SUMMARY
Cytoplasmic pattern recognition receptors detect non-self RNAs during virus infections and initiate antiviral signaling. One receptor, MDA5, possesses essential signaling domains, but weak RNA binding. A second receptor, LGP2, rapidly detects diverse dsRNA species, but lacks signaling domains. Accumulating evidence suggests LGP2 and MDA5 work together to detect viral RNA and generate a complete antiviral response, but the basis for their cooperation has been elusive. Experiments presented here address this gap in antiviral signaling, revealing that LGP2 assists MDA5-RNA interactions leading to enhanced MDA5-mediated antiviral signaling. LGP2 increases the initial rate of MDA5-RNA interaction and regulates MDA5 filament assembly, resulting in the formation of more numerous, shorter MDA5 filaments that are shown to generate equivalent or greater signaling activity in vivo than the longer filaments containing only MDA5. These findings provide a mechanism for LGP2 co-activation of MDA5 and a biological context for MDA5-RNA filaments in antiviral responses.
Keywords: LGP2, MDA5, RIG-I-like receptors, RNA recognition, innate immunity
INTRODUCTION
Mammalian cells have the ability to detect and respond to virus infections. Viral RNAs that accumulate in the cytoplasm of infected cells can be detected by cytosolic pattern recognition proteins of the RIG-I-like receptor (RLR) family (Goubau et al., 2013; Ramos and Gale, 2011). These sentinel proteins recognize viral RNA and activate downstream signal transduction pathways that culminate in the transcription of diverse direct and indirect antiviral effectors, resulting in the establishment of a broadly effective antiviral state, a primary barrier for virus replication (Berke et al., 2013; Bruns and Horvath, 2012; Kowalinski et al., 2011).
The three RLR proteins, RIG-I, MDA5, and LGP2 share homologous DECH-box helicase regions that have intrinsic dsRNA binding and ATP hydrolysis functions, and a C-terminal domain that has been implicated in binding to RNA termini. RIG-I and MDA5 also contain N-terminal CARD domains that are not involved in RNA binding, but are essential for mediating interactions with upstream and downstream regulatory machinery. Detailed studies have produced a canonical paradigm of RLR signaling. Interaction with non-self ligand RNAs leads to exposure of active RLR CARDs, enabling association with the CARD region of an essential mitochondrial antiviral signaling protein known as IPS-1 or MAVS (Kawai et al., 2005; Kumar et al., 2006; Seth et al., 2005; Sun et al., 2006). MAVS acts as a polymeric scaffold that mediates interactions with downstream signaling proteins culminating in the activation of the master transcription regulators, IRF3 and NFκB to induce expression of IFNβ, the primary antiviral cytokine, and diverse antiviral target genes (Freaney et al., 2013; Honda et al., 2006; Liu et al., 2013).
The specific role of LGP2 in this model of RLR signaling has been more difficult to illuminate. LGP2 shares sequence conservation within the helicase domain and the C-terminus, but lacks a CARD region entirely. The absence of a known signaling or interaction domain has made it difficult to generalize LGP2’s function in the RLR pathway, and three separate strains of LGP2-deficient mice were reported with disparate conclusions (Satoh et al., 2010; Suthar et al., 2012; Venkataraman et al., 2007). LGP2 is present at low levels in the uninfected cell, but accumulates in response to virus infection or treatment with antiviral mediators including poly(I:C) and IFN (Komuro and Horvath, 2006; Rothenfusser et al., 2005; Saito et al., 2007). This behavior, and the observation that expression of LGP2 protein from plasmid vectors inhibits RLR signaling are consistent with a feedback regulator of RLR signaling (Komuro and Horvath, 2006; Rothenfusser et al., 2005; Saito et al., 2007; Yoneyama et al., 2005).
In contrast, growing evidence indicates a role for LGP2 as a positive regulator of antiviral responses. For example, mice with a targeted disruption in the LGP2 locus are more susceptible to infection by the picornaviruses, encephalomyocarditis virus (EMCV) and poliovirus, two viruses thought to be recognized by MDA5 (Kato et al., 2006) rather than RIG-I (Satoh et al., 2010; Venkataraman et al., 2007). Experiments in LGP2-deficient cells also revealed a synergistic signal transduction activity resulting from co-expression of LGP2 with MDA5 that was not attributable to either RLR alone (Satoh et al., 2010), suggesting that LGP2 may work together with MDA5 to promote efficient signal transduction. Replacing LGP2 with an enzymatically inactive mutant did not reconstitute defective positive signaling responses in vivo, indicating the importance of ATP hydrolysis in LGP2-mediated positive regulation of antiviral signaling. This requirement distinguishes positive regulation by LGP2 from negative regulation, which is independent of enzymatic activity, suggesting that LGP2 may be a bifunctional protein (Bamming and Horvath, 2009; Bruns et al., 2013). Additional evidence for LGP2 co-activation of MDA5 is accumulating from investigation of virus-RLR interactions. For example, an LGP2-associated EMCV RNA was found to act as a physiological agonist of MDA5-dependent signaling (Deddouche et al., 2014). Additionally, interferon antagonist proteins encoded by paramyxoviruses target both the MDA5 and LGP2, disrupting their ATP hydrolysis activity (Parisien et al., 2009). ATP hydrolysis activity is required to enable LGP2 to efficiently engage diverse dsRNA species, and is requisite for enhancement of MDA5 signaling (Bruns et al., 2013).
Electron microscopy has revealed that MDA5 forms filaments on dsRNA, with ring-like asymmetric units that form helical twists (Feng et al., 2012; Peisley et al., 2012; 2011; Wu et al., 2013). Evidence suggests initial contact between MDA5 and dsRNA occurs very slowly, and is followed by MDA5 assembly into head-to-tail filaments. These structures are clearly visible in the EM when captured in the presence of an ATP transition state analogue (ADP-AlF4) (Berke and Modis, 2012; Berke et al., 2012; Motz et al., 2013; Wu et al., 2013), but the MDA5 filaments are destabilized by physiological ATP hydrolysis (Berke and Modis, 2012; Berke et al., 2012). The presence of hydrolysable ATP promotes MDA5 dissociation from dsRNA, indicating that filament formation is a dynamic process in vivo. As such, the MDA5 filaments observed by electron microscopy likely represent a captured transition state for MDA5 signaling, subject to continual assembly and disassembly. Interestingly, RIG-I also forms filaments on RNA, albeit with mechanistic distinctions from MDA5 (Patel et al., 2013; Peisley et al., 2014; 2013), indicating that receptor oligomerization is a conserved feature of RLR antiviral signaling. Neither MDA5 nor RIG-I filaments have been observed in vivo, but prion-like aggregation of their downstream signaling partner, MAVS, has been observed both in vitro and in vivo (Hou et al., 2011; Xu et al., 2014). Structural analysis of activated MAVS revealed that CARD domain interactions organize a rod-like structure based on a three-stranded helix. This CARD organization is required for IFNβ gene expression, and it is reasonable to speculate that the filamentous assembly of MDA5 (or RIG-I) on dsRNA would suffice to provide aligned CARD domains to propagate MAVS filament formation and subsequent signal transduction. Curiously, while structural data indicate that as few as three to four aligned MDA5 (or RIG-I) molecules would be sufficient to organize or ‘seed’ CARD-mediated MAVS filament formation, much longer MDA5-dsRNA filaments have been observed in the EM. Greater knowledge of MDA5 RNA binding, filament initiation, and its regulation are required to fully appreciate how protein assembly influences signal transduction.
The role for LGP2 in the synergistic activation of MDA5 RNA interaction, filament assembly, and antiviral signal transduction has been examined in detail. Results indicate that LGP2 enhances signaling only by filament-competent MDA5, and does so by increasing the initial rate of MDA5-RNA interaction. In addition, although LGP2 itself does not form filamentous structures on dsRNA, results indicate that LGP2 has the ability to attenuate MDA5 filament length, resulting in the rapid generation of more numerous, shorter MDA5-RNA polymers. Despite their shorter length, these RNP complexes are able to stimulate antiviral signaling better than longer MDA5 filaments. Together, these results reveal that LGP2 facilitates and modulates MDA5-RNA interactions to increase the specific activity of MDA5 signal transduction.
