Abstract
Legionella dumoffii is one of the common causes of Legionnaires' disease and is capable of replicating in macrophages. To understand the mechanism of survival within macrophages, transposon mutagenesis was employed to isolate the genes necessary for intracellular growth. We identified four defective mutants after screening 790 transposon insertion mutants. Two transposon insertions were in genes homologous to icmB or dotC, within dot/icm loci, required for intracellular multiplication of L. pneumophila. The third was in a gene whose product is homologous to the 17-kDa antigen forming part of the VirB/VirD4 type IV secretion system of Bartonella henselae. The fourth was in the djlA (for “dnaj-like A”) gene. DjlA is a member of the DnaJ/Hsp40 family. Transcomplementation of the djlA mutant restored the parental phenotype in J774 macrophages, A549 human alveolar epithelial cells, and the amoeba Acanthamoeba culbertsoni. Using confocal laser-scanning microscopy and transmission electron microscopy, we revealed that in contrast to the wild-type strain, L. dumoffii djlA mutant-containing phagosomes were unable to inhibit phagosome-lysosome fusion. Transmission electron microscopy also showed that in contrast to the virulent parental strain, the djlA mutant was not able to recruit host cell rough endoplasmic reticulum. Furthermore, the stationary-phase L. dumoffii djlA mutants were more susceptible to H2O2, high osmolarity, high temperature, and low pH than was their parental strain. These results indicate that DjlA is required for intracellular growth and organelle trafficking, as well as bacterial resistance to environmental stress. This is the first report demonstrating that a single DjlA-deficient mutant exhibits a distinct phenotype.
Legionella dumoffii was first isolated from cooling-tower water in 1979 (18) and later from a postmortem lung specimen in the same year (40) as an atypical Legionella-like organism. It was later classified by Brenner (11) as a new species, L. dumoffii. Legionella species are gram-negative, facultative intracellular parasites of freshwater amoebae in nature and are capable of growing within alveolar macrophages and epithelial cells after being accidentally transmitted to humans (22). The most common human pathogen in the genus Legionella is L. pneumophila, the causative agent of Legionnaires' disease (71). Humans contract the disease from contaminated environmental sources, primarily by aspiration of aerosolized water sources (22). After internalization by alveolar macrophages, L. pneumophila-containing phagosomes do not acidify (34) or fuse with lysosomes (33). Instead, the mitochondria, smooth vesicles, and rough endoplasmic reticula (RER) near these L. pneumophila-containing vacuoles are recruited, and L. pneumophila begins to multiply in this unique niche (32). This altered endocytic pathway is considered to be controlled by the Dot/Icm type IV protein secretion system (5, 17, 48, 55, 56, 74). The dot/icm genes are essential for the intracellular growth of L. pneumophila (5, 51, 60). The presence of the dot/icm loci in several species of Legionella was shown by Southern or PCR analysis (4, 36, 43); however, the contributions of these loci to the pathogenesis of other species have yet to be investigated.
L. dumoffii is the fourth or fifth most common pathogen causing Legionnaires' disease (8, 71). Some of proteins or factors which may promote L. pneumophila pathogenesis, such as flagella, catalase, and gelatinase, are also present in L. dumoffii. Several putative virulence factors—lipase, oxidase, and a zinc metalloprotease—are absent in L. dumoffii (6, 11, 52). L. dumoffii is capable of infecting and replicating within Vero cells and the human lung alveolar epithelial cell line A549 in vitro (41, 42). To elucidate the molecular mechanisms of the intracellular growth of this organism, we attempted to isolate the mutants that exhibited defective growth phenotypes in J774 mouse macrophage-like cells and A549 human type II alveolar epithelial cells by using transposon mutagenesis. We isolated four clones attenuated in virulence within mammalian cells by screening 790 derivatives with Tn903dIIlacZ insertions. Two of four genes flanking the transposon insertions encode the proteins homologous to L. pneumophila IcmB and DotC (5, 51, 60), respectively. One gene has similarity to virB5 (17-kDa antigen) in the VirB/VirD4 type IV secretion system of Bartonella henselae (14, 49, 59). The deduced protein encoded by a fourth gene showed homology to DjlA proteins (16). The DjlA homologue, a member of the DnaJ/Hsp40 family, was originally identified in Escherichia coli as a product of a hypothetical open reading frame (13, 72), and since then homologues have been identified in many other bacterial species, such as Coxiella burnetti (73), Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae, and Vibrio cholerae. DjlA carries the J-domain characteristic of the DnaJ/Hsp40 family and is essential for interaction with the Hsp70 homologue, DnaK, by increasing its ATPase activity (67). Overproduction of DjlA stimulates colanic acid production in E. coli (15, 16, 27, 73). Analysis of the DjlA null mutant demonstrated that the gene was not essential for viability (16). Although DjlA homologue is present in L. pneumophila (10), the role of this gene in pathogenesis has yet to be determined.
In this study, we investigated the role of the djlA gene in avoidance of fusion with lysosomes and its role in organelle trafficking within macrophages and in bacterial resistance to environmental stresses such as oxidative products, high temperature, high salt concentrations, and acidic pH.
MATERIALS AND METHODS
Bacterial Strains, plasmids, and media.
The bacterial strains and plasmids used in this work are described in Tables 1 and 2. The L. dumoffii Tex-KL strain and its derivatives were grown on buffered charcoal-yeast extract (BCYE) agar plates or in buffered yeast extract (BYE) broth. BYE broth was based on the formation of BCYE, but the charcoal and agar were omitted. E. coli DH5α (Toyobo Co., Ltd., Osaka, Japan) was used for the majority of the cloning experiments. As required, antibiotics were used at the following concentrations: kanamycin (KM), 30 μg/ml; chloramphenicol (CM), 5 or 20 μg/ml (for L. dumoffii); KM, 30 μg/ml; ampicillin (AMP), 50 μg/ml; CM, 20 μg/ml (for E. coli).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F−endA1 hsdR17 supE44D thi-1 recA1Δ(argF-lacZYA)U169(φ80δ lacZM15) gyrA96λ− | 30 |
| VCS257 | DP50 sup F[supE44 supF58 hsd53(rB mB) dapD8 lacY1 glnV44 Δ(gal-uvrB)47 tyrT58 gyrA29 tonA53 Δ(thyA57)] | Stratagene |
| L. dumoffii Tex-KL | ATCC 33343 | |
| HOLD254 | Tex-KL djlA::Tn903dIIlacZ | This study |
| HOLD491 | Tex-KL icmB(dotO)::Tn903dIIlacZ | This study |
| HMLD4001 | Tex-KL 17-kDa antigen::Tn903dIIlacZ | This study |
| HMLD4002 | Tex-KL dotC::Tn903dIIlacZ | This study |
| HOLD254-1 | Tex-KL djlA::Tn903dIIlacZ/pHRO18 | This study |
| HOLD254-2 | Tex-KL djlA::Tn903dIIlacZ/pHRO25 | This study |
| Plasmids | ||
| pGEM-T Easy | Ampr, lacZ, general cloning vector | Promega |
| pUC19 | Ampr, parental cloning vector | 70 |
| pBR322 | oriR (ColE1); Ampr Tcr | New England Biolabs |
| pHC79 | Wide-host-range pBR322 origin cosmid vector; Ampr Tcr | 31 |
| pLAW317 | rpsL MCSaoriT(RK2) CmrloxP oriR(ColE1) AmprloxP | 68 |
| pLAW330 | pLAW317::Tn903dIIlacZ tnpA(Tn903) oriR(f1) | 68 |
| pMMB207 | RSF1010 derivative, lncQ lac Iq Cmr Ptac oriT | 47 |
| pMMB207c | pMMB207 with 8-bp insertion in mobA; Mob | 45 |
| pHRO1 | Tn903dIIlacZ-containing HindIII fragment from HOLD254 in pBR322 | This study |
| pHRO2 | Tn903dIIlacZ-containing BamHI fragment from HOLD491 in pBR322 | This study |
| pHRO3 | Tn903dIIlacZ-containing HindIII fragment from HMLD4001 in pBR322 | This study |
| pHRO4 | Tn903dIIlacZ-containing HindIII fragment from HMLD4002 in pBR322 | This study |
| pHRO17 | Ampr; 4-kbp ScaI-EcoRI fragment containing djlA gene in pUC19 | This study |
| pHRO18 | 4-kbp Pst-EcoRI fragment containing djlA from pHRO17 in pMMB207c | This study |
| pHRO24 | PCR fragment of djlA cloned into pGEM-T Easy vector | This study |
| pHRO25 | EcoRI-PstI fragment (1,155 bp) containing djlA from pHRO24 cloned into pMMB207c | This study |
MCS, multiple-cloning site.
