SUMMARY
Signaling in the ancestral branch of the unfolded protein response (UPR) is initiated by unconventional splicing of HAC1/XBP1 mRNA during endoplasmic reticulum (ER) stress. In mammals, IRE1α has been known to cleave the XBP1 intron. However, the enzyme responsible for ligation of two XBP1 exons remains unknown. Using an XBP1 splicing-based synthetic circuit, we identify RtcB as the primary UPR RNA ligase. In RtcB knockout cells, XBP1 mRNA splicing is defective during ER stress. Genetic rescue and in vitro splicing show that the RNA ligase activity of RtcB is directly required for the splicing of XBP1 mRNA. Taken together, these data demonstrate that RtcB is the long sought RNA ligase that catalyzes unconventional RNA splicing during the mammalian UPR.
INTRODUCTION
Elaborate regulatory networks have evolved for maintaining homeostasis of the protein-folding environment in the endoplasmic reticulum (ER). These networks, collectively known as the Unfolded Protein Response (UPR), consist of three signaling branches, each eliciting distinct downstream effects through an ER-resident sensor - ATF6, IRE1, or PERK (Walter and Ron, 2011). When ER functions are perturbed, these UPR pathways work in synergy to mitigate ER stress by increasing ER chaperones, inhibiting protein synthesis, and enhancing ER-associated protein degradation (Walter and Ron, 2011). However, if cells cannot reestablish ER homeostasis through these adaptive responses, prolonged activation of the UPR components such as IRE1α and CHOP induce apoptosis (Tabas and Ron, 2011). Therefore, under physiological and pathological conditions that perturb ER homeostasis, different signaling outputs of the individual UPR branches ultimately influence cell fate by promoting either adaption or apoptosis (Lin et al., 2007).
The most conserved UPR branch is defined by IRE1, an ER transmembrane kinase/endoribonuclease (Cox et al., 1993). Upon sensing unfolded proteins through its luminal domain, IRE1 undergoes oligomerization, trans-autophosphorylation and nuclease activation[for review, see (Hetz and Glimcher, 2009; Walter and Ron, 2011)]. Activated IRE1, a sequence-specific endoribonuclease, initiates a two-step unconventional splicing reaction. In yeast cells, Ire1p cleaves the unspliced HAC1 mRNA at two RNA stem-loops and excises an intervening intron in the first step (Sidrauski and Walter, 1997). The tRNA ligase Trl1p joins the two exons and 2′ phosphotransferase Tpt1p removes the junction 2′ phosphate in the second step (Schwer et al., 2004; Sidrauski et al., 1996). Similar to yeast Ire1p, mammalian IRE1α removes a 26-nt intron from the unspliced XBP1u mRNA to generate a mature mRNA for the production of XBP1s (Calfon et al., 2002; Tirasophon et al., 1998; Yoshida et al., 2001). The second step of XBP1 splicing is accomplished by an as-yet-unknown RNA ligase in higher organisms. In both species, Hac1p and XBP1s, the primary outputs of IRE1 signaling, function as potent transcription activators that induce the UPR transcription program to restore ER homeostasis in stressed cells (Acosta-Alvear et al., 2007; Lee et al., 2003; Travers et al., 2000; Yoshida et al., 2001).
At the biochemical level, yeast Ire1p and mammalian IRE1α act similarly in the splicing reaction, resulting in a 2′, 3′-cyclic phosphate bond at the 3′-end of the 5′-exon and a hydroxyl group at the 5′-end of the 3′ exon (Gonzalez et al., 1999; Shinya et al., 2011). However, the subsequent step - ligation of the two exons differs mechanistically between yeast and mammals. In yeast, Trl1pacts through a 5′-PO4/3′-OH ligation pathway involving multiple steps to join two exon halves, leaving a 2′ phosphate at the splice junction (Gonzalez et al., 1999). The splicing reaction is then completed by Tpt1p to remove the 2′ phosphate (Schwer et al., 2004; Steiger et al., 2005). By contrast, in vitro reconstitution experiments have suggested that the splicing of XBP1u mRNA is likely via a single-step 3′-PO4/5′-OHligation pathway in mammalian cells (Shinya et al., 2011). Consistent with this view, mice deficient for Trpt1, the homolog of yeast Tpt1p, have no defect in the UPR-induced XBP1splicing (Harding et al., 2008). RtcB, a 3′-PO4/5′-OH RNA ligase, has recently been identified in bacteria, archaea, and mammals (Englert et al., 2011; Popow et al., 2011; Tanaka and Shuman, 2011). Interestingly, overexpression of bacteria RtcB rescued the Hac1 splicing defect in Trl mutant yeast (Tanaka et al., 2011)whereas mammalian RtcB failed to do so (Popow et al., 2012). A global survey of mRNA-binding proteins suggested that RtcB was associated with XBP1 mRNA(Baltz et al., 2012); however, RtcB knockdown by RNAi was unable to show a role of RtcB in the UPR (Iwawaki and Tokuda, 2011). Despite significant efforts to identify such enzymes over the past decade, the mammalian RNA ligases involved in unconventional XBP1 splicing remain elusive.
In this study, we aimed to identify RNA ligases that control the UPR in mammalian cells. As a first step, we designed a synthetic genetic circuit using an XBP1 splicing-based UPR sensor. The cells integrated with this circuit exhibited a synthetic apoptotic phenotype in response to an acute UPR stimulus. We performed a genome-wide RNAi screen to isolate suppressors of this synthetic phenotype. Using this synthetic biology approach we identified mouse RtcB(RNA 2′, 3′-cyclic phosphate and 5′-OH ligase) as the primary RNA ligase for the mammalian UPR pathway.
RESULTS
A genetic circuit creates a synthetic apoptotic phenotype in response to the UPR
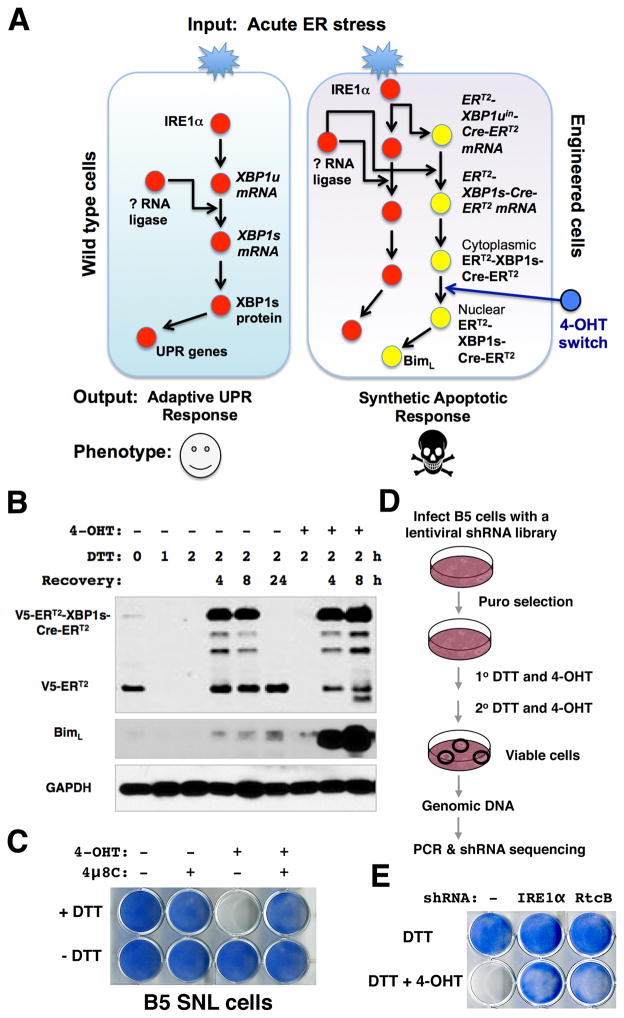
The unconventional splicing of XBP1 mRNA is the primary signaling output for the IRE1 branch of the UPR in mammalian cells (Walter and Ron, 2011). The XBP1 splicing-based fluorescent protein indicators have been previously developed to monitor the UPR activation in vivo (Back et al., 2005; Iwawaki et al., 2004; Sone et al., 2013). To couple the UPR-sensing mechanism of XBP1u mRNA with a synthetic signaling output, we designed an XBP1umRNA sensor that consists of a 4-hydroxytamoxifen (4-OHT)-inducible chimeric Cre (ERT2-Cre-ERT2)(Feil et al., 1997; Metzger et al., 1995)(Figure S1A–B). The sensor mRNA encodes an N-terminal V5-tagged estrogen receptor ligand-binding domain (ERT2), followed by a 120-nt XBP1 exon-intron-exon junction and a Cre-ERT2 fusion protein. Upon removal of the 26-nt XBP1 intron in response to ER stress, a full-length ERT2-XBP1s-Cre-ERT2 protein is translated from the spliced sensor mRNA (Figure S1C). By design, the activity of ERT2-XBP1s-Cre-ERT2 is tightly controlled by the UPR-induced splicing and 4-OHT-induced nuclear translocation (Figure S1D). We generated HEK293 cell lines that stably expressed both ERT2-XBP1uin-Cre-ERT2 and the GFP reporter (PB-CAG-loxP-RFP-loxP-GFP) (Figure S1E). As expected, the cells elicited robust GFP expression when treated with a combination of 4-OHT and UPR inducers. Exposure to acute ER stress signals did not affect the viability of these cells. Furthermore, transient expression and activation of ERT2-XBP1s-Cre-ERT2 permanently marked nearly all the cells that had experienced ER stress. Therefore, by coupling Cre activity with XBP1 splicing, we have constructed a UPR sensor that can alter the signaling output of the IRE1α pathway.
