Abstract
CRISPR drives prokaryotic adaptation to invasive nucleic acids such as phages and plasmids using an RNA-mediated interference mechanism. Interference in Type I CRISPR-Cas systems requires a targeting Cascade complex and a degradation machine Cas3, which contains both nuclease and helicase activities. Here we report the crystal structures of Cas3 bound to ss-DNA substrate and show that it is an obligated 3′-to-5′ ss-DNase preferentially accepting substrate directly from the helicase moiety. Conserved residues in the HD-type nuclease coordinate two irons for ss-DNA cleavage. ATP coordination and conformational flexibility are revealed for the SF2-type helicase moiety. Cas3 is specifically guided towards Cascade-bound target DNA with a correct PAM sequence, through physical interactions to both the non-target substrate strand and the CasA protein. The cascade of recognition events ensures a well-controlled DNA targeting and degradation of alien DNA by Cascade and Cas3.
Keywords: CRISPR, Cas, nuclease, helicase, Cas3
Introduction
CRISPR (Clustered Regularly Interspaced Palindromic Repeats) drives adaptation to invasive nucleic acids such as phages, conjugative plasmids, and transposable elements using an RNA-mediated interference mechanism that has fundamental similarities to our innate and adaptive immune responses 1–3. This RNA-based adaptive immunity system involves a noncoding CRISPR array and a nearby cas (CRISPR-associated) operon4–6. The encoded Cas proteins are involved in three key molecular events: CRISPR (Clustered Regularly Interspaced Palindromic Repeats)-Cas systems mediate three key molecular events: (1) adaptation through the insertion of short segments of “spacer DNA” derived from foreign genetic elements into the CRISPR array; (2) transcription of the CRISPR array and the endoribonucleolytic processing of it into crRNA; and (3) crRNA-guided degradation of the foreign DNA7 containing spacer-complementary sequences1–3 (RNAs are targeted in Type III-B CRISPR systems, as exemplified in Sulfolobus solfataricus and Pyrococcus furiosus8).
Based on the mutually exclusive presence of signature proteins Cas3, Cas9, and Cas10 in cas operons, known CRISPR-Cas systems can be further divided into Type I, II, and III classes, respectively, each achieve crRNA-guided nucleic acid degradation through a distinct mechanism9. The three major types can be further divided into subtypes. For example, the most wide-spread Type I system, accounting for 95% of all CRISPR systems, can be classified into six different subtypes (I-A to I-F), each encoding a unique set of subtype-specific Cas proteins9. Focusing on the most prevalent Type I systems, the emerging theme is that crRNA-guided ds-DNA degradation requires a targeting complex, the ~400 kDa Cascade (CRISPR associated complex for anti-viral defense), and a DNA unwinding and degradation enzyme, the Cas3 protein10,11. The seahorse-shaped Cascade promotes the invasion of crRNA into ds-DNA to selectively base-pair with the target strand DNA while looping out the non-target strand, generating the “R-loop” structure12–14. Subsequently, Cas3 is recruited by Cascade to degrade the target DNA, preferentially from the non-target strand12,14–16.
Being the signature gene and an essential factor in all Type I CRISPR systems, cas3 typically encodes an N-terminal HD nuclease and a C-terminal Type A (3′-to-5′) Superfamily 2 (SF2) helicase. Such nuclease/ribonuclease and helicase fusion proteins can be found in DNA replication/repair and RNA processing or interference systems17–20. Different modes of mechanistic coupling may exist depending on the architecture of the fusion enzyme. The HD nuclease has been characterized as a metal-dependent exo- and/or endo-nuclease, and the apo crystal structures revealed the presence of one or two divalent metals chelated by the invariable HD-motif at the active site 15,16,21–23. The coordination and function of these metal ions in DNA binding and catalysis was not convincingly defined since the available structures lacked the bound DNA substrate. The Type A SF2 helicase in Cas3 was shown to consume ATP and unwind a DNA duplex by displacing the nicked strand in a 3′-to-5′ direction22,23. Studies further reveal that Cas3 is activated at the Cascade-marked R-loop region, and it preferentially cleaves the non-target strand DNA ~12-nt into the R-loop region, then moves 3′-to-5′ driven by ATP hydrolysis, which is then followed by a similar degradation action on the target strand15,16. A recent study suggests that recruitment of Cas3 involves interaction with the CasA component of the Cascade complex24.
To fully understand the Cascade-activated DNA unwinding and degradation mechanism, we determined the crystal structure of the Cas3 protein bound to a ss-DNA substrate and biochemically defined its physical interactions with the Cascade complex. The catalysis mechanism of the HD nuclease was revealed with the snapshot of the ss-DNA substrate coordinated by two catalytic irons in the active site. The SF2 helicase was captured at open conformation, with and without an ATP molecule bound, providing hints about the ATP hydrolysis driven conformational switching cycle. The functions of the Cas3-specific structure features were revealed with the CRISPR interference assays and biochemical reconstitutions. We showed that Cas3 was specifically guided towards Cascade-bound target DNA in the presence of an optimal Protospacer Adjacent Motif (PAM) sequence, and through physical interactions with the CasA component of the Cascade and the non-complementary strand of the ds-DNA substrate. The stringent set of recognition events ensures a well-controlled DNA targeting and degradation of alien DNA in the Type I CRISPR-Cas system.