TABLE 2.
Strains of Legionella species used and their clinical relevance
| Legionella strain | Source | Clinical relevance |
|---|---|---|
| L. pneumophila serogroup1 (ATCC 33153) | Human | Yes |
| L. pneumophila serogroup6 (ATCC 33215) | Human | Yes |
| L. dumoffii (ATCC 33343) | Human | Yes |
| L. longbeachae (ATCC 33462) | Human | Yes |
| L. micdadei (ATCC 33218) | Human | Yes |
| L. bozemanii (ATCC 33217) | Human | Yes |
| L. feelei (ATCC 35849) | Human | Yes |
| L. gormanii (ATCC 33297) | Soil | Yes |
| L. jordanis (ATCC 33623) | Water | Yes |
| L. quinlivanii (ATCC 43830) | Water | No |
| L. moravica (ATCC 43877) | Water | No |
| L. gratiana (ATCC 49413) | Water | No |
| L. geestiana (ATCC 49504) | Water | No |
| L. rubrilucens (ATCC 35304) | Water | No |
| L. worsleiensis (ATCC 49508) | Water | No |
| L. jamestowniensis (ATCC 35298) | Soil | No |
| L. adelaidensis (ATCC 49625) | Water | No |
Cell culture.
J774A.1 macrophages (JCRB9108), refered to as J774 in this paper, were derived from mouse macrophage-like cells. The cell line A549 (JCRB0076) was donated by the Health Science Research Resources Bank, Osaka, Japan. The cells were established from a human alveolar epithelial carcinoma and have characteristics of well-differentiated type II pneumocytes. J774 cells and A549 cells were cultured in RPMI 1640 medium (GIBCO, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (FBS; Dainippon Pharmaceutical, Osaka, Japan). Acanthamoeba culbertsoni (44) was propagated at 28°C in 25-cm2 flasks (Falcon) containing 8 ml of peptone yeast extract glucose (PYG) and AC buffer (PYG + AC) (9, 46).
DNA manipulation.
Restriction enzymes and T4 DNA polymerase were purchased from Takara Shuzo Co., Ltd. (Kyoto, Japan) and Toyobo Co., Ltd. (Osaka, Japan). Calf intestine alkaline phosphatase was purchased from New England Biolabs Inc. (Beverly, Mass.). PCR amplification was performed by using The Ready To Go PCR-Beads (Amersham Pharmacia Biotech, Piscataway, N.J.) or Ex-Taq polymerase (Takara, Kyoto, Japan). Oligonucleotides used for PCR amplification were purchased from Japan Flour Co., Ltd. (Tokyo, Japan). Plasmid DNA was isolated from E. coli and L. dumoffii by using the Wizard Plus Mini Prep (Promega, Madison, Wis.) or the alkaline lysis method (58). Chromosomal DNA of L. dumoffii was purified using the Genomic Prep cells and tissue DNA isolation kit (Amersham Pharmacia Biotech). Electroporations were performed with a Bio-Rad Gene Pulser, as recommended by the manufacturer. Purification of DNA fragments from agarose gels for subcloning or labeling was carried out with a GFX PCR DNA and gel band purification kit (Amersham Pharmacia Biotech).
Transposon mutagenesis and construction of a bank of mutants.
L. dumoffii was mutated with the Tn903 derivative Tn903dIIlacZ, as described previously (68). Tn903dIIlacZ confers KM resistance (Kmr) and contains a 5′-truncated lacZ gene. Briefly, after electroporation of plasmid pLAW330, containing Tn903dIIlacZ, into L. dumoffii Tex-KL, bacteria were incubated in BYE broth for 5 h at 37°C and plated onto BCYE-KM plates. Kmr transformants containing β-galactosidase activity were identified as blue colonies after the the plates were overlaid with 0.8% agar containing 0.6 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) per ml. Kmr Cms colonies were saved as simple Tn903dIIlacZ insertion mutants of L. dumoffii.
Southern hybridization.
Chromosomal DNA from L. dumoffii strains was digested with HindIII, resolved on a 0.7% agarose gel in TBE buffer, and blotted onto a nylon membrane. DNA probes were prepared by random-primed labeling with digoxigenin-1 1-dUTP. The methods for prehybridization and hybridization and the washing conditions were the same as described previously (58), and the procedure for colorimetric detection of hybridized DNA was performed using the digoxigenin system (Roche Diagnostic Co., Indianapolis, Ind.).
Cloning and sequencing of the chromosomal junction of Tn903dIIlacZ insertion in the mutants.
Genomic DNA from the L. dumoffii mutants was digested with HindIII or BamHI and ligated to HindIII- or BamHI-digested pBR322. The ligation was used to transform DH5α, and the transformation mixture was plated on Luria-Bertani agar plates containing KM and AMP. Plasmid DNA was extracted, and the regions flanking Tn903dIIlacZ were sequenced with the lacZ primer (5′-CCCAGTCACGACGTTG-3′) and the Kmr primer (5′-AATTTAATCGCGGCCTCGAG-3′), corresponding to the 5′ and 3′ ends, respectively, of Tn903dIIlacZ.
Construction of plasmids for complementation.
For wild-type L. dumoffii genomic library construction, the genomic DNA was isolated from L. dumoffii and partially digested with Sau3 AI, and fragments of about 40 kb were purified. The fragments were ligated to the BamHI-digested, calf intestinal alkaline phosphatase-treated cosmid vecter pHC79 (31). The ligation products were packaged, in vitro, using the GigapackII Gold packaging system (Stratagene). Packaged hybrid cosmids were then used to infect E. coli strain VCS257. Recombinant clones were screened for the presence of a 1,085-bp PCR product (254-45), amplified using primers 254-4 (5′-GCTTCTTCCTTTCCACCATAA-3′) and 254-5 (5′-AGGTAGGCCTTGGGCAATTA-3′), by colony hybridization techniques. The probes used for colony hybridization were labeled with the digoxigienin random-primed DNA-labeling system (Roche). About 1,000 recombinant clones from the library were plated on the Luria-Bertani-plus-AMP plates for screening. Several positive cosmid clones were identified. The 4-kb ScaI-EcoRI fragment containing 254-45 from one of these cosmid clones was cloned into HincII-EcoRI-digested pUC19 to generate pHRO17. The recombinant clone was confirmed to contain 254-45 by Southern blot hybridization. The 4-kb PstI-EcoRI fragment from pHRO17 was cloned into shuttle vecter pMMB207c digested with PstI and EcoRI to generate pHRO18. pMMB207c is a nonmobilizable derivative of pMMB207 containing an 8-bp insertion within the mobA gene (at base 3325) and replicates stably in Legionella spp. (45). pHRO18 was electroporated into HOLD254 to yield the complemented strain HOLD254-1. The DNA fragment containing the djlA gene was PCR amplified from plasmid pHRO17 by using primer pair djlA-1-EcoRI (5′-GGGAATTCGAGTAGATACGAAGCAGGGT-3′) and djlA-2-PstI (5′-GGCTGCAGTTCCACCATAAACGGACTACA-3′). EcoRI and PstI sites (underlined sequences) were incorporated into these primers, respectively. The 1,155-bp PCR product that was generated contained 158 bp upstream of the ATG codon of djlA and 72 bp downstream of the stop codon of djlA. This PCR product was ligated into the pGEM-T Easy vector (Promega), resulting in pHRO24. The 1,155-bp EcoRI-PstI fragment from pHRO24 was then cloned into EcoRI-PstI-digested pMMB207c, creating pHRO25. The djlA mutant of L. dumoffii, HOLD254, was transformed with pHRO25 by electroporation. One of the transformants containing the desired plasmid was designated HOLD254-2. The cloned djlA gene was sequenced by using the primer within pMMB207c (pMMB207c-1; 5′-GTGTGGAATTGTGAGCGGAT-3′) and the primer within the djlA gene (254-3; 5′-GCTGATGGGCTGGATAGCAA-3′).
DNA sequence analysis of the region surrounding the djlA gene.
