It has been over two decades since the first preimplantation genetic diagnosis (PGD) for a monogenic disorder was performed [1], and methods have evolved to include a wide variety of techniques [2]. Among the most important advances was the incorporation of genotyping of linked informative markers near the mutation in order to avoid misdiagnosis from a phenomenon known as allele drop out (ADO). ADO occurs when two alleles are present, but the PCR-based test only detects one of the two, which can result in misdiagnosis of a monogenic disorder. However, by evaluating nearby linked informative polymorphisms, this type of error can be avoided since it is less likely to occur twice in the same test [2, 3].
The most common type of polymorphism used as a linked marker in the PGD setting is the short tandem repeat (STR). One advantage of the STR is that it is often multi-allelic, providing a high likelihood of being informative for a given family. However, a potential disadvantage is the relatively low frequency of STRs throughout the human genome [4, 5]. This becomes an important issue as genotypes of markers too far away from the mutation could be misinterpreted as a result of recombination. Specifically, if recombination occurred between the marker and the mutation, the genotypes could be misinterpreted as consistent with an ADO event at one of the two loci. This is even more of a concern when genes near telomeres are evaluated since the recombination frequency is considerably higher than other regions on the chromosome [6, 7].
In contrast to the STR, the single nucleotide polymorphism (SNP) is the most common polymorphism in the human genome, and therefore more likely to provide a marker within 1 Mb of the mutation, as recommended by the European Society for Human Reproduction and Embryology (ESHRE) PGD Consortium [2]. We recently reported the use of TaqMan PCR based allelic discrimination to genotype embryos for a single gene disorder in parallel with comprehensive chromosome screening [8]. This approach provides an opportunity to genotype SNPs as informative markers, instead of STRs, using a quantitative real time (q)PCR-based approach. This case report illustrates the particular advantage of qPCR, by identifying STR-based misdiagnoses due to recombination near the mutation.
Methods
This case involved a couple indicated for PGD since they were both carriers of the R1264H mutation in the Regulator of Telomere Length 1 (RTEL1) gene. They discovered their carrier status after the birth of their first and only child in 2009, who was homozygous for the mutation and was affected with Dyskeratosis Congenita. This disorder affects multiple organ systems, and can result in bone marrow failure, aplastic anemia, thrombocytopenia, osteoporosis, and liver and pulmonary fibrosis [9, 10]. Their daughter had been hospitalized since she was 6 months old and passed away at 2 and half years old. The female partner was 29 year old and the male partner was a 37 year old at the time of IVF for PGD.
The couple underwent routine controlled ovarian hyperstimulation through an antagonist protocol with intracytoplasmic sperm injection. Of the 46 oocytes retrieved, 17 made it to the blastocyst stage. Each embryo was biopsied twice on day 6. The first biopsy was used to perform comprehensive chromosome screening (CCS) using quantitative real-time PCR as previously described [11]. A second biopsy was used to diagnose Dyskeratosis Congenita at a reference laboratory using conventional methods of STR fragment size and Sanger sequencing as previously described [12]. After biopsies were performed all the embryos were cryopreserved to allow time for the reference laboratory to complete single gene disorder (SGD) analysis and provide a report. Upon receipt of the SGD report with unusually high rates of ADO and no results, the excess DNA from the CCS procedure was used to evaluate linked informative SNPs near the mutation, which were identified using NspI SNP arrays (Affymetrix Inc., Santa Clara, CA) on the couple. Phase was established using TaqMan allelic discrimination (Life Technologies Inc., Foster City, CA) of the informative SNPs on DNA from the couple’s affected daughter. A TaqMan assay was also developed to directly test the mutation through allelic discrimination in parallel as previously described [13]. The TaqMan assays for the linked markers and the mutation were used in a multiplex preamplification PCR reaction (Life Technologies Inc.) with the excess CCS DNA as template. Individual reactions with each individual primer set were performed using qPCR on the preamplified DNA as previously described [8]. Each Taqman assay allele specific probe was labeled with either a FAM or VIC dye in order to detect the major and minor SNP allele, respectively, and genotypes were designated as such in the results tables and figures.
This study was conducted under IRB approval and with patient consent.
