Abstract
During peptide bond formation on the ribosome the α-amine of an aminoacyl-tRNA attacks the ester carbonyl carbon of a peptidyl-tRNA to yield a peptide lengthened by one amino acid. Although the ribosome's contribution to catalysis is predominantly entropic, the lack of high-resolution structural data for the complete active site in complex with full-length ligands has made it difficult to assess how the ribosome might influence the pathway of the reaction. Here, we present crystal structures of pre-attack and post-catalysis complexes of the Thermus thermophilus 70S ribosome at ∼2.6 Å resolution. These structures reveal a network of hydrogen bonds along which proton transfer could take place to ensure the concerted, rate-limiting formation of a tetrahedral intermediate. Unlike earlier models, we propose that the ribosome and the A-site tRNA facilitate the deprotonation of the nucleophile through the activation of a water molecule.
Introduction
The ribosome is a large ribonucleoprotein complex responsible for synthesizing proteins in all living organisms. In bacteria, ribosomes are composed of large (50S) and small (30S) subunits, comprising approximately two-thirds rRNA and one-third protein. The active site where peptide bond formation takes place on the ribosome – also known as the peptidyl transferase center (PTC) – is located within the 50S subunit and consists mainly of RNA, with the N-terminal tail of the nearest ribosomal protein, L27, only approaching within 8-10 Å of the reaction center1. Thus, it has been concluded that the ribosome is a ribozyme, with RNA being the major contributor to catalysis2.
Peptide bond formation is an aminolysis reaction in which the α-amine of an aminoacyl-tRNA attacks the ester carbonyl carbon of a peptidyl-tRNA, resulting in the transfer of the peptidyl moiety to the aminoacyl-tRNA bound in the A-site. For each amino acid that is added to the nascent polypeptide chain, at least three protons must move within the PTC. Since primary amines in solution at near-neutral pH exist in the protonated ammonium form, it is necessary to first deprotonate the α-NH3+ group of the A-site tRNA in order to obtain the nucleophilic α-NH2 group that will initiate the reaction. In addition, a proton must be removed from this amine either during or following nucleophilic attack. Finally, a proton must be provided to the 3′-oxygen of the terminal P-site tRNA ribose in order to ensure the departure of the leaving group. As a facilitator of peptide bond formation, the ribosome could provide a path for the fast and efficient movement of protons around the PTC (i.e. through the ordering of water molecules and (or) the positioning of rRNA and tRNA residues), directly promote proton movement through acid-base catalysis, or do both. To understand how polypeptide chains are synthesized in vivo, one must therefore understand the role that the ribosome plays in facilitating proton transfer.
Models seeking to explain the movement of protons during peptide bond formation are typically evaluated on the basis of their agreement with high-resolution structures of the Haloarcula marismortui (Hma) 50S subunit in complex with minimal substrate, transition state or product analogs3-5. Although these structures have greatly advanced our understanding of ribosomal protein synthesis, they nevertheless restrict the interpretation of the available experimental and computational data to the context of the isolated large ribosomal subunit. In order to extend this structural framework to the full ribosome, we determined the structures of pre-attack and post-catalysis complexes of Thermus thermophilus (Tth) 70S ribosomes containing full-size tRNA ligands at resolutions of 2.6 Å and 2.55 Å, respectively (Fig. 1 and Supplementary Fig. 1), as well as two pre-reaction complexes featuring a short A-site substrate analog and a full-length P-site tRNA (Supplementary Fig. 2).
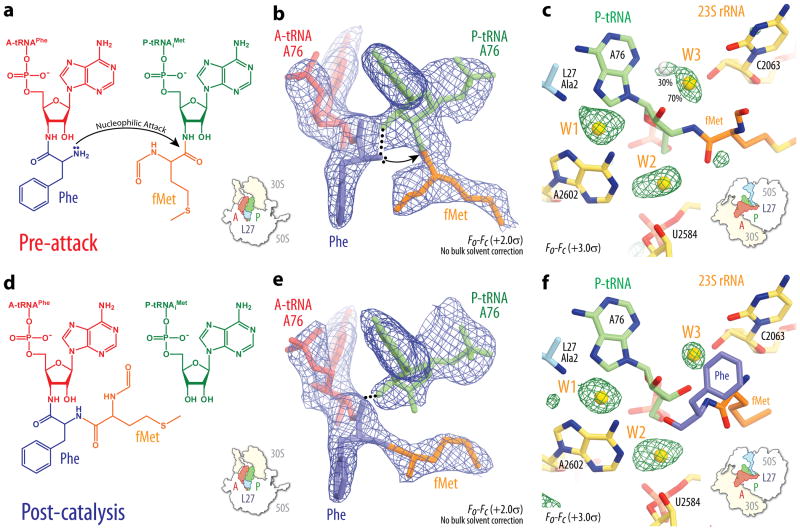
Figure 1. Chemical diagrams, electron density and water molecules in the peptidyl transferase center.
a, d, The chemical structures of Phe-NH-tRNAPhe and fMet-NH-tRNAiMet (a), and fMet-Phe-NH-tRNAPhe and tRNAiMet (d). b, e, Unbiased Fo-Fc electron density maps from co-crystallization experiments with the respective substrates: Phe-NH-tRNAPhe and fMet-NH-tRNAiMet at 2.6 Å resolution (+2.0σ) (b), and fMet-Phe-NH-tRNAPhe and tRNAiMet at 2.55 Å resolution (+2.0σ) (e). Bulk solvent correction was not applied to the PTC region, as explained in the legend for Supplementary Fig. 1 and online methods. In all cases the A-site tRNA and amino acid are shown in red and blue, respectively, while the P-site tRNA and amino acid are in green and orange, respectively. c, f, Unbiased Fo-Fc electron density maps of the Tth 70S pre-attack complex at 2.6 Å resolution (+2.5σ) showing the positions of PTC water molecules W1, W2 and W3 for pre-attack (c) and post-catalysis (f) states. The coloring scheme for the A- and P-site substrates is the same as in panels (b) and (e), 23S rRNA is colored in yellow, protein L27 is in blue, and a hydrated magnesium ion is in dark green. Alternate positions for W3 in the pre-attack state are indicated.
While these are not the first structures of the 70S ribosome to contain full-length aminoacylated tRNA ligands1, the 0.7-0.9 Å increase in resolution over similar structures that has been achieved here provides a complete description of the ribosomal PTC with bound tRNA substrates or products, ions and water molecules (Figs. 1 and 2). Importantly, these structures now allow us to perform a much more comprehensive structure-based assessment of the possible mechanisms for ribosome-catalyzed peptide bond formation.