Primer pair djlA-3 (5′-AAGGATGGTAACTCTGACTCT-3′) and pHC79-2 (5′-TTGGAGCCACTATCGACTAC-3′) within the djlA gene and pHC79, respectively, were used to amplify the flanking region of the djlA gene from the cosmid clone containing djlA gene. This 4-kb PCR product and the 4-kb plasmid DNA within pHRO17 were sequenced using a primer walking technique. DNA-sequencing reactions were performed on plasmid templates with the CEQ DTCS-Quick Start kit (Beckman Coulter, Inc., Fullerton, Calif.) and the CEQ DNA analysis system (Beckman Coulter, Inc.). The nucleotide sequences and deduced amino acid sequences were compared to the GenBank database by using the programs BLASTX and BLASTP and also to the incomplete genomic database of L. pneumophilaPhiladelphia I (http://genome3.cpmc.columbia.edu/∼legion/ngnp1033033). Motif searches were carried out using the Prosite program.
Intracellular growth assay.
Growth of L. dumoffii in J774 cells and A549 cells was determined by using a previously described standard intracellular growth assay (43, 74). L. dumoffii strains were grown in BYE broth to the early stationary phase. Approximately 2 × 109 bacteria were pelleted, resuspended, and diluted (1:1,000) in RPMI 1640 tissue culture medium. The bacteria were then added to J774 cells and A549 cells (2 × 105 per well) in 24-well dishes to give a multiplicity of infection (MOI) of about 10. The infected cells were incubated at 37°C under 5% CO2-air for 1.5 h and washed three times with phosphate-buffered saline (PBS) to remove extracellular bacteria. To measure bacterial internalization, 1 ml of sterile distilled H2O was added to the wells to release intracellular bacteria from the host cells, and CFU were determined by plating dilutions on BCYE agar plates. To each of the remaining wells, 0.5 ml of fresh tissue culture medium was added. At 24-h intervals, the intracellular and extracellular bacteria in each well were combined, and the total CFU was determined by plating the dilutions onto BCYE agar plates. Infection of A. culbertsoni was carried out in an almost identical manner, except that bacteria were suspended in AC buffer and 0.05% Triton X-100 was added to release intracellular bacteria.
Assessment of phagosome-lysosome fusion by confocal microscopy.
L. dumoffii strains were grown overnight to saturation at 37°C in BYE broth. They were added at an MOI of 25 to 50 to 8 × 104 J774 cells on glass coverslips in 24-well tissue culture plates. The plates were centrifuged at 150 × g for 5 min at room temperature and incubated for 20 min in 5% CO2-air at 37°C. Extracellular bacteria were removed by washing three times with PBS, and fresh tissue culture medium was added to each well. The plates were returned to the incubator for 4 h. Cells were fixed for 15 min at room temperature in P-PFA (4% paraformaldehyde in 1 × PBS [pH 7.4]) (43, 74). Coverslips were immersed in PBS-0.1% Saponin for 5 min to permeabilize the cells and blocked with 5% FBS in PBS for 5 min. Lysosomes and late endosomes were stained with rat monoclonal antibody 1 D4B (1:100) specific for LAMP-1 or Ab1 93 (1:100) specific for LAMP-2, and the bacteria were stained with rabbit anti-L. dumoffii polyclonal antibody (1: 10,000) for 1 h. The cells were washed with blocking solution three times and incubated for 30 min with Cy3-labeled goat anti-rat secondary antibody (1:300) and Alexa488-labeled goat anti-rabbit secondary antibody (1:300). The coverslips were then washed three times with blocking solution. All antibody dilutions were performed with PBS containing 0.5% FBS and 0.1% Saponin. Coverslips were inverted onto 1 μl of mounting medium (50% glycerol) on glass slides (39). Fluorescence was viewed using a Radiance 2100 MP confocal microscope (Bio-Rad Laboratories, Richmond, Calif.). Alexa488- and Cy3-labeled secondary antibodies were purchased from Molecular Probes (Eugene, Oreg.). Rat monoclonal antibodies to LGP107 (mouse LAMP-1) and LGP96 (mouse LAMP-2) were purified from mouse liver lysosomal membranes, as described previously (23).
Quantification of phagosome-lysosome fusion by electron microscopy.
To label cell lysosomes, J774 macrophages were incubated with bovine serum albumin (BSA)-conjugated colloidal 15-nm-diameter gold particles (BSA-gold) overnight, chased for 3 h, and pulsed with stationary-phase L. dumoffii strains at an MOI of 50 (19, 33). At 4 h postinfection, the cells were fixed and processed for electron microscopy as previously described (66). Briefly, infected macrophages were fixed with 2% glutaraldehyde and then with 1% OsO4, dehydrated with ethanol, and embedded in Epon. Ultrathin sections were stained with uranyl acetate followed by lead citrate and examined by electron microscopy in a JEM 2000EX instrument (JEOL, Ltd., Tokyo, Japan).
Examination of RER recruitment by transmission electron microscopy.
J774 cells were plated in 90-mm-diameter petri dishes (2 × 105 cells/ml) and infected with stationary-phase L. dumoffii strains at an MOI of 20 for 8 and 24 h (32). Ultrathin sections were prepared as described above.
Assays for survival under stress conditions.
L. dumoffii strains were grown for 2 to 3 days on BCYE agar plates and used to inoculate 4 ml of BYE medium. The bacteria were then grown at 37°C with aeration for at least 16 h. The initial CFU count was about 1010 per ml. Cells were divided into aliquots, centrifuged, and resuspended in equal volumes of 1 × M63 salts [22.0 mM KH2PO4, 40.2 mM K2HPO4, 14.6 mM (NH4)2SO4 500 nM FeSO4 (pH6.5)]. One aliquot was used for measuring the untreated CFU. For heat shock, aliquots were transferred to 48°C and incubated for 60 min. For oxidative stress, aliquots were exposed to 10 mM H2O2 for 30 min. For osmotic shock, aliquots were exposed to 5 M sodium chloride for 30 min. For acid shock, aliquots were resuspended in 0.1 M citric acid (pH 3) for 5 min. Except for heat stress, the cells were incubated in a 37°C heat block. At the indicated time points, the cells were washed with 1 × M63 salts and serially diluted to determine the CFU on BCYE agar plates (29).
Detection of a djlA gene in other Legionella spp.
The presence of djlA in 17 different strains of Legionella spp. was examined by PCR with the primer pair djlA-cons-1 (5′-ATAACAACCTGGTGGGGAAA-3′) and djlA-cons-2 (5′-TGGGCAATTAATTTATCTGGATG-3′), located in the transmembrane domain (TMD) and J domain within the djlA gene, respectively, which gave a 791-bp product. PCR was carried out by using chromosomal DNA from BCYE plate-grown bacteria as a template.
RESULTS
Isolation of intracellular growth mutants.
Wild-type L. dumoffii Tex-KL was mutagenized with Tn903dIIlacZ as described previously (57, 68). Plasmid pLAW330 containing Tn903dIIlacZ was introduced into L. dumoffii, and 790 Kmr Cms mutants of L. dumoffii (HOLD strains 1 to 656 and HMLD strains 4004 to 4044 and 4048 to 4140) with various levels of β-galactosidase activity were isolated. The 790 mutants were individually screened for their ability to kill mouse macrophage-like J774 cells and human alveolar epithelial A549 cells. The mutants were grown for 2 days in 96-well tissue culture plates containing BYE medium. Then 5-μl samples of 2-day-old cultures of mutants were transferred to another 96-well tissue culture plate containing J774 cells or A549 cells. At each 24-h time point after infection, the monolayers were visually examined to determine the extent of killing of both J774 cells and A549 cells. From several assays, we isolated five mutants, based on their reproducible phenotypes. Southern blot analysis of the HindIII-digested genomic DNA of each of the five mutants probed with pLAW330 showed that four of them contained a single copy of the Tn903dIIlacZ insertion and that these insertions were distributed in distinct locations within the chromosome of L. dumoffii (data not shown). For reasons not yet understood, one of the mutants showed no hybridization. Therefore, the four strains were chosen for further analysis. In vitro, the growth of these four mutants in BYE broth and on BCYE agar plates was similar to that of the wild-type strain (data not shown).
Intracellular growth phenotype of the mutants within J774 macrophages and alveolar epithelial cells.
