, our studies have demonstrated that the LPS mutant AL251 is still highly resistant to the bactericidal action of complement in chicken serum, indicating that a completely wild-type LPS structure is not required for the bacteria to have full serum resistance. This is not surprising, since the waaQPM mutant is still encapsulated and the hyaluronic acid capsule of P. multocida serotype A strains confers serum resistance (6).
Although it is likely that the observed attenuation of the waaQPM mutant was due directly to the truncated LPS, we cannot completely exclude the possibility that pleiotropic effects, such as changes to the outer membrane structure, may play a role. However, a similar E. coli LPS mutant retained near-wild-type levels of outer membrane stability, as assessed by sensitivity to SDS and novobiocin (45).
In conclusion, we have characterized a P. multocida waaQPM mutant that expresses a severely truncated LPS. We have determined the inner-core structure for both the wild-type P. multocida LPS molecule and the waaQPM mutant molecule and demonstrated that a functional waaQPM gene is required for the addition of the third heptose residue to the inner core of the LPS. Thus, the waaQPM product has been identified as a heptosyltransferase III, and through virulence trials we have demonstrated that its activity is required for bacterial virulence in chickens, the natural host for this P. multocida strain. The inactivation of waaQPM leads to the complete absence of the third heptose in the inner core and, as a result, to the fully wild-type glycoforms. The majority of glycoforms identified from the mutant LPS lack PCho residues that in LPS of other gram-negative mucosal pathogens play a key role in bacterial virulence. Future work will focus on identifying the specific residues present on the LPS that are required for bacterial virulence.
Acknowledgments
This work was funded in part by a grant from the Australian Research Council.
We acknowledge Perry Fleming for cell growth, Suzon Larocque for NMR spectroscopy, and Lisa Morisson and Jianjun Li for mass spectrometry. We also thank the staff at Veterinary Pathology, University of Queensland, for their valuable assistance with the chicken experiments and Vicki Vallance and Ian McPherson for their excellent technical assistance.
Editor: V. J. DiRita
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, M., E. Garrido, M. Llagostera, A. M. P. de Rozas, I. Badiola, and J. Barbe. 2002. Pasteurella multocida exbB, exbD and tonB genes are physically linked but independently transcribed. FEMS Microbiol. Lett. 210:201-208. [DOI] [PubMed] [Google Scholar]
- 4.Boyce, J. D., and B. Adler. 2000. The capsu0le is a virulence determinant in the pathogenesis of Pasteurella multocida M1404 (B:2). Infect. Immun. 68:3463-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brogden, K. A., and P. A. Rebers. 1978. Serologic examination of the Westphal-type lipopolysaccharides of Pasteurella multocida. Am. J. Vet. Res. 39:1680-1682. [PubMed] [Google Scholar]
- 6.Chung, J. Y., I. Wilkie, J. D. Boyce, K. M. Townsend, A. J. Frost, M. Ghoddusi, and B. Adler. 2001. Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect. Immun. 69:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciucanu, I., and F. Kerek. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131:209-217. [Google Scholar]
- 8.Cox, A. D., M. D. Howard, J. R. Brisson, M. van der Zwan, P. Thibault, M. B. Perry, and T. J. Inzana. 1998. Structural analysis of the phase-variable lipooligosaccharide from Haemophilus somnus strain 738. Eur. J. Biochem. 253:507-516. [DOI] [PubMed] [Google Scholar]
- 9.Cox, A. D., H. Masoud, P. Thibault, J. R. Brisson, M. van der Zwan, M. B. Perry, and J. C. Richards. 2001. Structural analysis of the lipopolysaccharide from the nontypable Haemophilus influenzae strain SB 33. Eur. J. Biochem. 268:5278-5286. [DOI] [PubMed] [Google Scholar]