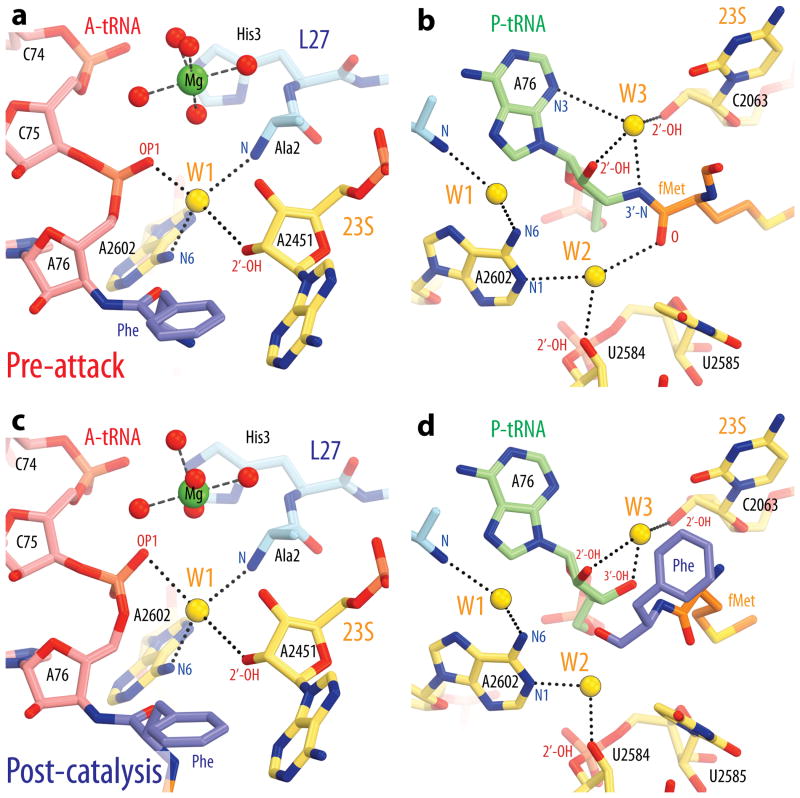
Figure 2. Coordination of water molecules in the active site.
a, c, Water molecule W1 is coordinated by the 2′-OH of 23S rRNA residue A2451, the phosphate oxygen of the A-site tRNA residue A76, the N-terminal amino group of ribosomal protein L27 and the N6 atom of 23S rRNA residue A2602 in both the pre-attack (a) and post-catalysis complex structures (c). b, d, Water molecule W2 is coordinated by the ester carbonyl oxygen of the peptidyl-tRNA, the N1 atom of 23S rRNA residue A2602 and the 2′-OH of U2585 in the pre-attack complex structure, while W3 is hydrogen bonded to the 2′-OH, 3′-OH, and N3 atoms of the P-site tRNA residue A76, and to the 2′-OH of C2063 (b). In the post-catalysis complex structure (d), contacts between W2 and the peptide are lost, while the hydrogen bond between W3 and the P-site A76 ribose shifts towards the 3′-OH of the deacylated tRNA.
Results
The structure of the PTC remains rigid throughout catalysis
In the Tth 70S ribosome pre-attack complex, unreacted, non-hydrolyzable fMet-NH-tRNAiMet occupies the P-site, while non-hydrolyzable Phe-NH-tRNAPhe is bound to the A-site (Fig. 1a,b). A comparison of the PTC seen in this structure with that of the lower resolution structure of a Tth 70S ribosome pre-attack complex1 and the structures of an Hma 50S subunit pre-attack complex with the A- and P-site substrate mimics cytidine-cytidine-hydroxypuromycin (CChPmn) and cytidine-cytidine-adenosine-phenylalanine-caproic acid-biotin (CCApcb)4 indicates that the 23S rRNA adopts similar conformations in all cases (Supplementary Fig. 3a,c). Nearly identical 23S rRNA conformations are also observed in our structures of Tth 70S ribosome in complex with fMet-NH-tRNAiMet in the P-site and the substrate analogs cytidine-puromycin (CPmn) or cytidine-cytidine-puromycin (CCPmn) in the A-site (Supplementary Fig. 3e). This is in contrast with earlier studies showing that the closely related analog cytidine-hydroxypuromycin (ChPmn) bound to Hma 50S fails to trigger the induced state, which is characterized by the rearrangement of residues U2506, G2583, U2584 and U2585 that results from proper binding of the substrate to the A-site4 (Supplementary Fig. 3f). Thus, all of the pre-attack structures presented here correspond to the induced state, with a fully accommodated A-site nucleophile poised to react with the P-site substrate.
In the Tth 70S ribosome post-catalysis complex, deacylated tRNAiMet occupies the P-site and dipeptidyl-tRNA obtained in situ by reaction of Phe-NH-tRNAPhe with fMet-tRNAiMet is found in the A-site (Fig. 1d,e), providing direct evidence that the ribosomes crystallized in our study are catalytically active under the buffer conditions used for complex formation. Strikingly, major differences are observed between the structure of the Tth 70S ribosome post-catalysis complex and that of an Hma 50S subunit complex with a product analog in the A-site5 (Supplementary Fig. 3b). While the PTC in the latter structure is in a conformation that resembles the uninduced state, our structure is very similar to the fully induced pre-attack state. Moreover the base of A2602 in the Hma 50S subunit product structure is rotated by ∼180° relative to its orientation in our structure. Since the rotated conformation of this base would result in a steric clash with the full-length P-site tRNA, we conclude that our Tth 70S ribosome post-catalysis structure is the correct structure of the PTC following peptide bond formation. The conformation of the 23S rRNA seen here is also very similar to that reported in the lower resolution structure of a Tth 70S ribosome complex mimicking the post-peptidyl transfer state1 (Supplementary Fig. 3d). However, this earlier structure featured an aminoacyl-tRNA instead of a dipeptidyl-tRNA in the A-site, making it unclear whether it represented the true post-catalysis state. From the nearly identical conformations of the 23S rRNA in the pre-attack and post-catalysis structures presented here and in earlier structures of the 50S subunit in complex with transition state analogs, we thus conclude that the PTC remains relatively rigid throughout catalysis.
Three ordered water molecules are trapped within the PTC
Due to the higher resolution of the current Tth 70S ribosome in complex with tRNAs compared with previous structures, peaks of positive density corresponding to three water molecules (W1-3) are visible in unbiased Fo-Fc maps of the pre-attack and post-catalysis Tth 70S ribosome complexes (Fig. 1c,f). W1 lies within a cavity that is formed by residues A2602 and A2451 of the 23S rRNA, the 3′-end of the A-site tRNA and the N-terminus of protein L27 (Fig. 2a,c). Although density for this water is absent from the Hma 50S subunit pre-attack structure4, it could nevertheless be modeled as a water molecule in all of our structures based on the following criteria: (i) comparison with the structures of the Hma 50S subunit in complex with the transition state mimics DAA3 or DCA4, which exhibited density in a similar location that could not be replaced by soaking of the crystals with high concentrations of Mn2+, K+ or Rb+ 3; (ii) tetrahedral coordination and distances from its ligands; (iii) strength of the electron density.