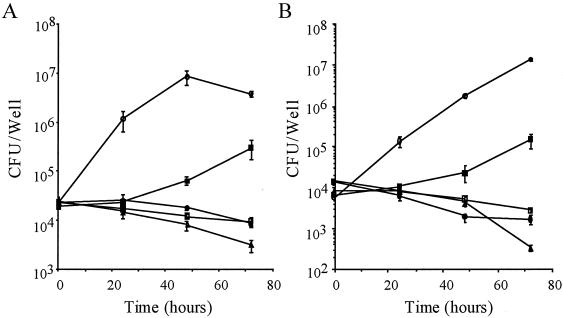
We examined the four candidates for their capacity to survive and to replicate within J774 macrophages and A549 epithelial cells. Bacterial CFU were determined over 3 days. The wild-type strain multiplied over 100-fold during the 3-day incubation period within J774 macrophages (Fig. 1A). HOLD254 showed a 1-log-unit increase after 3 days of incubation, whereas HOLD491, HMLD4001, and HMLD4002 did not grow during the incubation period in J774 cells. Within A549 epithelial cells (Fig. 1B), the wild-type strain increased approximately 1,000-fold over the 3-day period, while there was a 10-fold increase in the number of intracellular bacteria of HOLD254 over 3 days. For HOLD491 and HMLD4001, the number of CFU after 3 days of infection decreased 1 log unit to the initial number of CFU, and HMLD4002 was severely defective in intracellular survival (Fig. 1B).
FIG. 1.
Intracellular growth of L. dumoffii strains within J774 mouse macrophages (A) and A549 human epithelial cells (B). The formation of colonies (CFU per milliliter) was determined at the times indicated, in triplicate, for at least two independent experiments. Error bars indicate the standard deviations determined from samples taken from one experiment. Symbols: ○, L. dumoffii wild-type strain; ▪, HOLD254; □, HOLD491; •, HMLD4001; ▵, HMLD4002.
Sequence analysis of the junctions of Tn903dIIlacZ insertions.
We cloned the HindIII fragment containing the Tn903dIIlacZ insert and the flanking sequences of the mutants (HOLD254, HOLD491, HMLD4001, and HMLD4002). Using the primer within Tn903dIIlacZ, we partially sequenced and analyzed them to identify the genes responsible for intracellular multiplication. The results are summarized in Table 3. Sequence homology searches against the Gen Bank database were done with these genes and corresponding proteins. HOLD254, HOLD491, and HMLD4002 contain insertions within the genes homologous to known L. pneumophila genes. The gene disrupted in HOLD254 is the djlA (for “dnaJ-like A”) gene, encoding a member of the Hsp40 protein family, which has not been characterized in L. pneumophila. HOLD491 and HMLD4002 had a transposon insertion in their sequences similar to icmB (dotO) and dotC, respectively, identified as genes essential for intracellular growth in L. pneumophila (5, 51). HMLD4001 had an insertion within a gene whose product showed amino acid similarity to the 17-kDa antigen, VirB5, of B. henselae; the gene is located within the virB locus, which encodes a putative type IV secretion system together with the downstream virD4 gene (14, 49, 59). Recently, Schulein and Dehio (59) also showed that VirB4 and VirD4, encoded by the virB and virD4 loci of B. tribucorum, were required for establishing intraerythrocytic bacteremia.
TABLE 3.
Sequence similarities of L. dumoffii genes responsible for intracellular multiplicationa
| Mutant strain | Homologous gene | Organism | % Identity | % Positive |
|---|---|---|---|---|
| HOLD254 | djlA | Legionella pneumophila | 61 | 73 |
| HOLD491 | icmB/dotO | Legionella pneumophila | 89 | 95 |
| HMLD4001 | 17-kDa antigen gene | Bartonella henselae | 26 | 43 |
| HMLD4002 | dotC | Legionella pneumophila | 85 | 92 |
The values are taken from a Basic Local Alignment Search Tool for amino acid comparison (BLASTX program).
Complementation of an L. dumoffii djlA mutant.
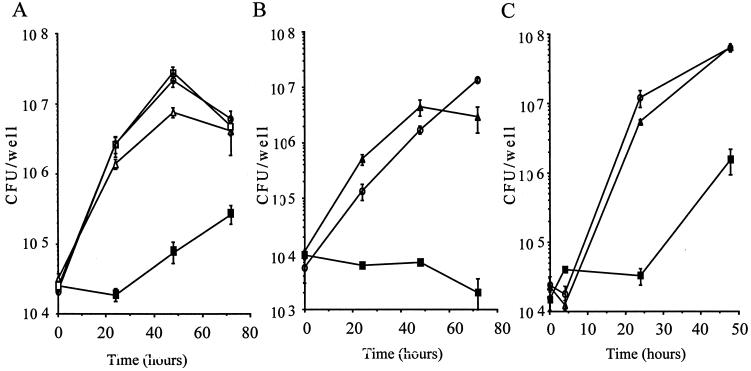
DjlA is known to be a heat shock protein DnaJ/Hsp40 homologue. The virulence of the djlA mutant was compared with that of the wild-type strain and the djlA-complemented mutant in J774 macrophages, A549 epithelial cells, and A. culbertsoni. The djlA mutant showed only a 100-fold increase in intracellular replication within A. culbertsoni (Fig. 2C). As shown in Fig. 2, bacterial growth was fully restored in the complemented strains HOLD254-1 and HOLD254-2. The restoration of the wild-type-level of multiplication of the djlA mutant within these cells, achieved after complementation in trans with the cloned djlA gene, is proof of the important role of djlA in the intracellular growth of L. dumoffii.
FIG. 2.
Complementation of intracellular growth defects of djlA mutant HOLD254 in J774 macrophages (A), in A549 epithelial cells (B), and in A. culbertsoni (C). Growth was measured over 72 h (A and B) or 48 h (C). The data points and error bars represent the mean CFU/well for triplicate samples from a typical experiment (performed at least twice) and their standard deviations. Symbols: ○, L. dumoffii wild-type strain; ▪, HOLD254; ▵, HOLD254-1 (djlA/pHRO18); □, HOLD254-2 (djlA/pHRO25).
Complete sequence and genetic structure of djlA.
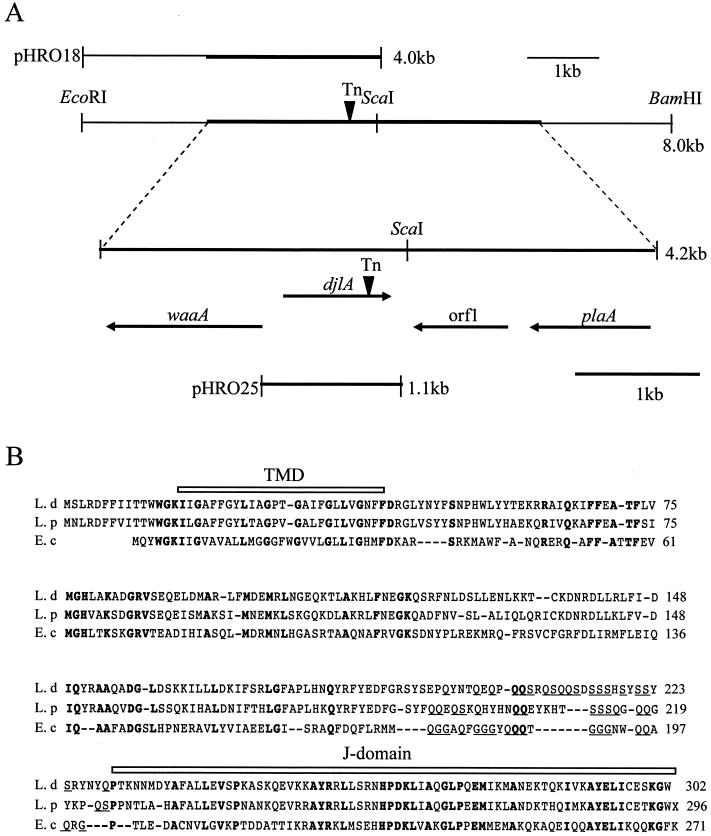
Figure 3A shows the organization around the djlA gene and the location of the Tn903dIIlacZ insertion. The transposon insertion (Tn) was located in the J domain at the C terminus of the predicted protein, which was the defined feature of the DnaJ family of molecular chaperones (16, 27). Since the two genes (waaA and orf1) which flanked djlA were both oriented in the opposite direction from the djlA, we consider the djlA to be transcribed monocistronically, and this transposon has no polar effect. The deduced amino acid sequence of L. dumoffii DjlA, together with L. pneumophila DjlA and E. coli DjlA, is presented in Fig. 3B. The putative L. dumoffii djlA gene encodes a protein of 302 amino acids with a predicted molecular mass of 35.33 kDa and an isoelectric point of 9.65. The protein size is similar to that of the L. pneumophila (296 amino acids) and E. coli (271 amino acids) proteins. L. dumoffii DjlA has 61% identity to L. pneumophila DjlA and 32% identity to E. coli DjlA (10, 16, 73). A potential TMD at the N terminus contains six glycines, spaced through the TMD at every three to five residues, which is similar to the structure of the TMD of E. coli (15, 16). There is a remarkable difference in the N terminus of DjlA protein between E. coli and Legionella spp. Clarke et al. (16) have demonstrated that E. coli DjlA is localized to the inner membrane and has a rare type III topology (i.e. N-out, C-in), with the N-terminal 6 to 8 residues located in the periplasm. Legionella spp. have longer stretches (15 residues) before the TMD structure, which are probably exposed in the periplasm. Another unique feature of Legionella DjlA is a glutamate-serine (QS)-rich spacer located before the J domain, instead of the glutamate-glycine (QG)-rich spacer of E. coli DjlA (Fig. 3B) (16). The cellular role of these QS- or QG-rich regions remain to be elucidated.