Results
Overall structure of the ss-DNA bound T. fusca Cas3 at 2.65 Å resolution
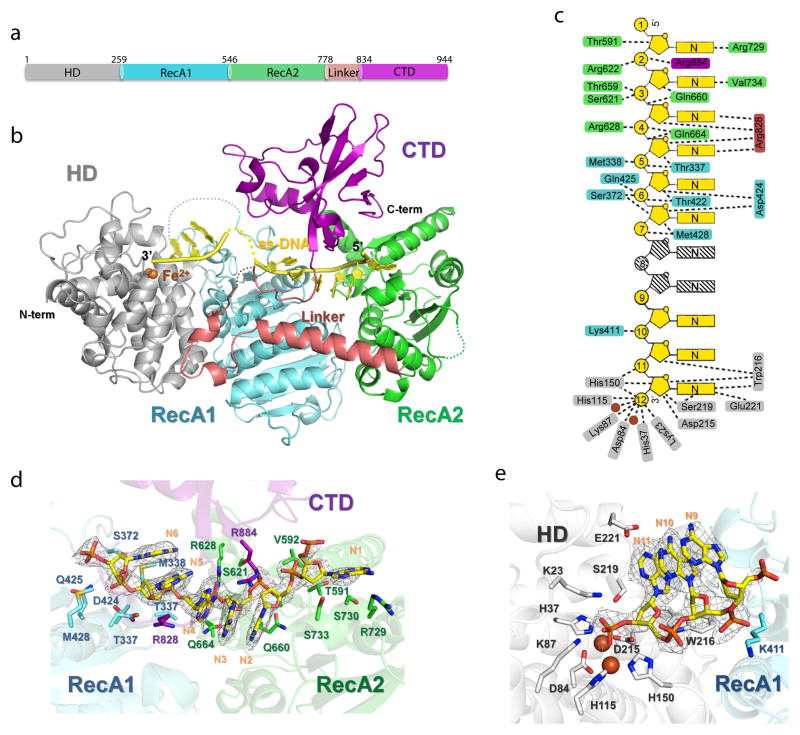
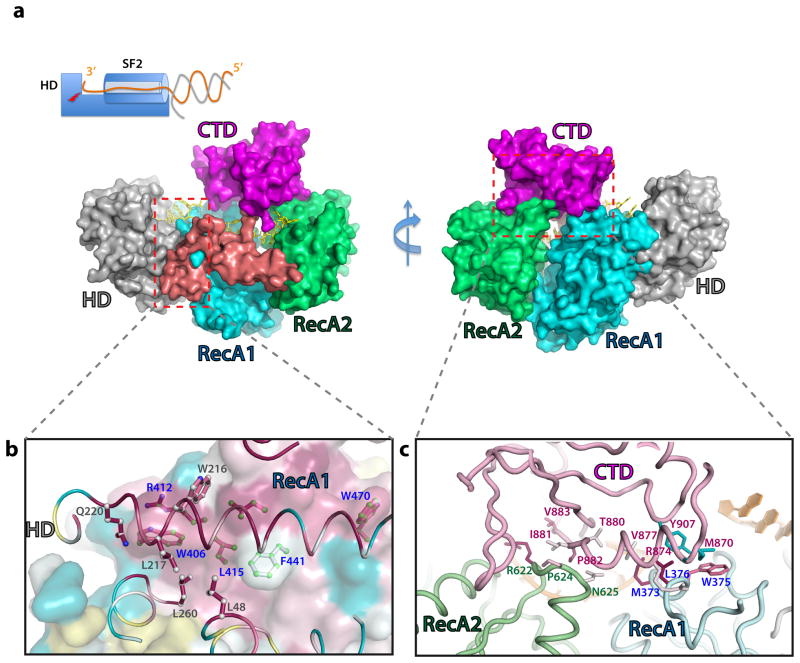
The crystal structure of the Cas3 protein in the Thermobifida fusca CRISPR-Cas Type I-E system was determined at 2.65 Å resolution, with a 12-nt endogenous ss-DNA substrate bound (Fig. 1; Table 1). The structure provides a snapshot of Cas3 where two enzymatic activities are combined to unwind and degrade its DNA substrate (Fig. 1a, b). The SF2 helicase has a classic arrangement of two juxtaposed RecA domains, followed by Cas3-specific structure features, including a long linker helix and an accessory C-terminal domain (CTD) spanning the top. The HD nuclease domain packs against the first RecA-like domain (RecA1) of the helicase through a large, conserved ~4200 Å2 hydrophobic interface. The key interface residues, including W216, L217, and L260 from HD and W406, R412, L415, F441, and W470 from RecA1, are highly conserved (Figs. 2a, S1). The RecA1 and RecA2 at the helicase core are separated by a cleft, where the ATP binding/hydrolysis induced conformational changes are expected to take place17,25. Following RecA2, a horizontally packed linker helix spans the entire helicase back to the HD domain. This is followed by a flexible linker projecting towards CTD, wrapping one side of the DNA-binding platform. The CTD contacts conserved surface loops in each of the RecA-like domains on the opposite side of the platform (Fig. 2b), burying a total surface area of ~2020 Å2, and leading to the formation of a closed ss-DNA threading channel.
Figure 1. Overall structure of ss-DNA-bound T. fusca Cas3 protein.
(a). Domain organization and (b) overall structure. HD, RecA1, RecA2, linker, and CTD domains and the bound ss-DNA substrate are colored in silver, cyan, green, pink, magenta and yellow, respectively. This coloring scheme is followed throughout figures. (c) DNA recognition diagram. Contacts from Cas3 residues to base, ribose, or phosphate of ss-DNA are illustrated in dashed lines. Two disordered nucleotides are shown in stripes. Views of the Cas3-DNA interaction in the helicase (d) and HD nuclease (e) regions. 2Fo-Fc ss-DNA electron densities (1.5σ) in light grey, protein in cartoon, and DNA-contacting side chains in sticks.
Table 1.
Data collection and refinement statistics (molecular replacement)
| Native | ATP | ADP | AMPPNP | Se-Met | Ta6Br12 soak | |
|---|---|---|---|---|---|---|
| Data collection# | ||||||
| Space group | P21 | P21 | P21 | P21 | P21 | P21 |
| Cell dimensions | ||||||
| a, b, c (Å) | 85.79, 218.54, 123.75 | 87.25, 222.81, 125.09 | 87.21, 222.61, 124.85 | 86.92, 222.06, 124.90 | 86.41, 218.85, 124.03 | 86.45, 218.08, 122.90 |
| α, β, γ(°) | 90.00, 90.00, 105.00 | 90.00, 90.00, 104.10 | 90.00, 90.00, 104.07 | 90.00, 90.00, 104.30 | 90.00, 90.00, 104.45 | 90.00, 90.00, 104.18 |
| Resolution (Å) | 2.65 | 3.34 | 3.12 | 2.93 | 3.5 | 4.8 |
| Rmerge * | 5.8 (37.8) | 6.4 (41.2) | 11.0 (48.8) | 8.2 (39.0) | 12.1 (33.9) | 10.8 (41.2) |
| I / σI | 18.1 (2.3)) | 9.3 (2.6) | 9.3 (3.2) | 9.4 (3.0) | 5.7 (1.5) | 14.7 (3.1) |
| Completeness (%) | 92.1 (86.8) | 99.2 (99.6) | 98.5 (98.3) | 99.1 (99.8) | 94.0 (86.9) | 98.6 (91.7) |
| Redundancy | 2.2 (1.9) | 2.2 (2.2) | 3.0 (2.9) | 2.6 (2.6) | 2.7 (2.0) | 3.8 (3.5) |
| Refinement | ||||||
| Resolution (Å) | 2.66 | 3.34 | 3.12 | 2.93 | 3.5 | 4.8 |
| No. reflections | 115323 | 66466 | 80395 | 97476 | ||
| Rwork / Rfree | 17.3/22.6 | 17.2/22.7 | 17.0/23.6 | 18.2/23.4 | ||
| No. atoms | ||||||
| Protein | 28027 | 27906 | 27922 | 28051 | ||
| DNA | 788 | 788 | 788 | 788 | ||
| Ligand/ion | 0/8 | 124/8 | 108/8 | 124/8 | ||
| Water | 215 | 0 | 0 | 0 | ||
| B factors | ||||||
| Protein | 57.9 | 107.2 | 63.4 | 35.0 | ||
| DNA | 66.2 | 118.4 | 80.6 | 52.3 | ||
| Ligand/ion | 36.9 | 123.2 | 92.2 | 86.7 | ||
| Water | 51.0 | 76.6 | 40.8 | 16.0 | ||
| r.m.s. deviations | ||||||
| Bond lengths (Å) | 0.020 | 0.015 | 0.015 | 0.019 | ||
| Bond angles (°) | 1.41 | 1.56 | 1.51 | 1.49 | ||
A single crystal was used for each structure listed.
