Abstract
The NF-κB family of inducible transcription factors is activated in response to a variety of stimuli. Amongst the best-characterized inducers of NF-κB are members of the TNF family of cytokines. Research on NF-κB and TNF have been tightly intertwined for more than 25 years. Perhaps the most compelling examples of the interconnectedness of NF-κB and the TNF have come from analysis of knock-out mice that are unable to activate NF-kB. Such mice die embryonically, however, deletion of TNF or TNFR1 can rescue the lethality thereby illustrating the important role of NF-κB as the key regulator of transcriptional responses to TNF. The physiological connections between NF-κB and TNF cytokines are numerous and best explored in articles focusing on a single TNF family member. Instead, in this review, we explore general mechanisms of TNF cytokine signaling, with a focus on the upstream signaling events leading to activation of the socalled canonical and noncanonical NF-κB pathways by TNFR1 and CD40 respectively.
Keywords: NF-kappaB, TNF, CD40 IKK, TRAF, RIP, signaling, inflammation
1. Introduction
1.1. A Brief Introduction to NF-κB
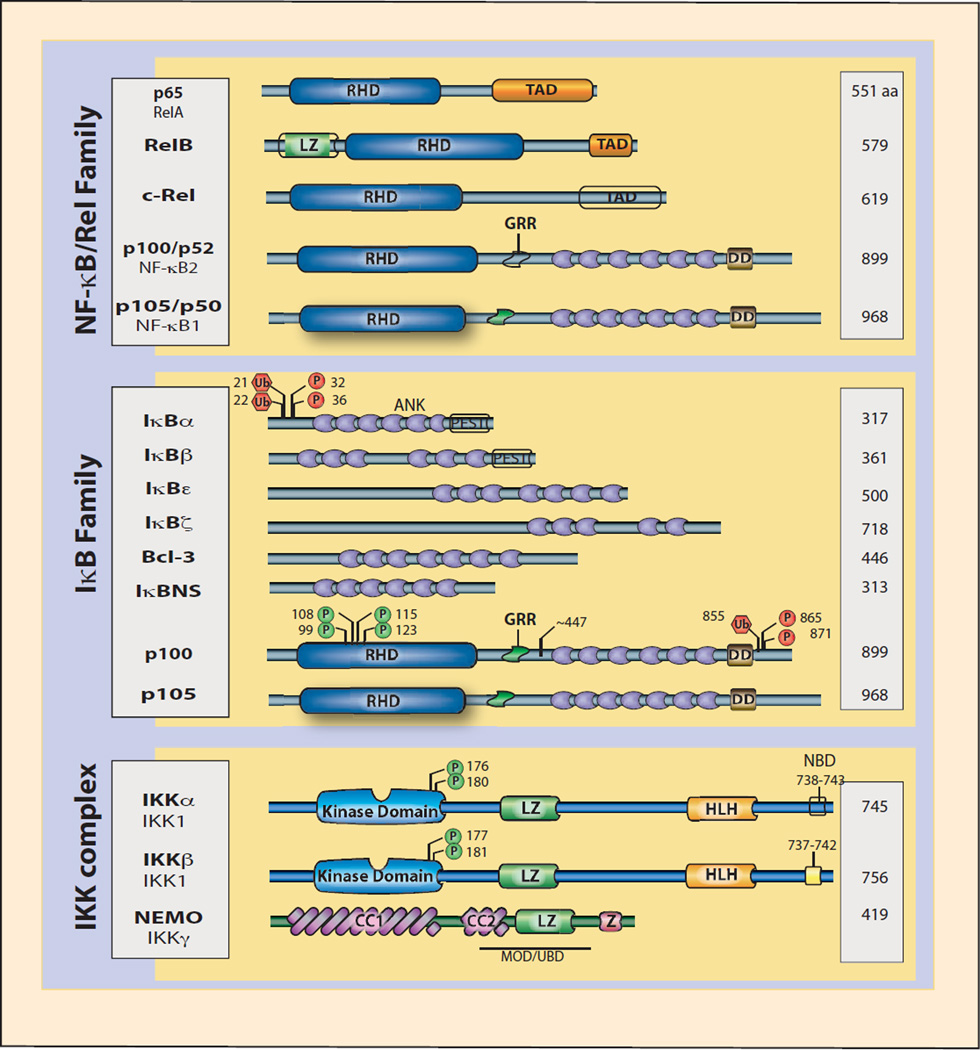
NF-κB is a family of inducible transcription factors that play a variety of evolutionarily conserved roles in the immune system [1, 2]. Cytokines belonging to the TNF family induce rapid transcription of genes regulating inflammation, cell survival, proliferation and differentiation, primarily through activation of the NF-κB pathway. The NF-κB family consists of five related proteins, p50 (NF-κB1) and p52 (NF-κB2), p65 (RelA), RelB and c-Rel (Rel), that share an approximately 300 amino acid long N-terminal Rel homology domain (RHD) (Figure 1). NF-κB proteins exist in cells as dimers, either homo or heterodimers, that are capable of binding to DNA. The RHD makes direct contact with DNA, while distinct protein domains mediate both positive and negative effects on target gene transcription through the recruitment of co-activators and co-repressors, respectively. The NF-κB proteins p65, c-Rel and RelB possess a transactivation domain allowing them to initiate transcription through co-activator recruitment. The p50 and p52 proteins do not have transactivation domains and therefore can affect transcription either through heterodimerization with p65, c-Rel, or RelB, through competition for binding to κB sites, or through heterotypic interaction with non-Rel transcription factors including certain IκB proteins.
Figure 1. Schematic representation of NF-κB, IκB and IKK proteins.
Key features of members of the NF-κB and IκB protein families and components of the IKK complex are shown. The number of amino acids in each protein is indicated on the right. Phosphorylation (P) and ubiquitination (U) sites on p100 and IκBα proteins that mediate proteasomal degradation and activation of the noncanonical and canonical NF-κB pathways are indicated. Presumed site of p100 cleavage (aa 447) is shown. Phosphorylation sites that mediate IKK kinase activation are indicated. α, β-helical domain; CC, coiled-coil domain; GRR, glycine-rich region; HLH, helix-loop-helix domain; LZ, leucine zipper domain; NBD, NEMO binding domain; RHD, rel homology domain; TAD, transactivation domain; Z, zinc finger domain.
In general the ‘resting’ state cytosolic NF-κB dimers are prevented from binding target sites through association with inhibitor of κB (IκB) proteins [3]. There are eight IκB proteins (Figure 1), IκBα, IκBβ, IκBε, IκBζ, BCL-3, IκBns, and the precursor proteins p100 (NF-κB2) and p105 (NF-κB1), which are characterized by the presence of multiple ankyrin repeat domain and the ability to bind NF-κB dimers. The function of IκB protein as regulators of NF-κB is highly variable: IκB family members can bind NF-κB dimers in the cytoplasm and nucleus and can both inhibit and augment transcriptional responses. For the purposes of this review, we will limit our discussion largely to the IκBα and p100 proteins, key regulators of the canonical and noncanonical NF-κB pathways, respectively. The functions of other IκB family members have recently been reviewed elsewhere [4, 5]. IκBα is the prototypical member of the IκB family. Although IκBα can regulate different NF-κB dimers, the primary target is the canonical p65:p50 heterodimer complex. IκBα enforces cytoplasmic localization of p65:p50 dimers through a robust nuclear export sequence as well as by masking the nuclear localization sequence of p65. In addition, rapid, signal-induced degration of IκBα via the proteasome [6–8] and concomitant release of NF-κB [9, 10] [11–13] is a hallmark of canonical NF-κB pathway. Therefore not only do IκBs negatively regulate NF-κB, they are also essential for the inducibility of the pathway: if all three traditional IκBs, IκBα, IκBε, and IκBβ, are removed, inducible NF-κB activation by TNF is completely abolished [14]. IκBα degradation is triggered upon phosphorylation of two serine residues within the so-called destruction box motif (DSGXXS) that is shared by all IκB proteins. The phosphorylated destruction box is recognized βTrCP leading to degradative, K48-linked polyubiquitination by the SCF/UbcH5 ubiquitin ligase complex [15].
Removal of IκB proteins allows the unbound NF-κB dimers to translocate to the nucleus and bind to accessible κB sites bearing the following sequence motif: 5’ GGGRNWYYCC 3’ (N - any base; R – purine; W – adenine or thymine; and Y – pyrimidine). The degenerate nature of the κB sequence and the ability of NF-κB to form heterotypic complexes with other transcription factors, allows NF-κB to regulate the expression of a wide variety of target genes [16]. NF-κB dimers can bind to both promoter and enhancer elements and have been shown to regulate both transcriptional responses and chromatin remodeling [17]. The activity of nuclear NF-κB dimers is further regulated by a variety of post-translational modifications that alter DNA and co-factor binding [18].
Cytokines of the TNF family trigger a variety of NF-κB-dependent responses that can be specific to both cell type and signaling pathway. It is not possible to provide in one article a detailed description of signaling mechanisms triggered by each individual TNF family member. Instead we will focus on two general categories of NF-κB signaling and explore the regulation of these pathways in the context of TNF family signaling. For didactic purposes it is useful to divide NF-κB signaling into two types: canonical and noncanonical. The “canonical” pathway, also termed the “classical” pathway, is typified by the inducible degradation of IκBα resulting from TNF ligation of TNFR1. As this pathway has been intensively studied, TNFR1 signaling will be the primary focus of our discussion of the regulation of canonical NF-κB by the TNF family. The “noncanonical”, or “alternative” pathway, is defined by the inducible processing of p100 (NF-κB1) bound to RelB to yield active p52:RelB heterodimers. Several members of the TNF cytokine family induce noncanonical NF-κB activation, however for this article we will focus primarily on signaling by CD40L through CD40 receptor as a model of noncanonical NF-κB activation.
All phosphorylation of IκB proteins on the conserved, destruction box serines are carried out by one of two highly related IκB kinases (IKK): IKKα (IKK1) or IKKβ (IKK2) (Figure 1). IKKα and IKKβ, along with a regulatory subunit called NEMO (NF-κB essential modulator, also known as IKKγ), exist in a large, 700–900kDa molecular weight, cytoplasmic complex [3]. All inducible signaling to NF-κB requires one of these two IκB kinases. The dependency on IKKα and IKKα is often used as a means of discriminating between canonical and noncanonical signaling pathways. While noncanonical signaling leading to inducible processing of p100 to p52 depends on IKKα, recent work has demonstrated that canonical signaling does not depend exclusively on IKKβ. A better discriminator appears to be NEMO, in that NEMO is always required for activation of the canonical pathway, while noncanonical signaling is NEMO independent. NEMO is essential for targeting of the IKK complex to IκBα and is, therefore, essential for the canonical pathway and inducible IκBα degradation [19]. In contrast, phosphorylation of p100 occurs normally in the absence of NEMO. In addition to phosphorylation of IκB it is important to note that IKKα and IKKβ can also regulate NF-κB responses through phosphorylation of NF-κB proteins, as well as mediate crosstalk with heterologous signaling pathways [20].
1.2. General Features of TNF Family Signaling to NF-κB
A variety of immunologically relevant ligands, and their receptors can activate the NF-κB pathway. These include, but are not limited to, the TNF receptor (TNFR), Toll-like receptor (TLR), IL-1 receptor (IL-1R), and antigen receptor families. One striking feature of all of these receptors, including TNF family receptors, is the lack of receptor enzymatic activity. Consequently, the physical act of ligand binding must be transmitted across a lipid bilayer and translated by adapter proteins into kinase activation. Current models hold that ligand binding promotes the formation of oligomeric receptor/ligand complexes. TNF family signaling to NF-κB involves binding of a series of adapter proteins to the ligand bound receptor complex, which in turn recruits and activates IKK. These adapter proteins possess well described protein:protein interaction domains that participate in assembly of the highly oligomeric protein complexes that bridge the ligand/receptor complex to IKK. The adapter protein interaction domains that are essential for TNF family signaling to NF-κB include the death fold domains (DD), RIP (Receptor Interacting Protein) homotypic interaction motifs (RHIM), and TRAF (TNF Receptor Associated Factor) domains. These adapter proteins mediate signaling to numerous downstream pathways. Here we will only discuss the roles of these proteins in NF-κB signaling. Altered receptor conformation, localization and/or increased avidity induced by ligand binding facilitates adapter protein binding through DD or TRAF binding motifs in the receptor cytoplasmic tail. The result is the recruitment of TRAF and RIP family members into large protein complexes. Oligomerization of these complexes is supported both by receptor crosslinking by ligand multimers, as well as the ability of protein:protein interaction domains, such as TRAF domains, to form trimers and higher order oligomers. These complexes recruit both upstream kinases as well as the IKK complexes. The upstream kinases leading to activation of IKK and NF-κB signaling by TNF family cytokines are NIK (NF-κB inducing kinase) and TAK1 (transforming growth factor-β activated kinase; MAP3K7). NIK mediates activation of IKKα and the noncanonical pathway. TAK1 functions in most NEMO dependent canonical pathways, although the requirement for TAK1 in all TNF family signaling to canonical NF-κB is not yet established. Once activated, these IKK kinases (IKK-K) can instigate traditional kinase cascades within and outside the NF-κB pathway.