Next, we introduced both ERT2-XBP1uin-Cre-ERT2and Cre-inducible Bim transgenes into mouse SNL cells (Figure S2A). In doing so, we coupled XBP1 slicing with the downstream effector Bim that can elicit a robust apoptotic response during the normally adaptive phase of the UPR (Figure 1A). Upon treatment with DTT and 4-OHT, robust Bim expression was rapidly induced in the engineered B5-SNL cells (Figure 1B), triggering massive apoptosis (Figure 1C, Figure S2B). 4μ8C, an IRE1α endonuclease inhibitor (Cross et al., 2012), efficiently blocked this synthetic phenotype (Figure 1C, Figure S2B). Thus, the induced apoptosis depends upon the activation of upstream IRE1α. Taken together, the data demonstrate that by coupling a synthetic ERT2-XBP1uin-Cre-ERT2 genetic circuit with the endogenous UPR pathway, we are able to switch the adaptive UPR response to a synthetic apoptotic response in the genetically engineered cells.
Figure 1. A synthetic circuit identifies RtcB as a candidate UPR RNA ligase.
A. A synthetic genetic circuit creates an apoptotic phenotype in response to acute ER stress. By coupling of an XBP1-splicing dependent Cre sensor with endogenous IRE1α, the 4-OHT-regulated Cre activates Bim expression and triggers a robust apoptotic response in the genetically engineered SNL cells. B. The UPR-induced splicing of V5-ERT2-XBP1uin-Cre- ERT2sensor and the induction of Bim in B5 SNL cells. DTT in combination with 4-OHTinduced an accumulation of full-length V5-ERT2-XBP1s-Cre- ERT2proteins and a robust induction of Bim. Note that the truncated V5-ERT2 proteins were depleted in cells acutely treated with DTT without recovery. The B5 cells underwent massive cell death 24 hours after being incubated with DTT and 4-OHT and the sample was not included in this analysis. C. Images of methylene blue staining showed that a combination of DTT and 4-OHT treatments was sufficient to cause robust apoptosis. Inhibition of the endonuclease activity of IRE1α by 4μ8C completely blocked the synthetic apoptosis. D. Experimental design for a genome-wide RNAi screen to identify genetic suppressors of the synthetic apoptosis in B5 SNL cells. E. RtcB was identified as a candidate UPR RNA ligase. The methylene blue stains showed that unlike parental B5 cells, the B5 cells that stably expressed an shRNA against RtcB were viable after DTT and 4-OHT treatment. IRE1α shRNA served as a positive control.
An RNAi screen identifies RtcB as an essential component of the synthetic apoptosis circuit
The SNL cells engineered with the synthetic XBP1 splicing circuit exhibit a robust apoptotic response to acute ER stress. We estimated that less than 1/200,000 of the engineered cells might have escaped the induction of apoptosis by this synthetic circuit, making it feasible to isolate viable cells that fail to carry out XBP1mRNA splicing in a high-throughput format. We therefore carried out a genetic screen using a whole-genome shRNA library as outlined in Figure 1D. First, the engineered SNL cells were infected with a pool of lentiviral shRNAs (Su et al., 2011). With puromycin selection, we obtained approximately 1 million independent viral integration events, equivalent to 5-fold coverage of the complexity of the shRNA library. Next, DTT and 4-OHT were used in combination to induce the synthetic apoptosis in the shRNA-expressing B5 cells. To increase the stringency and specificity of the screen, the viability selection process was repeated once after viable cells recovered from the first round of selection. We isolated 124 viable clones from the whole-genome screen and we identified 86 different candidate targets by sequencing (Table S1). As expected, we isolated several target genes implicated in apoptosis (e.g., Casp3) and nuclear import (e.g., Ranbp3, Kpna2). The upstream IRE1α was not isolated from the screen, likely due to inefficient IRE1α knockdown by particular shRNAs in the library. An independently designed IRE1α shRNA suppressed the apoptosis in B5 cells. Among these targets, RtcB gene, also previously known as HSPC117, C22orf28 and FAAP in human and D10Wsu52e as in mice (Ding et al., 2010), has recently been identified as a component of human tRNA ligase complex (Popow et al., 2011). Therefore, we decided to verify whether RtcB knockdown was able to suppress the apoptosis in B5 cells using a different shRNA against RtcB. Like the positive control IRE1α shRNA, RtcB shRNA efficiently blocked the synthetic apoptosis induced by DTT and 4-OHT (Figure 1E). Thus, RtcB is required for the synthetic apoptotic UPR pathway.
Generation of RtcB inducible knockout ES cells
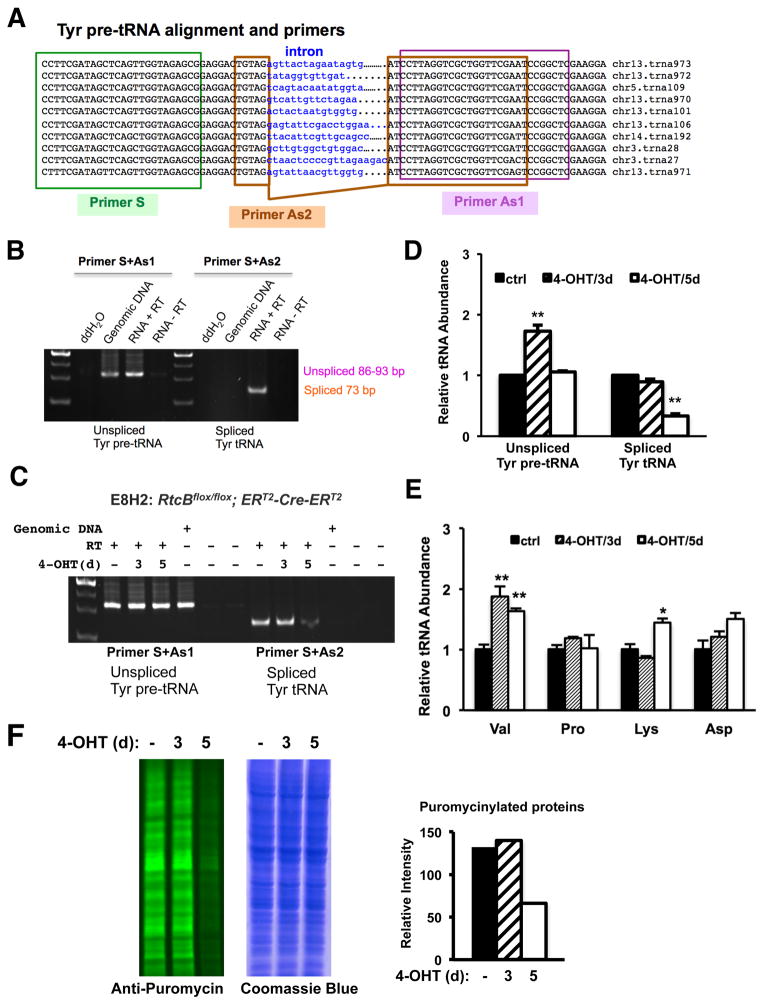
To investigate the in vivo function of RtcB in mammalian cells, we first generated a mouse ES cell line E8 that carries two homozygous RtcB conditional alleles (Figure S3A). We then introduced a PB-CAG-ERT2-Cre-ERT2 transgene into these cells to generate a conditional RtcB knockout (RtcBcKO) cell line E8H2 (Figure S3B). Upon Cre activation by 4-OHT, RtcB protein is depleted in E8H2 cells. We measured the growth rate of RtcB knockout cells and found that, in comparison with the untreated control or E8 ES cells without a Cre transgene, E8H2 ES cells deficient for RtcB began to grow significantly slower at day 4 after 4-OHT treatment (Figure S3C–D). RtcB has been shown to be a tRNA ligase in vitro and RtcB knockdown impairs the splicing of a tRNA reporter gene in human cells (Popow et al., 2011). Thus, we examined whether RtcBcKO cells are defective in the splicing of endogenous tRNAs. The mouse genome encodes 10 tyrosine tRNAs that all contain an intron (Figure 2A). Using a RT-PCR strategy that distinguish spliced Tyr tRNA and non-spliced Tyr pre-tRNA (Figure 2B), we showed that the level of spliced Tyr tRNAs was reduced by 70% at day 5 but not at day 3 after 4-OHT treatment (Figure 2C–D). Meanwhile, the levels of intron less tRNAs did not reduce in RtcB-depleted ES cells (Figure 2E). Interestingly, even though RtcB was completely depleted at day 3 after 4-OHT induction (Figure S3B), the level of spliced Tyr tRNAs remained unchanged, probably due to a compensatory up-regulation of unspliced Tyr pre-tRNA precursors seen at day 3(Figure 2D) and the stability and abundance of Tyr tRNA. Consistent with this observation, the global protein synthesis rates, as measured by transient labeling of puromycinylated proteins (Schmidt et al., 2009), did not change at day 3 but was significantly decreased at day 5 in E8H2 cells deficient for RtcB (Figure 2F). The reduction of spliced Tyr tRNA and the decrease of global protein synthesis coincided with the onset of proliferation defect of RtcB-deficient ES cells. Thus, the deficiency of tRNA splicing might contribute to the growth defect in RtcB knockout cells.