Values in parentheses are for highest-resolution shell.
Figure 2. Domain interfaces inside Cas3.
(a) Surface representation of Cas3 structure showing that CTD completely wraps the ss-DNA inside the helicase. Domains are colors as in Figure 1. Left inset: Schematics of DNA unwinding and degradation mechanism inside Cas3. The SF2 helicase feeds the 3′ DNA strand directly to the HD nuclease active site for degradation. (b) Conserved hydrophobic interactions at the HD-RecA1 interface, viewed from the HD side into RecA1. The two domains, HD in cartoon and RecA1 in surface representations, are colored according to the sequence conservation, dark red for the highly conserved residues, cyan for variable residues. The key interface residues are highlighted in sticks, colored according to their conservation. Carbon atoms from the HD and RecA1 side chains are colored in white and light green, respectively. (c) CTD-ReaA1 (in pink and cyan, respectively) and CTD-RecA2 (in pink and pale green) interfaces. Contacting residues are highlighted in sticks, colored according to their conservation.
The Path to degradation inside Cas3
A ss-DNA substrate spanning both enzymatic moieties is captured in the pre-cleavage state in the crystal structure, providing insights about the concerted DNA unwinding and degradation actions of Cas3. This substrate is endogenous DNA in origin, co-purified with the Cas3 protein (Fig. S2a), and was likely further processed during crystallization. Electron densities are visible for six nucleotides in the helicase region, three in the HD nuclease region, and two can be inferred in between (Fig. 1c–e). The DNA substrate traverses the most conserved surface regions in Cas3 (Fig. S3), and makes multiple sequence-nonspecific contacts along the path (Fig. 1c). The 5′-end of the DNA substrate enters from RecA2 side of Cas3. A short separation helix is inserted between nucleotide (nt-)1 and 2; the conserved R729 and V734 may play a role in committing nt-2 and beyond into the tunnel. The putative separation hairpin (aa715–727), conserved in many SF2 helicase to wedge into the ds-DNA11, is disordered in our structure. The sugar-phosphate backbones of nts-2–4 and 5–7 are contacted by RecA2 and RecA1, respectively, providing two anchoring points to enable the expected inchworm movement in the SF2 helicase (Fig. 1d). Contacts from RecA2 include salt bridge and hydrogen (H)-bond interactions from R622 and Q660 to nt-2, S621 and T659 to nt-3, and R628 and Q664 to nt-4. RecA1-DNA contacts are from T337, M338, and E424 to nt-5; S372, T422, and Q425 to nt-6; and M428 to nt-7. Emerging from the helicase channel, the nucleoside portion of nt-7 and nt-8 is difficult to trace due to lack of defined contacts, but density reappears towards the active site of the HD nuclease (nt-9~11 and phosphate of nt-12) after an ~90° twist at the sugar-phosphate backbone. Here, ss-DNA binding is assisted by Lys411(nt-10), Trp216(nt-10) as well as active site residues and catalytic metal ions (Fig. 1e). The electron density is poorly resolved beyond the labile phosphate of nt-12 at the HD active site, residual electron density suggests that the leaving nucleotide kinks 90° upward to avoid steric clashes (Fig. 1e). This sharp geometry distortion may form the basis for the HD nuclease to disfavor ds-DNA substrates. Overall, the path of ss-DNA substrate nicely demonstrates that Cas3 is an obligated 3′-to-5′ ss-DNase, preferentially accepting its substrate directly from the helicase moiety.
Metal coordination and catalysis mechanisms inside the HD nuclease
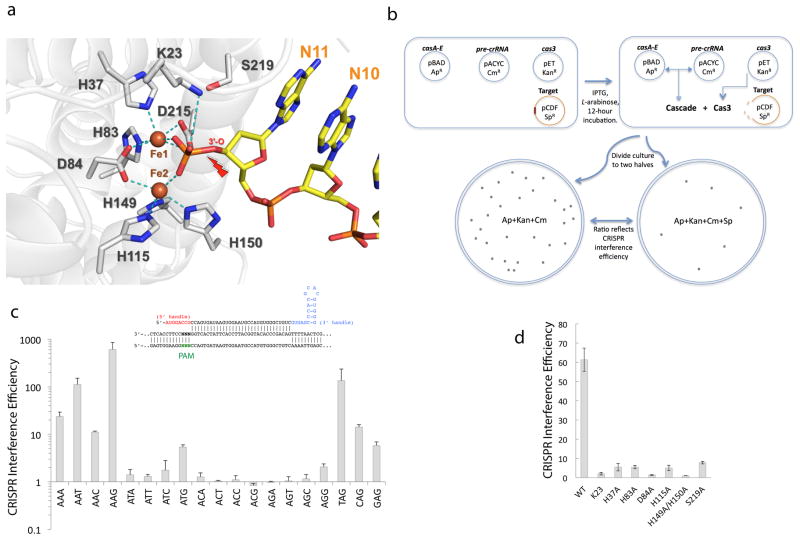
The HD nuclease has been characterized as a transition state metal-dependent single-stranded exo- and/or endo-nuclease15,16,21–23. The metal coordination scheme differs significantly among the published apo crystal structures 21–23 (Fig. S4). This may be the result of active site perturbation due to harsh purifications and/or the lack of ss-DNA substrate21–23. ss-DNase activity could be detected from T. fusca Cas3 with the supplement of Mg2+, Mn2+, or Co2+, and the cleavage product contained a 5′-PO43− and a 3′-OH (Fig. S2b–e). The substrate-bound Cas3 structure provides significantly more mechanistic insights regarding the substrate coordination and catalysis mechanism. The labile 3′-P bond spanning across the HD active site is trapped in the pre-cleavage state, and the two non-bridging oxygen atoms interact strongly with the two irons (most likely in FeII state) in the HD active site (Fig. 3a). The identity of the unexpected irons was subsequently verified using x-ray absorption fine-structure (XAFS) analysis and an anomalous difference map collected at the absorption edge of iron (Fig. S5). The strong affinity suggests that they are constitutive cofactors. The metal ions are coordinated by a cluster of highly conserved active site residues, including the invariable residues H83D84 (HD motif), H37, H115, H149, H150, and a less conserved residue D215. The soft nature of the iron chelating groups dictates the binding of transition state metal ions rather than Mg2+ in these two sites. A similar active site configuration was observed in a recently deposited apo putative HD nuclease domain structure (Fig. S4d), suggesting that the two-iron-architecture is likely conserved in a large percentage of Cas3 proteins. The strategic position of these two irons suggests their involvement in catalysis by i) coordinating a deprotonated water molecule between the two irons for nucleophilic attack, and ii) to stabilizing the negative charge together with K87 during the transition state26. Other HD nucleases likely coordinate transition metals at equivalent positions, although the exact configuration has not been fully resolved in the published apo structures possibly due to harsh purification conditions, weaker binding affinity, and/or the lack of ss-DNA substrate21–23.