RESULTS
LGP2 potentiation of MDA5 signaling requires ATP hydrolysis, RNA binding, and MDA5 oligomerization
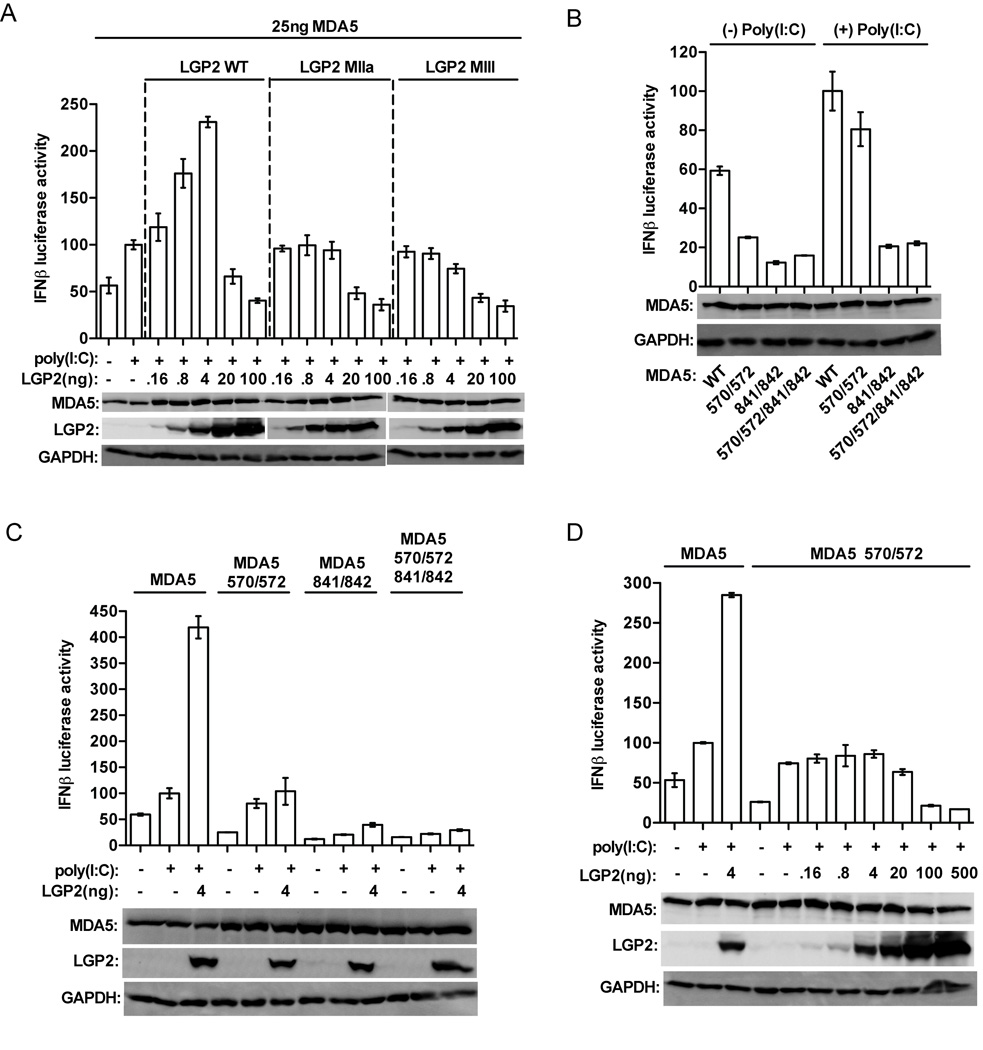
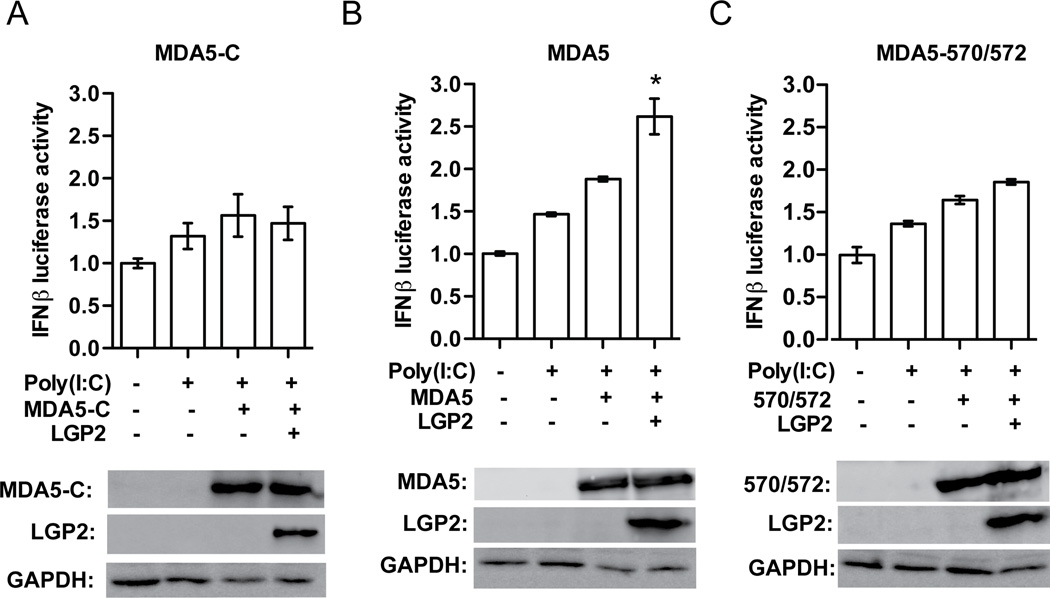
LGP2 has opposing effects on MDA5-mediated signaling that can be readily observed in the context of antiviral signal transduction assays (Figure 1). Expression of MDA5 in HEK293T cells is able to activate an IFNβ promoter driven luciferase reporter gene, and the potency of MDA5 can be further enhanced by transfection of poly(I:C). Titration of LGP2 by plasmid transfection reveals that low amounts of LGP2 result in enhanced MDA5-mediated signaling, while higher levels of LGP2 inhibit MDA5 signaling (Figure 1A) (Bruns et al., 2013; Pippig et al., 2009; Satoh et al., 2010). LGP2 with defective ATP hydrolysis and RNA binding, as with mutations to conserved helicase domain motif IIa (MIIa) or motif III (MIII) fail to enhance MDA5-mediated signaling. The mutated LGP2 proteins retain their ability to interfere with MDA5 signaling at higher concentrations. The dual actions of LGP2 differentially require intact ATP hydrolysis and RNA binding activities: both are required for positive effects on MDA5 signaling, but are dispensable for inhibition.
Figure 1. LGP2 enhancement of MDA5-mediated signaling requires ATP hydrolysis, RNA binding, and intact MDA5 oligomerization.
A. LGP2 ATP hydrolysis and RNA binding are required for MDA5 co-activation. HEK293T cells were transfected with −110 IFNβ-luciferase reporter gene, control Renilla luciferase plasmid, and expression vectors for MDA5, LGP2, or LGP2 mutants (MIIa and MIII). MDA5 was transfected at a constant 25ng plasmid/well, while the amount of LGP2 was titrated. After 24hr, cells were transfected with 5µg/ml poly(I:C) for 6hr prior to harvesting. In each plot, the activity of MDA5 stimulated by poly(I:C) is normalized to 100%. Expression levels of MDA5, LGP2, and GAPDH were analyzed by immunoblotting with specific antisera.
B. Signal transduction by MDA5 monomer interface mutants. IFNβ luciferase assays similar to A., but cells were transfected with 25ng of MDA5-WT or indicated MDA5 monomer-interface mutants: M570R/D572R, I841R/E842R, and M570R/D572R/I841R/E842R.
C. LGP2 does not augment signaling by MDA5 monomer interface mutants. IFNβ luciferase assay similar to A, but cells were transfected with 25ng of MDA5-WT or MDA5 monomer-interface mutants: 570/572, 841/842, and 570/572/841/842, and a single enhancing concentration of LGP2 (4ng).