Water molecules W2 and W3, which occupy pockets buried deep within the PTC (Fig. 2b,d), were previously observed in structures of Hma 50S subunits in complex with substrates or transition state analogs3,4, and were proposed to stabilize the oxyanion in the transition state6 and take part in proton shuttling3, respectively. However, the structures presented here exhibit all three water molecules at once, showing how they interact with both the ribosome and its tRNA substrates. Notably, no other solvent molecules are present within the PTC, including within the narrow cavity occupied by W3. This was not the case in many of the earlier structures of Hma 50S subunit complexes3,4, in which two water molecules were often modeled at this location and the active site was generally accessible to the bulk solvent. In our pre-attack structure, the unbiased difference density observed for W3 is elongated and could best be modeled as a single water molecule populating two alternate locations with relative occupancies of 0.7 and 0.3 (Fig. 1c). We hypothesize that the latter position corresponds to a minor sub-population in which the P-site tRNA is deacylated, thereby also explaining the relatively weak density observed for the fMet residue. For the purposes of our analysis we will therefore only consider the position of W3 modeled with an occupancy of 0.7.
A proton wire connects the attacking amine to W1
In all of the structures presented here, a short network of hydrogen bonds analogous to a proton wire7 connects W1 to the attacking amine via the 2′-OH of the P-site tRNA A76 and the 2′-OH of A2451 (Fig. 3a). The geometry of this network is well-suited for efficient proton transfer, with D–H…A distances of <3.1 Å in the Tth 70S ribosome pre-attack complex and the angle ω between three successive non-hydrogen atoms ranging from 107° to 121° and thus approximating the ideal value of 109.5° for a tetravalent atom (Supplementary Fig. 4a). This is in contrast with the hydrogen bond networks connecting the attacking amine to the 3′-O of the peptidyl-tRNA via the 2′-OH of the P-site tRNA A76, either through W3 (ω = 55°, 94°) (Supplementary Fig. 4b) or directly (ω = 61°) (Supplementary Fig. 4c). Moreover, W1 in the Tth 70S ribosome pre-attack complexes forms a strong (2.4 Å) hydrogen bond with the 5′-phosphate oxygen of A76 of the A-site tRNA, as well as additional hydrogen bonds with the N6 amino group of A2602 and the N-terminus of L27. As a result, this water molecule appears to be tightly coordinated from all sides and not likely to be exchanged with bulk solvent during catalysis.
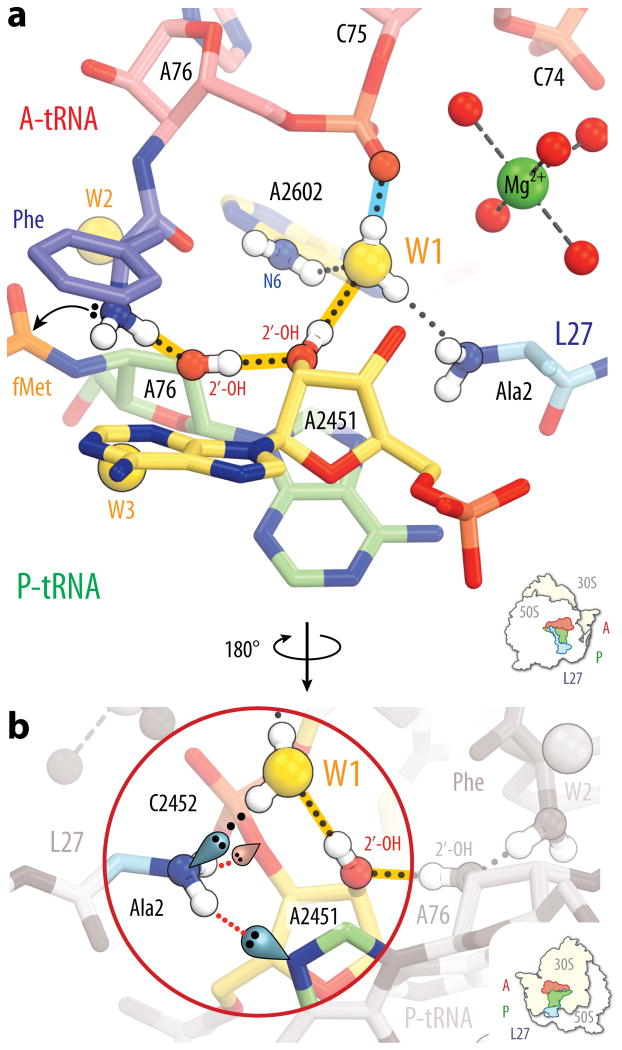
Figure 3. A proton wire in the peptidyl transferase center.
a, The hydrogen bond network connecting the α-amine of the A-site amino acid to water molecule W1 in the Tth 70S pre-attack complex is highlighted in gold. The hydrogen bond between water molecule W1 and the A-site tRNA A76 backbone phosphate is highlighted in blue. The coloring scheme is the same as in Figure 1. An arrow indicates nucleophilic attack by the nitrogen lone pair onto the P-site ester carbonyl carbon. Hydrogen atoms are for illustration purposes only and their positions were not refined by means of molecular simulations. b, The proton wire shown in panel (a) is seen from a view rotated by 180° around the vertical axis. Key hydrogen atoms, several lone pairs and hydrogen bonds are highlighted to indicate that proton transfer cannot be propagated beyond the N-terminal amino group of L27.
The N-terminus of L27 also interacts with two hydrogen bond acceptors: the N7 imino group of the P-site tRNA A76 and the 3′-O of A2451 (Fig. 3b). The nature of these interactions is such that proton exchange between the L27 terminus and the bulk solvent would require some backbone rearrangements to take place within the terminal residues of L27, resulting in the breaking of several hydrogen bonds. With the assistance of a nearby hydrated magnesium ion that closes the cavity containing W1, the N-terminus of L27 could therefore restrict the flow of protons between W1 and the bulk solvent. The conformation of the N-terminal segment of L27 in our current structures is in excellent agreement with that seen in earlier structures of the Tth 70S ribosome in complex with tRNAs1, yet importantly it is the resolution and quality of the electron density observed here that is key to revealing its interactions with W1.
Even though the resolution of our structures does not allow us to draw conclusions regarding the protonation state of the N-terminus of L27, computational simulations8 suggest that its α-amine is the ionizable ribosomal group with a pKa of ∼7.5-8.0 that is observed in the reaction with the A-site substrate puromycin9. The protonation state predicted by these studies for the reaction with tRNA substrates is more problematic however, given that they rely on a model of the PTC in which a water molecule that could in principle correspond to W1 makes a different set of interactions with components of the active site8. For the purposes of the discussion below, we will assume that the α-amine of L27 undergoes a downward pKa shift and is thus deprotonated in the physiological pH range.