2. TNFR1 Signaling to Canonical NF-κB
2.1. Introduction to TNF and NF-κB
The history and clinical significance of TNF and TNFR1 are reviewed elsewhere in this issue [21]. TNF and NF-κB research have long been closely associated, reflecting the biological relationship between the two pathways. TNF was first shown to activate NF-κB in the regulation of the HIV-1 LTR (long terminal repeat) and IL-2 receptor [22–24]. Shortly thereafter it was shown that NF-κB regulated TNF expression following stimulation of macrophages with LPS [25]. Since those early days it has increasingly been appreciated that many biological effects of TNF were actually mediated by NF-κB. Thus, in the absence of the canonical NF-κB subunit p65, transcriptional responses to TNF are severely blunted and induction of cell death is favored [26–28]. In the absence of p65, IKKβ, or NEMO, embryonic lethality occurs due to massive apoptosis of hepatocytes, and this lethality can be reversed by deletion of either TNFR1 [29, 30] or TNF [31–33].
TNF signals through two receptors, TNFR1 (p55; TNFR1a) and TNFR2 (p75; TNFR1b) [21]. However, because TNFR1, which exhibits ubiquitous and constitutive tissue expression, is thought to be the primary mediator of classical TNF functions in vivo, and has been the subject of more extensive research with regard to NF-κB, we limit our discussion here to TNFR1. In addition, although TNF can be expressed as both soluble and membrane bound forms, most of our knowledge of TNFR1 signaling derives from the use of soluble recombinant TNF, even though the pathways activated by soluble and cell surface TNF are not equivalent.
Ligation of TNFR1 by soluble TNF (Figure 2) induces activation of multiple signaling pathways, including NF-κB and MAPK, as well as apoptosis and necroptosis pathways [34, 35]. Activation of NF-κB is a key event in TNFR1 signaling, both for the induction of inflammatory gene expression as well as for the induction of anti-apoptotic transcriptional programs that counteract pro-apoptotic pathways [34–37]. As discussed above, TNF-induced activation of NF-κB is mediated by a series of intermediary adapters. The cytoplasmic tail of TNFR1 contains several protein binding domains, including a death domain, that mediates signaling following TNF binding. The DD of TNFR1 is essential for activation of apoptosis as well as induction of pro-survival and inflammatory pathways through activation of transcription factors such as NF-κB and AP-1.
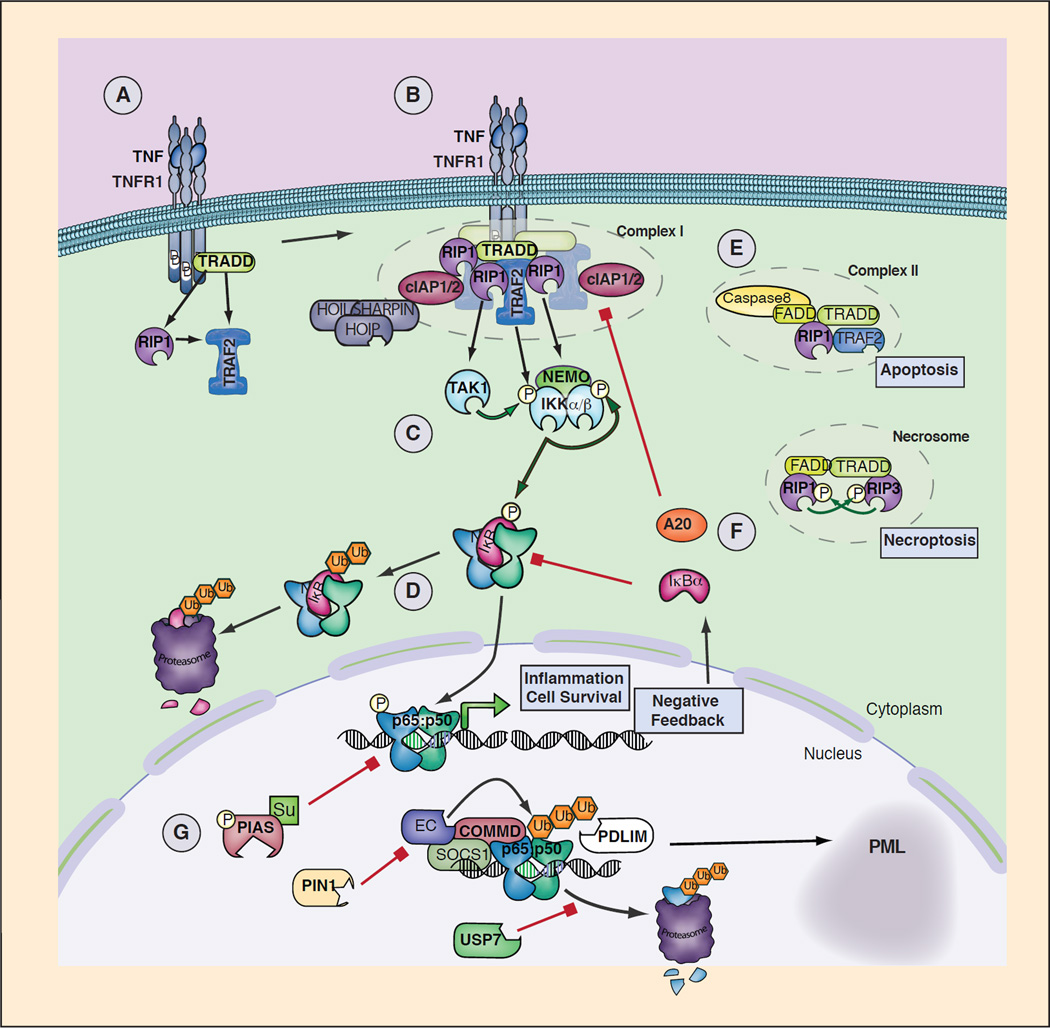
Figure 2. Model of TNFR1 Signaling to Canonical NF-κB.
A. Upon binding of TNF trimmers to TNFR1, oligomeric receptor complexes form resulting in recruitment of TRADD to the death domain (DD) of the receptors cytoplasmic tail. RIP1 is recruited through TRADD and TNFR1 through homotypic DD interactions. High affinity binding of TRAF2 trimers to TRADD is augmented by TRAF2/RIP1 interactions. B. TRAF2 trimers recruit cIAP1/2, which in turn recruit the LUBAC complex (HOIP, HOIL and SHARPIN), while RIP1 mediates recruitment of TAK1 and the IKK complex through NEMO. This oligomeric signaling complex is termed complex I and signals to NF-κB and AP-1 and MAPK pathways (not shown). C. Recruitment and induced proximity of TAK1 and the IKK complex supporting phosphorylation and activation of IKK. The ubiquitin ligase cIAP1/2 and LUBAC may facilitate TAK1 and IKK activation through production of linear and/or K63 linked ubiquitination. D. Activated IKK phosphorylates IκBα leading its ubiquitination and degradation and nuclear translocation of p65:p50 NF-κB complexes. DNA bound canonical NF-κB induces transcription of immune response genes as well as genes that protect the cell from TNF induced cell death. E. Cell death pathways are triggered by distinct signaling complexes triggering either apoptosis (complex I) or RIP1/RIP3 kinase dependent necroptosis (Necrosome). F. Negative feedback is mediated by NF-κB dependent resynthesis of IκB proteins as well as other factors including A20, which may facilitate complex I disassembly by disrupting TRAF2/cIAP binding. G. Multiple mechanisms, discussed in the text, act to induce degradation or displacement of DNA bound canonical NF-κB dimers.
2.2. TNFR1 Signaling to Canonical NF-κB
The initiating event in TNFR1 signaling pathways is the recruitment of the DD-containing adaptor protein TRADD (TNFR1 associated death domain protein) [38] through heterotypic interactions with the DD of TNFR1 (Figure 2). TRADD is required for activation of NF-κB, and as discussed above recruitment of TRAF proteins is essential for activation of the canonical pathway. TNFR1 lacks a TRAF interaction domain, however, TRADD, through its TRAF binding domain acts as an adapter for the recruitment of TRAF proteins. TRADD forms both a pro-apoptotic signaling complex through recruitment of FADD (Fas receptor-associated death domain), and pro-inflammatory/survival complex through recruitment of RIP1 and TRAF2 [39–41]. The requirement of TRADD for signaling to both these pathways has been confirmed in mouse knockout models [42–44] and deletion in human B cells [45]. The DD-containing kinase RIP1, can bind directly to the DD of ligand-bound TNFR1 [46]. However, it appears that RIP1 is more efficiently recruited to the receptor complex through TRADD [40, 41]. Consequently, TRADD is required for RIP1 recruitment in most cells. In macrophages, which express high levels of RIP1, RIP1 binds TNFR1 directly and mediates weak TNF induced IκBα degradation even in the absence of TRADD [44]. RIP1 also has a TRAF binding motif, and may contribute to TRAF2 recruitment under some circumstances [47], although it is generally thought that TRAF2 recruitment is primarily dependent on TRADD [41, 47–49]. Thus, upon TNF binding, TRADD binds the DD of TNFR1 and, in turn recruits RIP1. TRADD also brings TRAF2 to the receptor complex and RIP1 may also contribute to TRAF2 recruitment. This pro-inflammatory, multi-subunit signaling structure, is termed complex I, and spatially and functionally is distinct from the pro-cell death, FADD (Fas-associated protein with death domain) - caspase 8 containing complex II [50] and necrosome (Figure 2).
The high-affinity interaction between TRADD and TRAF2 in complex I leads to robust activation of NF-κB and AP-1 [51]. The crystal structure of TRAF2 interacting with TRADD has been solved and the structure predicts an interaction that is of much higher affinity than the interaction between TRAF2 and TNFR superfamily members with cytoplasmic TRAF interaction motifs [51]. The relative contribution, if any, of other TRAF proteins to NF-κB activation by TNFR1 is unclear. Confusion about the role of individual TRAFs in specific TNF family signaling pathways partly results from the ability of TRAF family members to compensate for one another in knockout models, as well as in overexpression experiments. This phenomenon has been described most extensively for TRAF2 and TRAF5 in TNFR1 signaling. TRADD shows a much higher affinity for TRAF2 than TRAF5 [51], and recruitment of TRAF2, but not TRAF5, to the TNFR1 complex is readily demonstrated in receptor pull-down experiments. Thus it is unlikely that TRAF5 is recruited by TRADD in wild-type cells. TRAF2 deficient mice have partly intact TNF signaling to NF-κB, but deficient AP-1 activation [52], whereas TRAF5 knockouts have normal TNF signaling to both NF-κB, and AP-1. TRAF2/5 double knockout cells however have markedly reduced TNF-induced NF-κB and AP-1 activation [52–54]. Furthermore, while TRAF2 knockout mice exhibit perinatal lethality that is rescued by TNF or TNFR1 deletion [52, 55], TRAF5 knockout mice develop normally [54]. In summary, these results suggest that while TRAF5, and likely other TRAFs, can compensate for the absence of TRAF2, in wild type cells, it is primarily TRAF2 that is recruited by TRADD to TNFR1 complex I.
Recruitment of the IKK complex, like recruitment of RIP1 and TRAF2, also appears to occur through multiple mechanisms. TRAF2 can recruit the IKK complex through direct binding to IKKα or IKKβ [56], although the IKK complex can be recruited and activated in TRAF2 knockout cells. RIP1 deficient cells, in contrast, are severely compromised in their ability to activate NF-κB [57]. Although recent work has indicated that there is significant residual TNF-induced IKK activation in some RIP1 knockout cells [58, 59], this residual NF-κB activation may be the result of a noncanonical pathway [60]. This residual activity could explain the NF-κB-independent contributions of RIP1 to pro-death signaling pathways [59] such as necroptosis [61], and the delayed lethality in RIP1 knockouts compared to IKKβ or p65 knockouts. However, while RIP1 is required for activation of IKK by TNFR1, at least in most cell types, RIP1 kinase activity is not [41, 62–64]. Indeed, kinase dead RIP1 knock-in mice are viable, fertile, and display normal TNFR1 signaling to NF-κB [65]. RIP1, therefore, functions as a scaffold for NF-κB activation [41, 62, 64] and contributes to IKK activation through an induced proximity mechanism [66, 67].
RIP1 mediates recruitment of IKK to TNFR1 complex by directly binding to NEMO [68]. Although recruitment of the IKK complex to TNFR1 can occur in RIP1 deficient cells, most likely through TRAF2 binding to IKKα/β, the IKK complex is not activated [56]. These data are consistent with the idea that RIP1 supports formation of a higher order structure through binding to both TRADD and TRAF2 trimers. Indeed, forced oligomerization of NEMO activates IKK [67, 69] and NEMO with a mutated oligomerization domain prevents IKK activation [70, 71]. However, such a model does not account for the requirement for TAK1, for TNF induced IKK activation [72, 73]. Interestingly, TAK1 is recruited to the receptor complex in a RIP1 dependent manner [74, 75], suggesting RIP1 may facilitate TNFR1 signaling to NF-κB activation through induced proximity of both the IKK complex and TAK1. However, the mechanism(s) of RIP1 mediated recruitment of IKK, through NEMO, and TAK1, through TAB2, remains controversial.