- 10.Cundell, D. R., N. P. Gerard, C. Gerard, I. Idanpaan-Heikkila, and E. I. Tuomanen. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435-438. [DOI] [PubMed] [Google Scholar]
- 11.Diallo, I. S., and A. J. Frost. 2000. Survival of avian strains of Pasteurella multocida in chicken serum. Vet. Microbiol. 72:153-161. [DOI] [PubMed] [Google Scholar]
- 12.Erler, W., H. Feist, K. D. Flossmann, B. Jacob, and A. Pilarski. 1981. Characterization of structural elements in lipopolysaccharides of Pasteurella multocida. Z. Allg. Mikrobiol. 21:507-517. (In German.) [DOI] [PubMed] [Google Scholar]
- 13.Erler, W., H. Feist, B. Jacob, and W. Schade. 1986. The heptose region of lipopolysaccharides of Pasteurella multocida. J. Basic Microbiol. 26:383-387. (In German.) [DOI] [PubMed] [Google Scholar]
- 14.Erler, W., H. Feist, and W. Schade. 1988. The structure of lipopolysaccharides from Pasteurella multocida. Arch. Exp. Veterinaermed. 42:32-40. (In German.) [PubMed] [Google Scholar]
- 15.Erridge, C., E. Bennett-Guerrero, and I. R. Poxton. 2002. Structure and function of lipopolysaccharides. Microbes Infect. 4:837-851. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez de Henestrosa, A. R., I. Badiola, M. Saco, A. M. Perez de Rozas, S. Campoy, and J. Barbe. 1997. Importance of the galE gene on the virulence of Pasteurella multocida. FEMS Microbiol. Lett. 154:311-316. [DOI] [PubMed] [Google Scholar]
- 17.Filiatrault, M. J., B. W. Gibson, B. Schilling, S. H. Sun, R. S. Munson, and A. A. Campagnari. 2000. Construction and characterization of Haemophilus ducreyi lipooligosaccharide (LOS) mutants defective in expression of heptosyltransferase III and β-1,4-glucosyltransferase: identification of LOS glycoforms containing lactosamine repeats. Infect. Immun. 68:3352-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foged, N. T., J. P. Nielsen, and S. E. Jorsal. 1989. Protection against progressive atrophic rhinitis by vaccination with Pasteurella multocida toxin purified by monoclonal antibodies. Vet. Rec. 125:7-11. [DOI] [PubMed] [Google Scholar]
- 19.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29:25-38. [DOI] [PubMed] [Google Scholar]
- 20.Galdiero, M., A. Folgore, I. Nuzzo, and E. Galdiero. 2000. Neutrophil adhesion and transmigration through bovine endothelial cells in vitro by protein H and LPS of Pasteurella multocida. Immunobiology 202:226-238. [DOI] [PubMed] [Google Scholar]
- 21.Ganfield, D. J., P. A. Rebers, and K. L. Heddleston. 1976. Immunogenic and toxic properties of a purified lipopolysaccharide-protein complex from Pasteurella multocida. Infect. Immun. 14:990-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper, M., J. D. Boyce, I. W. Wilkie, and B. Adler. 2003. Signature-tagged mutagenesis of Pasteurella multocida identifies mutants displaying differential virulence characteristics in mice and chickens. Infect. Immun. 71:5440-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homchampa, P., R. A. Strugnell, and B. Adler. 1997. Cross protective immunity conferred by a marker-free aroA mutant of Pasteurella multocida. Vaccine 15:203-208. [DOI] [PubMed] [Google Scholar]
- 24.Horadagoda, N. U., J. C. Hodgson, G. M. Moon, T. G. Wijewardana, and P. D. Eckersall. 2002. Development of a clinical syndrome resembling haemorrhagic septicaemia in the buffalo following intravenous inoculation of Pasteurella multocida serotype B:2 endotoxin and the role of tumour necrosis factor-alpha. Res. Vet. Sci. 72:194-200. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Masoud, H., E. R. Moxon, A. Martin, D. Krajcarski, and J. C. Richards. 1997. Structure of the variable and conserved lipopolysaccharide oligosaccharide epitopes expressed by Haemophilus influenzae serotype b strain Eagan. Biochemistry 36:2091-2103. [DOI] [PubMed] [Google Scholar]