D. LGP2 does not augment signaling by MDA5-570/572 mutant. IFNβ luciferase assay (as in A,B,C). HEK293T cells were transfected with 25ng of MDA5-WT or MDA5-570/572. The amount of LGP2 transfected was titrated at 0.16, 0.8, 4, 20, 100, and 500ng. No concentration of LGP2 was able to enhance MDA5-570572 mediated signaling.
A number of studies have demonstrated that MDA5 can assemble into tightly packed, ATP-sensitive filaments on dsRNA in vitro (Wu et al., 2013). Contacts between adjacent MDA5 molecules are thought to stabilize RNA-bound monomers and enable cooperative filament assembly. Mutations that disrupt the MDA5 monomer:monomer interface fail to support filament formation on long dsRNA (Wu et al., 2013). One of these MDA5 interface mutants, M570R/D572R, retains ≥ 70% of poly(I:C) stimulated signaling activity compared to wild-type MDA5, while the other mutants tested, I841R/E842R and M570R/D572R/I841R/E842R are only ~10–15% active (Wu et al., 2013) (Figure 1B). To ascertain if LGP2 co-activation is related to MDA5 filament formation, the monomer interface mutants were tested in the presence or absence of LGP2 (Figure 1C). None of the MDA5 mutants were enhanced by the addition LGP2. A full range titration of LGP2 demonstrated that MDA5-570/572 signaling ability is not enhanced at any concentration of LGP2 (Figure 1D). These data demonstrate that MDA5 oligomerization is required for LGP2-mediated enhancement. Together, the results indicate that both LGP2’s ATP-mediated RNA binding ability and MDA5’s RNA-mediated oligomerization ability are essential components of LGP2-mediated potentiation of MDA5 signaling.
LGP2 exponentially increases MDA5 interactions with dsRNA
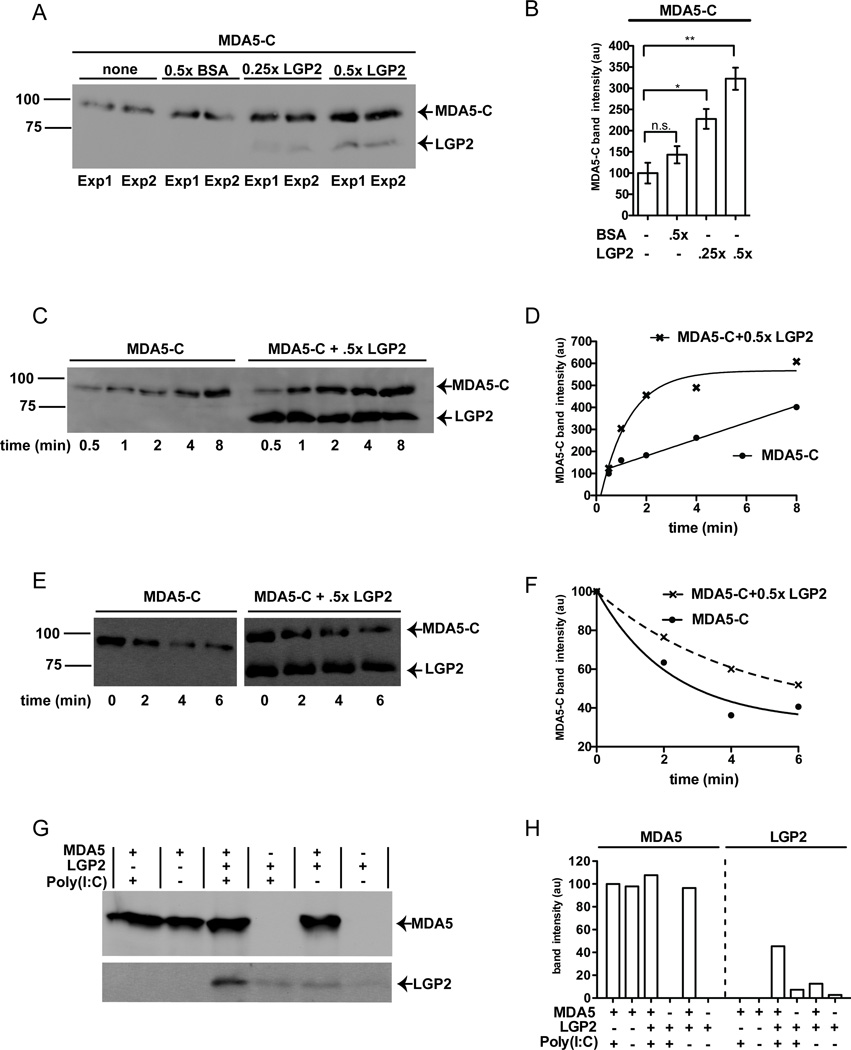
To directly test the hypothesis that LGP2-mediated enhancement of MDA5 signaling is related to MDA5 RNA interactions, the effect of LGP2 on MDA5-RNA interactions was investigated in solution using purified proteins and poly(I:C), a well characterized MDA5 agonist (Gitlin et al., 2006; Kato et al., 2008; Pichlmair et al., 2009).. To focus on the RNA-binding properties of MDA5 and avoid potential complications from the aggregation-prone CARDs, a truncated form of MDA5 lacking the CARDs was used in some experiments (MDA5-C). Purified, FLAG-tagged MDA5-C was incubated with biotinylated poly(I: C) and ATP, followed by ADP-AlF4 quenching to enable cooperative filament formation in vitro. Parallel samples were supplemented with purified FLAG-tagged LGP2 or a control non-specific protein (BSA), in empirically determined stoichiometric ratios. The biotinylated RNA and associated proteins were collected on streptavidin beads and washed extensively. Proteins were eluted with SDS, analyzed by anti-FLAG immunoblot (Figure 2A), and quantified (Figure 2B). MDA5 was found to bind to the dsRNA substrate in the absence of LGP2, but addition of LGP2 significantly increased the quantity of MDA5 bound to the RNA. The ability of LGP2 to enhance MDA5-dsRNA interaction was not observed in the absence of ATP/ADP-AlF4 (Figure S1). This is in agreement with both the observation that LGP2 mutants with defective ATP hydrolysis do not synergize with MDA5 (Figure 1A) and previous reports that ADP-AlF4 stabilizes MDA5-dsRNA interactions.
Figure 2. LGP2 enhances MDA5-dsRNA interactions by increasing the rate of MDA5 association.
A. LGP2 increases MDA5 binding to poly(I:C). MDA5-C was incubated with biotinylated-poly(I:C), in the presence of LGP2 or a control protein, BSA, in buffer containing ATP, and reactions were quenched with ADP-AlF4. RNA-bound proteins were analyzed by FLAG immunoblot. Duplicate experiments were carried out in parallel (Exp 1, Exp 2).
B. Quantification of MDA5-C band intensity from panel A, normalized to MDA5 alone. *, p=0.034; **, p=0.013.
C. LGP2 increases the rate of MDA5-C RNA interaction. Similar to (A), with changes to order of addition. Control (poly(I:C) alone) or LGP2-containing samples were incubated with ATP at 37°C for 1min, then MDA5-C was added to the reaction for the indicated times prior to quenching with ADP-AlF4.
D. Quantification of MDA5-C band intensity from panel C. MDA5-C binds to dsRNA with linear kinetics (R2=0.98) in the absence of LGP2, but LGP2 addition results in exponential MDA5 binding kinetics (R2=0.96 for exponential fit; R2=0.73 for linear fit).
E. LGP2 stabilizes MDA5-C association with poly(I:C). Similar to (C), but examining off-rate of MDA5. Biotinylated poly(I:C) was mixed with MDA5-C or MDA5-C and LGP2 and incubated with ATP for 5min, then 3x unlabeled competitor poly(I:C) was added for the indicated times prior to analysis.
F. Quantification of MDA5-C band intensity from panel E. MDA5-C dissociates from dsRNA with exponential kinetics in the absence (R2=0.97) and presence of LGP2 (R2=0.98), but the addition of LGP2 decreases the rate of MDA5-C dissociation from RNA.
G. RNA-mediated interaction of MDA5 and LGP2. Full-length MDA5 alone, LGP2 alone, or a mixture of LGP2 and MDA5 were incubated in the presence and absence of poly(I:C), and immunoprecipitated with MDA5-specific monoclonal antibody. Precipitates were washed, eluted, and analyzed by FLAG immunoblot.