Work over the past decade that has provided a large body of evidence suggesting a role for ubiquitination of RIP1 in TNF-induced NF-κB activation. TNF stimulation results in the appearance high molecular weight forms of RIP1 that are visible in western blotting [63] and are enriched when TNF is used to pull down active receptor complexes [76, 77]. However, even in these immunoprecipitated complexes, a minor fraction of RIP1 is ubiquitinated. Analyses by multiple groups have shown that this modified RIP1 is ubiquitinated either through K63 (lysine-63) [74, 75, 78, 79] or linear [77] ubiquitin linkages. It has been proposed that ubiquitin chains on RIP1 might mediate recruitment of IKK, through NEMO, and TAK1, through TAB2 (TAK1 binding protein 2, also known as MAP3KIP2) and TAB3, complexes. Ubiquitination of RIP1 depends on the presence of TRAF2 [63, 80]. Although it was initially thought TRAF2, which possesses an RING (really interesting new gene) domain, was directly responsible for RIP1 ubiquitination, it is now not considered to be true (discussed in [3]). Instead it has been shown that Inhibitor of apoptosis proteins (IAPs), cIAP1 and cIAP2, contribute to RIP1 ubiquitination [81]. IAP proteins are defined by the presence of a baculovirus IAP repeat (BIR) domain that mediates protein:protein interactions and cIAP1 and cIAP2 also have RING finger domains capable of acting as E3 ubiquitin ligases [82]. TRAF2 binds and recruits cIAP1 and cIAP2, which are essential for IKK activation [83–85], suggesting TRAF2 might mediate RIP1 ubiquitination through recruitment of cIAPs. The cIAPs can function as E3 ubiquitin ligases but are also responsible for the recruitment of the linear ubiquitin chain assembly complex, which is required for efficient activation of IKK and JNK pathways [77, 86–90]. In TNFR1 signaling, cIAP1 and cIAP2 are required for TRAF2-dependent K63-linked ubiquitination of RIP1 [81].
However, despite numerous studies demonstrating ubiquitination of a fraction of RIP1 during TNFR1 signaling, there is no convincing evidence that K63-linked polyubiquitination of RIP1 is required for IKK activation. In addition to studies in which RIP1 ubiquitination failed to correlate with IKK activation [91], there are now more direct studies demonstrating that K63 linked ubiquitination is not required for IKK activation by TNFR1 [92]. Furthermore, while TAK1 deficient cells demonstrate a profound defect in TNFR1 signaling, TAB2/3 double knockout MEFs display normal activation of the canonical NF-κB signaling pathway by TNF and, in B cells, normal responses to several other inducers of canonical NF-κB signaling [93]. These data suggest that if RIP1 acts in part through recruitment of TAK1, that this recruitment is independent of ubiquitin binding by TAB2 or TAB3. In summary, although a small portion of RIP1 undergoes K63-linked polyubiquitination during TNF signaling, current evidence does demonstrate that this ubiquitination is required for either recruitment of IKK or TAK1, or for activation of the NF-κB pathway.
NEMO is also ubiquitinated during TNFR1 signaling [86, 94, 95] and has recently been shown to be a substrate for linear ubiquitination [86, 96]. While most of ubiquitin chains involve linkage of the C terminal glycine of ubiquitin to lysine residues on the substrate linked ubiquitin, e.g. K48 (lysine 48) or K63, recent work has demonstrated the existence of head to tail linear ubiquitin chains (M1; methionine 1) assembled by a unique linear ubiquitin assembly complex (LUBAC) [97]. The LUBAC complex consists of heme-oxidized iron regulatory protein 2 ubiquitin ligase 1 (HOIL-1; also known as RBCK1) and HOIL-1 interacting protein (HOIP; also known as RNF31 and Zibra) [97]. Overexpression of the LUBAC complex activates NF-κB [97] and LUBAC co-purifies with TNFR1 complex I [98]. Initially it was shown that LUBAC stabilzes the TNFR1 signaling complex I, resulting in prolonged NF-κB activation [86, 87, 98]. More recently, SHARPIN has been identified as a third component of LUBAC [88–90]. SHARPIN deficient cells demonstrated variable defects in activation of IKK downstream of TNFR1, although the effects on TNF induced cell death appear more striking [88–90]. This discrepancy between NF-κB activation and TNFR1 induced cell death was also observed upon knockdown of the LUBAC complex [99]. These data suggest that LUBAC regulates TNF induced cell death independent of any role in NF-κB activation. Finally, the mild phenotype of SHARPIN deficient cpdm/cpdm (chronic proliferative dermatitis) mice is difficult to reconcile with the proposed requirement for TNFR1 signaling to NF-κB. Recent in vitro studies suggest that linear ubiquitination of NEMO, or linear ubiquitin binding by NEMO, may directly activate the IKK complex [100]. However generation of knock-in mice with point mutations in NEMO that block linear ubiquitination will be necessary to unequivocally demonstrate the importance of linear ubiquitination in NF-κB activation and cell death.
To summarize, it is not yet possible to definitively state whether regulatory ubiquitination plays an essential role in TNFR1 induced activation of NF-κB. While numerous studies have demonstrated K63, M1, and, most recently, mixed K63/M1 hybrid chains [101] on components of the TNFR1 signaling pathway, proof that these events cause, rather than result from, TNF signaling has not yet been provided. What remains clear is that TNF binding to TNFR1 induces the formation of a multiprotein signaling complex through the sequential binding of adapter proteins. Available data suggests that within this complex TRAF2 and RIP mediate recruitment of TAK1 and the IKK complexes, leading to induced proximity, and trans-autophosphorylation and TAK1-mediated activation of the IKK complex. While it is possible that ubiquitination of NEMO or ubquitin binding by TAK1 and IKK complexes contributes to IKK activation, several alternative scenarios, which we have discussed previously [3], appear equally consistent with available data.
IKK activation occurs extremely rapidly downstream of TNFR1, typically within three to five minutes [102]. Phosphorylation and degradation of IκBα is complete within 10 minutes and nuclear localization and DNA binding by NF-κB, as assessed by gel shift, is maximal by approximately thirty minutes [103]. While active IKK complex is capable of phosphorylating multiple IκB family members, IKK activation downstream of TNFR1 selectively results in rapid phosphorylation of IκBα (Figure 2). Phosphorylation of IκBβ occurs with delayed and prolonged kinetics, consistent with the demonstrated substrate preference of IKKβ for IκBα [104]. Rapid and selective phosphorylation of IκBα is also strongly enforced by selective IκBα binding by NEMO [19]. While as little as 5 minutes of TNF stimulation is sufficient to completely activate the canonical NF-κB pathway, the transcriptional response to TNF depends on repetitive activation of TNFR1 signaling [103]. Sustained TNF results in cyclic activation of the canonical NF-κB pathway with a periodicity of approximately 100 minutes [103]. Sustained activation of the pathway is necessary for the induction of many pro-inflammatory TNF target genes [105–107]. In addition to these kinetic signaling requirements, several intracellular pathways govern NF-κB activity downstream of IκBα. Multiple post-translational modifications regulate the ability of NF-κB to activate transcription of target genes. This complex area of NF-κB regulation is beyond the scope of the current discussion but has recently been thoroughly reviewed elsewhere [17, 108, 109]. In contrast to the positive regulation of canonical NF-κB, relatively little is known about the termination of NF-κB transcriptional activity. Therefore, we will briefly discuss some of the negative feedback loops involved in termination of TNFR1 signaling and describe some factors involved in terminating TNF induced canonical NF-κB transcriptional responses.
2.3. Shutting Down TNF-Induced NF-κB Signaling
Termination of NF-κB responses is an essential aspect of NF-κB regulation as elevated NF-κB activation is association with inflammation and oncogenesis [110, 111]. As recent reviews have discussed negative feedback loops activated by the NF-κB pathway [112], we limit the following discussion to feedback mechanisms that are particularly relevant for TNF family signaling, or are often overlooked. One of the primary means of signal termination is receptor downregulation. Because the TNF system produces a response that is proportional to the duration of stimulus, it is essential that signaling be terminated upon ligand withdrawal. TNF bound TNFR1 undergoes rapid internalization and trafficking to lysosomes where it is degraded (Reviewed in [113]).
Upon activation of NF-κB several negative feedback loops are engaged through NF-κB-dependent transcriptional responses. The primary negative feedback loop in NF-κB signaling is the induction of IκB transcription. IκBα is rapidly induced following NF-κB activation [114–117] and is crucial for terminating the NF-κB response [118, 119]. Mice lacking IκBα exhibit systemic inflammation resulting from a failure to appropriately terminate TNF dependent responses [118, 119]. Negative feedback resulting from TNF-induced expression of IκB proteins determines the periodicity and amplitude of the NF-κB responses occurring during sustained stimulation [103].
Another rapidly induced gene implicated in termination of the TNF response is A20 (TNFAIP3). A20 is of significant interest as it has been associated with several autoimmune and chronic inflammatory diseases through genome wide association studies (GWASs; [120]). A20 contains multiple zinc finger domains, and TRAF-binding, and deubiquitinating (DUB) OUT (ovarian tumor domain) domains. A20 is induced rapidly following TNFα induction [121–123]. Upon overexpression of A20, NF-κB activation by TNF is inhibited [124–126], although others have noted that A20 did not alter NF-κB nuclear localization [127]. Subsequently it was shown that A20 might function as an ubiquitin editing enzyme. The OTU domain was shown to mediate RIP1 K63 deubiquitination while the zinc finger domain induced K48-linked ubiquitination of RIP1, leading to RIP1 degradation [80]. Consistent with the idea that A20 is a an important negative feedback regulator of the NF-κB pathway, it was shown that mice lacking A20 exhibit spontaneous inflammation and modestly prolonged NF-κB activation in response to TNF [128]. The most notable alteration in A20 knockout cells was increased TNF induced cell death, consistent with the idea that A20 may regulate cell death independent of effects on NF-κB activation [129–131]. Nevertheless, in vivo, it appears that A20 is not, primarily a regulator of TNF responses, but rather of TLR and IL-1R signaling, as the inflammatory phenotype of the A20 deficient mice is rescued by deletion of MyD88 but not TNFR1 or TNF [132, 133]. Given that phenotypes resulting from hyperactivation of NF-κB in other genetic models are rescued by blocking or deleting TNF cytokine signaling [134, 135], these results suggest that A20 is not an important negative regulator of TNF or NF-κB signaling. Although it was shown that A20 instead may function to subtly limit the magnitude of late responses during sustained or repeated TNF stimulation [136], the mechanism for this function is unclear. Knock-in mice in which the OTU domain is inactivated do not exhibit any phenotype or alterations in NF-κB transcriptional responses (De et al., in press). Recently it was shown that zinc finger 7 of A20, which is not implicated in catalytic functions, may inhibit IKK activation by binding ubiquitinated NEMO and preventing IKK activation [137–139] or by dissociating TRAF2/3 from cIAP1/2 [140, 141]. As these experiments have relied on overexpression, these non-catalytic functions of A20 need to be confirmed in vivo. Given the subtle TNF signaling defect observed in A20 knockout cells and failure of deletion of TNF/TNFR1 to rescue the phenotype of A20 knockout mice, it remains unlikely that A20 has a major role as a negative regulator of canonical NF-κB pathway activated by TNF. However, given the genetic implication of A20 in autoimmunity and demonstrated role in some malignancies, further work to define the mechanisms of action of A20 is clearly warranted.
In addition to targeting signaling pathway components, inactivation of DNA-bound NF-κB is also essential for limiting the NF-κB response. Because the key role of IκB proteins in limiting the NF-κB response was recognized early, our understanding of other mechanisms that terminate NF-κB activity has continued to lag behind our understanding of signaling pathways leading to NF-κB activation. It is clear that IκBα resynthesis restricts the duration of the NF-κB dependent transcriptional response after TNF stimulation. However, it does not seem that IκBα acts directly on DNA-bound to terminate the transcriptional response. Indeed, even in cells lacking κB activity, NF-κB responses are restrained – emphasizing that additional factors mediate termination of the response while IκBs are essential for inducibility. Termination of NF-κB activation can be achieved by removal of co-activators, degradation of active NF-κB dimers, or removal of active dimers from DNA – none of these are directly mediated by IκB. Termination of transcriptional activation of DNA bound NF-κB dimers is achieved by either removal of co-activators, such as CBP/p300, or displacement of co-activators by co-repressors including histone deacetylases. Treatment of cells with the HDAC inhibitor TSA substantially augments the TNF induced transcriptional response [142], suggesting that HDACS actively repress NF-κB-dependent responses to TNF. Degradation of canonical NF-κB downstream of TNF stimulation is also implicated in restriction and termination of the transcriptional response.
Under some circumstances ubiquitination and proteasomal degradation of nuclear p65-containing heterodimers has been demonstrated [143]. Several ubiquitin ligases have been implicated in this process and we review them briefly here. Suppressor of cytokine signalling-1 (SOCS1) recruits protein substrates to ubiquitin ligase elongin-B–elongin-C–cullin-2–SOCS1 protein (ECS) complex [144] and decreases p65 stability [145, 146] and transcriptional activity when overexpressed [147, 148]. COMMD1 (copper metabolism (Murr1) domain containing 1), which also associates with the ECS ubiquitin ligase complex, facilitates p65 degradation by promoting binding of p65 and SOCS1, and SOCS1 and CUL2 [149, 150]. In the absence of COMMD1, p65 and target transcript levels are increased following TNF stimulation [150]. If re-synthesis of IκBα, which is enhanced due to the elevated p65 activity, is blocked then nuclear p65 is increased and prolonged in the absence of COMMD1 [150]. Therefore, COMMD1 targeting of a SOCS1-containing ubiquitin ligase complex to p65 is a significant contributor to the termination of the p65-mediated transcriptional response. There are 10 COMMD proteins, and several have been shown to have significant effects on TNF induced NF-κB activity [151]. Binding to other NF-κB family members has also been demonstrated [151], though the significance of these interactions is less clear. Binding of COMMD1 to p65, and recruitment of p65 to ECS, may be regulated by phosphorylation of serine 468 on p65 [152, 153]. IKKβ, inducible IKK (IKKi; also known as IKKε), and GSK-3β have been shown to phosphorylate S468 [154, 155]. TNF induces IKKi transcriptionally [156] and causes the nuclear accumulation and phosphorylation of p65 [157]. However, recent work demonstrates that phosphorylation of S468 occurs rapidly and is associated with increased binding to CBP/p300 [158], which promotes the p65-dependent transcriptional response. Thus induction of S468 phosphorylation of p65 alone is unlikely to promote COMMD1-dependent p65 degradation. Instead competition between CBP and COMMD proteins for binding to p65, additional post-translational modifications (reviewed in [108, 109]) or sub-nuclear localization may determine COMMD1 binding and termination of the p65 response.