Figure 3. Substrate recognition and catalysis in the Cas3 HD nuclease.
(a) View of HD nuclease active site in the pre-cleavage state. Two irons are coordinated by highly conserved HD residues and mediate inner sphere coordination to each of the non-bridging phosphoryl oxygen atoms in the leaving nucleotide (nt-12). Cleavage site is marked by a red arrow. (b) Schematics of the in vivo CRISPR interference assay. See online methods for details of the procedure. (c) Evaluation of PAM sequences using in vivo CRISPR interference assay. The consensus is 5′-WAK, and the strongest PAM 5′-AAG was used in subsequent assays. Averages and Standard Deviations (s.d.) from independent triplicates (n=3) are shown for data points here and in Fig. 3d and 4c. (d) HD active site mutagenesis using the same assay with shorter Cascade+Cas3 induction time (see supplemental methods for details).
Define the function of HD active site residues using in vivo CRISPR interference assay
An in vivo CRISPR interference assay was set up in E. coli to evaluate the function of HD active site residues (Fig. 3b). Induction of T. fusca Cascade and Cas3 expression led to efficient CRISPR interference when the target plasmid bears a protospacer adjacent to a strong Protospacer Adjacent Motif (PAM) sequence (5′-WAK; 5′-AAG being the most potent); interference dropped to background level with a weak PAM present (Fig. 3c). Knocking out either Cas3 HD nuclease or helicase activity by previously characterized mutations (D84A and D451A, respectively, and the D84A/D451A double mutant) reduced the crRNA-mediated plasmid degradation process to the background level, suggesting that both enzymatic activities are essential in CRISPR interference (Figs. 3d, 4d). Based on the new structural insight, HD residues involved in iron coordination (H37, H83, H115, and H149/H150), or ss-DNA binding (K23) were subjected to alanine scanning mutagenesis, and all resulted in dramatic loss-of-function phenotype similar to D84A mutant, confirming their essential role in the HD nuclease (Fig. 3a). The conserved residues S219 is strategically located to the 3′ leaving oxygen of the substrate and may be involved in its protonation. S219A mutation led to a significant loss-of-function in the in vivo assay (Fig. 3c).
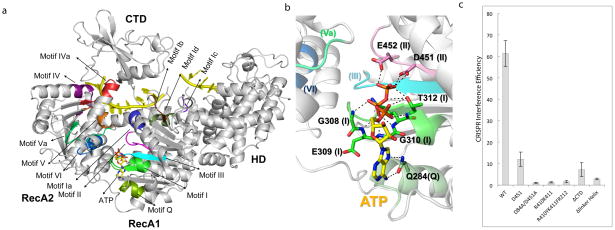
Figure 4. SF2 Helicase dissected.
(a) Mapping of the common SF2 motifs and Cas3-specific motifs onto the ATP-bound Cas3 structure. (b) Close-up of the ATP-binding interactions. Motifs are shown in parenthesis. More details are in Fig. S8. (c) Helicase region mutagenesis done using the same procedure as in Fig. 2c.
Essential features and ATP binding mode inside the Type A SF2 helicase of Cas3
Our crystal structure reveals that the nine of the twelve typical sequence commonly found in SF2 helicases are well conserved in Cas3 motifs (I-VI, Ia-d, IVa, Q)17,25,27 (Fig. 4a). Binding of ss-DNA involves a subset of these motifs (Ia, 1b, 1d, III, IV, IVa, and V) (Fig. S6a). Motif Ic is uniquely found in Cas3 and is essential in coordinating the HD and helicase activities (see below). Other Cas3-specific structure features include a sideway parked linker helix and the CTD domain that follow the helicase core, both were shown to be functionally important for Cas3 function (see below).
ATP, its non-hydrolyzable analog AMPPNP, and ADP were soaked into the Cas3 crystals and the resulting structures were determined at 3.1, 2.9, and 3.1 Å resolutions, respectively (Figs. 4b, S6b–d; Table 1). Their binding did not lead to full-scale conformational changes28 due to crystal lattice trapping (Fig. S6e–f). Recognition of the adenosine moiety is identical among three structures (Figs. 4c, S6b–d): it involves bidentate H-bonds to the Hoogsteen edge of adenosine from Q284 (Motif Q), base stacking from L277, and a H-bond to 2′-OH from E313. The first interaction disfavors binding of GTP to the same pocket, and the last one disfavors the binding of dATP. A second stacking interaction to the bottom of the adenine ring, observed in some helicase structures, is noticeably missing, likely from uncompleted conformational changes28. The triphosphate moiety is accommodated by a hairpin loop in Motif I (Walker A) through multiple electrostatic mainchain contacts from G308, E309, and G310. In the ATP-bound structure, the triphosphate moiety adopts an extended conformation, pointing towards D451 and E452 in Motif II (DEAH, Walker B motif). Mg2+ was supplemented in substoichiometric amounts and was not observed near the beta and gamma phosphates. However, residues responsible for positioning the Mg2+, such as T312 and D451 in Motifs I and II, as well as E452, the residue responsible for coordinating the attacking water molecule, are well conserved in space, suggesting that Cas3 would function as a typical SF2 helicase.