FIG. 3.
Chromosomal arrangement of the region surrounding the djlA gene and sequence alignment of DjlA proteins. (A) At the top is a plasmid used for complementation studies (pHRO18) and an 8-kb region of the L. dumoffii cosmid clone including the djlA gene, along with the location of relevant restriction enzyme sites. The thick line represents the DNA region that we sequenced. Below these diagrams, the distance between the djlA gene and neighboring genes and the orientation and size of the transcribed genes are delineated by the arrows below the 4.2-kb sequenced region. Another plasmid used for complementation studies (pHRO25) is also shown. The site of the Tn903dIIlacZ insertion (Tn) is indicated by the inverted arrowhead. The full names of the gene mapped are as follows: waaA, Kdo transferase gene; djlA, dnaJ-like A gene; plaA, lysophospholipase A gene. Orf1 is a putative open reading frame which showed no homology to known genes. (B) Sequence similarity of the predicted Dj1A protein of L. dumoffii (L.d, top line), L. pneumophila (L.p, middle line) and E. coli (E.c, bottom line). Amino acid residues conserved in the three sequences, appear in bold type. Gaps marked by dashes are introduced to reveal the maximal similarity among the sequences. The C-terminal J-domain and the N-terminal TMD are shown schematically above the sequences.
Quantification of endocytic maturation.
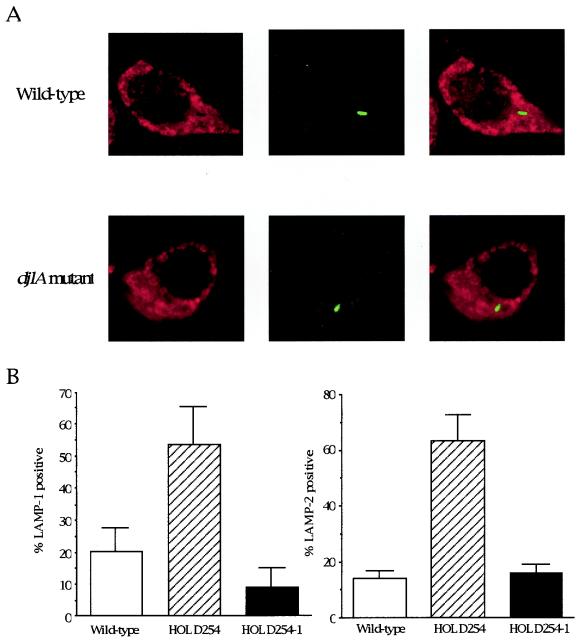
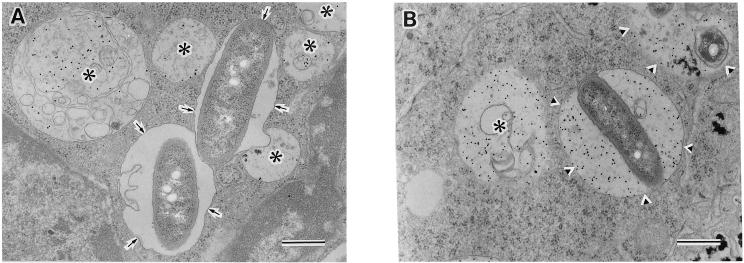
To determine whether the L. dumoffii strains were able to inhibit endocytic maturation, we measured the colocalization of L. dumoffii phagosomes with endocytic markers LAMP-1 and LAMP-2. J774 macrophages were infected with postexponential phase L. dumoffii strains for 4 h (Fig. 4). The permeabilized cells were stained with monoclonal antibody 1D4B or Abl 93, specific for late endosomal and lysosomal proteins, LAMP-1 or LAMP-2. The djlA mutant was found in phagosomes that contained LAMP-1 (Fig. 4A), indicating that these vacuoles had fused with late endosomes, whereas, phagosomes containing wild-type L. dumoffii did not colocalize with LAMP-1 (Fig. 4A). When each L. dumoffii strain found in the phagosomes was scored for fusion with the late endosomal/lysosomal markers LAMP-1 and LAMP-2, approximately 80% of the wild-type bacteria were found in LAMP-1- and LAMP-2-negative phagosomes while 50 to 60% of the HOLD254 was found in LAMP-1- and LAMP-2-positive compartments (Fig. 4B). We also performed the same analysis for HOLD4002, the dotC mutant, and found that this mutant followed the same endocytic pathway as HOLD254, with 60 to 70% LAMP-1- and LAMP-2-positive (data not shown). We also conducted an assay of phagosome-lysosome fusion, at the ultrastructural level, using electron microscopy. BSA-gold was used as a pinocytic, fluid-phase marker of the endosomal-lysosomal pathway. BSA-gold was accumulated mainly in lysosomes after endocytosis of the conjugate-containing medium overnight at 37°C, followed by a chase period of 3 h at 37°C in conjugate-free medium as previously described (33). After a pulse with L. dumoffii strains and another chase for 4 h, electron microscopy counting of L. dumoffii-containing phagosomes that fused with BSA-gold-labeled lysosomes was performed to assess fusion (Fig. 5). Wild-type-strain-containing phagosomes did not fuse with BSA-gold-marked lysosomes (Fig. 5A). Quantitation showed that only 11.4% (24 of 210) of the phagosomes containing the wild-type strain fused with BSA-gold-marked lysosomes. On the other hand, 85% (187 of 220) of the phagosomes containing the djlA mutant strain accumulated BSA-gold (Fig. 5B). Thus, the djlA mutant was not able to evade phagosome-lysosome fusion.
FIG. 4.
Colocalization of the intracellular growth mutant with late endosomal/lysosomal marker LAMP-1 or LAMP-2 in J774 mouse macrophage cells by confocal laser-scanning microscopy. J774 macrophages were incubated with the L. dumoffii mutant or wild-type strain for 4 h. (A) Late endosomes and lysosomes stained with rat monoclonal antibody 1D4B, specific for LAMP-1, and Cy3-labeled anti-rat secondary antibody (red) are shown on the left. Bacteria stained with rabbit polyclonal antibody specific for L. dumoffii Tex-KL and Alexa488-labeled anti-rabbit secondary antibody (green) are shown in the middle. Merged images showing LAMP-1-positive bacteria (yellow) and LAMP-1-negative bacteria (green) are shown on the right. (B) Data were collected from about 100 intracellular bacteria in total. The percentage that is LAMP-1 or LAMP-2 positive was calculated by dividing the number of colocalizing intracellular bacteria by the total number of intracellular bacteria scored. The average and standard deviation described here were calculated from three coverslips per strain in two independent experiments.
FIG. 5.
Distribution of a lysosomal marker, BSA-gold, in phagosomes containing the wild-type strain or the djlA mutant strain. To label the lysosomal compartment, J774 cells were incubated with 15-nm BSA-gold overnight, washed, and then chased for 3 h. Cells were then infected with wild-type strain (A) or djlA mutant strain (B). At 4 h postinfection, the cells were fixed and processed for electron microscopy. Arrows in panel A indicate phagosomes containing no detectable gold; arrowheads in panel B indicate phagolysosomes containing BSA-gold; asterisks indicate, lysosomes containing BSA-gold. Bar, 0.5 μm.
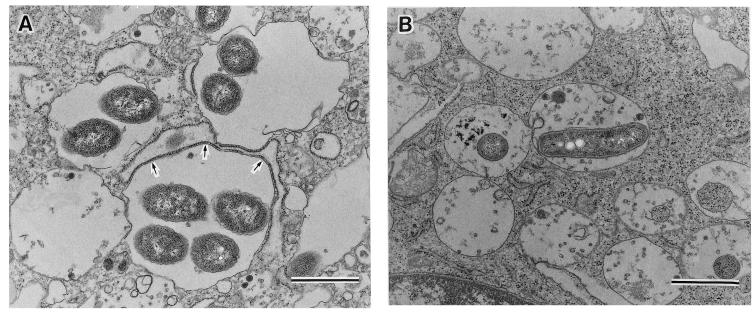
Recruitment of the RER.