Recruitment of p65 to the ECS may also be actively inhibited. Expression of the peptidyl-prolyl cis/trans isomerase Pin1 (protein NIMA-interacting 1) correlates with NF-κB activation in breast cancer, and loss of Pin1 prevents activation of NF-κB in response to TNF in MEFs and hepatocytes [146]. Pin1 competes with SOCS1 for binding to p65 and thus to prevents ECS-induced ubiquitination and degradation of p65-containing NF-κB dimers. Finally, the deubiquitinase USP7 promotes TNF induced transcriptional responses and nuclear p65 stability by deubiquitinating DNA bound p65 [159].
Both degradation and relocalization of NF-κB complexes are potential mechanism of transcription termination. The nuclear ubiquitin ligases PDLIM2 (PDZ and LIM domain 2; also known as SLIM and Mystique), and the SUMO ligases PIASy (protein inhibitor of activated STAT y) and PIAS1 have been implicated in regulation of nuclear p65. PDLIM2 induces p65 ubiquitination and relocalization of p65-containing dimers to PML nuclear bodies and loss of PDLIM2 in mice results in systemic inflammation [160]. PDLIM2 has been suggested to be regulated through cytoplasmic sequestration [161], at present, it is not known whether the function of PDLIM2 is regulated during TNF signaling. Indeed, it is not clear that PDLIM2 restricts TNF-induced NF-κB activation. Knockdown experiments demonstrate very modest reduction in TNF induced NF-κB transcriptional activity [162]. Furthermore, PDLIM2 function is not specific to NF-κB. STAT3 and STAT4 activity is also altered and contributes to the pro-inflammatory phenotype of the knockout mice [163–165].
The SUMO E3 ligases PIAS1 and PIASy, although initially characterized as STAT inhibitors, have both been shown to regulate NF-κB transcriptional responses [166]. PIAS1 inhibits the DNA binding of canonical NF-κB complexes [167] and PIAS1 knockout mice have increased sensitivity to LPS-induced shock[168]. PIAS1 acts early to limit immediate NF-κB dependent transcription [167], suggesting that its function may be more in establishing a threshold for p65-dependent DNA binding, than in termination of the NF-κB response. Deletion of PIASy in mice also results in hypersensitivity to LPS induced shock [169, 170] and increased p65 DNA binding in a stimulus-dependent manner [170]. Despite similar phenotypes of the knock-out mice, PIAS1 and PIASy regulate distinct sets of TNF-induced genes. For example, loss of PIAS1 has a marked effect on TNF-induced IκBα and IL-1β expression, whereas loss of PIASy does not. In contrast, TNFα-induced CXCL1 is affected by PIASy, but not by PIAS1 [167, 170]. PIAS1 and PIASy interact directly with p65 [171] but repression of p65 transcriptional functions is not dependent on binding only. Mutagenesis experiments indicate that PIAS1 SUMO ligase activity and IKKα mediated phosphorylation are required for inhibition of p65 function [171].
3. CD40 Signaling to NF-κB
Several members of the TNF family can induce activation of the IKKα-dependent noncanonical NF-κB pathway. The TNFR superfamily members CD40 and LTβR are the best-characterized mediators of noncanonical signaling, although signaling from Fn14, CD27, BAFF-R, TNFR2 and RANK, also lead to activation of noncanonical NF-κB [172]. Here we will focus on activation of the noncanonical pathway by CD40L. CD40L is expressed by activated CD4+ TH cells and is essential for the induction of B cell immunoglobulin class switch recombination in response to T-dependent antigens [173]. Consequently, most work on the biological role of CD40 has been done in the context of the humoral immune response. However, CD40 can be expressed on several hematopoietic cells including dendritic cells, macrophages, and T cells. Indeed, CD40 is also expressed on non-hematopoietic cells including epithelial and endothelial cells. The biological role of CD40 on these cell types is less well understood.
CD40 was originally identified as a surface marker on B cells, and most work has focused on the role of CD40 in B cell biology [174]. CD40 knockout B cells fail to undergo class switch recombination and exhibit defects in proliferation and germinal center formation [175]. B cells lacking CD40 also do not exhibit T cell induced co-stimulatory molecule upregulation and are, consequently, poor antigen presenting cells [176]. The requirement for the alternative NF-κB pathway in B-ells is not completely clear as B cells from relB−/− mice class switch normally [177] and defects in class switching in p52−/− mice are not B cell intrinsic [178]. B cells from rela−/− mice exhibit diminished class switching [28], which is augmented by deletion of p50 [179, 180]. c-Rel-deficient mice also indicate a requirement for c-Rel in class switch recombination [181–183]. Therefore, analyses of knockout animals suggest that the canonical NF-κB pathway likely has a role in maturation of the B-cell response in addition to directly mediating proliferative responses following BCR ligation. In addition to B cells, there has been considerable interest in the role of CD40 signaling in dendritic cells. However, it appears that most biological functions of CD40 in DCs are attributable to activation of the canonical signaling pathway. T cell CD40L expression also promotes dendritic cell survival, though this is likely through canonical pathway activation [184, 185]. Differentiation of effective dendritic cells from monocytes in vitro depends on activation of RelB complexes [186, 187], however these complexes consist of RelB:p50 [187, 188].
3.1. CD40 Signaling
CD40L, which is a type II transmembrane protein, is expressed on the cell surface of T cells, from which it engages CD40 on B cells and DCs. CD40L, like TNF, exists as a trimer [189], and therefore induces oligomerization of CD40 upon binding. In contrast to TNFR1, the crystal structure of CD40 bound to CD40L indicates that each CD40L trimer binds not two, but three, receptors [190]. However, given that CD40 forms ligand-independent homodimers [191], trimers of CD40L may bind two dimers of CD40, and induce formation of oligomeric receptor ligand complexes (Figure 3). Upon CD40 ligation by CD40L, CD40 binds to TRAF proteins. Although CD40 can trigger signaling resulting in activation of NF-κB, MAPK, JAK/STAT, and PI3K pathways (reviewed in [174]), we will focus on TRAF-dependent signaling to NF-κB. TRAF proteins exhibit different affinities for the CD40 cytoplasmic tail, with TRAF2 having the highest affinity, and it appears that receptor avidity of different TRAFs may govern their relative ability to participate in signaling [192]. Thus, the stoichiometry of the ligand receptor complex may be an important determinant of signaling outcomes.
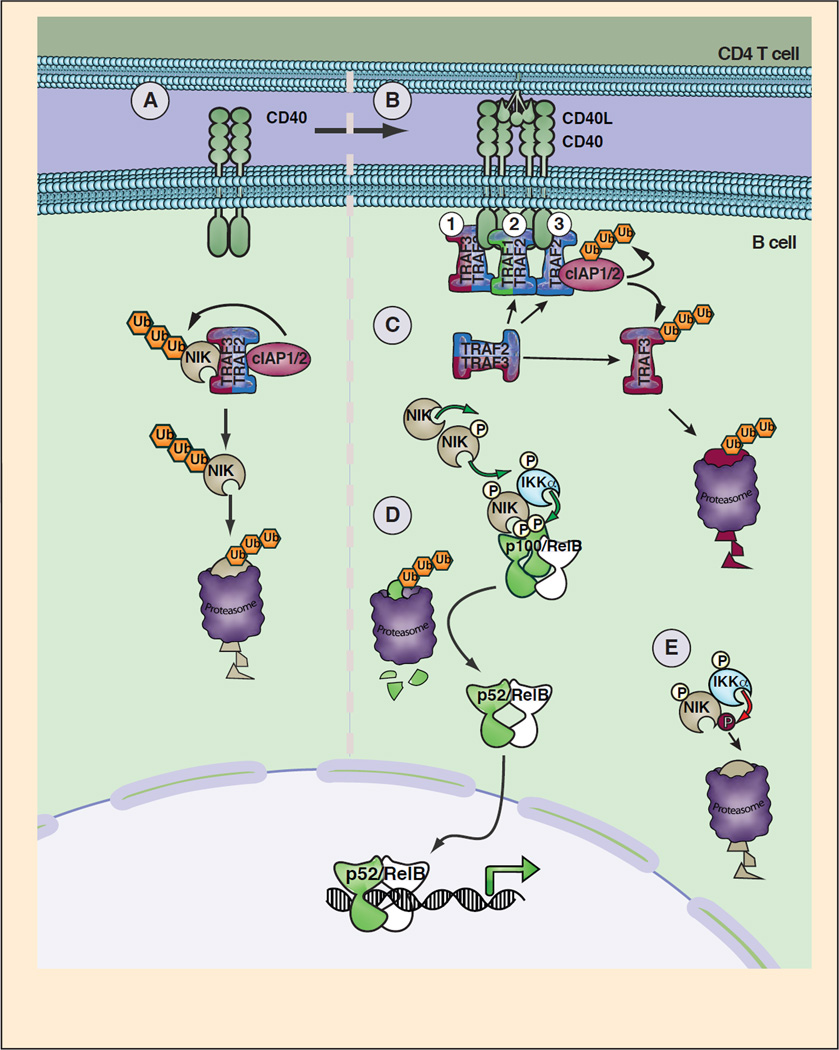
Figure 3. Model of CD40 signaling to Noncanonical NF-κB.
A. In resting B cells, a TRAF2/TRAF3 heterotrimer mediates constitutive NIK degradation by binding to NIK and the E3 ubiquitin ligases cIAP1/2. B. Upon encountering CD40L expressing CD4 T cells, CD40 dimers are oligomerized by CD40L trimmers resulting in altered affinity and avidity for TRAF proteins. C. Three non-exclusive mechanisms of NIK stabilization are shown. 1. Binding of TRAF2/3 trimers to the CD40 cytoplasmic tail results in decreased affinity for NIK. 2. TRAF3 is displaced from TRAF2 by preferential binding of TRAF1/2 heterotrimers to CD40 and cIAP1/2, resulting in CIAP1/2 mediated TRAF3 ubiquitination and degradation. 3. High affinity binding between TRAF2 and CD40 receptor displaces TRAF3 and alters cIAP1/2 function resulting in cIAP1/2, altered cIAP1/2 function and TRAF3 ubiquitination and degradation. D. Degradation or displacement from TRAF3 results in accumulation of newly synthesized NIK, which undergoes autophosphorylation and activation. NIK phosphorylates IKKα resulting in IKKα activation and phosphorylation of destruction box serines on p100 associated with RelB. Phosphorylated p100 is ubiquitinated and undergoes non-degradative processing by the proteasome yielding p52:RelB dimers that can mediate transcription by binding to DNA κB sites. E. IKKα induced phosphorylation of COOH-terminal serines on NIK may act as a negative feedback mechanism by inducing TRAF3-independent proteosomal degradation of NIK.
Unlike TNFR1, which lacks a TRAF binding motif and recruits TRAF2 through TRADD, CD40 possesses three well-characterized TRAF binding motifs in the cytoplasmic tail of the receptor (Figure 3). CD40 interacts with TRAFs 1, 2, 3, 5 and 6 [193–196]. CD40 binds to TRAF1, TRAF2 and TRAF3 using a prototypical TRAF binding site towards the COOH-terminus of the cytoplasmic tail [193]. The structure of TRAF2 bound to this site has been solved, [192] and reveals the expected trimer of TRAF2 that is capable of interacting with three receptor cytoplasmic tails. A second binding site that is selective for TRAF2 is located at the immediate COOH-terminus. The third TRAF binding site is located closer to the plasma membrane and binds to TRAF6 [194]. However, loss of this site does not abrogate TRAF6 recruitment, as TRAF6 can be recruited with TRAF2 [197]. Thus the relative contribution of each TRAF binding site and TRAF protein to CD40 signaling is complex. Therefore, we will touch only briefly on signaling to noncanonical NF-κB, as TRAF2 signaling to the canonical pathway has been discussed above, before moving on to discuss TRAF mediated activation of the alternative pathway downstream of CD40.
3.2. CD40 Signaling to Noncanonical NF-κB
The noncanonical NF-κB pathway culminates in inducible processing of p100 to p52 and is independent of IKKβ and NEMO. Instead, noncanonical signaling requires IKKα, which is phosphorylated and activated by the NF-κB inducing kinase (NIK). NIK is a member of the MAP3K family of kinases that can induce the activation of both the canonical and noncanonical NF-κB pathway [198]. Upon overexpression, NIK can activate canonical signaling through IKKβ [198–202], however the phenotype of NIK−/− and aly/aly mice, which have a point mutation in the NIK kinase domain, demonstrate that the physiological function of NIK is restricted to noncanonical signaling [203]. Although these mice were not initially characterized in terms of CD40 signaling to p100, it was subsequently observed that NIK deficient mice exhibited cell autonomous defects in B cell function, suggesting that CD40 signaling could be affected [204, 205]. Based on these observations, the role of NIK in p100 processing downstream of CD40 was studied and it was found that CD40L induces activation of the noncanonical pathway through NIK [206]. While we focused primarily on the mechanisms leading to recruitment of the IKK complex through the regulatory subunit NEMO during canonical signaling, we will focus on the receptor proximal events that lead to stabilization of NIK and phosphorylation IKKα.