Figure 2. RtcBcKO cells exhibit a defect in endogenous Tyr pre-tRNA splicing.
A. Sequence alignment of all 10 tyrosine pre-tRNA genes from the mouse genome. Primers used in RT-PCR analysis are highlighted. B. Verification of indicated primers for detecting unspliced and spliced Tyr tRNA. Primer S and Primer AS1 predominantly detect unspliced Tyr pre-tRNA in an RT-PCR assay as similar products were amplified from both genomic DNA and cDNA samples. Primer S and Primer As2 specifically detect a 73-bp spliced product only from the cDNA. C. RT-PCR analysis of Tyr tRNA splicing in RtcBcKO ES cells. At day 5 after 4-OHT treatment, the level of spliced Tyr tRNA was decreased in RtcB-depleted cells. D. RT-qPCR analysis of the relative abundance of unspliced Tyr pre-tRNA and spliced Tyr tRNA in RtcBcKO cells. GAPDH was used as an internal control. In comparison with the control, only 30% of spliced Tyr tRNA remained 5 days after 4-OHT induction. E. RT-qPCR analysis of the relative abundance of 4 intronless tRNAs in RtcBcKO cells. F. Quantitative western blot analysis of puromycin-labeled total cellular proteins in RtcBcKO cells. A Coomassieblue stained duplicate gel served as the loading control. Quantification of the intensity of fluorescence signal for each lane was plotted. * p< 0.05, ** p< 0.01, *** p< 0.0001, compared to the control groups.
Unconventional XBP1 mRNA splicing is severely defective in RtcB knockout cells
The synthetic apoptosis of B5 cells depends upon the splicing of XBP1 sensor. The finding that RtcB knockdown impairs this phenotype suggests a role of RtcB as a UPR RNA ligase that catalyzes XBP1u splicing. However, a previous report had directly argued against this possibility, both in vivo and in vitro (Iwawaki and Tokuda, 2011).
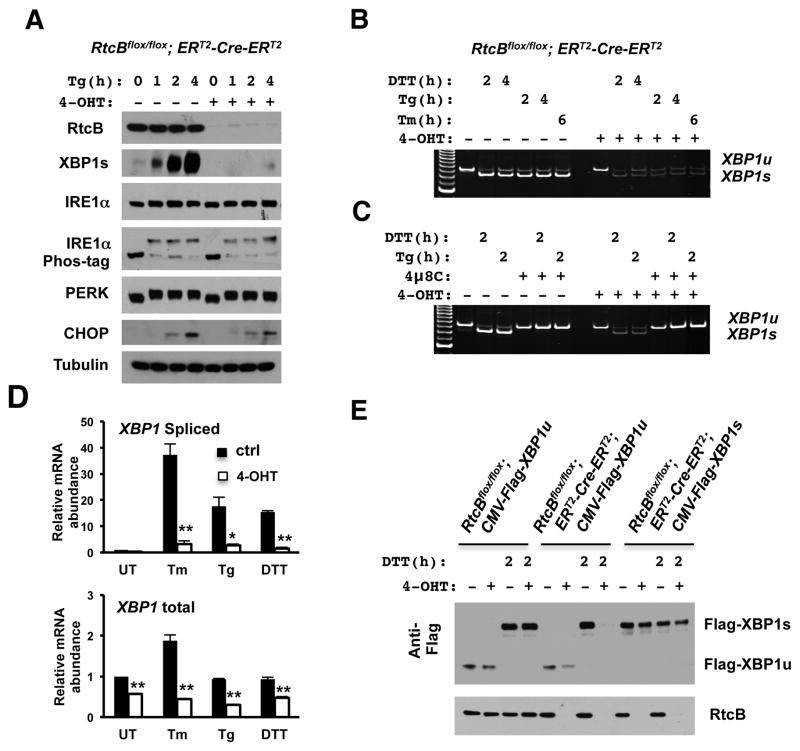
To examine whether RtcB functions in the XBP1u mRNA splicing pathway, we compared levels of UPR-induced XBP1s proteins in RtcBcKO ES cells at day 3 after 4-OHT treatment, prior to manifestation of at RNA splicing defect. Upon RtcB deletion, a dramatic reduction of XBP1s proteins was observed with diverse agents that induce the UPR (Figure 3A, Figure S4A–C), including thapsigargin (Tg), tunicamycin (Tm) and DTT. RT-qPCR showed that previously identified XBP1s target genes, such as Erdj4 and EDEM, were down-regulated as expected (Lee et al., 2003)(Figure S5). Meanwhile, we showed that the upstream IRE1α activation, measured by a reduction of mobility on Phos-tag™ SDS-PAGE, was not affected by RtcB knockout (Figure 3A, Figure S4A–C). In addition, other known ER stress induced signaling events, including PERK autophosphorylation, ATF6 proteolysis, and CHOP induction, remained unchanged in RtcB deficient cells (Figure 3A, Figure S4A–D). Thus, similar levels of ER stress were provoked in RtcB null and control cells and reduction of XBP1s was likely a direct consequence of RtcB deletion.
Figure 3. Unconventional XBP1 mRNA splicing is defective in RtcB deficient cells.
A. Western blot analysis showed that the accumulation of XBP1s proteins was blocked almost completely in response to Tg in RtcB deficient cells. Note that under the same condition, IRE1α phosphorylation, PERK phosphorylation and CHOP induction remained intact in RtcB deficient cells. B-C. The splicing of XBP1umRNA is defective in RtcBcKO cells. RT-PCR showed that both spliced and unsplicedXBP1 mRNAs were significantly reduced when RtcB were depleted (B). The accumulation of spliced XBP1s mRNA was blocked by IRE1α specific inhibitor 4μ8C in both control and RtcB-deficient cells (C). D. RT-qPCR analysis showed that the relative abundance of XBP1s mRNA in 4-OHT treated cells was drastically reduced in response to ER stress compared to that in untreated control cells. To induce ER stress, the cells were treated with Tm for 4 h, Tg for 2 h and DTT for 2h, respectively. In RtcB deficient cells, the levels of total XBP1 mRNA was also decreased but to a lesser extent. GAPDH served as an internal control in the experiments. * p< 0.05, ** p< 0.01, *** p< 0.0001, compared to the control groups. E. The splicing of Flag-tagged full-length XBP1u reporter in RtcB deficient cells. A constitutively expressed XBP1ucDNA expression vector was stably transfected into RtcBcKO ES cells, providing a reporter cell line lacking the endogenous XBP1 mediated feedback regulation. In E8H2 cells expressing Flag-XBP1ucDNA, XBP1s protein was not induced by DTT after 4-OHT treatment. Note that RtcB knockout had no direct effect on the expression of XBP1s encoded by a spliced Flag-XBP1scDNA transgene.
Next, we directly examined the splicing of XBP1u mRNA using an RT-PCR assay. In RtcB deficient cells, levels of both XBP1u and XBP1s mRNAs were significantly reduced compared to RtcB positive cells (Figure 3B). As expected, inhibition of IRE1α endonuclease activity by 4μ8C completely blocked the XBP1usplicing (Cross et al., 2012)(Figure 3C). Quantitative analysis of the relative abundance of spliced and total XBP1 mRNAs confirmed that the accumulation of spliced XBP1 was dramatically reduced when RtcB was deleted (Figure 3D, upper). Meanwhile, we observed a reduction of total XBP1 transcripts in RtcB null cells under both basal and stressed conditions (Figure 3D, lower). The reduction of total XBP1 mRNA can be in part explained by IRE1α-mediated cleavage and subsequent degradation of cleaved XBP1u mRNA. In addition, XBP1s is a potent transcription factor that activates its own expression through a positive feedback loop (Lee et al., 2003; Yoshida et al., 2006). Thus, reduction of XBP1s protein may also contribute to decreased levels of total XBP1 transcription in RtcB null cells. To evaluate the possibility that the effect of RtcB on XBP1u splicing was overstated by this positive feedback regulation, we generated ES cell lines that expressed XBP1u mRNA under the control of a constitutive CMV promoter. Like endogenous XBP1s protein, RtcB knockout cells failed to express Flag-tagged XBP1s encoded by CMV-driven XBP1u transgene (Figure 3E). As an additional control, a CMV-driven XBP1s transgene showed that Flag-XBP1s was equally expressed in the presence or absence of RtcB (Figure 3E), excluding the possibility that RtcB somehow influenced the stability of XBP1s protein or XBP1s mRNA. Taken together, the genetic evidence unequivocally demonstrates that RtcB is required for unconventional XBP1 splicing during the UPR.