In mammalian macrophages and protozoa, L. pneumophila replicates intracellularly in specialized vacuoles surrounded by the RER of the host cells (25, 32). To determine the intracellular location of L. dumoffii, we examined J774 macrophages infected with wild-type and djlA mutant L. dumoffii by using transmission electron microscopy. At 8 h postinfection, the RER around 61 (37.2%) of 164 phagosomes containing wild-type strains were recruited (Fig. 6A) whereas we could not find any phagosomes containing the djlA mutant surrounded by RER or attached directly by ribosomes (0 of 153 phagosomes). This was also the case at 24 h (Fig. 6B and data not shown). Phagosomes containing djlA mutant cells appeared to harbor much debris, resulting from fusing lysosomes with these vacuoles, while phagosomes containing wild-type cells did not have any contents other than replicating L. dumoffii cells (Fig. 6). At 24 h postinfection, many phagosomes containing wild-type cells were broken and their inhabiting macrophages were lysed (data not shown).
FIG. 6.
Transmission electron micrographs of J774 mouse macrophages infected by the wild-type L. dumoffii (A) and the djlA mutant HOLD254 (B) at 8 h after infection. (A) Wild-type L. dumoffii-containing phagosomes were surrounded by RER (arrows). (B) HOLD254-containing phagosomes appeared to harbor much debris resulting from fusing lysosomes. Bar, 1.0 μm.
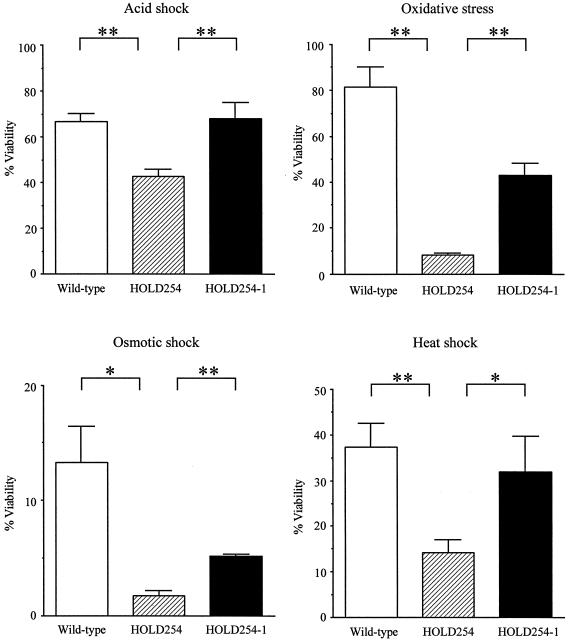
Susceptibility of the djlA mutant to stress stimuli.
In eukaryotic host cells, intracellular pathogens encounter hostile conditions such as toxic oxygen or nitrogen derivatives, intraphagosomal acidification, and harsh degradative enzymes (54, 62). As mentioned above, djlA is essential for intracellular growth of L. dumoffii. Thus, we examined whether the djlA mutant has an increased susceptibility to different environmental stresses. Since previous publications (12, 29) had demonstrated that L. pneumophila induces stress resistance in the stationary phase, L. dumoffii strains were grown to the stationary phase in BYE medium and subjected to acid shock, oxidative stress, osmotic stress, and heat shock (pH 3 for 5 min, 10 mM H2O2 for 30 min, 5 M sodium chloride for 30 min, and 48°C for 60 min, respectively). Compared to the wild-type strain, there was an elevated susceptibility to all stress conditions of the djlA mutant strain. There was an increase in the sensitivity of the mutant of 9.8-, 7.4-, 2.6-, and 1.6-fold on exposure to oxidative stress, osmotic stress, heat shock, and acid shock, respectively (Fig. 7). These results suggest that DjlA participated in the protection of L. dumoffii on exposure to environmental stress. In the djlA-complemented strain, in contrast, resistance to all stress stimuli was restored. The variability in the degree of complementation may result from the different expression of genes from the plasmid and the chromosome.
FIG. 7.
Sensitivities of in vitro-grown stationary-phase wild-type L. dumoffii (open bars), the djlA mutant strain (hatched bars), and the djlA complemented strain (solid bars) to oxidative stress, osmotic stress, acid stress, and heat shock (10 mM hydrogen peroxide for 30 min, 5 M sodium chloride for 30 min, pH 3 for 5 min, and 48°C for 60 min, respectively). Stationary-phase cultures were exposed to each stress as described in Materials and Methods. The percentage of viable bacteria was calculated by dividing the CFU obtained from plating the bacteria onto BCYE agar plates following exposure to the indicated stress by the CFU of the bacteria obtained from plating the bacteria onto BCYE agar plates prior to exposure to the stress and multiplying by 100. Experiments were performed at least three times, and the results represent the mean and standard deviation. Results were analyzed for significance by analysis of variance and by a two-tailed, unpaired t test. Asterisks indicate significant differences between the djlA mutant and two other strains. (*, P < 0.01; **, P < 0.001).
Presence of djlA in other Legionella spp.
To determine whether djlA is also present in nonpathogenic Legionella species, PCR amplification with primers in the djlA gene was performed for 17 different Legionella strains. All the strains used in this experiment are listed in Table 2. The expected 790-bp band was observed in all Legionella strains tested except L. jordanis and L. adelaidensis, irrespective of whether the strain was pathogenic (data not shown). Thus, djlA is not unique to particular Legionella strains.
DISCUSSION
Legionella spp. are facultative intracellular bacteria that overcome host cell defenses. Although many studies have been undertaken to understand the intracellular life cycle of L. pneumophila, very few species other than L. pneumophila have been examined phenotypically. The aim of this study was to uncover how L. dumoffii survives and replicates in mammalian cells and to identify the genes of L. dumoffii needed for intracellular growth. We isolated 4 mutants that were defective in intracellular growth in macrophages and alveolar epithelial cells among 790 independently derived Tn903dIIlacZ mutants of L. dumoffii. The defect in intracellular growth of these four mutants cannot be attributed to a defect in adherence or entry, because almost equal numbers of mutants and wild-type cells were present within mammalian cells at 0 h postinfection. Two of the four mutants had a transposon insertion in either the dotC or icmB homologues (5, 51, 60). The dot/icm genes are required for intracellular multiplication of L. pneumophila (5, 51, 60). Our results suggest that the dotC and icmB genes of L. dumoffii and L. pneumophila appear to perform similar functions. We propose that the dot/icm genes are involved in the pathogenesis of most Legionella species, since these genes are important in the intracellular growth of these distinct Legionella species.
One of the mutants defective in intracellular growth was shown to have a transposon insertion in the gene which had sequence similarity to the djlA gene (16). Cloning and sequence analysis of this gene revealed that the primary structure of L. dumoffii DjlA showed homology to other bacterial DjlA proteins (10, 16, 73). DjlA is the third DnaK cochaperone of E. coli, containing a J domain highly conserved in the DnaJ/Hsp40 family of molecular chaperones, including DnaJ and CbpA (16, 27, 65). CbpA is 39% identical to DnaJ along its entire length (64), while DjlA does not have any sequence similarity other than the J domain to DnaJ and CbpA in E. coli (26, 37). DjlA is unique in its structure and location in the DnaJ family. The J domain resides in the C terminus of DjlA but in the N terminus of other DnaJ family proteins. The N terminus of DjlA is integrated into the inner membrane through the single TMD, and the C-terminal J domain is located in the cytoplasm (16), while the whole of DnaJ and CbpA is localized in the cytoplasm. Moderate overproduction of djlA can trigger the synthesis of the colanic acid capsule in E. coli, mediated by the two-component regulatory system RcsC-RcsB, cooperating with DnaK and GrpE, but not DnaJ (15, 27, 37, 73). Unlike CbpA, DjlA could not adequately complement bacteriophage λ growth in a DnaJ-null background or restore bacterial growth above 40°C or below 16°C in the dnaJ cbpA null background in E. coli (15, 26, 37). The DjlA deletion mutant exhibits no apparent growth phenotype in E. coli (15, 16, 26). Thus, the true role of DjlA has been unclear.