H. Quantification of MDA5 (left) or LGP2 (right) band intensities, normalized to the signal of MDA5+poly(I:C) (100%). ≥3x more LGP2 co-precipitates with MDA5 in the presence of poly(I:C). See also Figure S1.
Single-molecule measurements of LGP2 interaction with a 25bp dsRNA have demonstrated that LGP2 has a fast on-rate (~0.086 sec−1, measured at 80nM), and that ATP hydrolysis dramatically increases LGP2’s ability to recognize dsRNAs (Bruns et al., 2013). Similar single molecule analysis was conducted to measure the on and off rates of MDA5-C, and it was found on the 25bp dsRNA MDA5-C has a very slow on rate (~0.039 sec−1, measured at 300nM) compared to LGP2 (Table 1). This slow on-rate for monomeric MDA5-dsRNA interactions accounts for the reported observation that MDA5 filament formation has a lag phase for nucleation in the range of 1–7 minutes (Peisley et al., 2012). MDA5-C displayed a faster on rate with a 44bp dsRNA (~0.063 sec−1), which is long enough to accommodate two MDA5 molecules, supporting the idea that protein-protein interactions between MDA5 monomers stabilize MDA5-RNA interactions. LGP2 displayed no significant difference in binding to the 25bp and 44bp substrates (Table 1) indicating that protein-protein interactions likely do not play a role in LGP2-RNA interactions. Considering these data along with the observed RNA binding enhancement suggested the hypothesis that LGP2 may function by altering the kinetics of MDA5-RNA interactions. To test this idea, an RNA binding time course was carried out. Consistent with its slow on-rate, MDA5-C was found to accumulate on RNA over time (Figure 2C). Incubation of the RNA with LGP2 for 1 min prior to MDA5 addition resulted in significantly faster MDA5 accumulation than in the absence of LGP2 (Figure 2C). Quantification of the MDA5-C band intensity revealed that the addition of LGP2 changed the rate of MDA5-C association with poly(I:C) from linear to exponential fit (Figure 2D), demonstrating that LGP2 increases the rate of MDA5 interaction with dsRNA. To investigate the effect of LGP2 on MDA5 disassembly from RNA, MDA5-C/RNA or MDA5-C/LGP2/RNA complexes were assembled using biotinylated poly(I:C) in the presence of ATP for 5 minutes. Three times excess of competitor unlabeled poly(I:C) was added for the indicated times, the reaction was quenched with ADP-AlF4, and proteins remaining bound to biotinylated poly(I:C) were analyzed by immunoblot (Figure 2E). Quantification revealed that while MDA5 displayed exponential off rate kinetics in both the presence and absence of LGP2, the presence of LGP2 stabilized MDA5-RNA interactions (Figure 2F).
Table I.
MDA5-C and LGP2 Binding Kinetics with 25bp and 44bp dsRNA
| MDA5-C | ||||
|---|---|---|---|---|
| dsRNA | Buffer | kon (sec−1) | koff (sec−1) | Kd (nM) |
| 25bp | No ATP | 0.039±0.009 | 0.059±0.011 | 452±24 |
| 500µM ATP | 0.025±0.017 | 0.093±0.018 | 1116±188 | |
| 44bp | No ATP | 0.063±0.01 | 0.071±0.009 | 326±138 |
| 500µM ATP | 0.04±0.008 | 0.098±0.017 | 771±207 | |
| LGP2 | ||||
| 25bp | No ATP | 0.086±0.007 | 0.053±0.006 | 49.8±8 |
| 44bp | No ATP | 0.084±0.015 | 0.059±0.004 | 56.1±10 |
Values are the average of at least three independent experiments shown plus or minus the standard deviation. Measurements made using 300nM MDA5-C or 80nM LGP2.
To determine if MDA5 and LGP2 directly interact, purified FLAG-tagged full length MDA5 and LGP2 proteins were combined and incubated in the presence or absence of poly(I:C), followed by immunoprecipitation with an MDA5-specific antibody and anti-FLAG immunoblotting (Figure 2G). A trace amount of LGP2 was detected non-specifically in the absence of MDA5, either in the presence or absence of poly(I:C). Incubation of MDA5 and LGP2 in the absence of poly(I:C) slightly increased the level of co-precipitated LGP2, but the presence of poly(I:C) increased the amount of co-precipitated LGP2 by over three fold (Figure 2H). These results indicate a low intrinsic ability of MDA5 and LGP2 to interact in solution, but their association is greatly enhanced via RNA interaction, consistent with the reported co-precipitation of LGP2 with MDA5 specifically in virus-infected cells (Komuro and Horvath, 2006).
MDA5 monomer-interface mutant is insensitive to LGP2-enhanced RNA binding
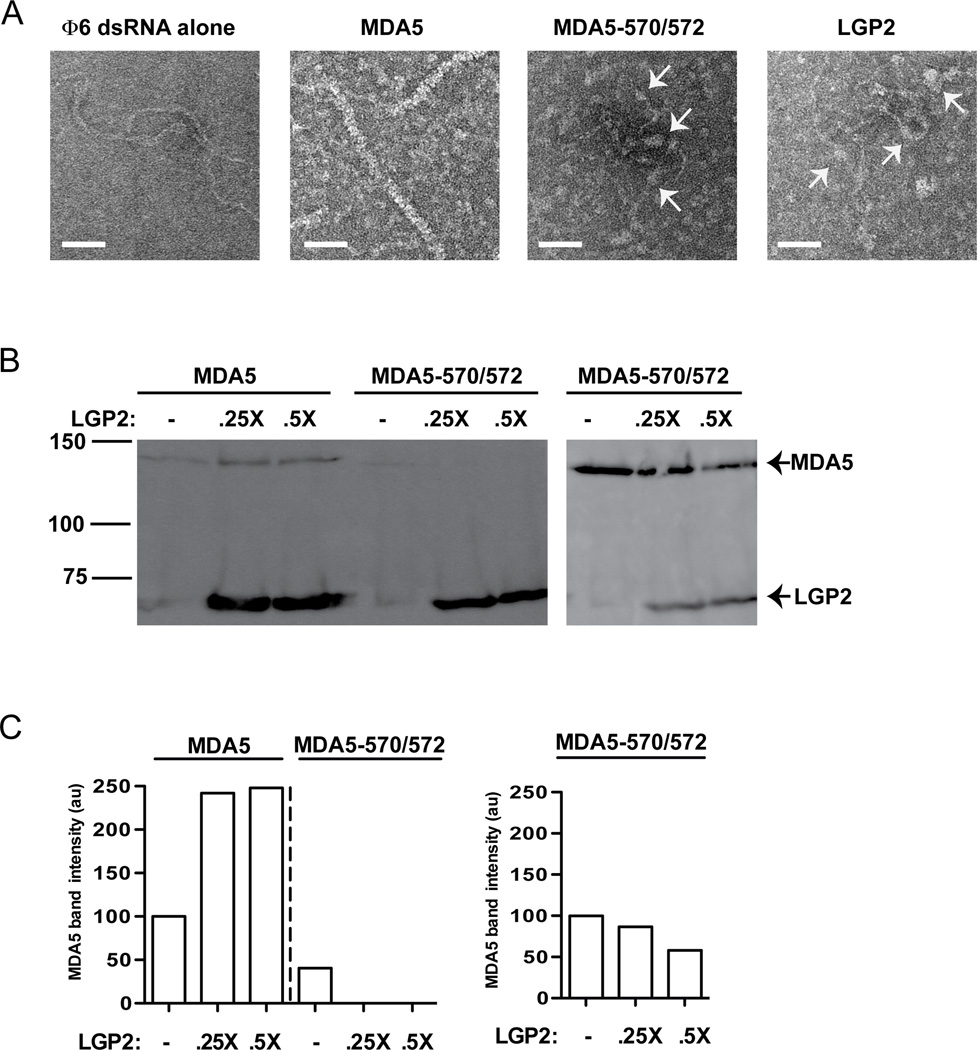
MDA5 assembly onto RNA results in filament formation, and mutations to the MDA5 monomeric interface prevent LGP2 co-activation. To directly determine the ability of the MDA5-570/572 mutant to form filaments on dsRNA, full length purified proteins were incubated with dsRNA and analyzed by negative stain electron microscopy. Full length MDA5 protein (containing the CARDs) was observed to form helical filaments along dsRNA, thoroughly coating most of the RNA surface (Figure 3A), consistent with prior reports. In contrast, long filaments were not observed with MDA5-570/572, and RNA interactions were limited to dispersed aggregates (see arrows, Figure 3A, third panel). It is noted that the MDA5-570/572 mutant retains the ability to form limited RNP assemblies at higher concentration. However, these quasi-filaments do not achieve the length found with WT MDA5 (Figure S2). As this MDA5 variant preserves much of its signal transduction capacity, this result indicates that the long polymers observed in vitro are not formally required for MDA5 signal transduction. LGP2 was not observed to form long filamentous structures on dsRNA under any conditions tested, but was also found to contact the dsRNA as dispersed aggregates (see arrows, Figure 3A, fourth panel).