Functional importance of the Cas3-specific helicase features
Since these SF2 motifs have been well-defined, we focus the mutagenesis on Cas3-specific structure features. Motif Ic is uniquely found in Cas3 and its strategic position suggests an involvement in coupling the helicase and nuclease activities. Indeed, R410A/K411A and R410Y/K411F/R412A targeting Motif Ic both led to strong loss-of-function phenotype similar to that of HD active site mutants (Fig. 4c). Replacing the linker helix (aa777-815) with a Gly/Ser linker was also more detrimental than the helicase null mutant, suggesting an essential function for this structure feature (Fig. 4c). Deletion of CTD (aa819-924) reduced CRISPR interference to the helicase null mutant level (Fig. 4c).
Biochemical reconstitution of the Cascade-Cas3 interaction
A missing link in Type I CRISPR system is the definition of specific interactions that lead to the Cas3 recruitment by a target DNA-bound Cascade. To address this, we reconstituted the Cascade-Cas3 interaction using electrophoretic mobility shift assay (EMSA) for deeper mechanistic dissections. Unlike the E. coli Cascade, the purified T. fusca Cascade mostly dissociated into free CasA and a crRNA-containing CasB-E sub-complex in the absence of a proper substrate (Figs. 5a–c); intact Cascade was only stable when bound to a protospacer and PAM containing ds-DNA, a bubbled substrate, or a nicked substrate mimicking an R-loop intermediate (Figs. 5d, 6a). The affinity of Cas3 alone for various DNA substrates was rather weak, and the rate-limiting step appeared to be at the substrate binding/exchange step (Fig. 6a, right 3 lanes). The nicked R-loop mimic supported the most stable Cas3-Cascade interaction, and Cas3 was specifically recruited to the Cascade when a series of conditions are met: 1) an intact Cascade where CasA was stably bound, 2) an optimal PAM sequence in the substrate; PAM substitution in either or both strands of the DNA substrate disrupted binding, presumably due to CasA dissociation (Fig. 6a, lanes 10–18), and 3) a 3′-overhang of at least 10-nt long in the non-target strand DNA; no Cas3 binding was detected when a 5-nt overhang was used (Fig. 6a, lanes 1–9). The 10-nt overhang requirement is roughly consist with the 11-nt ss-DNA inside our Cas3 crystal structure, suggesting that stable Cas3-Cascade association likely requires the non-target DNA strand to thread through the helicase to reach the HD active site in Cas3 (Fig. 1). Inclusion of AMPPNP or use of D451A helicase mutant strengthened Cas3-Cascade interaction, whereas ATP incubation led to much weaker interaction, presumably because the elevated helicase activity allowed Cas3 to clear the Cascade (Fig. 6b).
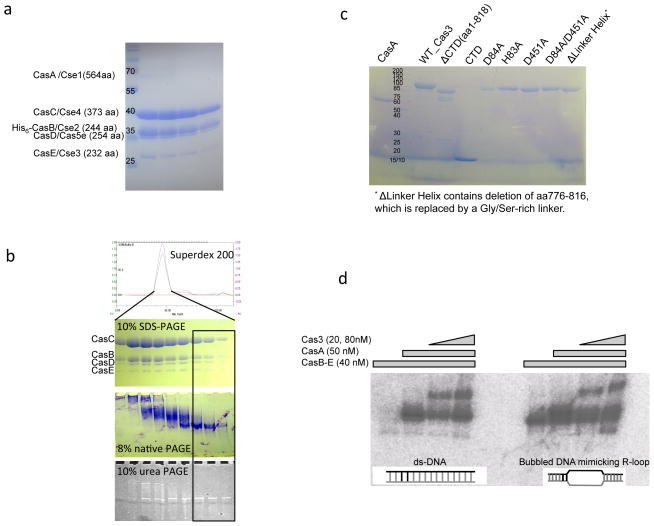
Figure 5. Reconstitution of Cas3-Cascade/R-loop interaction in vitro from purified components.
(a) Cascade (CasA-E) purification. SDS-PAGE of the Size Exclusion Chromatography (SEC) peak containing the co-expressed T. fusca Cascade. The protein identity in each band was verified by mass spectrometry. The expected stoichiometry of CasA-E and crRNA is 1:2:6:1:1:1. CasA is significantly sub-stoichiometric. (b) CasB-E Cascade sub-complex purification. Protein and RNA qualities were checked on the SEC elusion profile, 10% SDS-PAGE, 6% native PAGE, and 10% urea-PAGE. Note that even though the subunit stoichiometry was roughly the same across the SEC peak, the boxed half of the SEC peak contained monomeric CasB-E/crRNA complex, whereas the front half of the peak contained various aggregated forms. (c) Quality of CasA and Cas3 wild type and mutant proteins used in the biochemical studies on SDS-PAGE. (d) Reconstitution of Cascade/ R-loop and Cas3/Cascade/R-loop complexes. Substrate in the EMSA is either a perfectly base-paired ds-DNA or a bubbled DNA substrate mimicking an R-loop opened at the spacer region. R-loop formation with the former substrate requires CasA present. Cas3 interacts with the programmed Cascade robustly only when CasA is present.
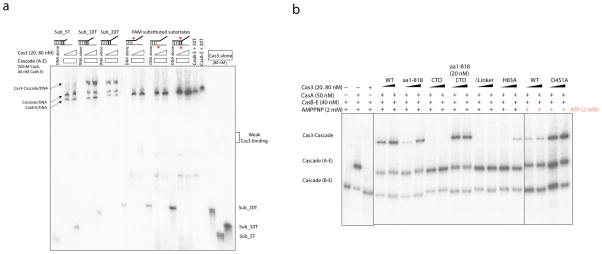
Figure 6. Cascade-Cas3 interaction reconstituted in vitro.
(a) 6% native PAGE showing stable Cas3-Cascade association. The bottom half of the gel is 12% to resolve free DNA. Substrates mimic a nicked R-loop structure. Non-target strand (in black) is 5′ 32P-labeled. Wrong PAM (red asterisk) in either or bother strands of DNA dissociates CasA, and Cas3 binding is lost in turn. (b) Cas3-specific structure features critically influence Cascade-Cas3 interaction. Sub_10T was used in all assays. Lanes 1–3: Cas3-Cascade binding requires CasA present. Lanes 4–15: The linker helix deletion in Cas3 disrupts Cascade binding. CTD deletion and HD active site mutation weaken the interaction. Lanes 16–19: ATP presence weakens Cas3-Cascade interaction, helicase null D451A restores the effect.