In unstimulated cells NIK is constitutively active, but is also subject to rapid, constitutive degradation thus leading to very low levels of the protein (Figure 3A). TRAF3 binds constitutively to NIK and is responsible for the ubiquitination of NIK and its degradation [207]. Consequently loss of TRAF3 results in stabilization and accumulation of NIK, and resulting hyperactivation of the noncanonical pathway [208]. NIK was initially identified as a TRAF2 binding factor, although it binds with higher affinity to TRAF3 [198, 207]. However loss of TRAF2 also results in constitutive activation of the noncanonical pathway [208–210]. This suggests that both TRAF2 and TRAF3 can affect constitutive downregulation of NIK. Indeed, loss of either TRAF2 [211] or TRAF3 [212], results in accumulation of NIK independent of stimulation. Although TRAF proteins have reported E3 ligase activity, it does not appear that TRAF3 can directly mediate NIK ubiquitination [172]. Instead, studies using cIAP inhibitors and knockout cells revealed that cIAP1/2 are responsible for constitutive degradation of NIK in unstimulated cells [213, 214]. TRAF3 binds with high affinity to the N-terminus of NIK [207], thus, the complex is formed by TRAF3 binding NIK and TRAF2 serving as a scaffold by bringing together cIAP1/2 and TRAF3 [208, 215, 216].
The preponderance of evidence suggests that NIK is constitutively active (reviewed in [172]). Activation of NIK is, therefore, dependent on inactivation of the TRAF/cIAP complex. As discussed above, TRAF2 is recruited to CD40 upon ligation by CD40L. Upon CD40 ligation, TRAF3 is targeted for proteasomal degradation [207]. Despite the role of TRAF2 and cIAP1/2 in NIK degradation, both are also required for inducible accumulation of NIK (Figure 3C). TRAF2 knockout cells fail to induce TRAF3 degradation downstream of CD40 [215]. It was shown that cIAP1/2 undergoes TRAF2 dependent ubiquitination through an unknown mechanism that leads to cIAP1/2 dependent TRAF3 ubiquitination and degradation [215]. TRAF1, which is also recruited to the CD40 complex [217], has been proposed to alter the function of TRAF2/cIAP1/2 complex [172, 218]. However, multiple studies have yielded conflicting results in terms of the effects of TRAF1 in canonical and noncanonical NF-κB signaling by TNF cytokines [217, 219–221]. An alternative proposal for the regulation of NIK stabilization, the “allosteric model,” considers that CD40 could stabilize NIK by binding TRAF2/3 and competitively preventing binding of newly synthesized NIK to the TRAF/cIAP complex [222]. Under this model, which is compatible with the role of TRAF2/cIAP in TRAF3 degradation, binding of the TRAF3/TRAF2/cIAP1/2 complex to CD40L bound CD40 prevents NIK binding to TRAF3, because the binding of TRAF3 to CD40 and NIK uses the same surface, and results in cIAP1/2 ubiquitin ligase activity being focused on TRAF3[222]. Whether due solely to degradation of TRAF3, or inability of receptor bound TRAF3 to bind NIK, recruitment of the TRAF/cIAP complex to the receptor results in accumulation of newly synthesized NIK.
One of the first groups to discover IKKα did so by using a yeast two hybrid screen to identify NIK binding proteins [199]. As, such, from the beginning, it has been known that NIK can bind and phosphorylate IKKα [199, 223–226]. NIK is a potent kinase for IKKs (IKK-K), and on overexpression robustly activates both canonical and noncanonical NF-κB pathways. NIK phosphorylation of IKKα leads phosphorylation of p100 in p100:RelB heterodimers, and processing to p52 [227]. In addition to activating IKKα, NIK has also been shown facilitate phosphorylation of p100 through kinase independent mechanisms [228]. Unlike canonical signaling to IκBα degradation, it was shown that the NIK- and IKKα-dependent pathway leading to p100 processing to p52 does not require NEMO [229, 230] or IKKβ [227]. However it is not yet clear whether IKKα that mediates p100 phosphorylation exists in a separate IKK complex. Given that NEMO has recently been shown to target the IKK complex to IκBα [19], it may be that only IKKα that is free of NEMO can target p100.
In unstimulated cells, p100 undergoes little constitutive processing to p52 [231, 232]. The C-terminal DD of p100, and the flanking ankyrin repeat, inhibits processing to p52 [233, 234]. However, the mutants that undergo constitutive processing appear to be processed by a distinct mechanism that requires nuclear shuttling [233], and, perhaps, cleavage of DNA bound p100 [235]. In contrast, NIK-induced processing of p100 likely occurs in the cytoplasm [236]. NIK and IKKα induce phosphorylation p100 on two COOH-terminal serine residues, 866 and 870 [228, 234, 237]. Although phosphorylation of these two serines depends on IKKα, direct phosphorylation of Serines 866 and 870 by IKKα has not yet been demonstrated [172]. Similar to IκBα, phosphorylation of p100 leads to recognition by βTrCP [237]. Subsequently, polyubiquitination of Lys855 in a region with sequence homology to Lys22 of IκBα, is mediated by the SCF complex leading to subsequent preoteasomal processing to p52 [237–239]. The exact mechanism that leads to processing of p100 to p52 remains unclear. Several factors have been proposed to function in this process including the S9 subunit of the proteasome. [240]. Another structural element of p100, that is also involved in p105 processing to p50, that may contribute to selective processing is a glycine rich repeat (GRR; Figure 1) that is thought to prevent complete degradation and promote release of active p52:NF-κB complexes [231, 241]. Indeed, p100 lacking the GRR undergoes NIK induced degradation rather than processing to p52 [236].
Although p100 can interact with several NF-κB dimers, it is believed that p100 interacts preferentially with RelB [242]. Processing of p100 therefore results in the release RelB:p52 dimers that translocate to the nucleus and induces transcription of a subset of target genes [243]. In this [243], and other studies, it has generally been assumed that deletion of IKKα or NIK reveals the subset of physiological functions governed by the noncanonical pathway. However, it should be emphasized that NIK is capable of activating IKKβ and inducing IκBα degradation, as discussed above. Furthermore, for TNF family receptors that activate the noncanonical pathway by inducing NIK accumulation, including CD40, activation of the canonical pathway has been shown to be variably dependent on NIK and IKKα [244]. Finally, NIK activation can be terminated through restoration of the TRAF2/3/cIAP1/2 complex, leading to NIK degradation. In addition, IKKα has recently been reported to phosphorylate NIK resulting in degradation that appears independent of the TRAF2/3/cIAP1/2 complex [245].
4. Conclusions
In the current review we have limited our discussion to signaling by TNF to the canonical NF-κB pathway, and by CD40 to the noncanonical NF-κB pathway. While several of the concepts discussed apply broadly to signaling by TNF family cytokines, it is clear that TNF family receptors diverge significantly in their signaling mechanisms. Nevertheless, the role or RIP1, TAK1, NIK, and other factors discussed above have not yet been rigorously evaluated in many TNF family signaling pathways. As is readily apparent from the discussion above, our understanding of the contribution of individual TRAF family members to specific signaling pathways is particularly inadequate. While genetic models are important, it is clear that for TRAF proteins, compensation leads to aberrant TRAF utilization and hence the conclusions that may not reflect the biology. More elegant and precise methods of interrogating TRAF function are needed. For example, careful biochemistry, using unmanipulated, and biologically relevant cells is required to map the role of individual TRAF proteins in signaling by individual TNF family members. Atypical mechanisms of TNF family receptor signaling to NF-κB have also not been broadly investigated. For example, TACI (TNFRSF13B), which is a receptor for the TNF family cytokines BAFF and APRIL, requires binding and signaling through MyD88 in order to drive B-cell class switch recombination [246]. While it is tempting to dismiss signaling pathways that fail to conform to established mechanisms for a receptor class, such differences likely underlie the need for 29 different receptors in the TNF family. In support of the biological significance of such pathways, a subsequent report demonstrated that a patient with a mutation in the MyD88 binding region of TACI displayed hyper-gammaglobulinemia and an inability to class switch in response to APRIL [247]. In addition to illuminating interesting biology, such differences may hold the key to developing targeted means of inhibiting the NF-κB pathway. Thus, there remains much to understand both ligand/receptor specific and cell type specific signaling by TNF family receptors to NF-κB.
Finally, we would like to note the controversy surrounding the function of regulatory ubiquitination in NF-κB signaling. We often do a disservice by too confidently presenting schematics of signaling pathways with little discussion of caveats and unknowns. Both in TNF signaling, and NF-κB signaling in general, considerable work remains to be done to clarify the role of regulatory ubiquitination. This is an area of active research, where yesterday’s models are invalid while today’s appear unquestionable. The once clear requirement for K63 linked ubiquitination, TRAF2 as E3 ubiquitin ligase, and many other aspects of the role of regulatory ubiquitin seem to have fallen out of favor. A20, a K63 DUB and negative regulator of NF-κB signaling seemed to provide the clearest evidence of the importance of K63 in TNF induced NF-κB. However, as discussed, knock-in mice produced by our lab (S.G.) and others disprove the requirement for A20’s deubiquitinating activity in regulating NF-κB. As the field now shifts its focus towards linear ubiquitin, we urge caution against overstating the case before all of the evidence, in particular from appropriate genetic models, is in.
Highlights.
TNF family cytokines activate both canonical and noncanonical NF-κB signaling pathways
TNF/TNFR1 signaling serves as a model of the canonical NF-κB pathway
CD40L/CD40 signaling serves as a model of the noncanonical NF-κB pathway
The molecular mechanisms linking ligation of TNFR1 or CD40 to activation of the canonical or noncanonical NF-κB signaling pathway remain unclear
Acknowledgments
This work was supported by institutional support from Columbia University and NIH grants RO1-AI068977 and RO1-AI 093985 (to S.G.).
