Abstract
We examined the role of Streptococcus pyogenes two-component response regulators (SptR) in expression of Mga and the Mga-regulated gene emm. Both serotype M6 and serotype M1 mutants in 12 of the 13 identified sptR genes exhibited levels of emm transcripts and Mga protein comparable to those of the wild type during exponential and stationary phases of growth. Thus, temporal control of these virulence genes does not require Spt response regulators.
To successfully colonize and persist within a variety of host tissues, pathogens have developed mechanisms by which they can respond to varying environments through coordinate changes in their patterns of gene expression. Bacteria often mediate such responses by two-component signal transduction systems, a family of phosphorelay proteins known to regulate a wide variety of cellular processes (9, 10). The typical two-component signal transduction system is composed of a membrane-bound sensor histidine kinase that detects the specific external signal and transfers a high-energy phosphate to a cognate cytoplasmic response regulator that interacts directly with DNA to control expression of a defined set of genes.
The group A streptococcus (GAS) (Streptococcus pyogenes) is an important bacterial pathogen and the causative agent of numerous diseases in its human host (4). GAS has evolved mechanisms that allow it to persist in varying microenvironments by coordinately expressing virulence factors in response to its changing surroundings. Analysis of the published genomes of GAS (M1, M3, and M18) has identified an average of 13 potential S. pyogenes two-component systems (SPTs) (1, 6, 21). Furthermore, 12 of the 13 Spts found in the M1 genome can also be found in M18 and M3, indicating their potential importance for environmental regulation among all class I serotypes of GAS. However, only three of these Spts have been characterized to any degree at the molecular level. The covRS/csrRS system represses expression of virulence genes encoding capsule and several exotoxins, as well as influencing the transcription of as much as 15% of the M6 GAS genome (2, 5, 7, 8, 13). The fasBCA system is a growth-phase-regulated SPT containing two histidine kinase components that uses a fasX effector RNA to down-regulate genes involved in adhesion and to up-regulate those encoding aggressins during the transition from exponential- to stationary-phase growth (11). Finally, the ihk/irr Spt has been shown to allow survival of GAS following uptake by polymorphonuclear leukocytes (5, 23).
In addition to SPTs, GAS possesses “stand-alone” global regulators that control large sets of virulence genes in response to both temporal and environmental stimuli. One such regulator is Mga, a DNA-binding protein that activates the expression of virulence genes encoding molecules important for colonization and immune evasion, such as the M protein family (emm, mrp, and enn), C5a peptidase (scpA), and collagen-like protein 1 (scl1) (12). Maximal expression of the Mga regulon occurs during exponential-phase growth and is rapidly shut down upon entering stationary phase (16). The regulon is also up-regulated by growth in elevated CO2 and iron levels and temperature (3, 14, 19). However, the mechanism by which any of these different conditions regulates expression of mga and its regulon is not known. In this study, 12 of the 13 putative response regulator genes were inactivated in two different class I GAS strains to determine their possible role in the temporal regulation of this important virulence cascade.
KSM148, a derivative of the S. pyogenes serotype M6 strain JRS4 containing a single-copy Pemm-gusA transcriptional reporter (20), was used for insertional inactivation of the response regulator gene (sptR). Open reading frame internal fragments for 12 of the 13 putative sptR genes in the serotype M1 genome (Fig. 1), except for the essential sycF (spt3R) gene, were amplified from M1 SF370 (6) genomic DNA by high-fidelity PCR using the primers listed in Table 1. Each PCR fragment was cloned into pCR-TOPO Blunt II (Invitrogen), purified following BamHI-PstI digestion, and subsequently cloned into BamHI-PstI-digested pJRS233, a temperature-sensitive integration vector (18). The 12 mutagenic plasmids (p233-spt1R, -2R, and -4R to -13R) (Fig. 1) were verified by PCR and DNA sequence analysis and integrated into the chromosome of KSM148 as previously described (18). PCR was used to verify the presence of the plasmid backbone, each integrant junction, and the absence of the wild-type sptR gene product using the appropriate primers (Table 1).