Discussion
Currently, two major models have been proposed to account for the movement of protons around the active site during peptide bond formation3. Referred to as the six- and eight-membered proton shuttles, both of these models rely on the 2′-OH of the P-site tRNA A76 ribose to transfer a proton from the attacking amine to the leaving group, with the eight-membered shuttle further incorporating a water molecule corresponding to W3 from this study into the proton relay. Importantly, the structures that helped formulate these models were obtained using isolated 50S subunits and lacked full-length tRNAs, thus precluding the ordering of protein L27 (or rather, as discussed below, its functional equivalent in archaea) that is needed to render the PTC inaccessible to the bulk solvent. What's more, the water referred to here as W1 was only seen in complexes with transition state analogs that lacked the 2′-OH of the P-site A76, making it difficult at the time to incorporate this molecule into the proposed reaction models.
The question that the current work addresses is the following: given the structures of the expanded PTC presented here, can we propose alternate models for peptide bond formation that the earlier Hma 50S ribosome structures did not allow us to foresee? In doing so, we do not mean to supersede or disprove the existing models, but rather to expand the conceptual framework needed to probe the mechanistic aspects of peptide bond formation by biochemical, kinetic and (or) computational means. Like the earlier proton shuttle models, our proposed mechanism does not seek to explain the initial deprotonation of the A-site α-NH3+ to give a nucleophilic amine, as this event may well take place prior to or during the induced fit triggered by the binding of the A-site substrate. Instead, we focus our attention on the circumstances surrounding the deprotonation of the attacking amine and the formation of a tetrahedral intermediate.
The Tth 70S pre-attack structure presented in this work is the outcome of crystallographic experiments performed on a thermodynamically equilibrated sample containing unreactive substrate analogs. Consequently, the state that is visualized here may differ to some extent from the catalytically active state, that presumably occurs immediately after accommodation of the aminoacyl-tRNA into the A site. This problem is by no means unique to this particular system, yet it is important to stress that the state populated here may not be the kinetically relevant one, despite the fact that the structures of the Hma 50S ribosome in complex with transition state analogs mirror our findings. With this in mind, we assume that the Tth 70S pre-attack structure represents a reasonable approximation of the reactive state, which is suitable enough to allow mechanistic hypotheses to be formulated. Similar assumptions are made concerning the Tth 70S post-catalysis structure.
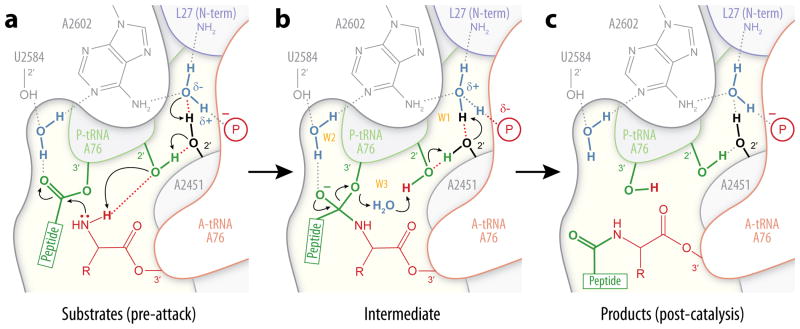
Superposition of the pre-attack and post-catalysis Tth 70S ribosome structures obtained here onto structures of the Hma 50S subunit in complex with various transition state analogs3,4 suggests at least one alternative model for proton transfer that differs substantially from the six- or eight-membered concerted proton shuttles3,10,11 (Fig. 4; Supplementary Video 1). Assuming that the α-amine of L27 is deprotonated, the presence of this basic group and of the negatively charged 5′-phosphate oxygen of the A-site A76 in the immediate vicinity of W1 could facilitate the formation of a partial negative charge on the water oxygen (Fig. 4a). Shortening of the O–H…O– distance between W1 and the 5′-phosphate along the reaction coordinate to form the low-barrier hydrogen bond suggested by our structures could, in turn, cause an upward shift of the pKa for the protonated form of W1 from its value in solution of -1.74. As the nucleophilic attack proceeds, the extremely high pKa of the α-amine would decrease substantially, ultimately triggering a concerted proton transfer from the attacking nucleophile to W1 via the proton wire formed by the 2′-OH of the P-site A76 ribose and the 2′-OH of A2451. This would result in the negatively charged tetrahedral intermediate (T–) described by recent kinetic isotope effect (KIE) experiments12, which might additionally receive a proton from the adjoining water molecule W2 to yield a neutral intermediate (T0)6 (Fig. 4b). Positive and negative charges would then be unable to propagate further due to a lack of neighboring groups suitable for proton transfer and would be transiently retained within the closed pockets containing W1 and W2, respectively.
Figure 4. A possible alternate pathway for peptide bond formation.
a, Attack by the α-amine of the aminoacyl-tRNA onto the ester carbonyl carbon of the peptidyl-tRNA is concerted with the deprotonation of the nucleophile in a single rate-limiting step. b, A tetrahedral intermediate is formed, with positive and negative charges becoming separated in space and delocalized over the pockets containing W1 and W2, respectively. Water molecules are labeled in gold. c, A partial reversal of the proton transfer event results in intermediate breakdown, yielding peptidyl-tRNA in the A-site and deacylated tRNA in the P-site.
In our model, a proton wire that consists of components provided by the ribosome and both tRNAs is formed only after the aminoacyl-tRNA is fully accommodated into the A-site, which ensures that the nucleophilic attack and deprotonation of the α-amine are coordinated in a single rate-limiting step. Although we propose that the 5′-phosphate oxygen of the A-site A76 aided by the α-amine of the L27 N-terminus could assist in the deprotonation of the nucleophile, further biochemical and kinetic studies will need to be carried out to establish whether this is true for all or perhaps only certain incoming amino acids. Studies on the effect of L27 on peptide bond formation performed to date have been rather limited, with the deletion of the first 3 residues of L27 resulting in only a modest decrease in the rate of peptide bond formation as measured by the standard puromycin assay13.
From a structural perspective, our model could be favored over the six- or eight-membered proton shuttles proposed earlier3. Indeed, the efficiency of proton transfer through an sp3 hybridized atom X is impaired by substantial deviations of the ω angle from the ideal value of 109.5° 14. While the proton wire described in this study is characterized by ω angles approximating this value, the hydrogen bond networks at the heart of the eight- and six-membered proton shuttles have a much less favorable geometry (Supplementary Fig. 4). In addition, the alternate positions observed for W3 in the pre-attack structure render this water molecule much less likely than W1 to be a part of the proton relay involved in deprotonating the attacking amine. Although the proton wire linking the attacking amine to W1 in the pre-attack Tth 70S ribosome structure appears to be the most geometrically favorable path along which a proton could be abstracted from the nucleophile, quantitative quantum mechanical calculations of the type performed for the six- and eight-membered proton shuttles11 could be performed to evaluate this prediction.