The function of the Cas3-specific structure features became apparent in the Cascade-Cas3 interaction assay (Fig. 6b). Cas3 lacking CTD (aa1-818) exhibited weakened affinity for Cascade. Complementing this truncation with CTD in trans restored its Cascade binding affinity to the wild type level, which is consistent with our structure model suggesting that the CTD domain regulates HD nuclease activity by functioning as a substrate filter. More importantly, Cascade binding was completely lost when the linker region in Cas3 was replaced by a flexible linker, pointing to a strong involvement of this region in Cascade interaction. Finally, HD nuclease mutant H83A also exhibited weaker affinity for Cascade, consistent with an accessory function of the HD domain in stabilizing the Cascade-Cas3 interaction by grabbing tighter to the substrate.
Discussion
The most wide-spread Type I CRISPR-Cas system can be further classified into six different subtypes (I-A to I-F), each encoding a unique set of subtype-specific genes9. While the composition of the target-searching Cascade complex varies significantly among these subtypes, the effector gene cas3 is universally conserved in all Type I systems, underlining its functional importance. The analysis of the Cas3 crystal structure in this study reveals the special arrangement of the nuclease and helicase activities inside the ~100 kDa Cas3 protein, sheds light into its DNA unwinding and degradation mechanism. The biochemical reconstitutions further define the set of conditions leading to the Cas3 recruitment to the Cascade-bound DNA substrate.
While the HD nuclease inside Cas3 was correctly defined as a transition state metal dependent ss-DNase, its exact catalysis mechanism remained elusive15,16,22–24. Our Cas3 structure reveals the coordination of two constitutively bound catalytic iron cofactors and their interactions with the ss-DNA substrate. Questions remain about why the ss-DNA substrate was not cleaved inside the Cas3 crystal. A possibility is that only FeII, but not its oxidized FeIII form, is capable of supporting the HD nuclease activity, and that our T. fusca Cas3 may have been slowly inactivated during purification and crystallization due to FeII oxidation, leading to the observation of a trapped ss-DNA in its precursor form. Efforts are underway to investigate this possibility, and to provide further snapshots of Cas3 in various stages of the enzymatic cycle.
The structure features commonly found in SF2 helicases are well conserved in the Cas3 helicase region17,25,27. This suggests that Cas3 would function like a typical SF2 helicase to unwind ds-DNA17,25,27,17,25,27,18,25,27,17,24,26. SF1 and SF2 monomeric helicases has been shown to unwind ds-DNA using an inchworm mechanism17,25,27. That is, binding of ATP to RecA1 would induce a rotation and closing movement in RecA2 to orient important residues for ATP hydrolysis. This structure compression is then relieved after ATP hydrolysis and dissociation. The ATP induced conformational change, coupled with alternate tight-loose grip on ss-DNA from the two RecA-like domains, leading to the inchworm movement of the helicase on one-strand of ds-DNA to unwind ds-DNA. The compression of the helicase upon ATP binding was not captured in the Cas3 crystals presumably due to crystal lattice trapping, however, hints of such movement is shown when we compare the four Cas3 molecules in the asymmetric unit of the crystal lattice, since they undergo different extent of rigid-body movement in the crystal lattice (Fig. S6e). A hinge motion in the ATP-binding cleft results in a closing-in motion in molecule D, such that its HD domain shifts as much as 3 Å, causing the active site metal ions and ss-DNA substrate to move ~1.7 Å (Fig. S6f). Such movements give hints about the consequence of ATP binding/hydrolysis induced conformational changes in the Cas3 helicase region, and point to a concerted mechanism to couple the inchworm movement of the helicase with the substrate translocation in the HD nuclease active site.
Taken together, the Cas3 crystal structure enabled us to resolve some of the most important mechanistic questions in the Type I CRISPR-Cas system. The two enzymatic activities in Cas3 work in a concerted fashion at several levels. Apart from unwinding and feeding the substrate into the HD nuclease, the helicase moiety also initiates the recruitment of the latter activity through physical interactions with the CasA component of the R-loop presenting Cascade (Fig. 6b). Subsequently, the HD nuclease strengthens the Cascade-Cas3 interaction with its strong ss-DNA binding affinity at the active site (Fig. 6b). Cascade-Cas3 interaction only occurs when a cascade of conditions are met, reflecting an evolutionary pressure to tightly regulate the activation of Cas3-mediated DNA degradation.
In consideration of the initial steps involving Cascade-mediated recruitment and activation of Cas3 at the R-loop region, questions remain about how Cas3 is loaded to the non-target strand in the R-loop structure prior to the first cleavage event, as threading a looped ss-DNA without open 3′-end through the caged Cas3 helicase would be topologically challenging. Although it is possible that the HD nuclease makes the first cut bypassing the helicase, the existing data15,16 are more consistent with a model in which the CTD of Cas3 transiently dissociates, possibly triggered by interaction with the Cascade, to allow ss-DNA placement into the helicase. Such accessory domain movement is observed in other helicases, as exemplified in UvrD28. Following CTD re-association, the HD nuclease makes the first cut and converts Cas3 into the processive conformation observed in the crystal structure, and ATP hydrolysis further drives 3′-to-5′ unwinding and degradation. An alternative model to resolve the topological challenge involves the retraction of the long flexible linker sequence (aa 816–832) beyond the HD nuclease to expose the side of the ss-DNA recruitment channel along Cas3. Single molecule fluorescent resonance energy transfer (FRET) experiment and co-crystal structures of the Cas3 bound to the target presenting Cascade complex will be required to distinguish these two competing models.