Figure 3. MDA5 oligomerization is required for LGP2 enhanced RNA interactions.
A. Filament profiles of full-length MDA5, MDA5-570/572, and LGP2. Electron micrographs illustrating Φ6 genomic dsRNA alone, full-length MDA5-dsRNA filaments, and the more dispersed interactions between full-length MDA5-570/572 mutant or LGP2 with dsRNA. Arrows indicate protein-dsRNA interactions. Scale bar =50nm.
B. LGP2 enhances RNA binding by full-length MDA5 but not MDA5-570/572. Biotin-poly(I:C) pulldown of MDA5 and MDA5-570572 in the presence and absence of LGP2, similar to Figure 2A, 2B. Right panel shows a duplicate experiment using a 3x higher concentration of the mutant MDA5.
C. Quantification of panel B. Data normalized to the signal of MDA5 (or mutant) alone (100%). See also Figure S2.
These results were corroborated by a biotin-poly(I:C) pulldown experiment using full-length wild type MDA5 or mutant (570/572) in the presence or absence of LGP2 (Figure 3B). Similar to MDA5-C (Figure 2A), the full length MDA5 association with dsRNA was also detected in the absence of LGP2, but the amount of full length MDA5 bound to RNA was increased by the addition of LGP2. Lower amounts of the MDA5-570/572 protein were detected in association with the dsRNA, and LGP2 did not increase the amount of RNA association. Increasing the concentration of MDA5-570/572 resulted in more robust detection of RNA binding, but again LGP2 did not augment RNA interaction (Figure 3B). The fact that MDA5-570/572 signal transduction is not enhanced by LGP2 expression coupled with the lack of LGP2-mediated increase in RNA interaction supports the conclusion that LGP2-mediated enhancement of MDA5 signaling is a consequence of LGP2’s ability to increase MDA5-RNA interactions.
LGP2 attenuates MDA5 filament formation
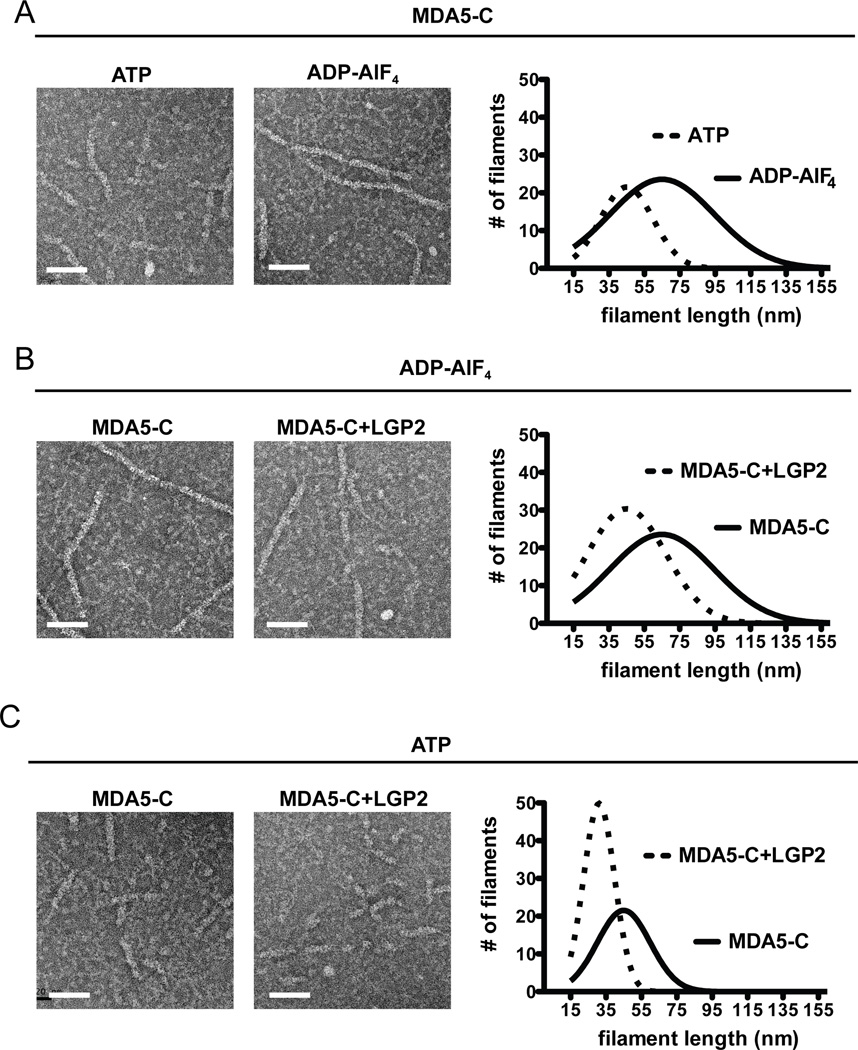
MDA5 filament formation initiates very slowly (Peisley et al., 2012) due to the low MDA5 on-rate (Table 1), and LGP2 apparently reduces this initial barrier to increase the extent of MDA5-dsRNA association (Figure 2D). Taking this into consideration along with the fact that signaling synergy between MDA5 and LGP2 is disrupted by mutations to the MDA5 monomer interface, suggested that LGP2 might function to improve rate-limiting MDA5 RNA recognition that is required to nucleate filament formation. To test this hypothesis, electron microscopy was used to investigate the effects of LGP2 on MDA5 filament formation. Purified MDA5-C proteins were incubated with dsRNA in the presence or absence of LGP2. Experiments were conducted in buffer containing only ADP-AlF4, or ATP quenched with ADP-AlF4 immediately prior to adsorption onto the EM grids. MDA5-C RNA filament formation was stabilized in the presence of ADP-AlF4, while incubation with ATP prior to ADP-AlF4 quenching resulted in quantifiably shorter filaments, with mean length of 45nm rather than 65nm (Figure 4A). Under filament stabilizing conditions (i.e. ADP-AlF4), the addition of LGP2 resulted in attenuation of filament length compared with MDA5-C alone, producing an average length of 45nm (Figure 4B). LGP2 had a more dramatic effect on MDA5-C filaments in the presence of ATP, resulting in the production of more numerous filaments of substantially shorter length (Figure 4C). The median size of these filaments is 28–34nm, and are estimated to be comprised of ~4–6 RLR molecules based on the 16–18bp footprint of MDA5 (Berke and Modis, 2012). The ability of LGP2 to attenuate MDA5 filament formation was also observed using LGP2 purified from mammalian cells (Figure S3). These data indicate that LGP2 not only promotes initial MDA5 association with dsRNA, but also regulates the extent of MDA5 filament formation on long dsRNA. In contrast to the dramatic reorganization of MDA5 filaments, the addition of LGP2 had no effect on MDA5-570/572 (Figure S2). As long MDA5 filaments are dispensable for much of its signaling ability, we conclude that co-activation of MDA5 by LGP2 is related to LGP2’s ability to increase the rate of MDA5 association with dsRNA, resulting in more rapid filament nucleation, and stabilization of these shorter MDA5-RNA structures.
Figure 4. LGP2 attenuates MDA5 filament formation.
Electron microscopy was used to assess MDA5-RNA complexes to evaluate effects of ATP and LGP2 on filament formation. “ATP” indicates incubation with ATP for 6min at 37°C, followed by quenching with ADP-AlF4 immediately before application to the EM grid. “ADP-AlF4” indicates incubation for 6min at 37°C with ADP-AlF4. Only filaments ≥18nm were quantified, a length estimated to bind at least a trimer of MDA5. Scale bar =50nm.