Genetic rescue shows that RtcB acts as the primary RNA ligase in XBP1 mRNA splicing
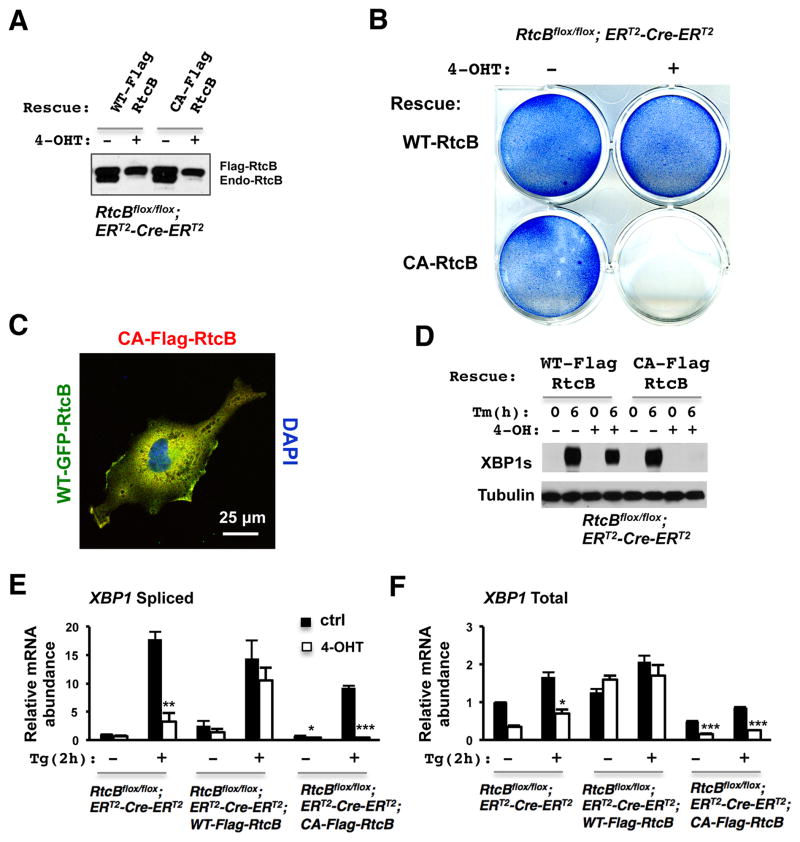
RtcB belongs to a subfamily of RNA ligases that are conserved in bacteria, archea and mammals (Englert et al., 2011; Popow et al., 2012; Tanaka and Shuman, 2011). It has recently been shown that RtcB/HSPC117 proteins, chromatographically purified from in Pyrobaculumaerophilum and cultured human cells, can function as 3′-5′ RNA ligases during tRNA maturation (Englert et al., 2011; Popow et al., 2011). Based on the structural studies on bacterial RtcB protein (Okada et al., 2006; Tanaka et al., 2011), a conserved cysteine residue at position 122 of mammalian RtcB has been proposed to participate in metal coordination at the active center of the enzyme. The point mutation C122A in human HSPC1117/RtcB abolished its tRNA ligase activity in vitro (Popow et al., 2011); therefore, we carried out genetic rescue experiments to test whether the RNA ligase activity of mouse RtcB is essential for its function in the UPR (Figure 4A). While wild type RtcB rescued the 4-OHT-induced growth defect, C122A mutant form of RtcB failed to do so (Figure 4B). Western blot analysis and immunofluorescence labeling showed that both wild type and mutant RtcB proteins were expressed at a similar level and had overlapping subcellular localization (Figure 4A/C). However, RtcBC122A-expressing mutant cells failed to express any detectable XBP1s (Figure 4D). Consistent with this observation, RT-qPCR showed that the induction of XBP1s mRNA by thapsigargin was completely abolished in RtcBC122A -expressing cells. Thus, RtcBC122A -expressing mutant cells displayed an even more dramatic XBP1 splicing defect than RtcB null cells (Figure 4E–F). Taken together, these data demonstrate that RtcB functions as an RNA ligase for XBP1 splicing during the UPR and ligase-dead RtcB may have a dominant/negative effect.
Figure 4. Genetic rescue shows that RtcB acts as an RNA ligase for XBP1 splicing.
A. The RtcBcKO cells were stably transfected with Flag-RtcBWT or Flag-RtcBC122A. Flag-RtcB proteins were expressed at a level similar to endogenous RtcB. B. Methylene blue stains showed that like RtcB null cells (Fig. S3D), RtcBC122A-expressing ES cells exhibited a severe growth defect after deleting endogenous RtcB. C. A confocal immunofluorescence image showing that GFP-RtcBWT and Flag-RtcBC122A have overlapping subcellular localization in transfected COS7 cells. D. In 4-OHT treated Flag-RtcBC122A-expressing E8H2 cells, ligase-dead RtcB failed to rescue the XBP1u splicing defect. No XBP1s protein was detected after Tm treatment. E–F. Similar to the experiment described in Fig. 3D, RT-qPCR showed that wild type RtcB rescued the splicing defect of XBP1u whereas C122A mutant did not. The basal level of XBP1s was significantly reduced and the induction of XBP1s by Tg was abolished in RtcBC122A -expressing cells. Similarly, under both basal and induced conditions, the levels of total XBP1 mRNA were decreased significantly in RtcBC122A-rescued cells.* p< 0.05, ** p< 0.01, *** p< 0.0001, compared to the control groups.
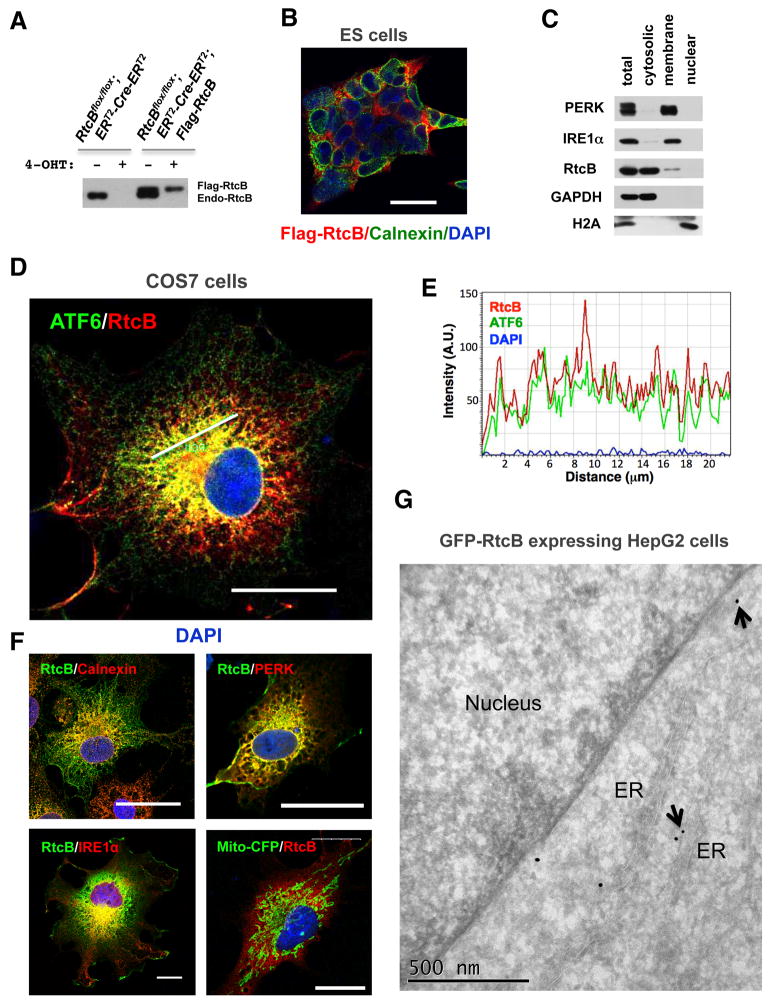
A subset of RtcB is associated with the endoplasmic reticulum
Because IRE1α is located on the ER, XBP1u splicing likely occurs on or near the ER. Consistent with this notion, it has been shown that XBP1u mRNA is bound to the ER membrane in an IRE1α independent manner (Yanagitani et al., 2009; Yanagitani et al., 2011). To analyze the subcellular localization of RtcB, we first carried out immunofluorescence labeling using a Flag-RtcB-rescued RtcBcKO cell line, in which Flag-RtcB was expressed at a comparable level to endogenous RtcB (Figure 5A). Flag-RtcB was perinuclear in the rescued ES cells (Figure 5B). To test whether RtcB is associated with the ER, we fractionated HEK293T cells using a combination of digit on inpermeabilization and NP-40 extraction, showing that a portion of endogenous RtcB co-fractionated with the ER proteins IRE1α and PERK (Figure 5C).