We demonstrated that the djlA mutant of L. dumoffii exhibited a defective growth phenotype in mammalian cells and protozoan hosts. Phagosomes containing wild-type L. dumoffii excluded the late endosomal/lysosomal markers LAMP-1 and LAMP-2 and a lysosomal marker, BSA-gold, and were surrounded by RER in J774 macrophages, while djlA mutant-bearing phagosomes contained LAMP-1, LAMP-2, and BSA-gold and were not surrounded by RER (Fig. 4 to 6). It has been reported that L. pneumophila is targeted into RER-surrounding phagosomes that do not fuse with lysosomes in mammalian cells (25, 33), while L. micdadei is targeted into RER-free phagosomes that are thought to fuse to lysosomes in mammalian cells (3, 36). Doyle et al. (20) reported that virulent L. longbeachae-containing phagosomes were surrounded by RER but avirulent L. longbeachae-containing phagosomes did not have RER. Our observations suggest that L. dumoffii might replicate in phagosomes which have not fused with lysosomes and are able to recruit host cell organelles, similar to that reported for L. pneumophila. The djlA mutant seemed to be intact (Fig. 5B), and no loss of CFU was observed during the infection (Fig. 1A and 2A). It is possible that the mutant bacteria are in either a late endosomal or a nondegradative lysosomal compartment, as described by Joshi et al. (35). The frequency of recruitment of L. dumoffii RER at 8 h is lower than that reported for L. pneumophila (32). We suspect that association with ER and avoidance of lysosomes by L. dumoffii is temporary, as shown for L. pneumophila (63).
Although the precise function of DjlA is unclear, it does not seem to play a direct role in intracellular trafficking. DjlA might contribute to folding or transportation of the proteins, such as Dot/Icm proteins, which play an important role in intracellular survival and growth. Most of the Dot/Icm proteins are located in the bacterial membranes, where they may associate to form a large transport complex, the type IV secretion apparatus (17, 43, 51, 60, 61). DjlA might cooperate with Dot/Icm proteins through their interaction in the membranes, since the N-terminal portion of DjlA is located in the cytoplastic membrane (16, 37). It has been reported that the two-component regulatory system, PhoP-PhoQ, of Salmonella enterica serovar Typhimurium plays an essential role in survival within macrophages (28). It is possible that DjlA promotes L. dumoffii to adapt to intracellular environments and to coordinate with the two-component signal transduction systems. In vitro, DjlA-deficient mutants showed an increased susceptibility to several stresses, including oxidative stress, that might be encountered by bacteria in mammalian cells. DjlA might protect the genes or proteins, including Dot/Icm and catalase-peroxidase (7), that are important for intracellular growth, from harmful stress in a direct or indirect manner. Several lines of evidence for the important role of stress proteins in intracellular growth and virulence have been reported for intracellular pathogens; these include DnaK of Brucella suis (38), ClpC and ClpP of Listeria monocytogenes (24, 54), Lon of B. abortus (53), and GsrA of Yersinia enterocolitica (69). In L. pneumophila, at least 30 proteins are included during the intracellular infection of macrophages and at least 13 of these proteins, including GroEL (Hsp60), GroES, and GspA, are also induced by several stress conditions in vitro (1, 2, 21). Recently, Pedersen et al. (50) demonstrated direct evidence for the role of the stress protein of L. pneumophila, HtrA, during intracellular growth in mammalian cells but not in protozoan cells. Our data indicated that DjlA plays an important role during intracellular growth in both mammalian and protozoan cells. Besides Dot/Icm proteins, stress proteins or molecular chaperones might play an important role in the intracellular growth of the Legionella species.
In conclusion, we showed the essential role of L. dumoffii Dot/Icm homologues and DjlA during the intracellular infection of mammalian cells and protozoa. The precise mechanism of DjlA involvement in intracellular multiplication, including interaction with DnaK, remains to be elucidated. Further investigation of specific substrates with which DjlA interacts will lead to a better understanding of the intracellular survival mechanism in the Legionella species.
Acknowledgments
We acknowledge H. A. Shuman for his generous gifts of plasmids pLAW330 and the pMMB207c. We thank H. Nakayama and C. C. Sze for scientific discussion. We also thank H. Fujita, K. Iida, and H. Kajiwara for technical assistance. We thank L. Saza for manuscript preparation.
This work was supported by grants-in-aid for scientific research (B)(2)14370094, (B)(1)12490009, and (C)(2)15590391 from the Ministry of Education, Science, Culture and Sports of Japan. This work was also supported by Health and Labour Sciences research grants (H15-Ganyobou-095) from the Ministry of Health, Labour and Welfare.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Abu Kwaik, Y., B. I. Eisenstein, and N. C. Engleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 61:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y., L. Y. Gao, O. S. Harb, and B. J. Stone. 1997. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol. Microbiol. 24:629-642. [DOI] [PubMed] [Google Scholar]
- 3.Abu Kwaik, Y., C. Venkataraman, O. S. Harb, and L. Y. Gao. 1998. Signal transduction in the protozoan host Hartmannella vermiformis upon attachment and invasion by Legionella micdadei. Appl. Environ. Microbiol. 64:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alli, O. A., S. Zink, N. K. Von Lackum, and Y. Abu-Kwaik. 2003. Comparative assessment of virulence traits in Legionella spp. Microbiology 149:631-641. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, H. L., J. P. Vogel, and R. R. Isberg. 1998. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baine, W. B. 1985. Cytolytic and phospholipase C activity in Legionella species. J. Gen. Microbiol. 131:1383-1391. [DOI] [PubMed] [Google Scholar]
- 7.Bandyopadhyay, P., B. Byrne, Y. Chan, M. S. Swanson, and H. M. Steinman. 2003. Legionella pneumophila catalase-peroxidases are required for proper trafficking and growth in primary macrophages. Infect. Immun. 71:4526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benin, A. L., R. F. Benson, and R. E. Besser. 2002. Trends in legionnaires disease, 1980-1998: declining mortality and new patterns of diagnosis. Clin. Infect. Dis. 35:1039-1046. [DOI] [PubMed] [Google Scholar]
- 9.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64:668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brabetz, W., C. E. Schirmer, and H. Brade. 2000. 3-Deoxy-d-manno-oct-2-ulosonic acid (Kdo) transferase of Legionella pneumophila transfers two Kdo residues to a structurally different lipid A precursor of Escherichia coli. J. Bacteriol. 182:4654-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner, D. J. 1985. The new species of Legionella. Int. J. Syst. Bacteriol. 35:50-59. [Google Scholar]
- 12.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casaregola, S., M. Chen, N. Bouquin, V. Norris, A. Jacq, M. Goldberg, S. Margarson, M. Tempete, S. McKenna, H. Sweetman, et al. 1991. Analysis of a myosin-like protein and the role of calcium in the E. coli cell cycle. Res. Microbiol. 142:201-207. [DOI] [PubMed] [Google Scholar]
- 14.Chen, L., Y. Chen, D. W. Wood, and E. W. Nester. 2002. A new type IV secretion system promotes conjugal transfer in Agrobacterium tumefaciens. J. Bacteriol. 184:4838-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke, D. J., L. B. Holland, and A. Jacq. 1997. Point mutations in the transmembrane domain of DjlA, a membrane-linked DnaJ-like protein, abolish its function in promoting colanic acid production via the Res signal transduction pathway. Mol. Microbiol. 25:933-944. [DOI] [PubMed] [Google Scholar]
- 16.Clarke, D. J., A. Jacq, and I. B. Holland. 1996. A novel DnaJ-like protein in Escherichia coli inserts into the cytoplasmic membrane with a type III topology. Mol. Microbiol. 20:1273-1286. [DOI] [PubMed] [Google Scholar]
- 17.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38:719-736. [DOI] [PubMed] [Google Scholar]
- 18.Cordes, L. G., H. W. Wilkinson, G. W. Gorman, B. J. Fikes, and D. W. Fraser. 1979. Atypical Legionella-like organisms: fastidious water-associated bacteria pathogenic for man. Lancet ii:927-930. [DOI] [PubMed] [Google Scholar]