FIG. 1.
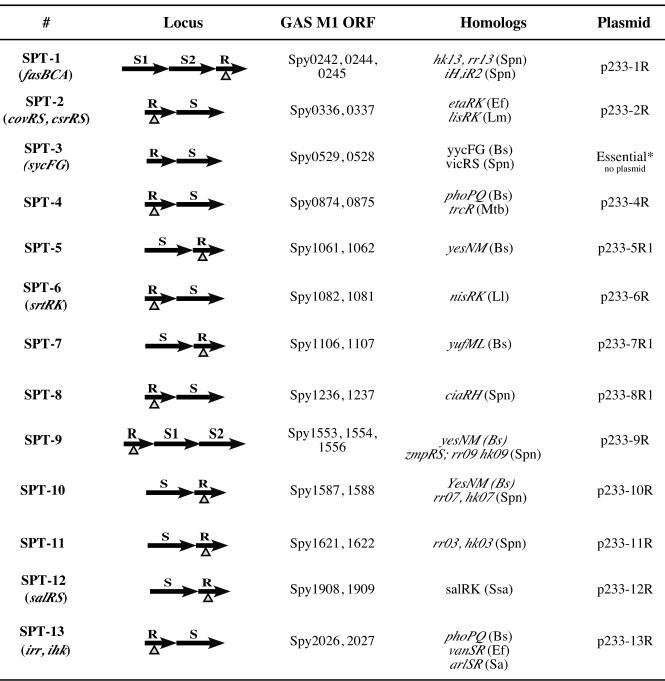
Thirteen loci encoding known or putative S. pyogenes SPTs in the serotype M1 SF370 genome. The SPT loci were numbered based on their order of appearance in the M1 genome sequence, with any previously assigned names provided in parentheses. A schematic representation of the gene positions for each SPT loci (R, response regulator; S, sensor histidine kinase) is shown, along with their respective open reading frame numbers (Spy) based on the original annotation by Ferretti et al. (6). Each insertion-duplication mutation within each sptR gene is indicated (▵). The homologs provided for each SPT locus represent those found in other gram-positive genomes (Bs, Bacillus subtilis; Spn, Streptococcus pneumoniae; Ef, Enterococcus faecalis; Lm, Listeria monocytogenes; Mtb, Mycobacterium tuberculosis; Ll, Lactococcus lactis; Ssa, Streptococcus salivarus; Sa, Staphylococcus aureus) that exhibit the highest similarity at the amino acid level to the respective locus. The names of the temperature-sensitive mutagenic plasmids used in the study to inactivate each sptR gene are listed on the right of the figure, excluding one for the essential spt3R locus.
TABLE 1.
Primers used in this study
| Target | Primera | Sequence (5′-3′) | Reference |
|---|---|---|---|
| spt1R (fasA) | spt1R-L | TACCATGTAATCAGCTTGAA | This study |
| spt1R-R | AACAAACTTTGTGAGGAGT | This study | |
| fasA-R1* | GGGATTGATTGCTCGATAAA | This study | |
| fasA-L1 | AGCACAAAAACCAATCGTGT | This study | |
| spt2R (covR) | covR-L | TAGTGAGAGAAATCTCATCG | 5 |
| covR-R | TATGAAGTCATTGTTGAGGT | 5 | |
| covR-R1* | AGGCAATCAGTGTAAAGGCA | This study | |
| covR-L1 | AATCCTTTTGCTAGCTTGCA | This study | |
| spt4R | spt4R-L | AAGGCCATTAATTTACCTTC | This study |
| spt4R-R | CGGTCAAGAGGCAATTGATA | This study | |
| spt4R-R1* | GCATCTTGTGATGTTACCAT | This study | |
| spt4R-L1 | CGGTTAATTTCTTGTTGACT | This study | |
| spt5R | spt5R-L1 | AAGAAGGAGCTGTTTTAATC | This study |
| spt5R-R1 | CCTGATTAATCCGATATTGA | This study | |
| spt5R-R2* | GCCATTTGCGTTTGAGTTTT | This study | |
| spt5R-L2 | TTCGGGAATTGATGCTCATC | This study | |
| spt6R | spt6R-L | ATTCTGAAGCTTATGAAGAC | This study |
| spt6R-R | TGACGTGACAGTAATTCTAA | This study | |
| spt6R-L2 | AGAGATCTTGAGTGATAGCA | This study | |
| spt6R-R2* | TAAACATCCCATTTATAACC | This study | |
| spt7R | spt7R-L1 | ACAAGCGGAAGCTAATTCTT | This study |
| spt7R-R1 | ATCCAGCGACTCTATGAAAG | This study | |
| spt7R-R2 | TCTCAGTGACAACACCTTAT | This study | |