The concerted formation of a tetrahedral intermediate implied by our model is consistent with the lack of positive charge buildup on the attacking amine in the transition state implied by the near-zero Brønsted coefficient15 and with KIE data suggesting that the ribosome alters the pathway of the uncatalyzed reaction by means of a pre-arranged proton transfer network12. The proposed pathway for proton transfer is in agreement with the rate-limiting formation of at least three hydrogen bonds in the transition state (i.e. from the attacking amine to the P-site A76 2′-OH to the A2451 2′-OH to W1), as suggested by kinetic solvent isotope effect (KSIE) studies performed with puromycin as the A-site substrate10. Moreover, the trajectory for the departing proton is supported by biochemical data showing that deletion of the 2′-OH of the A76 ribose of the P-site tRNA or substitution of the 2′-OH of A2451 with groups including -H or -OCH3 decrease the rate of peptide bond formation by at least 100-fold16-18 or 10 to 50-fold19,20, respectively. It is worth noting that the controversy surrounding the role of the 2′-OH of the P-site tRNA A76 ribose during peptide bond formation16-18 stems from the fact that the structural consequences of mutations at this position remain unknown. The values quoted above may thus be largely underestimated given that the high-resolution structures that would reveal compensatory rearrangements within the PTC16,21 and (or) functional complementation by ordered solvent molecules19 have not been observed.
Finally, the effect of base substitutions at position 2602 of the 23S rRNA on the rate of peptide bond formation in vivo are easily reconciled with our proposed model22. Indeed, when the minimal A-site substrate puromycin is used together with ribosomes mutated at position A2602, the rate constant for the reaction is 30-320 times slower than with wild-type ribosomes, whereas the rate remains unchanged when the reaction is performed with a full-length aminoacyl-tRNA. Modeled base substitutions at this position in the Tth 70S ribosome pre-attack structure suggest that hydrogen bonding between residue 2602 and water molecules W1 and W2 is preserved in all cases, in agreement with the rate constants observed for full-length substrates. Moreover, the mild reduction in rate that is measured upon complete deletion of A2602 could be explained by the fact that W1 remains tightly bound via three hydrogen bonds to the mutant ribosome pre-attack complex. Even though A2602 may seem dispensable at first sight, the effect of cumulative mutations within the W1 binding pocket on the rate of peptide bond formation must be measured in order to assess its full contribution to the reaction. A possible involvement of A2602 in coordinating the reaction should also be considered given that this residue acts as a bridge between the two proposed catalytic waters W1 and W2.
A key question raised by our model concerns the possible involvement of the ribosome in catalyzing the conversion of the tetrahedral intermediate into products. Increasing the rate of breakdown of the intermediate may indeed be necessary to drive the reaction forward by preventing the intermediate from reverting back to substrates. Under such circumstances, the ester bond of the peptidyl-tRNA would become exposed to hydrolysis, which in turn could result in the premature termination of protein synthesis. Based on the structures obtained here, an attractive possibility would be that the positive charge contained within the W1 pocket is used to break down the intermediate via the same proton wire that was used to create it. Indeed, the fast, concerted transfer of a proton from W1 to W3 to form a hydronium ion would increase the ability of this buried solvent molecule to catalyze the hydrolysis of the intermediate into products. A role for the N-terminal amino group of L27 in delaying the loss of a proton from W1 to the bulk solvent long enough to allow protonation of W3 could explain why shortening this highly defined tail by three or more residues causes growth defects and decreases the overall yield of protein synthesis13. The conserved length of the L27 N-terminal extension ensures that the entire proton wire and water molecule W2 form a shielded system that can only exchange a proton with the bulk solvent through the 2′-OH of the P-site A76 ribose. According to our model, deletion of the N-terminus of L27 could alter both the rates of intermediate formation and conversion of the intermediate into products, even though the effects on the overall reaction rate could be rather modest depending on the nature of the incoming amino acid in the A-site.
The lack of pH-sensitivity for peptide bond formation with either full-length tRNAs or with the short A-site substrate analogs CPmn or CCPmn23 may also be an indication that the PTC is closed and that protons involved in the reaction are not exchanged with the bulk solvent. This is in agreement with the three pre-attack structures presented here, which exhibit nearly identical PTC conformations (Supplementary Fig. 3e) and a shielded W1 cavity. On the other hand, the existence of an ionizable group with a pKa of ∼7.5 in the puromycin reaction9 is consistent with the N-terminus of L27 acting as a general base or acid once the PTC cavity is open to the solvent and the A76 phosphate oxygen of the A-site tRNA is no longer available to initiate the proton transfer. Our model does not exclude the possibility that the N-terminal amine of L27 also becomes protonated during the reaction with the full-length substrates. Such a protonation event would remain undetected when analyzing the pH profile of the reaction since the breakdown of the intermediate is likely to occur faster than the exchange of a proton with the bulk solvent. In light of our proposed mechanism, we therefore suggest that mutations which render the pocket containing W1 accessible to the bulk solvent, such as the deletion of residue A260224 or alterations to the N-terminus of L2724,25, will need to be re-evaluated in terms of their effect on the pH sensitivity of the reaction.
No homolog of L27 exists in archaea or in the cytoplasmic ribosomes of eukaryotes. In archaea L27 is replaced by protein L10e, which has an internal loop that is disordered in the Hma 50S subunit structure25, but may nevertheless extend towards the peptidyl transferase center in a 70S ribosome with bound full-length tRNA substrates. Indeed, a cryo-EM model of the wheat germ ribosome in complex with a P-site tRNA shows that the eukaryotic L10e homolog (RPL10) possesses a loop that reaches towards the PTC in a manner that is similar to the N-terminus of L2726. Although mutations within this highly conserved loop are lethal27, a high-resolution structure of the archaeal or eukaryotic ribosome in complex with aminoacylated tRNA substrates and kinetic data for the reaction by ribosomes with L27, L10e or RPL10 mutant proteins will be necessary to establish whether L10e or RPL10 could play a role comparable to that proposed here for L27.
In summary, we have obtained markedly improved snapshots of the PTC prior to and following catalysis. These structures have revealed a previously unseen network of hydrogen bonds that could play a catalytic role by ensuring that nucleophilic attack and deprotonation of the α-amine are concerted in the rate-limiting step. Though by no means definitive, the model put forward here is intended to steer research on the mechanism of peptide bond formation in directions that could not be foreseen when, nearly a decade ago, structures of the 50S ribosomal subunit in complex with small tRNA mimics provided us with high-resolution pictures of the PTC in different functional states. Thus, our data provide a robust, expanded structural framework on the basis of which alternate reaction mechanisms can be formulated and tested using kinetic, biochemical and computational approaches.
Online Methods
1. Purification of Thermus thermophilus 70S ribosomes
Tth 70S ribosomes were prepared as described previously28. The final 70S pellets were resuspended at a concentration of approximately 500 A260 units/mL in a buffer containing 5 mM HEPES-KOH (pH 7.6), 50 mM KCl, 10 mM NH4Cl, 10 mM Mg(CH3COO)2, 6 mM β-mercaptoethanol, flash frozen in liquid nitrogen and stored in small aliquots at -80°C until used in crystallization experiments.