- 27.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melaugh, W., N. J. Phillips, A. A. Campagnari, M. V. Tullius, and B. W. Gibson. 1994. Structure of the major oligosaccharide from the lipooligosaccharide of Haemophilus ducreyi strain 35000 and evidence for additional glycoforms. Biochemistry 33:13070-13078. [DOI] [PubMed] [Google Scholar]
- 29.Mendes, S., K. P. Carmichael, J. C. Nunnally, J. R. Glisson, I. H. Cheng, and B. G. Harmon. 1994. Lesions resulting from attempted Shwartzman reaction in turkey poults inoculated with Pasteurella multocida lipopolysaccharide. Avian Dis. 38:790-796. [PubMed] [Google Scholar]
- 30.Phillips, N. J., M. A. Apicella, J. M. Griffiss, and B. W. Gibson. 1992. Structural characterization of the cell surface lipooligosaccharides from a nontypable strain of Haemophilus influenzae. Biochemistry 31:4515-4526. [DOI] [PubMed] [Google Scholar]
- 31.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramdani, and B. Adler. 1991. Opsonic monoclonal antibodies against lipopolysaccharide (LPS) antigens of Pasteurella multocida and the role of LPS in immunity. Vet. Microbiol. 26:335-347. [DOI] [PubMed] [Google Scholar]
- 33.Rebers, P. A., M. Phillips, R. Rimler, R. A. Boykins, and K. R. Rhoades. 1980. Immunizing properties of Westphal lipopolysaccharide from an avian strain of Pasteurella multocida. Am. J. Vet. Res. 41:1650-1654. [Google Scholar]
- 34.Rhoades, K. R., and R. B. Rimler. 1987. Effects of Pasteurella multocida endotoxins on turkey poults. Avian Dis. 31:523-526. [PubMed] [Google Scholar]
- 35.Sawardeker, D. G., J. H. Sloneker, and A. Jeanes. 1965. Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal. Chem. 37:1602-1604. [Google Scholar]
- 36.Schenkein, H. A., S. E. Barbour, C. R. Berry, B. Kipps, and J. G. Tew. 2000. Invasion of human vascular endothelial cells by Actinobacillus actinomycetemcomitans via the receptor for platelet-activating factor. Infect. Immun. 68:5416-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serino, L., and M. Virji. 2002. Genetic and functional analysis of the phosphorylcholine moiety of commensal Neisseria lipopolysaccharide. Mol. Microbiol. 43:437-448. [DOI] [PubMed] [Google Scholar]
- 38.Swords, W. E., B. A. Buscher, K. Ver Steeg Ii, A. Preston, W. A. Nichols, J. N. Weiser, B. W. Gibson, and M. A. Apicella. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13-27. [DOI] [PubMed] [Google Scholar]
- 39.Swords, W. E., M. R. Ketterer, J. Shao, C. A. Campbell, J. N. Weiser, and M. A. Apicella. 2001. Binding of the non-typeable Haemophilus influenzae lipooligosaccharide to the PAF receptor initiates host cell signalling. Cell Microbiol. 3:525-536. [DOI] [PubMed] [Google Scholar]
- 40.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver strain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 41.Weiser, J. N., N. Pan, K. L. McGowan, D. Musher, A. Martin, and J. Richards. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharide. Extraction with phenol water and further applications of the procedure. Methods Carbohydr. Chem. 5:88-91. [Google Scholar]
- 43.Wijewardana, T. G., C. F. Wilson, N. J. Gilmour, and I. R. Poxton. 1990. Production of mouse monoclonal antibodies to Pasteurella multocida type A and the immunological properties of a protective anti-lipopolysaccharide antibody. J. Med. Microbiol. 33:217-222. [DOI] [PubMed] [Google Scholar]
- 44.Wilkie, I. W., S. E. Grimes, D. O'Boyle, and A. J. Frost. 2000. The virulence and protective efficacy for chickens of Pasteurella multocida administered by different routes. Vet. Microbiol. 72:57-68. [DOI] [PubMed] [Google Scholar]
- 45.Yethon, J. A., D. E. Heinrichs, M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273:26310-26316. [DOI] [PubMed] [Google Scholar]