A. Effect of ATP on MDA5-C filaments. Representative images of MDA5-C filaments formed during incubation with ATP, and parallel samples incubated continuously in ADP-AlF4. Graph of filament size distributions is presented at right for 20 random images. For ATP, 137 filaments were measured with an average length of 45nm; for ADP-AlF4, 226 filaments were measured, with an average length of 65nm.
B. Effect of LGP2 on MDA5-C filament formation in ADP-AlF4 conditions. Similar to A, using only filament favoring ADP-AlF4 conditions to compare MDA5-C alone to MDA5-C with LGP2. Graph of filament size distributions is presented at right for 20 random images. For MDA5-C alone, 226 filaments were measured with an average length of 65nm. For the MDA5-C+LGP2 sample, 203 filaments were measured with an average length of 45nm.
C. Effect of LGP2 on MDA5 filament formation in ATP hydrolysis conditions. Similar to B, comparing MDA5-C alone to MDA5-C+LGP2 in the presence of ATP. Graph of filament size distributions is presented at right for 20 images. For MDA5-C alone, 137 filaments were measured with an average length of 45nm. For the MDA5-C+LGP2 sample, 158 filaments were measured with an average length of 31nm. See also Figure S3.
Short MDA5-LGP2-RNA complexes stimulate in vivo antiviral signaling
In addition to its positive contributions to MDA5 signaling, LGP2 overexpression has previously been found to interfere with RLR signaling (Komuro and Horvath, 2006; Rothenfusser et al., 2005; Saito et al., 2007). To determine if the MDA5-RNA complexes formed in the presence of LGP2 are capable of mediating antiviral signaling, these RNP complexes were introduced directly into HEK293T cells in the context of an IFNβ promoter-luciferase reporter gene assay (Lamartina et al., 1998; Sells et al., 1995; Tinsley et al., 1998; Zelphati et al., 2001). Cells expressing the IFNβ promoter-luciferase reporter were transfected with “naked” poly(I:C), or RNP complexes assembled in vitro from the same concentration of poly(I:C) along with MDA5 or its variants, with or without the addition of LGP2. All RNPS transfected contain the same amount of poly(I:C), and RNPs containing MDA5 protein have the same concentration of MDA5, MDA5-C, or MDA5-570/572 (Figure 5). MDA5-C, a negative control which lacks the CARD signaling domains did not induce the reporter gene activity greater than poly(I:C) alone, irrespective of LGP2 (Figure 5A). RNPs formed with full length MDA5 were able to induce signaling to the IFNβ promoter, resulting in reporter gene activity greater than that of naked poly(I:C) (Figure 5B). RNPs that contain both LGP2 and MDA5 retained signaling ability and resulted in equal or greater signaling than complexes formed from only MDA5 and poly(I:C), reflecting LGP2-mediated augmentation of MDA5 signaling. RNPs formed from the filament-defective MDA5-570/572 slightly increased signaling activity beyond that of poly(I:C) (Figure 5C). Importantly, the activity of MDA5-570/572 RNPs was not significantly stimulated by LGP2. The fact that RNP complex transfection is inefficient compared to naked poly(I:C), highlights the signaling potency of the MDA5+LGP2 RNA complexes (Figure S4). These results indicate that while long MDA5 filaments are able to induce antiviral signaling, the shorter MDA5 filaments formed in the presence of LGP2 are able to elicit an equivalent or slightly more potent signaling response.
Figure 5. LGP2 enhances the signal transduction activity of MDA5- RNA complexes.
Luciferase reporter gene assays were carried out as in Figure 1, but cells were stimulated by transfection with either 5µg/ml naked poly(I:C) or pre-formed RNP complexes composed of MDA5-C (panel A), WT MDA5 (panel B), or MDA5-570/572 (panel C) with or without LGP2 for 6hr before harvesting and luciferase measurement. Protein expression in lysates presented below each graph was analyzed with an MDA5-specific antibody, FLAG-specific antiserum, or a GAPDH-specific antibody. MDA5-C is incapable of signaling due to absence of the CARD domain, and serves as a negative control. MDA5 570/572 is insensitive to LGP2, but WT MDA5 signaling is increased by LGP2. *= p<0.01. See also Figure S4.
DISCUSSION
Many lines of evidence support the concept of synergy between the cytosolic RNA sensor, LGP2, and the signaling protein, MDA5. Experiments presented here provide a mechanistic basis for LGP2 enhancement of MDA5-mediated signaling by demonstrating that LGP2 modulates MDA5-RNA interactions. LGP2 both facilitates initial MDA5-RNA interactions, leading to the nucleation of MDA5 filament assembly, and regulates the extent of filament progression, resulting in the rapid formation of numerous shorter MDA5-RNA complexes. In this way, LGP2 increases the specific activity of MDA5 signaling by facilitating the formation of a greater number of more productive RNP complexes.
Both MDA5 and RIG-I have been shown to form filaments on dsRNA substrates (Peisley et al., 2013; Wu et al., 2013), resulting in close positioning of their CARDs. In turn, the aligned RLR CARDs interact with and stimulate the assembly of MAVS into a helical, three-stranded filament (Xu et al., 2014). The arrangement of CARDs in the MAVS triple helix suggests as few as three RLR CARD domains, aligned through dsRNA filament formation, would be sufficient to seed MAVS filament formation. Once seeded, MAVS polymerization is capable of self-propagation to form long signaling fibers (Hou et al., 2011; Xu et al., 2014). RIG-I filaments differ from MDA5 in several ways, including their limited propagation that results in multiple shorter filaments along the dsRNA (Patel et al., 2013; Peisley et al., 2013; 2014). RIG-I bound to dsRNA as short as 62bp, a length that can accommodate only 4–5 RIG-I molecules, represents the shortest RNP capable of activating MAVS oligomerization in vitro (Peisley et al., 2013). The fact that short filaments represent unit signaling activity suggests that the observed long MDA5 filaments containing dozens of side-by-side CARDs are far longer than is necessary to induce MAVS aggregation. MDA5 dissociation from dsRNA is inversely proportional to the filament length, and short MDA5 filaments are quite unstable compared to longer MDA5 filaments (Peisley et al., 2011). We demonstrate that LGP2 not only increases the rate of MDA5-RNA interactions, but also results in the formation of more numerous MDA5 filaments in vitro. The shorter RNPs are apparently stabilized by LGP2, enabling the formation of a greater number of signaling complexes that are sufficient to activate MAVS (Figure 4). Direct analysis of RNP complexes demonstrates that RNPs formed from MDA5 and LGP2 are slightly more active than complexes containing MDA5 alone (Figure 5). Filament formation itself, however, is apparently indispensable for LGP2 augmentation, as MDA5 monomer-interface mutants are not susceptible to signaling enhancement by LGP2.