Figure 5. Subcellular localization of RtcB.
A. Expression of Flag-RtcB proteins in genetically rescued RtcBcKO ES cells. B. A confocal imunofluorescence image showing that RtcB was enriched in the perinuclear region in ES cells. C. Cell fractionation showed that a portion of endogenous RtcB was associated with the membrane fraction. IRE1α and PERK are ER transmembrane proteins. GAPDH and Histone H2A are cytosolic and nuclear, respectively. D–F. Subcellular localization of RtcB with the ER resident proteins. COS7 cells were co-transfected with Flag-RtcB and indicated ER markers including GFP-ATF6 (in D), calnexin, IRE1αKA, PERK-ΔC-9E10 and a mitochondria marker mito-CFP (in F). Confocal images showed that RtcB overlapped partially with these ER markers. Note that kinase-dead IRE1α and C-terminal truncated PERK were used in the experiment to achieve higher levels of expression. Bars, 25 μm in B, D and F. In E, an intensity plot along the line drawn in the confocal image showed that RtcB was colocalized with ATF6 in this region. G. An immuno-gold labeled EM image showing that GFP-RtcB proteins were labeled on the ER in HepG2 cells. Gold particles on the ER membrane are marked by arrows.
Next, we chose COS7 cells to better visualize the colocalization of RtcB with the ER. Cells expressing tagged-RtcB and several ER proteins were analyzed by fluorescence microscopy. Confocal images show that a subset of RtcB is colocalized with the ER markers tested in the experiments, including ATF6, calnexin, IRE1α/β, and PERK(Figure 5D–F and data not shown). Using RtcBcKO cells, we showed that GFP-RtcB fully rescued the loss of endogenous RtcB (Figure S6A–B). Thus, we generated a stable HepG2 cell line expressing GFP-RtcB because the GFP epitope is suitable for immuno-gold electron microscopy. The EM images confirm that RtcB is partially localized on the ER membrane and nuclear envelope where IRE1α is located (Figure 5G, Figure S6C).
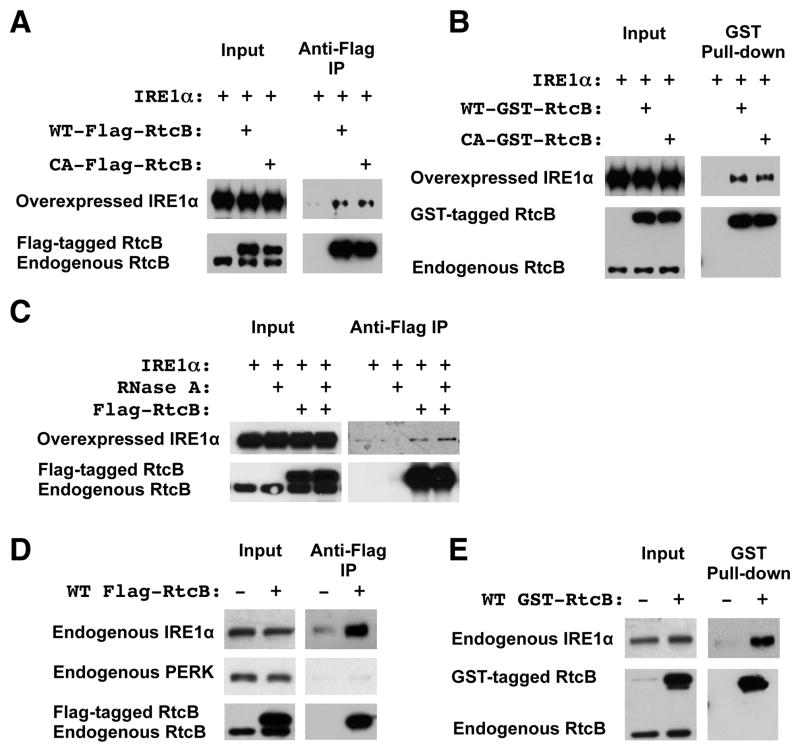
RtcB is associated with IRE1α
Because both RtcB and IRE1α are located to the ER, we investigated whether both proteins are physically associated with each other. HEK293T cells that stably expressed either wild type or C122A Flag-RtcB were transfected with IRE1α for co-immunoprecipitation (co-IP) analysis. Anti-Flag-IP showed that both wild type and ligase-dead RtcB proteins were associated with IRE1α (Figure 6A). We repeated a similar experiment using cells that expressed GST-tagged RtcB. GST-pull down experiments verified that IRE1α was present in RtcB-containing complexes (Figure 6B). In addition, we showed that the association of RtcB with IRE1α was insensitive to RNase A treatment (Figure 6C). Because overexpression of IRE1α induces the UPR and levels of IRE1α in these experiments were significantly higher than that of endogenous protein, we used two stable HEK293T cell lines that expressed Flag-RtcB or GST-RtcB to isolate RtcB protein complexes and assay endogenous IRE1α. In these cells, levels of Flag-RtcB and GST-RtcB were slightly higher or comparable to that of endogenous RtcB. In the absence of the UPR induction, co-IP showed that RtcB was in complex with endogenous IRE1α but not with a related ER-resident protein PERK (Figure 6D–E).
Figure 6. RtcB is in complex with IRE1α.
A. In HEK293T cells stably transfected with either Flag-RtcBWT or Flag-RtcBC122A, the level of Flag-RtcB was comparable to that of endogenous RtcB (lower panel, input). Anti-Flag co-IP showed that both wild type and ligase-dead RtcB proteins were associated with overexpressed IRE1α. Parental HEK293T cells that overexpressed IRE1α were used as a negative control. B. Using glutathione-Sepharose pull-down, a similar experiment was performed with GST-RtcB stably transfected HEK293T cells, showing that GST-RtcB interacted with overexpressed IRE1α. C. The association of RtcB with IRE1α was resistant to RNase A treatment. D–E. Endogenous IRE1α was associated with Flag-tagged or GST-tagged RtcB in two stable HEK293T cell lines that expressed Flag-RtcB and GST-RtcB, respectively.
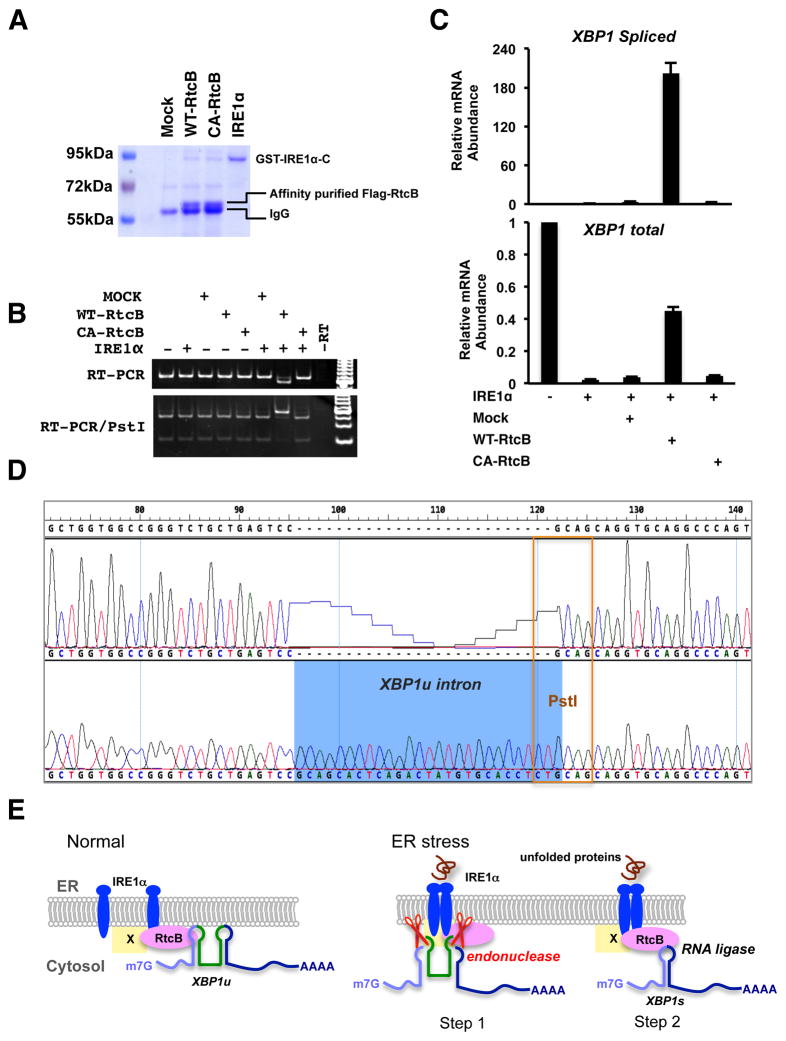
Reconstitution of XBP1u splicing in vitro
To further demonstrate the role of RtcB in XBP1u splicing, we performed a reconstitution experiment using purified RtcB and IRE1α. Because purified RtcB and IRE1α might be associated with endogenous XBP1u or XBP1s mRNA, we first designed an artificial XBP1u splicing reporter V5-XBP1uin-Rn luciferase (Figure S7A). Stable cell lines expressing this reporter were constructed using RtcBcKO and RtcB-rescued ES cells. Luciferase assays showed that this reporter was induced by different UPR stimuli in an RtcB-dependent manner (Figure S7B). We next transcribed an XBP1u intron-containing RNA substrate for in vitro splicing assaywithT7 RNA polymerase. Last, we reconstituted an XBP1u splicing reaction using baculovirus-expressed GST-IRE1αC that includes the cytoplasmic domain of human IRE1α and affinity-purified RtcB (Figure 7A). To exclude the possibility that other associated or contaminant proteins might contribute to the ligase activity, RtcBC122A was used as a negative control. As shown in Figure 7, when both wild type RtcB and IRE1α were incubated with the XBP1uin–containing mRNA substrate, was the spliced product detected by RT-PCR (Figure 7B). Quantitative RT-PCR analysis showed that over 95% of XBP1u substrates were cleaved by recombinant IRE1α and approximately 50% of the cleaved exons were rejoined during the in vitro splicing reaction (Figure 7C). Sequence analysis of the PCR product verified that the exact XBP1s exon-exon junction was generated in vitro (Figure 7D). Thus, the unconventional XBP1 mRNA splicing can be reconstituted by endonuclease IRE1α and its downstream RNA ligase RtcB in vitro.