Results
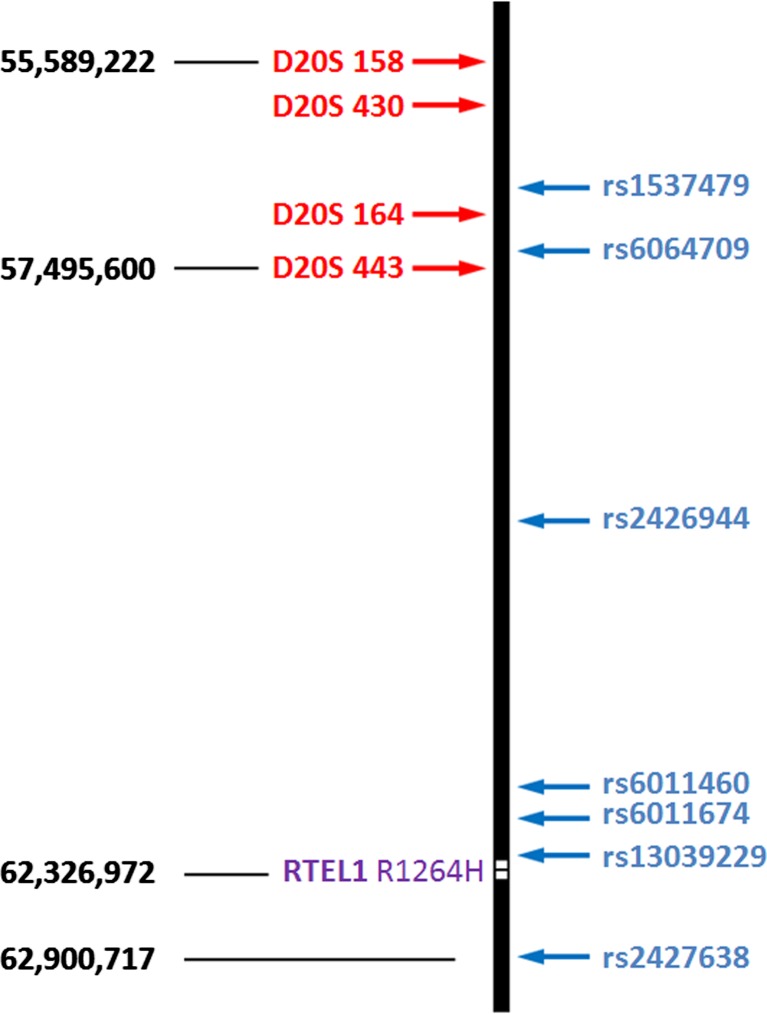
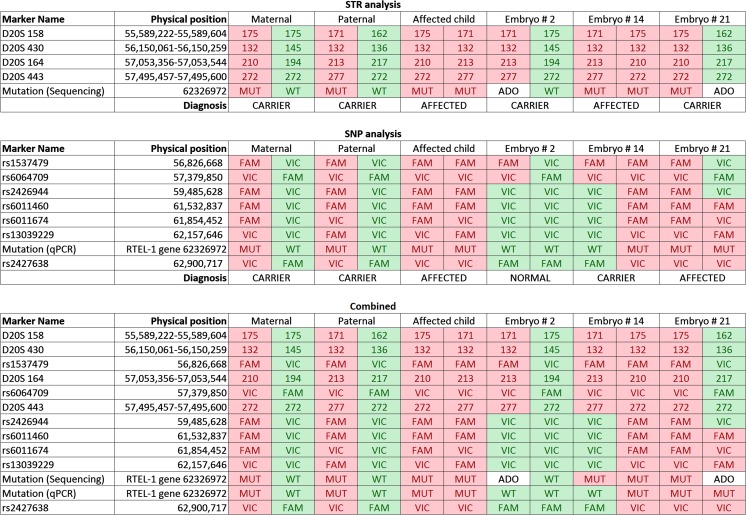
CCS indicated that 12/17 (70 %) of the embryos were euploid and potential candidates for transfer (Table 1). The PGD report from the reference laboratory using conventional methods of STR and Sanger sequence analysis indicated an ADO rate of 8 % (14/170) and a non-diagnosis rate of 18 % (3/17), despite having been performed on trophectoderm biopsies. Given the unusually high rates of ADO and no results, analysis of the SGD on the excess DNA from CCS was performed using qPCR for allelic discrimination of informative SNPs and the mutation. Seven informative SNPs were evaluated including 4 between the nearest STR marker (which was 4.8 Mb away from the mutation) and one on the telomeric side of the mutation (Fig. 1). In each of the 4 cases that the reference laboratory interpreted the mutation analysis as having been affected by ADO, the SNP based methodology demonstrated that recombination occurred between the nearest STR and the mutation (Supplementary Table 1). This led to a reference laboratory misdiagnosis rate of 21 % (3/14), including an embryo diagnosed as a carrier that was actually affected (Fig. 2). Interestingly, the recombination rate within the 7.3 Mb of interrogated sequence was 53 % (9/17). Fortunately, the patient had 4 embryos which were diagnosed as normal by both laboratories, one of which was selected for transfer and resulted in an ongoing pregnancy.