| spt7R-L2* | GTTTATGACAATTTAGGGTC | This study | |
| spt8R | spt8R-L1 | ATCACTATCGAATCCCCATA | This study |
| spt8R-R1 | TGAAGGTTTATACGAAGCAG | This study | |
| spt8R-L2 | AAACTACTGTCAACTGACGA | This study | |
| spt8R-R2* | GGCTATTAGCCTCTTAAATA | This study | |
| spt9R | spt9R-L | TTTTAATCAAGACGCTAAGG | This study |
| spt9R-R | GTTGATTTTAGCCAGTTCAA | This study | |
| spt9R-R1* | ATATGGAAGCTAAGGGCTAT | This study | |
| spt9R-L1 | TACGAGAACTCGTCTGGTAA | This study | |
| spt10R | spt10R-L | AGTGGATGTCATGATTTCAG | This study |
| spt10R-R | CGCTTTTGTAGATCATAGGT | This study | |
| spt10R-R1* | TTGGTAGCCAGTCACTGTCT | This study | |
| spt10R-L1 | TCAAGGCTTTAGATGAGACG | This study | |
| spt11R | spt11R-L | TCCGCATGGGACTCAAGAGT | This study |
| spt11R-R | GTCAGGGTGTTGATCATGCG | This study | |
| spt11R-R1* | AGGCATATAACCCGTATCTT | This study | |
| spt11R-L1 | TTGAAGATGATTGATGATGG | This study | |
| spt12R | spt12R-L | GCAAAGAGTATCCAACTGTT | This study |
| spt12R-R | TTCCTGGTCAGTTAAAGATT | This study | |
| spt12R-R1* | TAGCAACGACACAAGTAAAA | This study | |
| spt12R-L1 | TTCAAGAAACACTAGCAGCT | This study | |
| spt13R (irr) | irr-L | GGTGACGTTTTGCTAAATAA | 5 |
| irr-R | AAAGCGAATAACTATGATCC | 5 | |
| irr-R1* | TGTCTTTGGACTATTACCAG | This study | |
| irr-L1 | CTTCTTTGTCTTTGACTTTG | This study | |
| M13 | 1201 | AACAGCTATGACCATGATTACG | Clontech |
| 1211 | GTAAAACGACGGCCAGT | Clontech | |
| pWV01 ori | pLZ12ori-L | TTATATCCTGACTCAATTCC | This study |
| pLZ12ori-R | CTCAAACCATAATCTAAAGG | This study | |
| emm | OM6-35 | AACAGCAAATTAGCTGCTC | 15 |
| OM6-16 | GTTTCCTTCATTGGTGCT | 15 | |
| 23S rRNA | rRNA_23S-L | GGAAGGTAAGCCAAAGAGAG | 20 |
| rRNA_23S-R | TCCTAGTTTCTGTGCAACC | 20 |
Boldface indicates wild-type sptR gene pair. An asterisk indicates integrant junction primer.
To initially characterize each M6 SptR mutant strain, growth curves were determined in Todd-Hewitt broth at 37°C and compared to those of parental KSM148 and mga-inactivated KSM148.586 (20). Most of the mutants exhibited rates of growth identical to that of wild-type KSM148, while KSM148.spt5R actually grew slightly faster and KSM148.spt2R (covR) demonstrated an extended lag phase prior to reaching comparable growth rates (data not shown). The ability to obtain mutants for each of the 12 targeted sptR genes with no obvious effects on growth demonstrated that none of 12 two-component loci tested is essential for growth of GAS under normal laboratory conditions.