2. Preparation of mRNA, tRNAs and tRNA analogs
Synthetic mRNA with the sequence 5′-GGC AAG GAG GUA AAA AUG UUC UAA-3′ was obtained from Integrated DNA Technologies (Coralville, IA). This mRNA includes a Shine-Dalgarno sequence, an AUG start codon and a phenylalanine UUC codon followed by a stop codon.
Unmodified E.coli tRNAs were overexpressed and purified as described previously for tRNAiMet29 and for tRNAPhe 30. tRNAiMet was charged with methionine and formylated as described29 to yield fMet-tRNAiMet with a conventional 3′-ester linkage between the tRNA and the amino acid residue.
Modified aminoacyl-tRNAs – fMet-NH-tRNAiMet and Phe-NH-tRNAPhe – featuring non-hydrolyzable 3′-amide linkages between the tRNA and the amino acid moieties were prepared as previously described1 (with modifications). The ATP analog 3′-amino-3′-deoxyadenosine-5′-O-triphosphate was obtained from Axxora (Farmingdale, NY). The 3′-terminal AMP of tRNAPhe was removed by incubation with 170 μg/mL phosphodiesterase I (Worthington Biochemical Corporation, Lakewood, NJ) at 37°C for 2 hours in a solution containing 50 mM Glycine-NaOH (pH 9.2), 100 mM NaCl and 15 mM MgCl2. Following phenol/chloroform extraction and ethanol precipitation of the trimmed tRNA, 3′-amino-3′-deoxy-AMP was incorporated at the 3′-end of tRNAPhe by incubation with 1 mM of the modified nucleotide and 125 μg/mL E.coli CCA-adding enzyme in a buffer containing 50 mM Glycine-NaOH (pH 9.2), 30 mM KCl, 12 mM Mg(CH3COO)2, 100 μg/mL bovine serum albumin and 3 mM CTP (with modifications from 31). To perform a similar replacement of the terminal A76 for the tRNAiMet, CCA-adding enzyme alone was used. In the first step, the incorporation/hydrolysis equilibrium of the reaction catalyzed by CCA-adding enzyme was shifted towards the hydrolysis by addition of 250 μM Na2P2O7 in order to remove the terminal nucleotides. In the following step, the hydrolysis reaction was reversed by addition of 2 U/mL E.coli inorganic pyrophosphatase (NEB) and the resulting incorporation of the modified terminal residue was allowed to proceed for 1 hour at 37°C. The resulting full-length 3′-NH2-tRNAPhe and 3′-NH2-tRNAiMet were enzymatically acylated with phenylalanine31 or methionine29, respectively, to yield Phe-NH-tRNAPhe and Met-NH-tRNAiMet. The latter was then formylated as described29 to yield fMet-NH-tRNAiMet.
All obtained aminoacylated tRNA species were further purified by HPLC on a 20-mL C4 reversed phase column (Vydac) in a buffer containing 20 mM NH4CH3COO (pH 5.5), 400 mM NaCl and 10 mM MgCl2, using a 0-40% linear gradient of methanol for elution. The fractions from the C4 column that contained the desired tRNAs were pooled, concentrated, flash-frozen in liquid nitrogen in small aliquots and stored at -80°C until further use in crystallization experiments.
The aminoacyl-tRNA mimics – cytidine-puromycin (CPmn) and cytidine-cytidine-puromycin (CCPmn) – were obtained from Thermo Scientific.
3. Complex formation
Ribosome-mRNA-tRNA complexes were formed by programming of 5 μM 70S Tth ribosomes with 10 μM mRNA and incubation at 55°C for 10 minutes, followed by addition of 20 μM P- and A-site tRNA substrates (with minor changes from 1). Each of the last two steps was allowed to reach equilibrium for 10 minutes at 37°C. In the co-crystallization experiments with aminoacyl-tRNA analogs, both CPmn and CCPmn were used at final concentrations of 100 μM. Immediately prior to use in crystallization experiments, all complexes were briefly centrifuged. The final ionic conditions consisted of 5 mM HEPES-KOH (pH 7.6), 50 mM KCl, 10 mM NH4Cl and 10 mM Mg(CH3COO)2.
4. Crystallization
Initial crystalline needles were obtained by screening around previously published ribosome crystallization conditions28,32,33. Crystals were grown by vapor diffusion in sitting drop crystallization trays at 19°C. 2-3 μL of the mRNA-tRNA-ribosome complex were mixed with 3-4 μL of a reservoir solution containing 100 mM Tris-HCl (pH 7.6), 2.9% (w/v) PEG-20K, 7-12% (v/v) MPD, 100-200 mM Arginine, 0.5 mM β-mercaptoethanol. Crystals appeared within 2-3 days and grew up to 150 × 150 × 1600 μm in size within 7-8 days. Crystals were cryo-protected stepwise using a series of buffers with increasing MPD concentrations until reaching a final concentration of 40% (v/v) MPD, in which they were incubated overnight at 19°C. In addition to MPD, all stabilization buffers contained 100 mM Tris-HCl (pH 7.6), 2.9% (w/v) PEG-20K, 50 mM KCl, 10 mM NH4Cl, 10 mM Mg(CH3COO)2 and 6 mM β-mercaptoethanol. During crystal cryo-protection with aminoacyl-tRNA analogs, CPmn and CCPmn were added to all stabilization solutions at 50 μM or 20 μM, respectively. After stabilization, crystals were harvested and immediately flash frozen in a nitrogen cryo-stream at 80K.
5. Data Collection and Processing
Diffraction data were collected at beamline X25 at the Brookhaven National Laboratory (Upton, NY) and at beamline 24ID-C at the Advanced Photon Source (Argonne, IL). A complete dataset for each ribosome complex was collected at 100K from multiple regions of the same crystal using 0.2° oscillations and 0.9789 Å wavelength. The raw data were integrated and scaled using the XDS software package34. All of the crystals belonged to the primitive orthorhombic space group P212121 with approximate unit cell dimensions of 210Å × 450Å × 620Å and contained two copies of the 70S ribosome per asymmetric unit (Table 1). Each structure was solved by molecular replacement using PHASER from the CCP4 program suite35. The search model was generated from the previously published structure of Tth 70S ribosome with bound mRNA and tRNAs (PDB codes: 2WDG, 2WDI1). The mRNA, all three tRNAs as well as 23S rRNA residues comprising the peptidyl transferase center were excluded from the starting search model. The initial molecular replacement solutions were refined by rigid body refinement with the ribosome split into multiple domains, followed by 7 cycles of positional and individual B-factor refinement using PHENIX36. Non-crystallographic symmetry restraints were applied to 4 domains of the 30S ribosomal subunit (head, body, spur, helix 44), and 3 domains of the 50S subunit (body, L1-stalk, N-terminus of the L9 protein).