Online Methods
Expression, Purification, crystallization and Diffraction of Cas3 protein
The full-length T. fuaca cas3 (Accession No.:Q47PJ0, Gene name: Tfu_1593) fused to a N-terminal His6-tag was cloned into the pET28-b expression vector between NdeI and XhoI restriction sites. Escherichia coli BL21 (DE3) star cells containing the sequence-verified Cas3 construct were grown in LB medium at 37 °C to O.D.600 of 0.6, and the protein expression was induced by the addition of 0.5 mM isopropyl b-D-1-thiogalactopyranoside (IPTG), and the further culturing at 18 °C for 16 hours. The harvested cells were disrupted by sonication in buffer A (20 mM Tris-HCl pH 7.5, 300 mM NaCl, 10% glycerol, and 2 mM β-mercaptoethanol). The supernatant after centrifugation was applied onto the Ni-NTA column pre-equilibrated in Buffer A. Following 10–15 column volumes of wash in Buffer A plus 5 mM imidazole, Cas3 protein was eluted with Buffer A plus 250 mM imidazole. After dialysis into low-salt buffer, Cas3 was further purified on MonoS column, and the peak fractions were concentrated and purified on Superdex 200 column. The peak fractions were concentrated to 20 mg/ml and flash-frozen at 80 °C. A similar purification procedure was followed to obtain Se-methionine derived proteins. Cas3 crystals were grown using hanging drop vapor diffusion method by mixing 1 μl of protein solution with 1 μl of mother liquid (100 mM MES, pH 6.0, and 24% PEG 4000) at 18 °C. ATP, AMPPNP, and ADP bound structure was determined from crystals soaked in the well solution supplemented with 2 mM of each compound and 1 mM MgCl2 over night. Diffraction data were collected through mail-in data collection service at beam lines in APS NECAT, processed using Program HKL200029. Mutant Cas3 plasmids were constructed by site-directed mutagenesis.
Cas3 mutants were constructed using site-directed mutagenesis. Proteins were purified using Ni-NTA and MonoQ columns, concentrated to over 10 mg/ml, and stored at −20°C in 50% Glycerol.
Phasing, model building and structure refinement
The initial experimental phases were determined by single-wavelength anomalous dispersion (SAD) method using a data set collected at the tantalum L-III absorption edge from a crystal derivatized with Ta6Br12 cluster. The positions of 4 cluster sites were located using SHELXC/D30. Low-resolution SAD phasing was carried out at 6.0 Å using Phenix.autosol31, in which the four Ta6Br12 clusters were treated as superatoms. The resulting phases were then improved and extended to 3.0A using Phenix.autobuild32 by applying the noncrystallography symmetry (NCS) identified by Molrep33. The Ta cluster phases allowed the placement of some alpha-helices and beta-sheets into the electron density map. To reliably trace the entire Cas3 molecule, we switched to the 3.5 Å Se-SAD data set, where 51 different Se sites were identified from the anomalous difference electron density map by combining the Se-SAD anomalous diffraction data set with the Ta phases. These sites provided enough phasing power to generate a traceable electron density map and partial models for all four copies of Cas3 molecules using the automated experimental phasing and model building procedures in Phenix.autosol31 and Phenix.autobuild32. The phases were further improved by density modification using DM34, and the model was manually rebuilt into the electron density map using Coot35. The result model was refined using REFMAC536 and Phenix.refine37. The Rfree statistics of the final structure models are well below 30%, with good r.m.s. deviations in bond length and bond angle. The backbone conformations of ~93–96% of the protein residues are in the most favored region in the Ramachandran plot, ~4–7% in the allowed region, and none in the disallowed region. The geometry of coordinates was validated by MOLPROBITY38. All figures were generated using PyMOL (http://www.pymol.org).
X-ray absorption fine-structure (XAFS) analysis for metal ion identity
Energy-dispersive X-ray spectroscopy (EDS) was carried out using a silicon drift detector (model X-123SDD, Amptek Inc, USA) at NE-CAT 24ID-C beam line. The built-in multi-channel analyzer of X-123SDD was calibrated with known fluorescent emission lines of several metals. The gain of the detector was set to 75%, corresponding to an energy range of 0–16.7keV. EDS experiments were carried out with incident X-ray energy of 12.66keV, just above the K absorption edge of Selenium. First, EDS spectrum was recorded for 127.85 seconds with X-rays incident on the crystal in the cryo-loop (Fig. S5a). As a negative control experiment, EDS was also recorded for 190.7 seconds on the cryo-loop, but away from the crystal (Fig. S5b). To verify the presence of Iron in the crystal, near edge X-ray absorption fine structure spectroscopy (XAFS) was further carried out on the crystal (Fig. S5c) and away from the crystal (Fig. S5d) by tuning the incident X-ray energy close to K absorption edge of Iron. The anomalous-scattering components, f′ and f″ were derived from XAFS spectrum using the program CHOOCH39.
Expression and Purification of T. fusca Cascade complexes
T. fusca cascade operon encoding casA to CasE genes (Tfu_1592 to Tfu_1588) was PCR amplified from the genomic DNA and cloned into a pET expression vector. A His6 tag was inserted via mutagenesis into the N-terminus of casB for Ni-NTA purification. Co-expression of Tfu_Cascade with pre-crRNA expression vector led to a stable crRNA-containing CasB-E sub-complex, with CasA weakly associated. Subsequently, CasA was removed from the expression operon and recombinantly expressed separately as a His6-tagged protein, and purified into homogeneity using Ni-NTA and MonoQ columns. CasB-E sub-complex lacking CasA was purified using Ni-NTA, MonoQ, and Superdex 200 columns (Fig. 5). Even though the Superdex 200 SEC peak fractions contained CasB-E proteins at the correct stoichiometry, the higher molecular weight half of the peak contained contaminating nucleic acids, which resulted in various degree of aggregation as revealed by the 6% native gel (Fig. 5b). The fractions lacking these contaminating nucleic acids were pooled, concentrated to over 10 mg/ml, and flash-frozen in small aliquots in −80°C for biochemical assays.
In vivo Assay for T. fusca Cascade and Cas3 mediated target plasmid loss
We modified the assay from a published protocol14 to recombinantly express the T. fusca Cascade and T. fusca Cas3 for target plasmid destruction in the E. coli BL21_AI cell line, which does not express the E. coli CRISPR and cas operons. The experimental design and the list of plasmids used are detailed in Fig. 3b and Table S1, respectively. All Tfu_Cas3 constructs were pET28b (KanR) based. The Tfu_Cascade expression construct was generated by inserting the T. fusca CasA-E fragment into a pBAD (ApR) vector. The pre-crRNA expression cassette containing four identical CRISPR units under the control of T7 RNA polymerase promoter and terminator was synthesized by GenScript and cloned into the pACYC-Duet-1 (CmR) vector. A 332 bp fragment of λ phage genome containing a matching protospacer and a 3-bp Protospacer Adjacent Motif (PAM) was cloned into the pCDFDuet-1 (SmR) vector to serve as the target DNA. All plasmids were sequence verified. The pET28b_Cas3, pBAD_Cascade, pACYC_CRISPR and pCDF_target plasmids were transformed into BL21_AI competent cells, grown on LB plates containing kanamycin (50 μg/ml), ampicillin (100 μg/ml), streptomycin (50 μg/ml) and chloramphenicol (34 μg/ml).