Abbreviations
- AP-1
Activator Protein-1
- APRIL
a proliferation inducing ligand
- BAFF
B cell activating factor
- BCL-3
B cell lymphoma 3
- βTrCP
beta transducing repeat containing
- CD40
cluster of differentiation 40
- Fn14
fibroblast growth factor inducible 14
- IκB
inhibitor of kappa B
- IKK
IκB kinase
- JNK
c-Jun N-terminal kinase
- MAPK
Mitogen activate protein kinase
- MyD88
myeloid differentiation primary response gene 88
- NF-κB
nuclear factor interacting with the kappa light chain enhancer of activated B cells or, commonly, nuclear factor kappa B
- RANK
receptor activator of NF-κB
- SHARPIN
SHANK-Associated RH domain interacting protein
- SCF
Skp1-Culin-Roc1/Rbx1/Hrt-1-F-box
- STAT
signal transducer and activator of transcription
- SUMO
small ubiquitin like modifier
- TACI
transmembrane activator and CALM interactor
- TNF
tumor necrosis factor
- TNFAIP3
Tumor necrosis factor alpha induced protein 3
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gilmore TD, Wolenski FS. NF-kappaB: where did it come from and why? Immunological reviews. 2012;246:14–35. doi: 10.1111/j.1600-065X.2012.01096.x. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. NF-kappaB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh S, Hayden MS. Celebrating 25 years of NF-kappaB research. Immunological reviews. 2012;246:5–13. doi: 10.1111/j.1600-065X.2012.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinz M, Arslan SC, Scheidereit C. It takes two to tango: IkappaBs, the multifunctional partners of NF-kappaB. Immunological reviews. 2012;246:59–76. doi: 10.1111/j.1600-065X.2012.01102.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, et al. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 7.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 8.Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Stimulation-dependent I kappa B alpha phosphorylation marks the NF-kappa B inhibitor for degradation via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haskill S, Beg AA, Tompkins SM, Morris JS, Yurochko AD, Sampson-Johannes A, et al. Characterization of an immediate-early gene induced in adherent monocytes that encodes I kappa B-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh S, Baltimore D. Activation in vitro of NF-kappa B by phosphorylation of its inhibitor I kappa B. Nature. 1990;344:678–682. doi: 10.1038/344678a0. [DOI] [PubMed] [Google Scholar]
- 11.Lin YC, Brown K, Siebenlist U. Activation of NF-kappa B requires proteolysis of the inhibitor I kappa B-alpha: signal-induced phosphorylation of I kappa B-alpha alone does not release active NF-kappa B. Proc Natl Acad Sci U S A. 1995;92:552–556. doi: 10.1073/pnas.92.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henkel T, Machleidt T, Alkalay I, Kronke M, Ben-Neriah Y, Baeuerle PA. Rapid proteolysis of I kappa B-alpha is necessary for activation of transcription factor NFkappa B. Nature. 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- 13.Mellits KH, Hay RT, Goodbourn S. Proteolytic degradation of MAD3 (I kappa B alpha) and enhanced processing of the NF-kappa B precursor p105 are obligatory steps in the activation of NF-kappa B. Nucleic Acids Res. 1993;21:5059–5066. doi: 10.1093/nar/21.22.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tergaonkar V, Correa RG, Ikawa M, Verma IM. Distinct roles of IkappaB proteins in regulating constitutive NF-kappaB activity. Nature cell biology. 2005;7:921–923. doi: 10.1038/ncb1296. [DOI] [PubMed] [Google Scholar]
- 15.Kanarek N, Ben-Neriah Y. Regulation of NF-kappaB by ubiquitination and degradation of the IkappaBs. Immunological reviews. 2012;246:77–94. doi: 10.1111/j.1600-065X.2012.01098.x. [DOI] [PubMed] [Google Scholar]
- 16.Smale ST. Dimer-specific regulatory mechanisms within the NF-kappaB family of transcription factors. Immunological reviews. 2012;246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 17.Natoli G. NF-kappaB and chromatin: ten years on the path from basic mechanisms to candidate drugs. Immunological reviews. 2012;246:183–192. doi: 10.1111/j.1600-065X.2012.01103.x. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nature reviews Immunology. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 19.Polley S, Huang DB, Hauenstein AV, Fusco AJ, Zhong X, Vu D, et al. A structural basis for IkappaB kinase 2 activation via oligomerization-dependent trans auto-phosphorylation. PLoS biology. 2013;11:e1001581. doi: 10.1371/journal.pbio.1001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 21.Aggarwal BB, Gupta SC, Kim JH. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood. 2012;119:651–665. doi: 10.1182/blood-2011-04-325225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osborn L, Kunkel S, Nabel GJ. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappaB. Proc Natl Acad Sci U S A. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duh EJ, Maury WJ, Folks TM, Fauci AS, Rabson AB. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci U S A. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowenthal JW, Ballard DW, Bohnlein E, Greene WC. Tumor necrosis factor alpha induces proteins that bind specifically to kappa B-like enhancer elements and regulate interleukin 2 receptor alpha-chain gene expression in primary human T lymphocytes. Proc Natl Acad Sci U S A. 1989;86:2331–2335. doi: 10.1073/pnas.86.7.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shakhov AN, Collart MA, Vassalli P, Nedospasov SA, Jongeneel CV. Kappa B-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor alpha gene in primary macrophages. J Exp Med. 1990;171:35–47. doi: 10.1084/jem.171.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappaB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 27.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 28.Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y. NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J Exp Med. 1997;185:953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenfeld ME, Prichard L, Shiojiri N, Fausto N. Prevention of hepatic apoptosis and embryonic lethality in RelA/TNFR-1 double knockout mice. The American journal of pathology. 2000;156:997–1007. doi: 10.1016/S0002-9440(10)64967-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alcamo E, Mizgerd JP, Horwitz BH, Bronson R, Beg AA, Scott M, et al. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-kappa B in leukocyte recruitment. Journal of immunology. 2001;167:1592–1600. doi: 10.4049/jimmunol.167.3.1592. [DOI] [PubMed] [Google Scholar]
- 31.Doi TS, Marino MW, Takahashi T, Yoshida T, Sakakura T, Old LJ, et al. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proc Natl Acad Sci U S A. 1999;96:2994–2999. doi: 10.1073/pnas.96.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 33.Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, et al. The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berghe TV, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 35.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nature reviews Immunology. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 36.Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 37.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 38.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 39.Liu ZG, Hsu H, Goeddel DV, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 40.Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 41.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 42.Ermolaeva MA, Michallet MC, Papadopoulou N, Utermohlen O, Kranidioti K, Kollias G, et al. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol. 2008;9:1037–1046. doi: 10.1038/ni.1638. [DOI] [PubMed] [Google Scholar]
- 43.Chen NJ, Chio II, Lin WJ, Duncan G, Chau H, Katz D, et al. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proc Natl Acad Sci U S A. 2008;105:12429–12434. doi: 10.1073/pnas.0806585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pobezinskaya YL, Kim YS, Choksi S, Morgan MJ, Li T, Liu C, et al. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat Immunol. 2008;9:1047–1054. doi: 10.1038/ni.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider F, Neugebauer J, Griese J, Liefold N, Kutz H, Briseno C, et al. The viral oncoprotein LMP1 exploits TRADD for signaling by masking its apoptotic activity. PLoS biology. 2008;6:e8. doi: 10.1371/journal.pbio.0060008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng L, Bidere N, Staudt D, Cubre A, Orenstein J, Chan FK, et al. Competitive control of independent programs of tumor necrosis factor receptor-induced cell death by TRADD and RIP1. Mol Cell Biol. 2006;26:3505–3513. doi: 10.1128/MCB.26.9.3505-3513.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pobezinskaya YL, Kim YS, Choksi S, Morgan MJ, Li T, Liu C, et al. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat Immunol. 2008;9:1047–1054. doi: 10.1038/ni.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen NJ, Chio II, Lin WJ, Duncan G, Chau H, Katz D, et al. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12429–12434. doi: 10.1073/pnas.0806585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ermolaeva MA, Michallet MC, Papadopoulou N, Utermohlen O, Kranidioti K, Kollias G, et al. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol. 2008;9:1037–1046. doi: 10.1038/ni.1638. [DOI] [PubMed] [Google Scholar]
- 50.Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol. 2001;21:5299–5305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 52.Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 53.Tada K, Okazaki T, Sakon S, Kobarai T, Kurosawa K, Yamaoka S, et al. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-kappa B activation and protection from cell death. J Biol Chem. 2001;276:36530–36534. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- 54.Nakano H, Sakon S, Koseki H, Takemori T, Tada K, Matsumoto M, et al. Targeted disruption of Traf5 gene causes defects in CD40- and CD27-mediated lymphocyte activation. Proc Natl Acad Sci U S A. 1999;96:9803–9808. doi: 10.1073/pnas.96.17.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen LT, Duncan GS, Mirtsos C, Ng M, Speiser DE, Shahinian A, et al. TRAF2 deficiency results in hyperactivity of certain TNFR1 signals and impairment of CD40-mediated responses. Immunity. 1999;11:379–389. doi: 10.1016/s1074-7613(00)80113-2. [DOI] [PubMed] [Google Scholar]
- 56.Devin A, Lin Y, Yamaoka S, Li Z, Karin M, Liu Z. The alpha and beta subunits of IkappaB kinase (IKK) mediate TRAF2-dependent IKK recruitment to tumor necrosis factor (TNF) receptor 1 in response to TNF. Mol Cell Biol. 2001;21:3986–3994. doi: 10.1128/MCB.21.12.3986-3994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 58.Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J. RIPK1 is not essential for TNFR1-induced activation of NF-kappaB. Cell Death Differ. 2010;17:482–487. doi: 10.1038/cdd.2009.178. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 2011;471:373–376. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gentle IE, Wong WW, Evans JM, Bankovacki A, Cook WD, Khan NR, et al. In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8. The Journal of biological chemistry. 2011;286:13282–13291. doi: 10.1074/jbc.M110.216226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ting AT, Pimentel-Muinos FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. Embo J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 63.Lee TH, Shank J, Cusson N, Kelliher MA. The kinase activity of Rip1 is not required for tumor necrosis factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J Biol Chem. 2004;279:33185–33191. doi: 10.1074/jbc.M404206200. [DOI] [PubMed] [Google Scholar]
- 64.Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity. 2000;12:419–429. doi: 10.1016/s1074-7613(00)80194-6. [DOI] [PubMed] [Google Scholar]
- 65.Berger SB, Kasparcova V, Hoffman S, Swift B, Dare L, Schaeffer M, et al. Cutting Edge: RIP1 Kinase Activity Is Dispensable for Normal Development but Is a Key Regulator of Inflammation in SHARPIN-Deficient Mice. Journal of immunology. 2014 doi: 10.4049/jimmunol.1400499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delhase M, Hayakawa M, Chen Y, Karin M. Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science. 1999;284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 67.Inohara N, Koseki T, Lin J, del Peso L, Lucas PC, Chen FF, et al. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 68.Zhang SQ, Kovalenko A, Cantarella G, Wallach D. Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKgamma) upon receptor stimulation. Immunity. 2000;12:301–311. doi: 10.1016/s1074-7613(00)80183-1. [DOI] [PubMed] [Google Scholar]
- 69.Poyet JL, Srinivasula SM, Lin JH, Fernandes-Alnemri T, Yamaoka S, Tsichlis PN, et al. Activation of the Ikappa B kinases by RIP via IKKgamma /NEMO-mediated oligomerization. J Biol Chem. 2000;275:37966–37977. doi: 10.1074/jbc.M006643200. [DOI] [PubMed] [Google Scholar]
- 70.Tegethoff S, Behlke J, Scheidereit C. Tetrameric oligomerization of IkappaB kinase gamma (IKKgamma) is obligatory for IKK complex activity and NF-kappaB activation. Mol Cell Biol. 2003;23:2029–2041. doi: 10.1128/MCB.23.6.2029-2041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agou F, Traincard F, Vinolo E, Courtois G, Yamaoka S, Israel A, et al. The trimerization domain of NEMO is comprised of the interacting C-terminal CC2 and LZ coiled-coil subdomains. J Biol Chem. 2004 doi: 10.1074/jbc.M314278200. [DOI] [PubMed] [Google Scholar]
- 72.Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6:1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 74.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 75.Li H, Kobayashi M, Blonska M, You Y, Lin X. Ubiquitination of RIP is required for tumor necrosis factor alpha-induced NF-kappaB activation. J Biol Chem. 2006;281:13636–13643. doi: 10.1074/jbc.M600620200. [DOI] [PubMed] [Google Scholar]
- 76.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 77.Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Molecular cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 78.Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-{kappa}B activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem. 2005;280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- 79.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 80.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. Deubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 81.Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–10567. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Estornes Y, Bertrand MJ. IAPs, regulators of innate immunity and inflammation. Seminars in cell & developmental biology. 2014 doi: 10.1016/j.semcdb.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 83.Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11778–11783. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vince JE, Chau D, Callus B, Wong WW, Hawkins CJ, Schneider P, et al. TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1-TRAF2 complex to sensitize tumor cells to TNFalpha. The Journal of cell biology. 2008;182:171–184. doi: 10.1083/jcb.200801010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nature cell biology. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 87.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 88.Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 89.Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, et al. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 91.Zhang L, Blackwell K, Shi Z, Habelhah H. The RING domain of TRAF2 plays an essential role in the inhibition of TNFalpha-induced cell death but not in the activation of NF-kappaB. Journal of molecular biology. 2010;396:528–539. doi: 10.1016/j.jmb.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Molecular cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ori D, Kato H, Sanjo H, Tartey S, Mino T, Akira S, et al. Essential roles of K63-linked polyubiquitin-binding proteins TAB2 and TAB3 in B cell activation via MAPKs. Journal of immunology. 2013;190:4037–4045. doi: 10.4049/jimmunol.1300173. [DOI] [PubMed] [Google Scholar]