Table 1. Data collection and refinement statistics.
| 70S-tRNA Pre-Attack |
70S-tRNA Post-Catalysis |
70S-CPmn Pre-Attack |
70S-CCPmn Pre-Attack |
|
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 209.45, 448.85, 619.02 | 209.32, 450.06, 622.23 | 209.25, 448.46, 618.08 | 207.56, 444.23, 613.03 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 224-2.60 (2.67-2.60)a | 256-2.55 (2.62-2.55)c | 363-2.90 (2.98-2.90)e | 222-2.80 (2.62-2.80)g |
| Rmerge | 23.2 (225.0) | 13.1 (120.9) | 18.5 (124.4) | 18.4 (128.6) |
| I / σ I | 8.58 (0.97)b | 8.91 (1.02)d | 7.13 (1.08)f | 6.04 (1.02)h |
| Completeness (%) | 99.9 (99.8) | 95.9 (85.6) | 96.9 (98.8) | 98.0 (97.8) |
| Redundancy | 9.39 (8.99) | 3.90 (3.09) | 3.62 (3.48) | 3.79 (3.66) |
| Refinement | ||||
| Resolution (Å) | 2.60 | 2.55 | 2.90 | 2.80 |
| No. reflections | 1,760,252 | 1,803,654 | 1,229,259 | 1,345,662 |
| Rwork/Rfree | 22.3 / 26.4 | 23.2 / 27.9 | 23.1 / 28.6 | 23.3 / 28.0 |
| No. atoms | ||||
| Protein | 91,253 | 91,181 | 91,171 | 91,175 |
| Ligand/ion | 202,093 | 202,139 | 196,240 | 196,499 |
| Water | 3,022 | 3,062 | 2,071 | 2,367 |
| B factors | ||||
| Protein | 63.6 | 63.9 | 64.4 | 63.4 |
| Ligand/ion | 59.5 | 60.5 | 59.9 | 57.5 |
| Water | 44.4 | 45.5 | 40.1 | 41.0 |
| r.m.s. deviations | ||||
| Bond lengths (Å) | 0.005 | 0.005 | 0.005 | 0.005 |
| Bond angles (°) | 0.982 | 0.989 | 0.965 | 0.991 |
Values in parentheses are for highest-resolution shell.
Two crystals were used to obtain the structure
I/σI = 2 at 2.80Å resolution
Single crystal was used to obtain the structure
I/σI = 2 at 2.76Å resolution
Single crystal was used to obtain the structure
I/σI = 2 at 3.05Å resolution
Single crystal was used to obtain the structure
I/σI = 2 at 2.97Å resolution
After initial refinement, electron density corresponding to the mRNA, all three tRNAs, water molecules (W1, W2, W3) and amino acid moieties attached to the 3′-terminal residues of the A- and P-site tRNAs became evident in the unbiased Fo-Fc difference maps. To further improve the quality of the difference maps and to aid model building, we used an approach in which bulk solvent correction was omitted for the ribosome's active site region. To protect specific features of the density from being included into the solvent mask (Supplementary Fig. 1a), a sphere of evenly-spaced dummy atoms with zero occupancy (Supplementary Fig. 1b) was used to cover the active site region during mask generation, but was ultimately excluded from the refinement process. All dummy atoms in the sphere grid were spaced at 0.7 Å intervals as suggested by PHENIX developers36 and a sphere radius of 15Å was chosen to completely cover the active site. Any dummy atom located <2 Å away from any atom of the physical model was excluded. The resulting unbiased Fo-Fc maps revealed more prominent features and had continuous electron densities for the amino acid moieties (Supplementary Fig. 1c). Owing to the absence of bulk solvent correction, the scale of these maps within the active site region was shifted in a seemingly arbitrary fashion. An equivalent contour level was obtained by visual comparison of the active site densities between the bulk solvent corrected and uncorrected maps.
6. Model Building
The mRNA, full-length E-site tRNA and residues 1-75 of A- and P-site tRNA ligands were built into the unbiased difference density maps from the initial round of refinement, and the refinement scheme described above was performed after the addition of each ligand. A high resolution structure of unmodified tRNAPhe from E.coli (PDB code: 3L0U37) was used as a guide to build the A- and E-site tRNAs. All of the hydrogen bonds defining the tRNA secondary and tertiary structures, as well as those involved in the mRNA-tRNA and 23S-tRNA base-pairs, were included as distance restraints during refinement. The 3′-terminal A76 residues of A- and P-site tRNAs with amino acids attached were omitted until the remainder of the active site of the ribosome had been built and refined. They were then placed together with water molecules into the unbiased difference density maps using previous Tth 70S1 or Hma 50S4,5 structures as guides. Initial models for the 3′-terminal A76 residues of the A- and P-site tRNAs with the amino acids or with dipeptide attached via amide bonds were created de novo. The corresponding restraints needed for positional and B-factor refinement were generated manually using existing restraints from the ccp4-library for AMP, phenylalanine and formyl-methionine as templates. Geometric restraints for the refinement of aminoacyl-tRNA mimics, CPmn and CCPmn, were taken from CCP4-library.
The final models for each ribosome complex were generated by multiple rounds of model building in COOT38, followed by refinement in PHENIX36. The statistics of data collection and refinement for each complex are compiled in Table 1.
The 0.7-0.9 Å increase in resolution achieved over existing structures of similar 70S complexes1 enabled us to build a more complete and accurate model of the 70S ribosome than in previous structures. The markedly improved overall quality of the electron density maps allowed us to rectify some main-chain register problems and to improve the number of residues falling into the favored and allowed regions of the Ramachandran plot for many ribosomal proteins in our starting model.
Additionally, we found that the ligand coordinated by residues Cys9, Cys12, Cys26, and Cys31 of ribosomal protein S4 is a 4Fe-4S iron-sulfur cluster instead of a Zn2+-ion, as suggested by all previous structures of the Tth 70S ribosome. Initial refinement with a Zn2+-ion placed in the center of the positive Fo-Fc density peak adjacent to the protein S4 cysteine residues, which were connected to the Zn2+-ion by coordination bonds, demonstrated a significant distortion in the protein's backbone geometry and lingering strong peaks of positive density. This suggested that the ligand coordinated by cysteines of protein S4 should be larger in size, possess more electrons, and have the same tetrahedral coordination geometry as compared with the Zn2+-ion. The only ligand, which meets all these criteria, is known to be coordinated by cysteines and has biological significance for the Tth species is the cubic 4Fe-4S iron-sulfur cluster. Thus, all structures presented here were built and refined with a 4Fe-4S cluster coordinated by ribosomal protein S4 of the 30S subunit.
7. Figures and Movie
All figures showing atomic models and movie were generated using PYMOL (www.pymol.org).