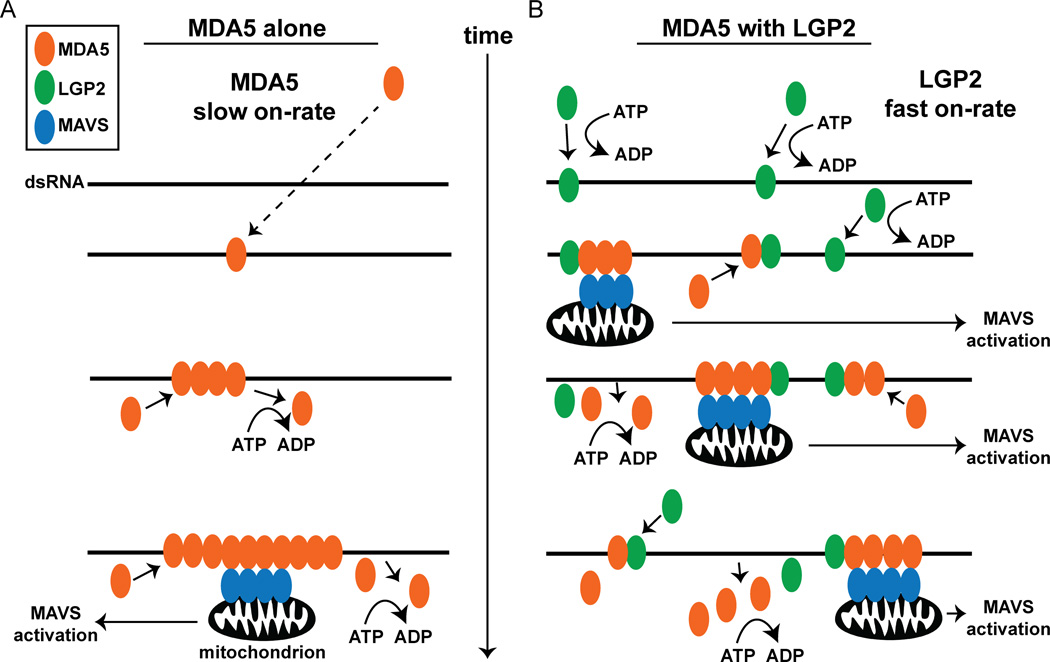
A conceptual model is presented to illustrate the central conclusions derived from this study in the context of prior knowledge of MDA5 and LGP2, emphasizing the primary features of RNA detection and antiviral signaling during the initial period of virus infection (Figure 6). In the absence of LGP2, MDA5 engages dsRNA templates slowly, with low affinity, and physiological ATP hydrolysis supports MDA5 dissociation from dsRNA. It is conceivable that alterations in MDA5 concentration, specific post-translational modifications, or interactions with additional signaling partners could modify the protein’s ability initiate filament assembly. In the presence of ADP-AlF4, filament formation is stabilized, resulting in the observed long RNP fibers mediated by essential monomer-monomer interactions. These long filaments are capable of activating downstream signaling, but data indicate they are not necessarily more active than shorter RNPs, such as the quasi-filaments formed by defective MDA5 monomer-interface mutants.
Figure 6. Mechanistic model for LGP2 co-activation of MDA5 signaling.
Representation of MDA5 dsRNA recognition, filament assembly, and signal transduction in the absence (A) or presence (B) of LGP2. The dsRNA is depicted as a black line, and MDA5, LGP2, and MAVS are illustrated as orange, green, and blue ovals, respectively. Time is indicated as an arrow from top to bottom.
A. MDA5 monomers bind to dsRNA with a slow on-rate (dashed line), and ATP hydrolysis mediates dynamic MDA5 filament assembly and disassembly in vivo. Filaments formed with sufficient aligned MDA5 CARDs can induce activation of MAVS assembly in the mitochondria (black) and subsequent signal transduction. Long filaments take longer to form and are limited in their ability to initiate MAVS activation over time.
B. LGP2 monomers rapidly bind to dsRNA utilizing ATP hydrolysis to enhance dsRNA recognition, enabling more rapid recognition of the dsRNA by MDA5, enabling more short filaments to assemble quickly. The dynamic, ATP-enhanced RNA binding activity of LGP2 allows MDA5 filament initiation at multiple sites over time. This rapid assembly produces a greater number of short, signaling-competent MDA5 filaments in the presence of LGP2, and a greater yield in MAVS activation over time versus longer filaments formed in the absence of LGP2.
In contrast, the presence of LGP2 alters the kinetics of MDA5-RNA interaction and regulates filament formation. LGP2 has a high affinity for dsRNA, and utilizes ATP hydrolysis to efficiently engage diverse dsRNA based substrates (Bruns et al., 2013). LGP2 rapidly detects dsRNA, enabling more efficient recruitment of MDA5 to the RNA, and this augmentation, as well as LGP2-mediated signaling enhancement, requires that MDA5 is competent for filament formation (Figure 3B,C). Results indicate that the LGP2-mediated RNA recognition enables MDA5 to nucleate filament assembly at more loci, resulting in more numerous MDA5 filaments formed over the same period of time. All MDA5 filaments are subject to ATP-dependent disassembly, but LGP2 enables them to form faster, stabilizes the formation of shorter structures, and allows them to accumulate to higher abundance, accounting for their greater signaling potency over time. The short MDA5 filaments assembled in the presence of LGP2 are sufficient to induce MAVS activation, resulting in greater signaling to the IFNβ promoter in the same period of time than the long MDA5 filaments formed in the absence of LGP2 (Figure 6).
The observation that longer filaments containing more CARD domains do not result in higher signaling capacity seems counter-intuitive. However, each of the short LGP2-containing filaments are capable of inducing MAVS polymer activation independently at many mitochondria surfaces within the cell. Longer filaments, once engaged, may be restricted in their ability to generate multiple sites of MAVS oligomerization. In the context of virus infection, MDA5 filament formation is almost certainly a dynamic process with filamentous structures constantly assembling and disassembling under physiological conditions of ATP hydrolysis. Because LGP2 is very efficient in dsRNA recognition and uses ATP hydrolysis to engage diverse dsRNA species, it is plausible that during virus infection LGP2 would recognize exposed dsRNA first, reducing energetic barriers for initial MDA5-RNA interactions, increasing the likelihood of productive signal assembly. LGP2 stabilizes the formation of shorter MDA5 filaments, resulting in numerous small bundles of aligned CARDs. As exposed dsRNA regions are thought to be a rarity during natural RNA virus infections, this mechanism may ensure MDA5 recognition of even short stretches of non-self dsRNA to result in productive signaling events. The ability of LGP2 to help MDA5 form active MAVS-stimulating complexes more rapidly and frequently ultimately leads to increased transcription of IFNβ and a fully productive antiviral response.
Additional studies will be required to determine if additional aspects of LGP2 action contribute to MDA5 signal co-activation, and if this synergy is modulated by infection-induced post-translational modification. Given the importance of a tightly regulated innate antiviral response, it is likely that additional layers of regulation modulate the extent of LGP2-MDA5 antiviral signaling synergy.
EXPERIMENTAL PROCEDURES
Plasmids and mutagenesis
Expression vectors for FLAG-tagged MDA5 and LGP2 described previously (Bruns et al., 2013), were used as templates for site-directed mutagenesis (QuickChangeXL, Stratagene). For recombinant baculovirus production FLAG-tagged MDA5 and MDA5-C (residues 295–1025) were subcloned into pBac2cp (Novagen), and FLAG-tagged LGP2 cDNA was cloned pVL-1393 (BD Biosciences).
Cell culture, luciferase, and immunoblotting
HEK293T cells were maintained in DMEM supplemented with 10% CCS and 1% Pen/Strep. For luciferase assays, cells were transfected using Lipofectamine 2000 (Invitrogen). The −110 IFNβ promoter-luciferase reporter is co-transfected with control Renilla luciferase for signal normalization. MDA5 plasmid was used at 25ng/well, and the amount of LGP2 plasmid was varied. The following day cells were transfected with 5µg/ml HMW poly(I:C) (Invivogen) for 6hr before harvesting. For RNP-activated luciferase, cells were similarly transfected and treated the following day by transfection of HMW poly(I:C) (3µg) or freshly-prepared RNP complexes containing 3µg HMW poly(I:C) and 2.5µg MDA5, MDA5-C, or MDA5-570/572, with or without 0.25µg LGP2 using Lipofectamine 2000 for 6hr before measuring luciferase activity (Dual Luciferase™, Promega), and are plotted as the average of triplicates ± S.D. Lysates were analyzed by immunoblot with a monoclonal antibody to MDA5 (Bruns et al., 2013) and with mouse anti-FLAGM2 to detect MDA5-C and LGP2 expression (Sigma), and with GAPDH as a loading control (Santa Cruz Biotechnologies).
Baculovirus, protein expression, and purification
To generate recombinant baculovirus expressing MDA5, MDA5-C, LGP2, and mutant proteins, transfer vectors containing the cDNA of interest were co-transfected with linearized baculovirus DNA (BD Biosciences) into Sf9 insect cells. Proteins were purified from infected Sf9 cells using FLAG immunoaffinity chromatography (Bruns et al., 2013). Proteins were quantified by Bradford assay, and purity assessed by Coomassie Blue staining. 3xFLAG-LGP2 was cloned into pSNAPf (NEB), and 20µg 3xFLAG-LGP2-SNAP plasmid was transfected by CaPO4 into each 20×10cm plates of 293T cells. Lysate was harvested in whole cell extract buffer, and FLAG purified.