- 94.Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 95.Zhou H, Wertz I, O'Rourke K, Ultsch M, Seshagiri S, Eby M, et al. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 96.Belgnaoui SM, Paz S, Samuel S, Goulet ML, Sun Q, Kikkert M, et al. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell host & microbe. 2012;12:211–222. doi: 10.1016/j.chom.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 97.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. The EMBO journal. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haas AL. Linear polyubiquitylation: the missing link in NF-kappaB signalling. Nature cell biology. 2009;11:116–118. doi: 10.1038/ncb0209-116. [DOI] [PubMed] [Google Scholar]
- 99.Vanlangenakker N, Bertrand MJ, Bogaert P, Vandenabeele P, Vanden Berghe T. TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell death & disease. 2011;2:e230. doi: 10.1038/cddis.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fujita H, Rahighi S, Akita M, Kato R, Sasaki Y, Wakatsuki S, et al. Mechanism underlying IkappaB kinase activation mediated by the linear ubiquitin chain assembly complex. Mol Cell Biol. 2014;34:1322–1335. doi: 10.1128/MCB.01538-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci U S A. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blackwell K, Zhang L, Workman LM, Ting AT, Iwai K, Habelhah H. Two coordinated mechanisms underlie tumor necrosis factor alpha-induced immediate and delayed IkappaB kinase activation. Mol Cell Biol. 2013;33:1901–1915. doi: 10.1128/MCB.01416-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 104.Wu C, Ghosh S. Differential phosphorylation of the signal-responsive domain of I kappa B alpha and I kappa B beta by I kappa B kinases. J Biol Chem. 2003;278:31980–31987. doi: 10.1074/jbc.M304278200. [DOI] [PubMed] [Google Scholar]
- 105.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hao S, Baltimore D. RNA splicing regulates the temporal order of TNF-induced gene expression. Proc Natl Acad Sci U S A. 2013;110:11934–11939. doi: 10.1073/pnas.1309990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bhatt D, Ghosh S. Regulation of the NF-kappaB-Mediated Transcription of Inflammatory Genes. Frontiers in immunology. 2014;5:71. doi: 10.3389/fimmu.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang H, Tang QZ, Wang AB, Chen M, Yan L, Liu C, et al. Tumor suppressor A20 protects against cardiac hypertrophy and fibrosis by blocking transforming growth factor-beta-activated kinase 1-dependent signaling. Hypertension. 2010;56:232–239. doi: 10.1161/HYPERTENSIONAHA.110.149963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baker RG, Hayden MS, Ghosh S. NF-kappaB, inflammation, and metabolic disease. Cell metabolism. 2011;13:11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.DiDonato JA, Mercurio F, Karin M. NF-kappaB and the link between inflammation and cancer. Immunological reviews. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 112.Ruland J. Return to homeostasis: downregulation of NF-kappaB responses. Nat Immunol. 2011;12:709–714. doi: 10.1038/ni.2055. [DOI] [PubMed] [Google Scholar]
- 113.Schutze S, Tchikov V, Schneider-Brachert W. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat Rev Mol Cell Biol. 2008;9:655–662. doi: 10.1038/nrm2430. [DOI] [PubMed] [Google Scholar]
- 114.Brown K, Park S, Kanno T, Franzoso G, Siebenlist U. Mutual regulation of the transcriptional activator NF-kappa B and its inhibitor, I kappa B-alpha. Proc Natl Acad Sci U S A. 1993;90:2532–2536. doi: 10.1073/pnas.90.6.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 116.de Martin R, Vanhove B, Cheng Q, Hofer E, Csizmadia V, Winkler H, et al. Cytokine-inducible expression in endothelial cells of an I kappa B alpha-like gene is regulated by NF kappaB. The EMBO journal. 1993;12:2773–2779. doi: 10.1002/j.1460-2075.1993.tb05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Le Bail O, Schmidt-Ullrich R, Israel A. Promoter analysis of the gene encoding the I kappa B-alpha/MAD3 inhibitor of NF-kappa B: positive regulation by members of the rel/NF-kappa B family. The EMBO journal. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Beg AA, Sha WC, Bronson RT, Baltimore D. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 119.Klement JF, Rice NR, Car BD, Abbondanzo SJ, Powers GD, Bhatt PH, et al. IkappaBalpha deficiency results in a sustained NF-kappaB response and severe widespread dermatitis in mice. Mol Cell Biol. 1996;16:2341–2349. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Majumdar I, Paul J. The deubiquitinase A20 in immunopathology of autoimmune diseases. Autoimmunity. 2014 doi: 10.3109/08916934.2014.900756. [DOI] [PubMed] [Google Scholar]
- 121.Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem. 1992;267:17971–17976. [PubMed] [Google Scholar]
- 122.Laherty CD, Hu HM, Opipari AW, Wang F, Dixit VM. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor kappaB. J Biol Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- 123.Evans PC, Ovaa H, Hamon M, Kilshaw PJ, Hamm S, Bauer S, et al. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J. 2004;378:727–734. doi: 10.1042/BJ20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jaattela M, Mouritzen H, Elling F, Bastholm L. A20 zinc finger protein inhibits TNF and IL-1 signaling. Journal of immunology. 1996;156:1166–1173. [PubMed] [Google Scholar]
- 125.Cooper JT, Stroka DM, Brostjan C, Palmetshofer A, Bach FH, Ferran C. A20 blocks endothelial cell activation through a NF-kappaB-dependent mechanism. J Biol Chem. 1996;271:18068–18073. doi: 10.1074/jbc.271.30.18068. [DOI] [PubMed] [Google Scholar]
- 126.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc Natl Acad Sci U S A. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zetoune FS, Murthy AR, Shao Z, Hlaing T, Zeidler MG, Li Y, et al. A20 inhibits NF-kappa B activation downstream of multiple Map3 kinases and interacts with the I kappa B signalosome. Cytokine. 2001;15:282–298. doi: 10.1006/cyto.2001.0921. [DOI] [PubMed] [Google Scholar]
- 128.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.He KL, Ting AT. A20 inhibits tumor necrosis factor (TNF) alpha-induced apoptosis by disrupting recruitment of TRADD and RIP to the TNF receptor 1 complex in Jurkat T cells. Mol Cell Biol. 2002;22:6034–6045. doi: 10.1128/MCB.22.17.6034-6045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 131.De Valck D, Jin DY, Heyninck K, Van de Craen M, Contreras R, Fiers W, et al. The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene. 1999;18:4182–4190. doi: 10.1038/sj.onc.1202787. [DOI] [PubMed] [Google Scholar]
- 132.Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, Lee B, et al. Homeostatic MyD88-dependent signals cause lethal inflamMation in the absence of A20. J Exp Med. 2008;205:451–464. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 134.Dong J, Jimi E, Zeiss C, Hayden MS, Ghosh S. Constitutively active NF-kappaB triggers systemic TNFalpha-dependent inflammation and localized TNFalpha-independent inflammatory disease. Genes Dev. 2010;24:1709–1717. doi: 10.1101/gad.1958410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rebholz B, Haase I, Eckelt B, Paxian S, Flaig MJ, Ghoreschi K, et al. Crosstalk between keratinocytes and adaptive immune cells in an IkappaBalpha protein-mediated inflammatory disease of the skin. Immunity. 2007;27:296–307. doi: 10.1016/j.immuni.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 136.Werner SL, Kearns JD, Zadorozhnaya V, Lynch C, O'Dea E, Boldin MP, et al. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22:2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Skaug B, Chen J, Du F, He J, Ma A, Chen ZJ. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Verhelst K, Carpentier I, Kreike M, Meloni L, Verstrepen L, Kensche T, et al. A20 inhibits LUBAC-mediated NF-kappaB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 2012;31:3845–3855. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tokunaga F, Nishimasu H, Ishitani R, Goto E, Noguchi T, Mio K, et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation. EMBO J. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yamaguchi N, Oyama M, Kozuka-Hata H, Inoue J. Involvement of A20 in the molecular switch that activates the non-canonical NF-small ka, CyrillicB pathway. Scientific reports. 2013;3:2568. doi: 10.1038/srep02568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Jr, Conaway RC, et al. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Strebovsky J, Walker P, Lang R, Dalpke AH. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25:863–874. doi: 10.1096/fj.10-170597. [DOI] [PubMed] [Google Scholar]
- 146.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, et al. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 147.Kinjyo I, Hanada T, Inagaki-Ohara K, Mori H, Aki D, Ohishi M, et al. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity. 2002;17:583–591. doi: 10.1016/s1074-7613(02)00446-6. [DOI] [PubMed] [Google Scholar]
- 148.Nakagawa R, Naka T, Tsutsui H, Fujimoto M, Kimura A, Abe T, et al. SOCS-1 participates in negative regulation of LPS responses. Immunity. 2002;17:677–687. doi: 10.1016/s1074-7613(02)00449-1. [DOI] [PubMed] [Google Scholar]
- 149.Ganesh L, Burstein E, Guha-Niyogi A, Louder MK, Mascola JR, Klomp LW, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- 150.Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. EMBO J. 2007;26:436–447. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, et al. COMMD proteins, a novel family of structural and functional homologs of MURR1. J Biol Chem. 2005;280:22222–22232. doi: 10.1074/jbc.M501928200. [DOI] [PubMed] [Google Scholar]
- 152.Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML. Phosphorylation of NF-kappaB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 2009;10:381–386. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, et al. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-kappaB/RelA. Genes Dev. 2009;23:849–861. doi: 10.1101/gad.1748409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mattioli I, Geng H, Sebald A, Hodel M, Bucher C, Kracht M, et al. Inducible phosphorylation of NF-kappa B p65 at serine 468 by T cell costimulation is mediated by IKK epsilon. J Biol Chem. 2006;281:6175–6183. doi: 10.1074/jbc.M508045200. [DOI] [PubMed] [Google Scholar]
- 155.Schwabe RF, Sakurai H. IKKbeta phosphorylates p65 at S468 in transactivaton domain 2. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19:1758–1760. doi: 10.1096/fj.05-3736fje. [DOI] [PubMed] [Google Scholar]
- 156.Chatterji D, Ogawa Y, Shimada T, Ishihama A. The role of the omega subunit of RNA polymerase in expression of the relA gene in Escherichia coli. FEMS microbiology letters. 2007;267:51–55. doi: 10.1111/j.1574-6968.2006.00532.x. [DOI] [PubMed] [Google Scholar]
- 157.Moreno R, Sobotzik JM, Schultz C, Schmitz ML. Specification of the NF-kappaB transcriptional response by p65 phosphorylation and TNF-induced nuclear translocation of IKK epsilon. Nucleic Acids Res. 2010;38:6029–6044. doi: 10.1093/nar/gkq439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mukherjee SP, Behar M, Birnbaum HA, Hoffmann A, Wright PE, Ghosh G. Analysis of the RelA:CBP/p300 interaction reveals its involvement in NF-kappaB-driven transcription. PLoS biology. 2013;11:e1001647. doi: 10.1371/journal.pbio.1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Colleran A, Collins PE, O'Carroll C, Ahmed A, Mao X, McManus B, et al. Deubiquitination of NF-kappaB by Ubiquitin-Specific Protease-7 promotes transcription. Proc Natl Acad Sci U S A. 2013;110:618–623. doi: 10.1073/pnas.1208446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8:584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 161.Healy NC, O'Connor R. Sequestration of PDLIM2 in the cytoplasm of monocytic/macrophage cells is associated with adhesion and increased nuclear activity of NF-kappaB. Journal of leukocyte biology. 2009;85:481–490. doi: 10.1189/jlb.0408238. [DOI] [PubMed] [Google Scholar]
- 162.Bowe RA, Cox OT, Ayllon V, Tresse E, Healy NC, Edmunds SJ, et al. PDLIM2 regulates transcription factor activity in epithelial-to-mesenchymal transition via the COP9 signalosome. Molecular biology of the cell. 2014;25:184–195. doi: 10.1091/mbc.E13-06-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tanaka T, Yamamoto Y, Muromoto R, Ikeda O, Sekine Y, Grusby MJ, et al. PDLIM2 inhibits T helper 17 cell development and granulomatous inflammation through degradation of STAT3. Science signaling. 2011;4:ra85. doi: 10.1126/scisignal.2001637. [DOI] [PubMed] [Google Scholar]
- 164.Tanaka T, Soriano MA, Grusby MJ. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22:729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 165.Guo H, Mi Z, Bowles DE, Bhattacharya SD, Kuo PC. Osteopontin and protein kinase C regulate PDLIM2 activation and STAT1 ubiquitination in LPS-treated murine macrophages. J Biol Chem. 2010;285:37787–37796. doi: 10.1074/jbc.M110.161869. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 166.Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006;16:196–202. doi: 10.1038/sj.cr.7310027. [DOI] [PubMed] [Google Scholar]
- 167.Liu B, Yang R, Wong KA, Getman C, Stein N, Teitell MA, et al. Negative regulation of NF-kappaB signaling by PIAS1. Mol Cell Biol. 2005;25:1113–1123. doi: 10.1128/MCB.25.3.1113-1123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu B, Mink S, Wong KA, Stein N, Getman C, Dempsey PW, et al. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol. 2004;5:891–898. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- 169.Wong KA, Kim R, Christofk H, Gao J, Lawson G, Wu H. Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol Cell Biol. 2004;24:5577–5586. doi: 10.1128/MCB.24.12.5577-5586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tahk S, Liu B, Chernishof V, Wong KA, Wu H, Shuai K. Control of specificity and magnitude of NF-kappa B and STAT1-mediated gene activation through PIASy and PIAS1 cooperation. Proc Natl Acad Sci U S A. 2007;104:11643–11648. doi: 10.1073/pnas.0701877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu B, Yang Y, Chernishof V, Loo RR, Jang H, Tahk S, et al. Proinflammatory stimuli induce IKKalpha-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129:903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- 172.Sun SC. The noncanonical NF-kappaB pathway. Immunological reviews. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Aruffo A, Farrington M, Hollenbaugh D, Li X, Milatovich A, Nonoyama S, et al. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72:291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- 174.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunological reviews. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, et al. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 176.Hollander GA, Castigli E, Kulbacki R, Su M, Burakoff SJ, Gutierrez-Ramos JC, et al. Induction of alloantigen-specific tolerance by B cells from CD40-deficient mice. Proc Natl Acad Sci U S A. 1996;93:4994–4998. doi: 10.1073/pnas.93.10.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Snapper CM, Rosas FR, Zelazowski P, Moorman MA, Kehry MR, Bravo R, et al. B cells lacking RelB are defective in proliferative responses, but undergo normal B cell maturation to Ig secretion and Ig class switching. J Exp Med. 1996;184:1537–1541. doi: 10.1084/jem.184.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Franzoso G, Carlson L, Poljak L, Shores EW, Epstein S, Leonardi A, et al. Mice deficient in nuclear factor (NF)-kappa B/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J Exp Med. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Horwitz BH, Zelazowski P, Shen Y, Wolcott KM, Scott ML, Baltimore D, et al. The p65 subunit of NF-kappa B is redundant with p50 during B cell proliferative responses, and is required for germline CH transcription and class switching to IgG3. Journal of immunology. 1999;162:1941–1946. [PubMed] [Google Scholar]