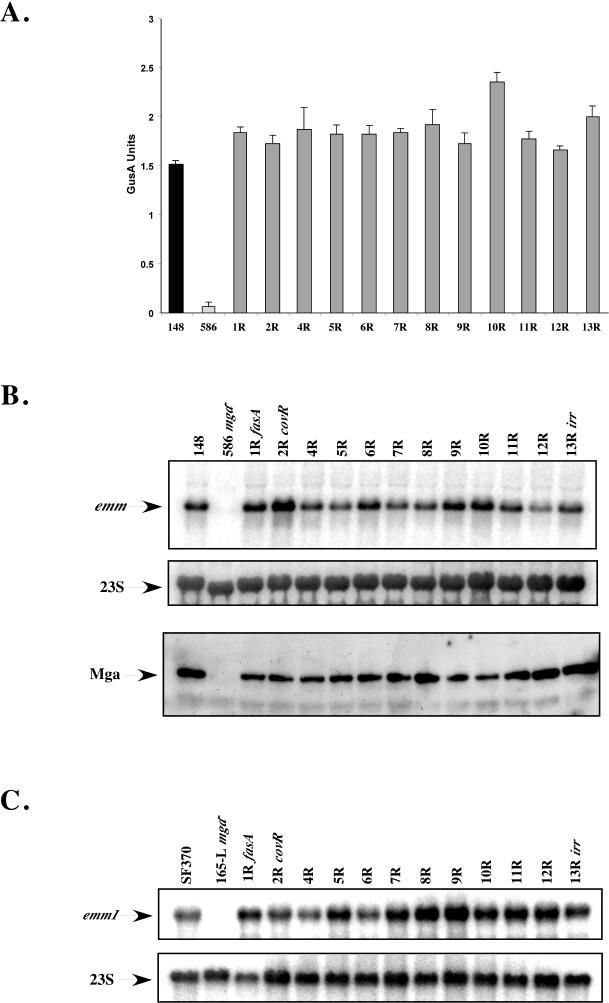
Since mga expression is optimally expressed during exponential phase and repressed during stationary phase, it was expected that the loss of an Spt necessary for temporal regulation would produce a significant decrease in expression of Mga-regulated emm during exponential phase and/or a dramatic increase in expression during stationary phase. Liquid GusA assays were performed, with each mutant strain isolated at exponential phase of growth and compared to both the wild-type KSM148 and mga-inactivated KSM148.586. All of the mutants exhibited Mga-regulated GusA activity equal to or slightly above the levels observed for wild-type KSM148 (Fig. 2A), with none showing decreased GusA activity during exponential-phase growth comparable to that of the mga-inactivated control, KSM148.586 (Fig. 2A). To verify that the levels of GusA activity observed reflected Mga-regulated gene expression, Northern blot analysis using an emm6 probe was carried out, and the results were normalized using a probe for 23S RNA as a control for loading (20). As observed in the GusA assays, each of the 12 SPTR mutant strains showed levels of Mga-regulated emm6 transcripts similar to those of wild-type KSM148 (Fig. 2B). In addition, the Mga protein was found in each mutant strain lysate at levels comparable to that for the wild type, as determined by Western blot analysis (Fig. 2B) (15). These data clearly demonstrate that none of the 12 sptR genes examined are necessary for exponential-phase expression of Mga or emm in the serotype M6 GAS strain KSM148.
FIG.2.