Figure 7. IRE1α and RtcB reconstitute XBP1 splicing in vitro.
A. A Coomassie blue stained gel showing the affinity-purified RtcB proteins and the recombinant cytoplasmic domain of IRE1α. Both Flag-RtcBWT and Flag-RtcBC122A proteins were purified from the HEK293T stable cell lines described in Fig. 6A. B. An XBP1u intron-containing RNA substrate described in Fig. S7 was generated by in vitro transcription. Indicated purified proteins were incubated with the RNA substrate. The RNA products were purified and analyzed by RT-PCR. Note that the PCR was carried at saturation to increase signal intensity. The size shift and Pst I digestion patterns showed that wild type RtcB and IRE1α were both required and sufficient for XBP1u splicing in vitro. C. RT-qPCR analysis of the in vitroXBP1 splicing. D. Sequence analysis of the PCR products confirmed that the reconstituted splicing reaction occurred at the exact exon-intron junction. E. A working model to demonstrate the role of RtcB during mammalian UPR. Under normal condition, a subset of RtcB is in complex with IRE1α while binding to XBP1u mRNA. Upon ER stress, IRE1α oligomerizes, trans-autophosphorylates, and activates its endonuclease activity to excise a 26-nt intron of XBP1u. The IRE1α-associated RtcB then ligates two XBP1 exons to generate XBP1s mRNA. By physical interaction between RtcB and IRE1α, the two sequential steps of unconventional XBP1 splicing occur on the ER membrane. At present, it is not known whether RtcB directly interacts with IRE1α or through an intermediate (marked as “X”).
DISCUSSION
The unfolded protein response (UPR), a well-known adaptive mechanism for cells to maintain ER homeostasis, has been implicated in the pathogenesis of a variety of human diseases [for review see (Wang and Kaufman, 2012)]. Recent studies have demonstrated that genetic deletion of the UPR signaling components can significantly influence the disease progression in mouse models for diabetes, hepatic steatosis, IBD, ALS and other diseases (Wang and Kaufman, 2012). Despite the importance of UPR, a number of mechanistic questions remain to be answered. For instance, the mammalian RNA ligases responsible for unconventional XBP-1 splicing have been sought for over a decade. As a step to identify this essential component, we constructed a synthetic circuit that consists of an IRE1α sensor and an inducible apoptotic effector. By pairing this circuit with endogenous UPR pathway, we show that the resulting cells exhibit rapid and massive apoptosis, in response to acute UPR stimuli that normally do not affect cell survival. Building upon this robust synthetic phenotype, we performed a genome-wide RNAi screen for genetic suppressors and we identified RtcB as the primary RNA ligase for the mammalian UPR.
Prior to this study, RtcB was identified as an essential component of tRNA ligase complex, but not as an RNA ligase for XBP1 splicing (Iwawaki and Tokuda, 2011; Popow et al., 2011). Subsequent studies on the bacterial and archaeal RtcB have also demonstrated the biochemical mechanism by which RtcB catalyzes 3′-PO4/5′-OH ligation reaction (Chakravarty and Shuman, 2012; Desai et al., 2013; Englert et al., 2012). Consistent with the previous finding (Popow et al., 2011), we find that RtcB is required for endogenous tyrosine tRNA splicing. In contrast to a previous report (Iwawaki and Tokuda, 2011), we demonstrate that RtcB is essential for XBP1u splicing during the mammalian UPR. The obvious difference might be due to an insufficient RtcB knockdown by RNAi. In our study, genetic rescue and in vitro splicing experiments further establish that RtcB’s RNA ligase activity is directly responsible for the splicing of XBP1u downstream of IRE1α. In RtcBcKO cells, a small portion of XBP1s was still produced in the absence of RtcB, suggesting that an additional RNA ligase might compensate for RtcB function. Unlike mammalian cells, yeastHAC1mRNA splicing is mediated by the 5′-PO4/3′-OHligation pathway utilizing tRNA ligase Trl1p and 2′-phosphotransferase Tpt1p (Gonzalez et al., 1999; Schwer et al., 2004; Sidrauski et al., 1996). Consistent with this observation, a similar RNA ligase activity has been detected biochemically in HeLa cell extract but the identity of such ligase remains unknown (Zillmann et al., 1991). Therefore, it is possible that an RNA ligase in the 5′-PO4/3′-OHpathway compensates for loss of RtcB ligase activity in RtcB null cells. Nevertheless, we have shown that the cells expressing ligase-dead RtcB exhibited a nearly complete loss of XBP1s protein and XBP1s mRNA, suggesting that RtcBC122A may act as a dominant/negative protein to prevent XBP1u substrates from being ligated by a compensatory RNA ligase. Taken together, these data demonstrate RtcB is the predominant UPR RNA ligase in the wild type cells.
The unconventional XBP1 mRNA splicing has been suggested to occur in the cytoplasm based on the observation that isolated cytoplasmic fraction contains an activity capable of ligating XBP1 mRNA in vitro (Shinya et al., 2011; Uemura et al., 2009). However, because the first-step endonuclease IRE1α is an ER- resident transmembrane protein, the subsequent ligation step of XBP1 splicing would be more efficient if located on or near the ER. Results from several studies favor the latter possibility. For instance, it has been shown that unsplicedXBP1u mRNA is mainly localized to the ER membrane (Yanagitani et al., 2009). The ER localization of XBP1u mRNA is independent of IRE1α. Instead, the C-terminal region of nascent XBP1u polypeptide contains a hydrophobic signal sequence to target XBP1u-translating mRNA to the ER (Yanagitani et al., 2011). In our study, we show that the primary UPR RNA ligase RtcB is partially localized on the ER membrane, supporting the idea that ligation is more likely on the ER. Lastly, genome-wide analyses of UV-crosslinked RNA targets for RNA-binding proteins have recently identified XBP1u mRNA as a target for RtcB/C22orf28(Baltz et al., 2012). RtcB has been shown to bind to two sites flanking the 26-nt intron in XBP1u mRNA. Therefore, two different mechanisms are involved to recruit XBP1u to the ER. First, XBP1u mRNA is localized on the ER by an RNA-protein interaction with the ER-bound RtcB [this study and (Baltz et al., 2012)]. Second, XBP1u mRNA being translated remains associated with the ER through the nascent polypeptide ofXBP1u (Yanagitani et al., 2011). Thus, accumulating evidence suggests that the splicing of XBP1 mRNA primarily occurs on the ER.
Higher order organization of IRE1 has been shown to be critical for the UPR response in yeast and mammalian cells (Ali et al., 2011; Korennykh et al., 2009; Lee et al., 2008). During ER stress, IRE1α undergoes oligomerization and forms dynamic foci (Aragon et al., 2009; Li et al., 2010). Meanwhile, the substrates HAC1 mRNA in yeast and XBP1u mRNA in mammals are both recruited to IRE1 foci, albeit through different mechanisms[(Aragon et al., 2009; Yanagitani et al., 2011) and see discussion above]. As a signaling center, these foci initiate downstream signaling events including the unconventional mRNA splicing. Here, we demonstrate that the downstream RNA ligase RtcB not only resides on the ER membrane, but also is in complex with IRE1α protein even when the UPR is not activated. While the co-IP experiments did not differentiate whether the interaction between IRE1α and RtcB is direct or through other protein partners, their association in a protein complex suggests that the two proteins are in proximity with each other on the ER. Based on the findings presented here and previous studies on IRE1(Walter and Ron, 2011), we propose a working model for the action of RtcB during the UPR (Figure 7E). In this model, prior to ER stress, a subset of RtcBis associated with IRE1α while bindingXBP1u mRNA. Upon ER stress, oligomerized IRE1α forms higher-order foci on the ER, activating its endonuclease activity to cleave the intron of XBP1u. The proximally located RNA ligase RtcB then joins the two cleaved XBP1 exons to produce XBP1s mRNA. By physical interaction between IRE1α and RtcB, the two sequential steps of XBP1 splicing can be coupled on the ER membrane to increase the efficiency of signaling outputs.