- 180.Snapper CM, Zelazowski P, Rosas FR, Kehry MR, Tian M, Baltimore D, et al. B cells from p50/NF-kappa B knockout mice have selective defects in proliferation, differentiation, germ-line CH transcription, and Ig class switching. Journal of immunology. 1996;156:183–191. [PubMed] [Google Scholar]
- 181.Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 182.Carrasco D, Cheng J, Lewin A, Warr G, Yang H, Rizzo C, et al. Multiple hemopoietic defects and lymphoid hyperplasia in mice lacking the transcriptional activation domain of the c-Rel protein. J Exp Med. 1998;187:973–984. doi: 10.1084/jem.187.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zelazowski P, Carrasco D, Rosas FR, Moorman MA, Bravo R, Snapper CM. B cells genetically deficient in the c-Rel transactivation domain have selective defects in germline CH transcription and Ig class switching. Journal of immunology. 1997;159:3133–3139. [PubMed] [Google Scholar]
- 184.Ouaaz F, Arron J, Zheng Y, Choi Y, Beg AA. Dendritic cell development and survival require distinct NF-kappaB subunits. Immunity. 2002;16:257–270. doi: 10.1016/s1074-7613(02)00272-8. [DOI] [PubMed] [Google Scholar]
- 185.Kriehuber E, Bauer W, Charbonnier AS, Winter D, Amatschek S, Tamandl D, et al. Balance between NF-kappaB and JNK/AP-1 activity controls dendritic cell life and death. Blood. 2005;106:175–183. doi: 10.1182/blood-2004-08-3072. [DOI] [PubMed] [Google Scholar]
- 186.O'Sullivan BJ, MacDonald KP, Pettit AR, Thomas R. RelB nuclear translocation regulates B cell MHC molecule, CD40 expression, and antigen-presenting cell function. Proc Natl Acad Sci U S A. 2000;97:11421–11426. doi: 10.1073/pnas.97.21.11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.O'Sullivan BJ, Thomas R. CD40 ligation conditions dendritic cell antigen-presenting function through sustained activation of NF-kappaB. Journal of immunology. 2002;168:5491–5498. doi: 10.4049/jimmunol.168.11.5491. [DOI] [PubMed] [Google Scholar]
- 188.Shih VF, Davis-Turak J, Macal M, Huang JQ, Ponomarenko J, Kearns JD, et al. Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-kappaB pathways. Nat Immunol. 2012;13:1162–1170. doi: 10.1038/ni.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Karpusas M, Hsu YM, Wang JH, Thompson J, Lederman S, Chess L, et al. 2 A crystal structure of an extracellular fragment of human CD40 ligand. Structure. 1995;3:1426. [PubMed] [Google Scholar]
- 190.An HJ, Kim YJ, Song DH, Park BS, Kim HM, Lee JD, et al. Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation. J Biol Chem. 2011;286:11226–11235. doi: 10.1074/jbc.M110.208215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Smulski CR, Beyrath J, Decossas M, Chekkat N, Wolff P, Estieu-Gionnet K, et al. Cysteine-rich domain 1 of CD40 mediates receptor self-assembly. J Biol Chem. 2013;288:10914–10922. doi: 10.1074/jbc.M112.427583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pullen SS, Labadia ME, Ingraham RH, McWhirter SM, Everdeen DS, Alber T, et al. High-affinity interactions of tumor necrosis factor receptor-associated factors (TRAFs) and CD40 require TRAF trimerization and CD40 multimerization. Biochemistry. 1999;38:10168–10177. doi: 10.1021/bi9909905. [DOI] [PubMed] [Google Scholar]
- 193.Pullen SS, Miller HG, Everdeen DS, Dang TT, Crute JJ, Kehry MR. CD40-tumor necrosis factor receptor-associated factor (TRAF) interactions: regulation of CD40 signaling through multiple TRAF binding sites and TRAF hetero-oligomerization. Biochemistry. 1998;37:11836–11845. doi: 10.1021/bi981067q. [DOI] [PubMed] [Google Scholar]
- 194.Ishida T, Mizushima S, Azuma S, Kobayashi N, Tojo T, Suzuki K, et al. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J Biol Chem. 1996;271:28745–28748. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- 195.Ishida TK, Tojo T, Aoki T, Kobayashi N, Ohishi T, Watanabe T, et al. TRAF5, a novel tumor necrosis factor receptor-associated factor family protein, mediates CD40 signaling. Proc Natl Acad Sci U S A. 1996;93:9437–9442. doi: 10.1073/pnas.93.18.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lu LF, Cook WJ, Lin LL, Noelle RJ. CD40 signaling through a newly identified tumor necrosis factor receptor-associated factor 2 (TRAF2) binding site. J Biol Chem. 2003;278:45414–45418. doi: 10.1074/jbc.M309601200. [DOI] [PubMed] [Google Scholar]
- 197.Rowland SL, Tremblay MM, Ellison JM, Stunz LL, Bishop GA, Hostager BS. A novel mechanism for TNFR-associated factor 6-dependent CD40 signaling. Journal of immunology. 2007;179:4645–4653. doi: 10.4049/jimmunol.179.7.4645. [DOI] [PubMed] [Google Scholar]
- 198.Malinin NL, Boldin MP, Kovalenko AV, Wallach D. MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 199.Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 200.Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 201.Lin X, Mu Y, Cunningham ET, Jr, Marcu KB, Geleziunas R, Greene WC. Molecular determinants of NF-kappaB-inducing kinase action. Mol Cell Biol. 1998;18:5899–5907. doi: 10.1128/mcb.18.10.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, et al. A physical and functional map of the human TNF-alpha/NF-kappaB signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 203.Shinkura R, Kitada K, Matsuda F, Tashiro K, Ikuta K, Suzuki M, et al. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat Genet. 1999;22:74–77. doi: 10.1038/8780. [DOI] [PubMed] [Google Scholar]
- 204.Karrer U, Althage A, Odermatt B, Hengartner H, Zinkernagel RM. Immunodeficiency of alymphoplasia mice (aly/aly) in vivo: structural defect of secondary lymphoid organs and functional B cell defect. European journal of immunology. 2000;30:2799–2807. doi: 10.1002/1521-4141(200010)30:10<2799::AID-IMMU2799>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 205.Yamada T, Mitani T, Yorita K, Uchida D, Matsushima A, Iwamasa K, et al. Abnormal immune function of hemopoietic cells from alymphoplasia (aly) mice, a natural strain with mutant NF-kappa B-inducing kinase. Journal of immunology. 2000;165:804–812. doi: 10.4049/jimmunol.165.2.804. [DOI] [PubMed] [Google Scholar]
- 206.Coope HJ, Atkinson PG, Huhse B, Belich M, Janzen J, Holman MJ, et al. CD40 regulates the processing of NF-kappaB2 p100 to p52. EMBO J. 2002;21:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 208.Gardam S, Sierro F, Basten A, Mackay F, Brink R. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity. 2008;28:391–401. doi: 10.1016/j.immuni.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 209.Grech AP, Amesbury M, Chan T, Gardam S, Basten A, Brink R. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-kappaB activation in mature B cells. Immunity. 2004;21:629–642. doi: 10.1016/j.immuni.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 210.Xia M, Ling W, Zhu H, Wang Q, Ma J, Hou M, et al. Anthocyanin prevents CD40-activated proinflammatory signaling in endothelial cells by regulating cholesterol distribution. Arteriosclerosis, thrombosis, and vascular biology. 2007;27:519–524. doi: 10.1161/01.ATV.0000254672.04573.2d. [DOI] [PubMed] [Google Scholar]
- 211.Zhang L, Blackwell K, Thomas GS, Sun S, Yeh WC, Habelhah H. TRAF2 suppresses basal IKK activity in resting cells and TNFalpha can activate IKK in TRAF2 and TRAF5 double knockout cells. J Mol Biol. 2009;389:495–510. doi: 10.1016/j.jmb.2009.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.He JQ, Zarnegar B, Oganesyan G, Saha SK, Yamazaki S, Doyle SE, et al. Rescue of TRAF3-null mice by p100 NF-kappa B deficiency. J Exp Med. 2006;203:2413–2418. doi: 10.1084/jem.20061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 214.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 215.Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Fotin-Mleczek M, Henkler F, Hausser A, Glauner H, Samel D, Graness A, et al. Tumor necrosis factor receptor-associated factor (TRAF) 1 regulates CD40-induced TRAF2-mediated NF-kappaB activation. J Biol Chem. 2004;279:677–685. doi: 10.1074/jbc.M310969200. [DOI] [PubMed] [Google Scholar]
- 218.Zheng C, Kabaleeswaran V, Wang Y, Cheng G, Wu H. Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation. Mol Cell. 2010;38:101–113. doi: 10.1016/j.molcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Guo F, Sun A, Wang W, He J, Hou J, Zhou P, et al. TRAF1 is involved in the classical NF-kappaB activation and CD30-induced alternative activity in Hodgkin's lymphoma cells. Mol Immunol. 2009;46:2441–2448. doi: 10.1016/j.molimm.2009.05.178. [DOI] [PubMed] [Google Scholar]
- 220.McPherson AJ, Snell LM, Mak TW, Watts TH. Opposing roles for TRAF1 in the alternative versus classical NF-kappaB pathway in T cells. J Biol Chem. 2012;287:23010–23019. doi: 10.1074/jbc.M112.350538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Xie P, Hostager BS, Munroe ME, Moore CR, Bishop GA. Cooperation between TNF receptor-associated factors 1 and 2 in CD40 signaling. Journal of immunology. 2006;176:5388–5400. doi: 10.4049/jimmunol.176.9.5388. [DOI] [PubMed] [Google Scholar]
- 222.Sanjo H, Zajonc DM, Braden R, Norris PS, Ware CF. Allosteric regulation of the ubiquitin:NIK and ubiquitin:TRAF3 E3 ligases by the lymphotoxin-beta receptor. The Journal of biological chemistry. 2010;285:17148–17155. doi: 10.1074/jbc.M110.105874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ling L, Cao Z, Goeddel DV. NF-kappaB-inducing kinase activates IKK-alpha by phosphorylation of Ser- 176. Proc Natl Acad Sci U S A. 1998;95:3792–3797. doi: 10.1073/pnas.95.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kashiwada M, Shirakata Y, Inoue JI, Nakano H, Okazaki K, Okumura K, et al. Tumor necrosis factor receptor-associated factor 6 (TRAF6) stimulates extracellular signal-regulated kinase (ERK) activity in CD40 signaling along a ras-independent pathway. J Exp Med. 1998;187:237–244. doi: 10.1084/jem.187.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Uhlik M, Good L, Xiao G, Harhaj EW, Zandi E, Karin M, et al. NF-kappaB-inducing kinase and IkappaB kinase participate in human T-cell leukemia virus I Tax-mediated NF-kappaB activation. J Biol Chem. 1998;273:21132–21136. doi: 10.1074/jbc.273.33.21132. [DOI] [PubMed] [Google Scholar]
- 226.Nemoto S, DiDonato JA, Lin A. Coordinate regulation of IkappaB kinases by mitogen-activated protein kinase kinase kinase 1 and NF-kappaB-inducing kinase. Mol Cell Biol. 1998;18:7336–7343. doi: 10.1128/mcb.18.12.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 228.Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IKKalpha to p100 and IKKalpha-mediated phosphorylation. J Biol Chem. 2004 doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 229.Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 230.Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 231.Heusch M, Lin L, Geleziunas R, Greene WC. The generation of nfkb2 p52: mechanism and efficiency. Oncogene. 1999;18:6201–6208. doi: 10.1038/sj.onc.1203022. [DOI] [PubMed] [Google Scholar]
- 232.Betts JC, Nabel GJ. Differential regulation of NF-kappaB2(p100) processing and control by amino-terminal sequences. Mol Cell Biol. 1996;16:6363–6371. doi: 10.1128/mcb.16.11.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Liao G, Sun SC. Regulation of NF-kappaB2/p100 processing by its nuclear shuttling. Oncogene. 2003;22:4868–4874. doi: 10.1038/sj.onc.1206761. [DOI] [PubMed] [Google Scholar]
- 234.Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 235.Qing G, Yan P, Qu Z, Liu H, Xiao G. Hsp90 regulates processing of NF-kappa B2 p100 involving protection of NF-kappa B-inducing kinase (NIK) from autophagy-mediated degradation. Cell Res. 2007;17:520–530. doi: 10.1038/cr.2007.47. [DOI] [PubMed] [Google Scholar]
- 236.Qing G, Qu Z, Xiao G. Stabilization of basally translated NF-kappaB-inducing kinase (NIK) protein functions as a molecular switch of processing of NF-kappaB2 p100. J Biol Chem. 2005;280:40578–40582. doi: 10.1074/jbc.M508776200. [DOI] [PubMed] [Google Scholar]
- 237.Fong A, Sun SC. Genetic evidence for the essential role of beta-transducin repeat-containing protein in the inducible processing of NF-kappa B2/p100. J Biol Chem. 2002;277:22111–22114. doi: 10.1074/jbc.C200151200. [DOI] [PubMed] [Google Scholar]
- 238.Amir RE, Haecker H, Karin M, Ciechanover A. Mechanism of processing of the NF-kappa B2 p100 precursor: identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCF(beta-TrCP) ubiquitin ligase. Oncogene. 2004;23:2540–2547. doi: 10.1038/sj.onc.1207366. [DOI] [PubMed] [Google Scholar]
- 239.Cohen S, Achbert-Weiner H, Ciechanover A. Dual effects of IkappaB kinase beta-mediated phosphorylation on p105 Fate: SCF(beta-TrCP)-dependent degradation and SCF(beta-TrCP)-independent processing. Mol Cell Biol. 2004;24:475–486. doi: 10.1128/MCB.24.1.475-486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Fong A, Zhang M, Neely J, Sun SC. S9, a 19 S proteasome subunit interacting with ubiquitinated NF-kappaB2/p100. J Biol Chem. 2002;277:40697–40702. doi: 10.1074/jbc.M205330200. [DOI] [PubMed] [Google Scholar]
- 241.Lin L, Ghosh S. A glycine-rich region in NF-kappaB p105 functions as a processing signal for the generation of the p50 subunit. Mol Cell Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV. RelB cellular regulation and transcriptional activity are regulated by p100. J Biol Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- 243.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 244.Ramakrishnan P, Wang W, Wallach D. Receptor-specific signaling for both the alternative and the canonical NF-kappaB activation pathways by NF-kappaB-inducing kinase. Immunity. 2004;21:477–489. doi: 10.1016/j.immuni.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 245.Razani B, Zarnegar B, Ytterberg AJ, Shiba T, Dempsey PW, Ware CF, et al. Negative feedback in noncanonical NF-kappaB signaling modulates NIK stability through IKKalpha-mediated phosphorylation. Science signaling. 2010;3:ra41. doi: 10.1126/scisignal.2000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.He B, Santamaria R, Xu W, Cols M, Chen K, Puga I, et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat Immunol. 2010;11:836–845. doi: 10.1038/ni.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Almejun MB, Cols M, Zelazko M, Oleastro M, Cerutti A, Oppezzo P, et al. Naturally occurring mutation affecting the MyD88-binding site of TNFRSF13B impairs triggering of class switch recombination. European journal of immunology. 2013;43:805–814. doi: 10.1002/eji.201242945. [DOI] [PMC free article] [PubMed] [Google Scholar]