Analysis of exponential-phase Mga-regulated gene expression in serotype M6 KSM148 sptR mutants. (A) GusA activity of whole-cell lysates from wild-type KSM148 (148), mga-inactivated KSM148.586 (586), and the 12 sptR mutants (1R, 2R, and 4R to 13R). Whole-cell lysates from exponential-phase cells were examined in a liquid GusA assay for the production of β-glucuronidase. (B) Northern analysis (1 μg of total RNA) of serotype M6 KSM148 sptR mutants for emm6 transcript levels (top). Northern blots were stripped and reprobed for 23S RNA as a control for loading. Mga protein levels (bottom) in each lysate were determined by Western blotting (7 μg of total protein), with an antibody to a peptide of Mga used as a probe. (C) Northern analysis (1 μg of total RNA) of serotype M1 SF370 sptR mutants for emm1 transcript levels (top). Northern blots were stripped and reprobed for 23S RNA as a control for loading (bottom).
To investigate whether the results obtained with the M6 strain were serotype specific, we inactivated the comparable sptR genes in the sequenced serotype M1 strain SF370 (6, 22) as described for KSM148. Growth curves for the M1 mutants were almost identical to those observed for the M6 KSM148 mutants (data not shown), demonstrating that these response regulators are not required for in vitro growth in rich medium with either serotype. Furthermore, inactivation of spt2R (covR/csrR) in SF370 and KSM148 resulted in a colony phenotype on agar plates that was highly mucoid compared to that of the parental strain, as expected (data not shown) (13). Since a Pemm-gusA reporter fusion is not available in the SF370 background, Northern blot analysis using an emm1 probe was performed, and the results were normalized using a probe for 23S RNA as a control for loading. Each of the 12 SPTR mutant strains showed levels of Mga-regulated emm1 transcripts comparable to those for wild-type SF370 and significantly higher than the levels seen for mga-inactivated KSM165-L (Fig. 2C). Therefore, there is no major requirement for SPTRs in exponential-phase expression of Mga or the Mga-regulated emm gene in either of the two class I strains of GAS.
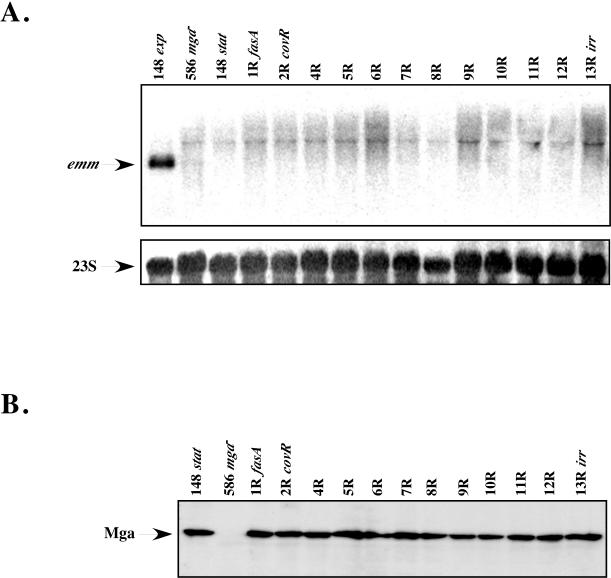
As GAS cells enter into stationary-phase growth, a dramatic decrease in the levels of mga and emm is observed that is mediated through the mga promoter (16, 17). To investigate whether SPTRs may be involved in negatively influencing expression of emm late in the growth cycle, Northern blot analysis using an emm6 probe was performed. Total RNA from the 12 M6 KSM148 sptR mutants, wild-type KSM148, and mga-inactivated KSM148.586 was isolated at 6 h after reaching stationary phase (90 to 100 Klett units), and results were normalized using a probe for 23S RNA as a control for loading. Comparison of emm transcript levels from previously isolated logarithmic-phase cells (1 μg of RNA) to levels in stationary-phase cells (5 μg of RNA) clearly shows the expected dramatic reduction in Mga-regulated gene expression following entry of GAS into the later phase of growth (Fig. 3A). Importantly, no detectable increase in emm transcripts during stationary-phase growth was observed for any of the sptR mutant strains compared to levels for wild-type KSM148 (Fig. 3A). Levels of Mga protein detected in the same stationary-phase lysates were similar to wild-type levels as assessed by Western blotting using an Mga-specific antibody (Fig. 3B). Interestingly, comparable levels of protein were observed in both exponential- and stationary-phase cells, indicating that Mga is stably maintained in the cell during times when transcription of Mga-regulated genes is absent (Fig. 3A). Taken together, these data strongly suggest that repression of Mga-regulated emm transcription upon entry into stationary phase does not require the involvement of the SPTR response regulators studied.