Single molecule RNA binding
Experiments conducted as in (Bruns et al., 2013) with Cy-3 labeled dsRNA alone or in combination with proteins visualized on a 37°C objective using a Olympus IX81 TIRF microscope, ImageEM EMCCD camera (Hammamatsu), and 561 nm (75 mW) laser (Melles Griot). Image acquisition was done using Slidebook software (Olympus). The dwell times were analyzed the presence and absence of ATP, using both a 25bp dsRNA (Bruns et al., 2013), and a 44bp dsRNA (IDT): 5’-CGAGCAGACUGGCAAUCCGGAUCUCCCAGGCCCGGCUUUCAAGC-/Cy3/-3’ 5’-/5AmMC6/-GCUUGAAAGCCGGGCCUGGGAGAUCCGGAUUGCCAGUCUGCUCG-/3Bio/-3’
Poly(I:C) binding assays
Biotinylated poly(I:C) (Invivogen; 4ng/µl) was incubated with proteins at 37°C in (20mM Hepes pH 7.5, 150mM NaCl, 3mM MgCl2, 1mM DTT). MDA5 (full length) or MDA5-C ranged from 0.05-0.2µM in each reaction, determined empirically based on the specific RNA-binding activity of each preparation. LGP2 was used in each experiment relative to MDA5, also determined empirically. Reactions were quenched for 1m at 4°C with 1000µM ADP-AlF4. For kinetic RNA binding analysis during filament assembly (Figure 2C,D), 60µg/mL heparin was added to the quenching buffer to sequester unbound proteins. In experiments analyzing off-rate, proteins were incubated with biotin-poly(I:C) and 500µM ATP for 5min at 37°C to allow for filament assembly, and for the indicated times 3X excess non-biotinylated poly(I:C) was added to the reaction, followed by quenching with ADP-AlF4. Biotin-poly(I:C) and associated proteins were collected with streptavidin magnetic beads (Invitrogen), washed 3X with Buffer A +0.1% NP-40, and eluted with PLB (0.3M Tris pH6.8, 10% SDS, 25% β-mercaptoethanol, 20% glycerol, 0.01% bromophenol blue). Eluates were probed with with FLAG-M2 antibody (Sigma). The intensity of bound MDA5 was quantified using ImageJ64 and normalized to the corresponding signal of MDA5 without LGP2.
Co-immunoprecipitation
For interaction analysis, 0.2µg MDA5 and 0.02µg LGP2 were incubated alone or in combination in Buffer A plus 500µM ADP-AlF4 in the presence and absence of 10µg poly(I:C). Pre-cleared supernatants were mixed with 20µl protein A/G agarose conjugated to 8µg of a monoclonal MDA5 antibody (Bruns et al., 2013) and incubated at 4°C for 1hr. Beads were washed 3X with 1ml buffer A, and eluted in PLB.
Electron Microscopy
For filament imaging, MDA5, MDA5-C (0.1µM), or MDA5-570/572 (0.1µM and 0.3µM) and Φ6 genomic dsRNA (Thermo Scientific) (10ng/µl) were incubated with buffer A for 6min to allow MDA5-RNA interactions. LGP2 (0.02µM) was added to the RNA in Buffer A prior to addition of MDA5 or MDA-C. “ADP-AlF4” indicates 6min incubation at 37°C in the presence of ADP-AlF4; “ATP” indicates incubation with ATP for 6min at 37°C, followed by immediate quenching with ADP-AlF4 and absorption onto 300 mesh copper grids covered with a thin carbon film that were glow-discharged immediately prior to use, stained with 1% uranyl formate, and visualized at 100kV with a JEOL 1230 microscope. Images were acquired with a Gatan 831 CCD camera. Only filaments ≥18nm were quantified. In Figs 3 and 4 filaments from 20 random images (n=130–230 per condition) were measured using ImageJ64, grouped into 10nm bins, and used to generate length averages and distributions. In Fig S2, filament-like structures of MDA5-570/572 were scarce, so images of fields containing quasi-filaments were specifically captured, and lengths quantified (n=60–80).
Supplementary Material
Highlights.
-
-
Antiviral synergy requires LGP2 ATPase and RNA binding, and MDA5 oligomerization.
-
-
LGP2 increases the rate of initial MDA5-dsRNA interaction.
-
-
LGP2 results in more numerous MDA5-dsRNA filaments of shorter length.
-
-
Shorter filaments formed in the presence of LGP2 retain their signaling capacity.
ACKNOWLEDGMENTS
We are grateful to John Marko for assistance with single molecule experiments, and members of the Horvath lab for helpful comments on this work and manuscript. Research on RLRs in the Horvath lab was supported by NIH grants AI073919 and AI50707 to CMH. AMB was supported by a predoctoral fellowship from the NIH Cellular and Molecular Basis of Disease Training Grant T32GM008061. GPL and RAL are a Research Specialist and an Investigator, respectively, of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
CMH and AMB conceived experiments.
AMB and GPL performed experiments.
RAL provided reagents and expertise.
CMH and AMB wrote the manuscript.
REFERENCES
- Bamming D, Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J Biol Chem. 2009;284:9700–9712. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. Embo J. 2012;31:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Li Y, Modis Y. Structural basis of innate immune recognition of viral RNA. Cell Microbiol. 2013;15:386–394. doi: 10.1111/cmi.12061. [DOI] [PubMed] [Google Scholar]
- Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci USA. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Horvath CM. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol. 2012;47:194–206. doi: 10.3109/10409238.2011.630974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Pollpeter D, Hadizadeh N, Myong S, Marko JF, Horvath CM. ATP hydrolysis enhances RNA recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (LGP2) J Biol Chem. 2013;288:938–946. doi: 10.1074/jbc.M112.424416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard PV, Borg A, Matthews N, Feng Q, van Kuppeveld FJM, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJM. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freaney JE, Kim R, Mandhana R, Horvath CM. Extensive cooperation of immune master regulators IRF3 and NFκB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 2013;4:959–973. doi: 10.1016/j.celrep.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell R, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic-polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, Deddouche S, Reis e, Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang Q-X, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamartina S, Roscilli G, Rinaudo D, Delmastro P, Toniatti C. Lipofection of purified adeno-associated virus Rep68 protein: toward a chromosome-targeting nonviral particle. J Virol. 1998;72:7653–7658. doi: 10.1128/jvi.72.9.7653-7658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen J, Cai X, Wu J, Chen X, Wu Y-T, Sun L, Chen ZJ. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2:e00785–e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann K-K, Hopfner K-P. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339:690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- Parisien J-P, Bamming D, Komuro A, Ramachandran A, Rodriguez JJ, Barber G, Wojahn RD, Horvath CM. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J Virol. 2009;83:7252–7260. doi: 10.1128/JVI.00153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JR, Jain A, Chou Y-Y, Baum A, Ha T, García-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Jo MH, Lin C, Wu B, Orme-Johnson M, Walz T, Hohng S, Hur S. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci USA. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci USA. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan C-P, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis E, Sousa C. Activation of MDA5 Requires Higher-Order RNA Structures Generated during Virus Infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner K-P. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos HJ, Gale M. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol. 2011;1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo Y-M, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Li J, Chernoff J. Delivery of protein into cells using polycationic liposomes. Biotechniques. 1995;19:72–6. [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea C-K, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu H-H, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, McMillan A, Diamond MS, Clark EA, Bevan MJ, et al. The RIG-I-like Receptor LGP2 Controls CD8(+) T Cell Survival and Fitness. Immunity. 2012;37:235–248. doi: 10.1016/j.immuni.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley JH, Hawker J, Yuan Y. Efficient protein transfection of cultured coronary venular endothelial cells. Am J Physiol. 1998;275:H1873–H1878. doi: 10.1152/ajpheart.1998.275.5.H1873. [DOI] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, la Cruz de MJ, Borkowski B, Zhang X, Chen ZJ, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. Elife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo Y-M, Gale M, Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Zelphati O, Wang Y, Kitada S, Reed JC, Felgner PL, Corbeil J. Intracellular delivery of proteins with a new lipid-mediated delivery system. J Biol Chem. 2001;276:35103–35110. doi: 10.1074/jbc.M104920200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.