- 19.Da Silva, T. R., J. R. De Freitas, Q. C. Silva, C. P. Figueira, E. Roxo, S. C. Leao, I. A. De Freitas, and P. S. Veras. 2002. Virulent Mycobacterium fortuitum restricts NO production by a gamma interferon-activated J774 cell line and phagosome-lysosome fusion. Infect. Immun. 70:5628-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyle, R. M., N. P. Cianciotto, S. Banvi, P. A. Manning, and M. W. Heuzenroeder. 2001. Comparison of virulence of Legionella longbeachae strains in guinea pigs and U937 macrophage-like cells. Infect. Immun. 69:5335-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez, R. C., S. M. Logan, S. H. Lee, and P. S. Hoffman. 1996. Elevated levels of Legionella pneumophila stress protein Hsp60 early in infection of human monocytes and L929 cells correlate with virulence. Infect. Immun. 64:1968-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 23.Furuno, K., T. Ishikawa, K. Akasaki, S. Yano, Y. Tanaka, Y. Yamaguchi, H. Tsuji, M. Himeno, and K. Kato. 1989. Morphological localization of a major lysosomal membrane glycoprotein in the endocytic membrane system. J. Biochem. (Tokyo) 106:708-716. [DOI] [PubMed] [Google Scholar]
- 24.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 25.Gao, L. Y., O. S. Harb, and Y. A. Kwaik. 1998. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect. Immun. 66:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genevaux, P., F. Schwager, C. Georgopoulos, and W. L. Kelley. 2001. The djlA gene acts synergistically with dnaJ in promoting Escherichia coli growth. J. Bacteriol. 183:5747-5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genevaux, P., A. Wawrzynow, M. Zylicz, C. Georgopoulos, and W. L. Kelley. 2001. DjlA is a third DnaK co-chaperone of Escherichia coli, and Dj1A-mediated induction of colanic acid capsule requires DjlA-DnaK interaction. J. Biol.Chem. 276:7906-7912. [DOI] [PubMed] [Google Scholar]
- 28.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 31.Hohn, B., and J. Collins. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291-298. [DOI] [PubMed] [Google Scholar]
- 32.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horwitz, M. A., and F. R. Maxfield. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi, A. D., S. Sturgill-Koszycki, and M. S. Swanson. 2001. Evidence that Dot-dependent and -independent factors isolate the Legionella pneumophila phagosome from the endocytic network in mouse macrophages. Cell. Microbiol. 3:99-114. [DOI] [PubMed] [Google Scholar]
- 36.Joshi, A. D., and M. S. Swanson. 1999. Comparative analysis of Legionella pneumophila and Legionella micdadei virulence traits. Infect. Immun. 67:4134-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley, W. L., and C. Georgopoulos. 1997. Positive control of the two-component RcsC/B signal transduction network by DjIA: a member of the DnaJ family of molecular chaperones in Escherichia coli. Mol. Microbiol. 25:913-931. [DOI] [PubMed] [Google Scholar]
- 38.Kohler, S., J. Teyssier, A. Cloeckaert, B. Rouot, and J. P. Liautard. 1996. Participation of the molecular chaperone DnaK in intracellular growth of Brucella suis within U937-derived phagocytes. Mol. Microbiol. 20:701-712. [DOI] [PubMed] [Google Scholar]
- 39.Kuronita, T., E. L. Eskelinen, H. Fujita, P. Saftig, M. Himeno, and Y. Tanaka. 2002. A role for the lysosomal membrane protein LGP85 in the biogenesis and maintenance of endosomal and lysosomal morphology. J. Cell Sci. 115:4117-4131. [DOI] [PubMed] [Google Scholar]
- 40.Lewallen, K. R., R. M. McKinney, D. J. Brenner, C. W. Moss, D. H. Dail, B. M. Thomason, and R. A. Bright. 1979. A newly identified bacterium phenotypically resembling, but genetically distinct from, Legionella pneumophila: an isolate in a case of pneumonia. Ann. Intern. Med. 91:831-834. [DOI] [PubMed] [Google Scholar]
- 41.Maruta, K., H. Miyamoto, T. Hamada, M. Ogawa, H. Taniguchi, and S. Yoshida. 1998. Entry and intracellular growth of Legionella dumoffii in alveolar epithelial cells. Am. J. Respir. Crit. Care Med. 157:1967-1974. [DOI] [PubMed] [Google Scholar]
- 42.Maruta, K., M. Ogawa, H. Miyamoto, K. Izu, and S. I. Yoshida. 1998. Entry and intracellular localization of Legionella dumoffii in Vero cells. Microb. Pathog. 24:65-73. [DOI] [PubMed] [Google Scholar]
- 43.Matthews, M., and C. R. Roy. 2000. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect. Immun. 68:3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamoto, H., H. Taniguchi, and S. Yoshida. 2003. A simple qualitative assay for intracellular growth of Legionella pneumophila within Acanthamoeba culbertsoni. Kansenshogaku Zasshi. 77:343-345. (In Japanese) [DOI] [PubMed] [Google Scholar]
- 45.Miyamoto, H., S. I. Yoshida, H. Taniguchi, and H. A. Shuman. 2003. Virulence conversion of Legionella pneumophila by conjugal transfer of chromosomal DNA. J. Bacteriol. 185:6712-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 48.Nagai, H., and C. R. Roy. 2001. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 20:5962-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Padmalayam, I., K. Karem, B. Baumstark, and R. Massung. 2000. The gene encoding the 17-kDa antigen of Bartonella henselae is located within a cluster of genes homologous to the virB virulence operon. DNA Cell Biol. 19:377-382. [DOI] [PubMed] [Google Scholar]
- 50.Pedersen, L. L., M. Radulic, M. Doric, and Y. Abu Kwaik. 2001. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect. Immun. 69:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell, M., and H. A. Shuman. 1998. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect. Immun. 66:2245-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinn, F. D., M. G. Keen, and L. S. Tompkins. 1989. Genetic, immunological, and cytotoxic comparisons of Legionella proteolytic activities. Infect. Immun. 57:2719-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson, G. T., M. E. Kovach, C. A. Allen, T. A. Ficht, and R. M. Roop, Jr. 2000. The Brucella abortus Lon functions as a generalized stress response protease and is required for wild-type virulence in BALB/c mice. Mol. Microbiol. 35:577-588. [DOI] [PubMed] [Google Scholar]
- 54.Rouquette, C., C. de Chastellier, S. Nair, and P. Berche. 1998. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 27:1235-1245. [DOI] [PubMed] [Google Scholar]
- 55.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 56.Roy, C. R., and L. G. Tilney. 2002. The road less traveled: transport of Legionella to the endoplasmic reticulum. J. Cell Biol. 158:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook, J., and W. J. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Schulein, R., and C. Dehio. 2002. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 46:1053-1067. [DOI] [PubMed] [Google Scholar]
- 60.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Segal, G., and H. A. Shuman. 1998. How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol. 6:253-255. [DOI] [PubMed] [Google Scholar]
- 62.Small, P. L., L. Ramakrishnan, and S. Falkow. 1994. Remodeling schemes of intracellular pathogens. Science 263:637-639. [DOI] [PubMed] [Google Scholar]
- 63.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ueguchi, C., M. Kakeda, H. Yamada, and T. Mizuno. 1994. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc. Natl. Acad. Sci. USA 91:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ueguchi, C., T. Shiozawa, M. Kakeda, H. Yamada, and T. Mizuno. 1995. A study of the double mutation of dnaJ and cbpA, whose gene products function as molecular chaperones in Escherichia coli. J. Bacteriol. 177:3894-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wall, D., M. Zylicz, and C. Georgopoulos. 1994. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J. Biol. Chem. 269:5446-5451. [PubMed] [Google Scholar]
- 68.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903 dIIlacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto, T., T. Hanawa, S. Ogata, and S. Kamiya. 1996. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect. Immun. 64:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp 18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 71.Yu, V. L., J. F. Plouffe, M. C. Pastoris, J. E. Stout, M. Schousboe, A. Widmer, J. Summersgill, T. File, C. M. Heath, D. L. Paterson, and A. Chereshsky. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186:127-128. [DOI] [PubMed] [Google Scholar]
- 72.Yura, T., H. Mori, H. Nagai, T. Nagata, A. Ishihama, N. Fujita, K. Isono, K. Mizobuchi, and A. Nakata. 1992. Systematic sequencing of the Escherichia coli genome: analysis of the 0-2.4 min region. Nucleic Acids Res. 20:3305-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zuber, M., T. A. Hoover, and D. L. Court. 1995. Analysis of a Coxiella burnetti gene product that activates capsule synthesis in Escherichia coli: requirement for the heat shock chaperone DnaK and the two-component regulator RcsC. J. Bacteriol. 177:4238-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuckman, D. M., J. B. Hung, and C. R. Roy. 1999. Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol. Microbiol. 32:990-1001. [DOI] [PubMed] [Google Scholar]