FIG. 3.
Analysis of stationary-phase Mga-regulated gene expression in serotype M6 KSM148 sptR mutants. (A) Northern analysis of wild-type KSM148 (148), mga-inactivated KSM148.586 (586), and the 12 sptR mutants (1R, 2R, and 4R to 13R). Total RNA (1 μg of total RNA for exponential-phase KSM148; 5 μg of total RNA for all stationary-phase samples) was probed for emm6 transcript levels. Northern blots were stripped and reprobed for 23S RNA as a control for loading. (B) Mga protein levels in each stationary-phase lysate were determined by Western blotting (7 μg of total protein), with an antibody to a peptide of Mga used as a probe.
Since spt3RS (sycFG) (Fig. 1) appears to be essential for growth in GAS, its role in Mga regulon expression was not investigated in these studies and we cannot rule out that this two-component locus may be involved in regulating expression of mga and emm in response to growth-phase signals. Additionally, the apparent lack of input from sptR components in the growth-phase regulation of mga expression does not rule out the possibility that Mga itself may directly interact with one of the sensor kinase components (sptS), resulting in a modification of Mga activity. Although half of our sptR mutations are likely to confer a polar effect on its downstream sptS gene (Fig. 1), we did not directly ask whether the cognate sptS genes in the remaining loci were required for Mga-regulated gene control. Therefore, the direct interaction of Mga with a sensor kinase may still represent a valid model for temporal regulation of the pathway. Finally, the potential role of SPTs in the response of the Mga pathway to other stimuli, such as CO2 levels and temperature, remains to be investigated.
Acknowledgments
We thank J. Ferretti and J. Scott for strains and plasmids used in this study.
This work was supported by a Public Health Service grant (R01-AI47928) from the NIH/NIAID awarded to K.S.M. D.A.R. was supported in part by an NIH/NIAID Molecular Microbiology Training Grant (5T32-AI0520).
Editor: V. J. DiRita
REFERENCES
- 1.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernish, B., and I. van de Rijn. 1999. Characterization of a two-component system in Streptococcus pyogenes, which is involved in regulation of hyaluronic acid production. J. Biol. Chem. 274:4786-4793. [DOI] [PubMed] [Google Scholar]
- 3.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 10.Hoch, J. A., and T. J. Silhavy (ed.). 1995. Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 11.Kreikemeyer, B., M. D. Boyle, B. A. Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 12.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 13.Levin, J. C., and M. R. Wessels. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol. Microbiol. 30:209-219. [DOI] [PubMed] [Google Scholar]
- 14.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIver, K. S., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 43:1591-1602. [DOI] [PubMed] [Google Scholar]
- 16.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIver, K. S., A. S. Thurman, and J. R. Scott. 1999. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J. Bacteriol. 181:5373-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podbielski, A., J. A. Peterson, and P. Cleary. 1992. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol. Microbiol. 6:2253-2265. [DOI] [PubMed] [Google Scholar]
- 20.Ribardo, D. A., and K. S. McIver. 2003. amrA encodes a putative membrane protein necessary for maximal exponential phase expression of the Mga virulence regulon in Streptococcus pyogenes. Mol. Microbiol. 50:673-685. [DOI] [PubMed] [Google Scholar]
- 21.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suvorov, A. N., and J. J. Ferretti. 1996. Physical and genetic chromosomal map of an M type 1 strain of Streptococcus pyogenes. J. Bacteriol. 178:5546-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voyich, J. M., D. E. Sturdevant, K. R. Braughton, S. D. Kobayashi, B. Lei, K. Virtaneva, D. W. Dorward, J. M. Musser, and F. R. DeLeo. 2003. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. USA 100:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]