Supplementary Material
Acknowledgments
We thank the staff at the Advanced Photon Source (beamline 24ID) and at the National Synchrotron Light Source (beamline X25) for help during data collection, and the staff at the Richards Center at Yale University for computational support. We also thank P.B. Moore, S.A. Strobel and D.A. Hiller for critical reading of the manuscript, R.L. Grodzicki for preparation of the unmodified tRNAs, J. Lin, C. Mackereth and D. Dupuy for discussions and advice, and members of the T.A.S. and C.A.I. laboratories for discussions. This work was supported by US National Institutes of Health grant GM022778 (T.A.S.) and by starting funds from the Fondation pour la Recherche Médicale, the Conseil Régional d'Aquitaine, the Institut National de la Santé et de la Recherche Médicale and the Centre National de la Recherche Scientifique (C.A.I.).
Footnotes
Accession Codes: Coordinates and structure factors were deposited in the RCSB Protein Data Bank with accession codes 4QCM, 4QCN, 4QCO, and 4QCP for the Tth 70S ribosome pre-attack complex; 4QCQ, 4QCR, 4QCS, and 4QCT for the Tth 70S ribosome post-catalysis complex; 4QCU, 4QCV, 4QCW, and 4QCX for the Tth 70S ribosome pre-attack complex with CPmn; and, 4QCY, 4QCZ, 4QD0, and 4QD1 for the Tth 70S ribosome pre-attack complex with CCPmn.
Author Contributions: C.A.I. and Y.S.P. devised experiments and prepared modified tRNA substrates, Y.S.P. crystallized ribosomal complexes and performed crystallographic data collection and processing, C.A.I. and Y.S.P. analyzed the data, and C.A.I., Y.S.P. and T.A.S. wrote the manuscript.
References
- 1.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 3.Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell. 2005;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 5.Schmeing TM, et al. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat Struct Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- 6.Carrasco N, Hiller DA, Strobel SA. Minimal transition state charge stabilization of the oxyanion during peptide bond formation by the ribosome. Biochemistry. 2011;50:10491–10498. doi: 10.1021/bi201290s. [DOI] [PubMed] [Google Scholar]
- 7.Nagle JF, Morowitz HJ. Molecular mechanisms for proton transport in membranes. Proc Natl Acad Sci USA. 1978;75:298–302. doi: 10.1073/pnas.75.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trobro S, Aqvist J. Role of ribosomal protein L27 in peptidyl transfer. Biochemistry. 2008;47:4898–4906. doi: 10.1021/bi8001874. [DOI] [PubMed] [Google Scholar]
- 9.Katunin VI, Muth GW, Strobel SA, Wintermeyer W, Rodnina MV. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol Cell. 2002;10:339–346. doi: 10.1016/s1097-2765(02)00566-x. [DOI] [PubMed] [Google Scholar]
- 10.Kuhlenkoetter S, Wintermeyer W, Rodnina MV. Different substrate-dependent transition states in the active site of the ribosome. Nature. 2011;476:351–354. doi: 10.1038/nature10247. [DOI] [PubMed] [Google Scholar]
- 11.Wallin G, Aqvist J. The transition state for peptide bond formation reveals the ribosome as a water trap. Proc Natl Acad Sci USA. 2010;107:1888–1893. doi: 10.1073/pnas.0914192107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiller DA, Singh V, Zhong M, Strobel SA. A two-step chemical mechanism for ribosome-catalysed peptide bond formation. Nature. 2011;476:236–239. doi: 10.1038/nature10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA. A protein component at the heart of an RNA machine: the importance of protein l27 for the function of the bacterial ribosome. Mol Cell. 2005;20:427–435. doi: 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Hammes-Schiffer S, Stuchebrukhov AA. Theory of coupled electron and proton transfer reactions. Chem Rev. 2010;110:6939–6960. doi: 10.1021/cr1001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingery DA, et al. An uncharged amine in the transition state of the ribosomal peptidyl transfer reaction. Chem Biol. 2008;15:493–500. doi: 10.1016/j.chembiol.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaher HS, Shaw JJ, Strobel SA, Green R. The 2′-OH group of the peptidyl-tRNA stabilizes an active conformation of the ribosomal PTC. EMBO J. 2011;30:2445–2453. doi: 10.1038/emboj.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang YW, Sprinzl M. Peptide Bond Formation on the Ribosome: The Role of the 2 ′-OH Group on the Terminal Adenosine of Peptidyl-tRNA and of the Length of Nascent Peptide Chain. Angew Chem Int Ed. 2011;50:7287–7289. doi: 10.1002/anie.201005245. [DOI] [PubMed] [Google Scholar]
- 18.Koch M, Huang Y, Sprinzl M. Peptide-bond synthesis on the ribosome: no free vicinal hydroxy group required on the terminal ribose residue of peptidyl-tRNA. Angew Chem Int Ed Engl. 2008;47:7242–5. doi: 10.1002/anie.200801511. [DOI] [PubMed] [Google Scholar]
- 19.Erlacher MD, et al. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2′-OH at A2451 of 23S rRNA. J Am Chem Soc. 2006;128:4453–4459. doi: 10.1021/ja0588454. [DOI] [PubMed] [Google Scholar]
- 20.Lang K, Erlacher M, Wilson DN, Micura R, Polacek N. The role of 23S ribosomal RNA residue A2451 in peptide bond synthesis revealed by atomic mutagenesis. Chem Biol. 2008;15:485–492. doi: 10.1016/j.chembiol.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 22.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 23.Bieling P, Beringer M, Adio S, Rodnina MV. Peptide bond formation does not involve acid-base catalysis by ribosomal residues. Nat Struct Mol Biol. 2006;13:423–428. doi: 10.1038/nsmb1091. [DOI] [PubMed] [Google Scholar]
- 24.Polacek N, et al. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol Cell. 2003;11:103–112. doi: 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- 25.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 26.Armache JP, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci USA. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofer A, Bussiere C, Johnson AW. Mutational analysis of the ribosomal protein Rpl10 from yeast. J Biol Chem. 2007;282:32630–32639. doi: 10.1074/jbc.M705057200. [DOI] [PubMed] [Google Scholar]
- 28.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt E, Blanquet S, Mechulam Y. Crystallization and preliminary X-ray analysis of Escherichia coli methionyl-tRNAMet(f) formyltransferase complexed with formyl-methionyl-tRNAMet(f) Acta Crystallogr D Biol Crystallogr. 1999;55:332–334. doi: 10.1107/S0907444998011780. [DOI] [PubMed] [Google Scholar]
- 30.Junemann R, et al. In vivo deuteration of transfer RNAs: overexpression and large-scale purification of deuterated specific tRNAs. Nucleic Acids Res. 1996;24:907–913. doi: 10.1093/nar/24.5.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraser TH, Rich A. Synthesis and aminoacylation of 3′-amino-3′-deoxy transfer RNA and its activity in ribosomal protein synthesis. Proc Natl Acad Sci USA. 1973;70:2671–2675. doi: 10.1073/pnas.70.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 33.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byrne RT, Konevega AL, Rodnina MV, Antson AA. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 2010;38:4154–4162. doi: 10.1093/nar/gkq133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.