EXPERIMENTAL PROCEDURES
Cell culture and induction of the UPR
HEK293T, COS7 and SNL cells were cultured in DMEM with 10% fetal bovine serum (FBS) and Penicillin-Streptomycin-Glutamine. HepG2 cells were cultured in Eagle’s MEM supplemented with 10% FBS. Mouse AB2.2 ES cells were cultured with Knockout™ DMEM with 15% FBS and mouse LIF with or without feeder cells. To induce ER stress, cells were treated with tunicamycin (2 μg/ml), thapsigargin (0.5 μg/ml) or DTT (2 mM) for indicated times before harvesting. To inhibit the endonuclease activity of IRE1α, cells were treated with 4μ8C (64 μM)30 min prior to the induction of ER stress (Cross et al., 2012). For cells to recover from the acute ER stress, treated cells were rinsed with warm culture media and incubated for an indicated recovery period without UPR inducers.
Construction of SNL sensor cells with a synthetic genetic circuit
To establish a stable SNL cell line with a UPR-inducible synthetic apoptosis phenotype, we transfected mouse SNL cells with a mixture of expression vectors including PB-CAG- ERT2-XBP1uin-Cre-ERT2:: PGK-Puro, PB-CAG-loxP-BSD-loxP-BimL and CAG-PBase (Su et al., 2009). Multiple stable cell lines were established after sequential drug-resistance selections with puromycin (4 μg/ml) and blasticidin (10 μg/ml). A cell line “B5” was used in the subsequent genetic screen.
RNAi based genetic suppressor screen
A pooled lentiviralpSIH-H1-PuroshRNA from System Biosciences were used in the screen as previously described (Su et al., 2011). The B5 SNL cells were infected with the lentiviral shRNA library and the infected cells were selected with puromycin (2 μg/ml). The PuroR SNL cells were treated with DTT (2 mM) for 2 hours to induce the UPR in the presence of 4-OHT (0.5 μM). After removing DTT, the cells were continuously treated with 4-OHT for 2 days. Robust cell death was observed 18–24 hours after adding 4-OHT and viable cells were cultured in non-selection media for 2 days. To increase the stringency and specificity of the RNAi screen, the selection by DTT and 4-OHT was repeated once as described above. After two rounds of DTT and 4-OHT selections, viable SNL clones were picked and expanded individually for isolating genomic DNA. Target shRNAs were identified by sequence analysis of PCR-amplified gene-specific shRNAs.
Gene targeting in ES cells
To generate RtcBcKO ES cells, we obtained an RtcB gene-targeting vector (PGS00036_A_C04) from the KOMP through BACPAC Resources (Skarnes et al., 2011). AB2.2 ES cells were electroporated with the linearized targeting vector and G418 resistant clones were selected. The 5′- and 3′- homologous recombination events were identified by long-range PCR(Skarnes et al., 2011). After the first RtcB allele was targeted, the Neo selection marker was removed by transient FLPe expression. Targeting of the second RtcB allele was repeated with the same targeting vector. Following G418 selection for the targeted allele, the second Neo marker was also deleted. The homozygous RtcBflox/flox ES cells were initially identified by long-range PCR with allele-specific primers. Using a previously described PB-CAG-ERT2-Cre-ERT2::PGK-Hygrovector (Su et al., 2009), we generated 4-OHT-inducible RtcB knockout ES cell lines. Knockout of RtcB in these cells were confirmed by deletion of RtcB exon 4 in RT-PCR assays and verified by a complete loss of RtcB protein in western blot analysis. The primer sequences for PCR screen and expression studies are listed in Table S2.
cDNA expression vectors
The original mouse RtcB/D10Wsu52e cDNA was obtained from Open Biosystems (IMAGE: 3583081). XBP1u, ATF6, IRE1a, IRE1β and PERK cDNA expression vectors were originally from Addgene. All cDNA expression vectors for stable transfection were constructed as PB vectors with the CAG promoter to express various genes of interest (Lu et al., 2009). Each PB vector consists of a drug resistance gene driven by PGK promoter. Mutant mouse RtcBC122A PB expression vector was constructed by an overlapping PCR strategy. All expression vectors were verified by sequencing analysis.
RNA isolation and quantitative PCR analysis
Total RNA was prepared using TRIzol (Invitrogen) followed by DNase I digestion. Reverse Transcription were performed with SuperScript™ III Reverse Transcriptase (Invitrogen) with oligo-dTor gene specific primers. After RT reactions, quantitative PCR were performed with an ABI Prism 7700 PCR instrument using SYBR green PCR Master Mix reagent kit (Applied Biosystems). The primers used in the experiments are listed in Table S2. In RT-PCR analysis of Tyr tRNA and Try pre-tRNA, primer S and primer AS1 only amplify pre-tRNA though the primers are designed to amplify both unspliced and spliced Tyr tRNA (Figure 2AB), perhaps due to the hairpin structure of spliced Tyr tRNA.
Measurement of the rate of global protein synthesis
A nonradioactive SUnSET method was used to measure the rate of protein synthesis (Schmidt et al., 2009). With or without 4-OHT, RtcBcKO cells were transiently labeled with puromycin at a final concentration of 2 μg/ml for 10 min before lysisin Laemmli buffer. Anti-puromycin antibody 3RH11 (KeraFAST) was used for quantitative western blot analysis. Quantification of puromycin-labeled total proteins was carried out using an Odyssey detection system.
Subcellular fractionation
HEK293T cells were fractionated into the cytosolic, membrane, and nuclear fractions using a differential detergent extraction method as previously described (Ramsby and Makowski, 2011). In brief, the cell pellet was permeabilized with cytosolic buffer (50 mMTris-HCl pH 7.4, 150 mMNaCl, 2 mM MgCl2, 5 mM EDTA, 1 mM PMSF, 100 μg/ml Digitonin) by incubation on ice for 10 min. The suspension was centrifuged at 2,000 RCF for 10 minutes and the supernatant was collected as the cytosolic fraction. After washing 2 times with cytosolic buffer to remove remaining digitonin extract, the pellet was resuspended in membrane buffer (50 mMTris-HCl pH 7.4, 150 mMNaCl, 2 mM MgCl2, 5 mM EDTA, 1 mM PMSF, 1% NP40) and incubated on ice for 30 minutes. The supernatant was collected as the membrane fraction after a 10-min centrifugation at 2,000 RCF. The resulting pellet was washed 2 times in membrane buffer and lysed in Laemmli buffer as the nuclear fraction.
Subcellular localization of RtcB by immunofluorescence
Mouse ES cells or COS7 cells were cultured on glass coverslips for immunofluorescence and confocal imaging analysis. Cells were washed with PBS followed by fixation in 2% paraformaldehyde (PFA) for 10 minutes. The following antibodies were used for staining: mouse anti-Myc (9E10, Sigma), mouse anti-Flag (M2, Sigma), rat-anti-GFP (MBL), rabbit anti-Calnexin (Abcam) and rabbit anti-GFP (Invitrogen). Double staining was visualized with DyLight488, DyLight 594- and Cy3-conjugated secondary antibodies (Jackson ImmunoResearch) and counterstained with DAPI. Images were captured with a Leica SP5 confocal microscope and fluorescence intensity plots were generated using the software Leica LAS AF lite.
Immunogold electron microscopy
HepG2 cells stably expressing GFP-RtcB were fixed in a fixative containing 2% paraformaldehyde and 0.1% glutaraldehyde in PBS (pH 7.4) for overnight at 4°C. After rinsing with PBS, cells were embedded with 10% gelatin, infused by sucrose, and cryosectioned. 80-nm sections were collected on electron microscopy grids. The grids were blocked with 1% cold water fish skin gelatin (Sigma) for 5 min, then incubated with rabbit anti-GFP antibody (Invitrogen) in blocking solution for 2 hour at room temperature. Following washing with PBS, a gold conjugated secondary antibody (18 nm Colloidal Gold-AffiniPure Goat Anti-Rabbit IgG, Jackson ImmunoReasearch) was incubated for 1 hour. The grids were fixed in 1% Glutaraldehyde for 5 min, washed with distilled water, contrasted and embedded in a mixture of 3% uranyl acetate and 2% methylcellulose (at a ratio of 1 to 9). All stained grids were examined under a Philips CM-12 electron microscope and photographed with a Gatan (4k × 2.7k) digital camera.