In the PAM sequence evaluation assays, the four-plasmid-containing BL21_AI cells from a single colony were cultured at 37 °C in non-selective LB media to O.D.600 of 0.3, at which point the expression of T. fusca Cascade, Cas3, and pre-crRNA was induced for 12 hour by the addition of 0.5% L-arabinose and 2.5 mM IPTG. Cell culture was then divided into two equal volumes and plated onto Kan+Ap+Cm LB plates (non-selective for pCDF_target) and Kan+Ap+Cm+Sm plates (selective for the target plasmid) in series of dilutions. The number of colonies on each plate was counted after overnight incubation at 37 °C. The CRISPR interference efficiency was reflected in the ratio of colony forming units on the non-selective versus selective plates. 10-fold titration series were routinely plated to capture the dynamic range. Each experiment was repeated three times to calculate the standard deviations.
All Cas3 mutants were evaluated following a slightly different protocol. The target plasmid contains the strongest PAM sequence, 5′-AAG. After induction of Cascade and Cas3 expression, the E. coli cells were incubated for 6 hours before plating, instead of 12 hours used in the PAM sequence assays. The shorter induction time led to slightly reduced dynamic range in CRISPR interference efficiency. However, the disadvantage is outweighed by a significantly reduced error spread, making the mutagenesis data more reliable for quantitative comparison.
in vitro reconstitution of Cas3-Cascade/R-loop interaction
DNA oligos (Table S1) were chemically synthesized and gel-purified. The non-target DNA strand was 32P-labeled and annealed with the target strand at 1:2 molar ratio by incubating at 65 °C for 10 minutes followed by on-bench cooling. The Cascade/R-loop complex (or mimics) were assembled by incubating the annealed DNA substrate with 40 nM CasB-E and 50 nM CasA at 25 °C for 20 minutes in a buffer containing 20 mM HEPES pH7.5, 100 mM NaCl, 10 mM MgCl2, 10 μM FeCl2, 2 mM AMPPNP (or ATP), and 5% Glycerol. Wild-type or mutant Cas3 protein were then introduced at two different concentrations (20 nM or 80 nM), and the reaction mixture was incubated at 37 °C for an additional 30 minutes. The reaction mixtures were then electrophoretically separated on a 6% Tris/Borate/EDTA (TBE) native PAGE in the cold room. The gel was exposed to a storage phosphor screen, and the radiograph signals were recorded by a Typhoon 9200 machine. All controls were done in the same buffer condition following the same incubation procedure.
Nuclease assay
Nucleic acid cleavage reactions were performed at 37 °C for 60 min in a buffer containing 10 mM Tris–HCl pH 8.0, 60 mM KCl, 10 mM MgCl2, 1mM dithiothreitol (DTT), where 0.1 μM 5′-Cy5-labeled ss-DNA was incubated with 1 μM of wild type or mutant Cas3 proteins. Reactions were initiated by the addition of protein, and stopped by the addition of 3X stop buffer [67.5 mM EDTA, 27% (v/v) glycerol, and 0.3% (w/v) SDS]. The reaction mixture was separated by electrophoresis on 10% (w/v) 8 M urea polyacrylamide gel (PAGE), and the Cy5 fluorescent signal was recorded using Typhoon 2900. The metal-dependency experiment was carried out in essentially the same condition, except the Mg2+ was substituted with 10 mM other metal ions.
Supplementary Material
Acknowledgments
This work is supported by NIH grants GM-086766 and GM-102543 to AK and Korean Postdoctoral Fellowship NRF-2010-357-C00106 to KHN. Use of NE-CAT and MACCHESS beamlines was supported by NIH grants GM103403 and GM103485, respectively. We thank Yujie Chen and Kay Perry for technical help, John van der Oost, Jason Grigg, Robert Hayes, and Ian Price for helpful discussions.
Footnotes
Database accession numbers
The coordinates for Cas3, Cas3-ATP, Cas3-AMPPNP, and Cas3-ADP have been deposited in the Protein Data Bank under accession numbers 4QQW, 4QQX, 4QQZ, and 4QQY, respectively.
Author Contributions Y.H, K.H.N., A.K., R.R., and I.K. collected diffraction data, K.H.N., L.W., and A.K. determined the structure. F.D., H.L., A.K., Y.X., Y.H., and S.Z. performed the biochemical analyses. A.K. designed the research and wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Current opinion in microbiology. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 3.Jore MM, Brouns SJ, van der Oost J. RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS computational biology. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biology direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 7.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nature reviews. Microbiology. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson RN, Lavin M, Carter J, Wiedenheft B. Fitting CRISPR-associated Cas3 into the Helicase Family Tree. Curr Opin Struct Biol. 2014;24C:106–114. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jore MM, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature structural & molecular biology. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 13.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Molecular cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinkunas T, et al. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. The EMBO journal. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulepati S, Bailey S. In Vitro Reconstitution of an Escherichia coli RNA-guided Immune System Reveals Unidirectional, ATP-dependent Degradation of DNA Target. The Journal of biological chemistry. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annual review of biochemistry. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 18.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 19.Taylor DW, et al. Substrate-specific structural rearrangements of human Dicer. Nature structural & molecular biology. 2013;20:662–670. doi: 10.1038/nsmb.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry JJ, et al. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nature structural & molecular biology. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- 21.Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) The Journal of biological chemistry. 2011;286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beloglazova N, et al. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. The EMBO journal. 2011;30:4616–4627. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinkunas T, et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. The EMBO journal. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochstrasser ML, et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W. Lessons learned from UvrD helicase: mechanism for directional movement. Annual review of biophysics. 2010;39:367–385. doi: 10.1146/annurev.biophys.093008.131415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W. Nucleases: diversity of structure, function and mechanism. Quarterly reviews of biophysics. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nature structural & molecular biology. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 28.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta crystallographica. Section D, Biological crystallography. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta crystallographica. Section D, Biological crystallography. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta crystallographica. Section D, Biological crystallography. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta crystallographica. Section D, Biological crystallography. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 34.Cowtan KD, Main P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta crystallographica. Section D, Biological crystallography. 1993;49:148–157. doi: 10.1107/S0907444992007698. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods in enzymology. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans GP, RF CHOOCH : a program for deriving anomalous-scattering factors from X-ray fluorescence spectra. J Appl Cryst. 2001;34:82–86. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.