Luciferase reporter assay
For luciferase assay to monitor the splicing reporter V5-XBP1uin-Renilla luciferase, RtcBcKO cells were stably transfected with the reporter using a Piggy-Bac transposon (Lu et al., 2009). Rn luciferase activity was assayed using a Promega luciferase system. The induction of XBP1u splicing is reflected by the relative ratio of Rn luciferase activities with ER stress versus those without. Results were presented as mean values ± SEM of triplicate samples.
Immunoprecipitation, GST pull-down and western blot
Immunoprecipitation of RtcB protein complexes was carried out as previously described (Su et al., 2011). Briefly, HEK293T cells expressing Flag-tagged RtcB were lysed in a buffer (50 mMTris, pH 7.5, 150 mMNaCl, 1.0% NP-40, 10% glycerol, 1 mM EDTA, 1 mM PMSF, and Roche protease inhibitors). The resulting lysates were used to purify RtcB protein complexes with Flag M2 affinity-gel (Sigma). Similarly, GST-RtcB complexes were purified with glutathione Sepharose 4B beads (GE Healthcare) under the same condition.
Antibodies used in western blot analysis were from the following sources: rabbit anti-RtcB/C22orf28 (ProteinTech); rabbit anti-IRE1α (14C10, Cell Signaling); rabbit anti-PERK (C33E10, Cell Signaling); rabbit anti-GAPDH (Cell Signaling); rabbit anti-Bim (Assay Designs); goat anti-H2A (Cell Signaling); mouse anti-β-tubulin (DSHB, E7); mouse anti-FLAG (Sigma); rabbit anti-FLAG (Sigma); mouse anti-myc (DSHB, 9E10); mouse anti-V5 (AbDSerotec); and HRP conjugated secondary antibodies (Santa Cruz and Bio-Rad).
Reconstitution of XBP1u splicing reaction in vitro
To reconstitute XBP1u splicing in vitro, we first cloned a cDNA fragment encodingV5-XBP1uin-Rn luciferase into a pBluescript vector downstream of T7 promoter. The splicing substrate V5-XBP1uin-Rn mRNA was prepared using a MAXIscript T7 kit (Life Technologies) at 37°C for 1 hour. In vitro transcripts were digested with RNase-free DNase I and purified with RNeasy kit (Qiagen). Flag-RtcB and Flag-RtcBC122A proteins were purified from stable HEK293T cell lines that expressed the two proteins, respectively. Cell pellets were lysed in a buffer containing 30 mMTris-HCl, pH 7.4, 150 mMNaCl, 2mM MgCl2, 0.5% Triton X-100, 5 mM β-mercaptoethanol, 1 mM PMSF, 1X Roche protease inhibitor cocktail and 1 X Sigma phosphatase inhibitor cocktail. Flag-tagged RtcB proteins were affinity-purified from the lysates with anti-Flag M2 affinity-gel (Sigma) followed by extensive washes. The human GST-IRE1a cytoplasmic domain was purified from Baculovirus-infected insect cells and was purchased from Sino Biological.
The reconstituted in vitroXBP1u splicing was carried out at 37°C for 2 hours in Kinase buffer (2 mM ATP, 2 mM GTP, 50 mMTris-HCl pH 7.4, 150 mMNaCl, 1 mM MgCl2, 1mM MnCl2, 5 mM β-mercaptoethanol). 10 ngV5-XBP1uin-Rn RNA (2 nM), 1.0μg GST-IRE1a protein and IP-purified Flag-RtcBon beads, were used for a 50-μl splicing reaction. The spliced products were column-purified with an RNA clean & Concentrator kit (Zymo Research) for RT-PCR analysis. The PCR products with and without Pst I digestion were resolved on a 10% polyacrymide gel. In parallel, the PCR products were cloned into pUC57 vector and 30 clones from each reaction were analyzed by clonal PCR and Pst I digestion. Corresponded clones were confirmed by sequencing analysis.
Statistical Analysis
All data are shown as means with standard error of the means. Student’s t test (two-tailed, unpaired) was used for comparisons between two experimental groups.
Supplementary Material
Highlights.
A synthetic apoptosis circuit enables the identification of a UPR RNA ligase.
RtcB catalyzes unconventional XBP1 mRNA splicing during ER stress.
A subset of RtcB is associated with the endoplasmic reticulum.
IRE1α and RtcB reconstitute XBP1 splicing in vitro.
Acknowledgments
We thank R. A. Holmgren, J. H. Brickner and R. I. Morimoto for comments on the manuscript and we thank W. Russin for the assistance of confocal imaging. This work was in part supported by grants from NIH (1R21CA161483 and 1R21NS079880) to X.W.
Footnotes
Supplemental Information includes 7 figures and 2 tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Molecular cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Ali MM, Bagratuni T, Davenport EL, Nowak PR, Silva-Santisteban MC, Hardcastle A, McAndrews C, Rowlands MG, Morgan GJ, Aherne W, et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. The EMBO journal. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods (San Diego, Calif) 2005;35:395–416. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Molecular cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chakravarty AK, Shuman S. The sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic acids research. 2012;40:8558–8567. doi: 10.1093/nar/gks558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E869–878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai KK, Bingman CA, Phillips GN, Jr, Raines RT. Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry. 2013;52:2518–2525. doi: 10.1021/bi4002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding NZ, He M, He CQ, Hu JS, Teng J, Chen J. Expression and regulation of FAAP in the mouse epididymis. Endocrine. 2010;38:188–193. doi: 10.1007/s12020-010-9371-z. [DOI] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Aslanian A, Yates JR, 3rd, Soll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Xia S, Okada C, Nakamura A, Tanavde V, Yao M, Eom SH, Konigsberg WH, Soll D, Wang J. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:15235–15240. doi: 10.1073/pnas.1213795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochemical and biophysical research communications. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dorfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. The EMBO journal. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Lackey JG, Hsu HC, Zhang Y, Deng J, Xu RM, Damha MJ, Ron D. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA (New York, NY) 2008;14:225–232. doi: 10.1261/rna.859908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Molecular cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nature medicine. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- Iwawaki T, Tokuda M. Function of yeast and amphioxus tRNA ligase in IRE1alpha-dependent XBP1 mRNA splicing. Biochemical and biophysical research communications. 2011;413:527–531. doi: 10.1016/j.bbrc.2011.08.129. [DOI] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Molecular and cellular biology. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Dey M, Neculai D, Cao C, Dever TE, Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16113–16118. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science (New York, NY) 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lin C, Wang X. PiggyBac transgenic strategies in the developing chicken spinal cord. Nucleic acids research. 2009;37:e141. doi: 10.1093/nar/gkp686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada C, Maegawa Y, Yao M, Tanaka I. Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold. Proteins. 2006;63:1119–1122. doi: 10.1002/prot.20912. [DOI] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science (New York, NY) 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- Popow J, Schleiffer A, Martinez J. Diversity and roles of (t)RNA ligases. Cellular and molecular life sciences: CMLS. 2012;69:2657–2670. doi: 10.1007/s00018-012-0944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsby M, Makowski G. Differential detergent fractionation of eukaryotic cells. Cold Spring Harbor protocols. 2011;2011:prot5592. doi: 10.1101/pdb.prot5592. [DOI] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nature methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Schwer B, Sawaya R, Ho CK, Shuman S. Portability and fidelity of RNA-repair systems. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2788–2793. doi: 10.1073/pnas.0305859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya S, Kadokura H, Imagawa Y, Inoue M, Yanagitani K, Kohno K. Reconstitution and characterization of the unconventional splicing of XBP1u mRNA in vitro. Nucleic acids research. 2011;39:5245–5254. doi: 10.1093/nar/gkr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M, Zeng X, Larese J, Ryoo HD. A modified UPR stress sensing system reveals a novel tissue distribution of IRE1/XBP1 activity during normal Drosophila development. Cell stress & chaperones. 2013;18:307–319. doi: 10.1007/s12192-012-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger MA, Jackman JE, Phizicky EM. Analysis of 2′-phosphotransferase (Tpt1p) from Saccharomyces cerevisiae: evidence for a conserved two-step reaction mechanism. RNA (New York, NY) 2005;11:99–106. doi: 10.1261/rna.7194605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Meng S, Lu Y, Trombly MI, Chen J, Lin C, Turk A, Wang X. Mammalian hyperplastic discs homolog EDD regulates miRNA-mediated gene silencing. Molecular cell. 2011;43:97–109. doi: 10.1016/j.molcel.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes & development. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nature cell biology. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Meineke B, Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. The Journal of biological chemistry. 2011;286:30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. The Journal of biological chemistry. 2011;286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes & development. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Uemura A, Oku M, Mori K, Yoshida H. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. Journal of cell science. 2009;122:2877–2886. doi: 10.1242/jcs.040584. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science (New York, NY) 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. The Journal of cell biology. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Imagawa Y, Iwawaki T, Hosoda A, Saito M, Kimata Y, Kohno K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Molecular cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science (New York, NY) 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. The Journal of cell biology. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M, Gorovsky MA, Phizicky EM. Conserved mechanism of tRNA splicing in eukaryotes. Molecular and cellular biology. 1991;11:5410–5416. doi: 10.1128/mcb.11.11.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.