Table 1.
Results of CCS, STR, SNP, and recombination analyses in embryos at risk of Dyskeratosis Congenita
| Embryo number | CCS | STR/sequencing analysis | SNP qPCR analysis | Recombination |
|---|---|---|---|---|
| 1 | 46, XY | Carrier | Carrier | No |
| 2 | 46, XY | Carriera | Normal | Yes |
| 3 | 46, XX | Normal | Normal | Yes |
| 4 | 45, XX,−16 | N/Ab | Carrier | No |
| 5 | 46, XX | Affected | Affected | No |
| 6 | 46, XX | Carrier | Carrier | Yes |
| 7 | 46, XX | Affected | Affected | No |
| 8 | 46, XY, +18,−22 | Normal | Normal | Yes |
| 9 | 45, XX,−11 | Normal | Normal | No |
| 10 | 46, XX | Affecteda | Carrier | Yes |
| 11 | 46, XY | Normal | Normal | No |
| 12 | 47, XY, +18 | Carriera | Affected | Yes |
| 13 | 46, XY | Carrier | Carrier | No |
| 14 | 46, XX | N/Ab | Normal | Yes |
| 15 | 46, XY | Affected | Affected | No |
| 16 | 46, XY | N/Ab | Carrier | Yes |
| 17 | 47, XY, +12 | Affected | Affected | Yes |
aMisdiagnosis, bNo result obtained
Fig. 1.
Locations of linked markers surrounding the RTEL1 gene locus on chromosome 20 (purple). STRs are shown in red, SNPs are shown in blue. Nucleotide positions are based on human genome version 18
Fig. 2.
Results of analysis using each approach for parents, affected child, and misdiagnosed embryos. MT- Mutant; WT- Wild type; ADO- Allele dropout
Conclusions
This case illustrates the particular problem of high rates of recombination near the telomeres of human chromosomes [14, 7] and the impact it can have when performing PGD with linked informative STR markers too far from the mutation. The exact recombination rates approaching the telomeric ends may not be available or reliable from published studies, and in this case the rates of the full region surrounding the gene were not. With the use of technologies which rely upon whole genome amplification and SNP array based analysis, the significant locus dropout from WGA may also prevent the identification of crossovers between the nearest available SNP marker and the mutation [15, 16]. In the case presented here, a misdiagnosis rate of 21 % was identified as a result of excessive STR marker distances, with respect to the mutation locus, failing to detect recombination and inappropriately assuming ADO at the mutation locus. The qPCR approach presented here overcomes these potential limitations allowing for simultaneous analysis of a large commercially available library of linked SNPs near the mutation, the mutation itself, and CCS within 4 hour of obtaining the sample for analysis.
Electronic supplementary material
(PDF 283 kb)
References
- 1.Handyside AH, Lesko JG, Tarin JJ, Winston RM, Hughes MR. Birth of a normal girl after in vitro fertilization and pre-implantation diagnostic testing for cystic fibrosis. N Engl J Med. 1992;327(13):905–909. doi: 10.1056/NEJM199209243271301. [DOI] [PubMed] [Google Scholar]
- 2.Harton GL, De Rycke M, Fiorentino F, Moutou C, SenGupta S, Traeger-Synodinos J, et al. ESHRE PGD consortium best practice guidelines for amplification-based PGD. Hum Reprod. 2010;26(1):33–40. doi: 10.1093/humrep/deq231. [DOI] [PubMed] [Google Scholar]
- 3.Wilton L, Thornhill A, Traeger-Synodinos J, Sermon KD, Harper JC. The causes of misdiagnosis and adverse outcomes in PGD. Hum Reprod. 2009;24(5):1221–1228. doi: 10.1093/humrep/den488. [DOI] [PubMed] [Google Scholar]
- 4.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev. 2004;5(6):435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 5.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachman MW. Variation in recombination rate across the genome: evidence and implications. Curr Opin Genet Dev. 2002;12:657–663. doi: 10.1016/S0959-437X(02)00358-1. [DOI] [PubMed] [Google Scholar]
- 8.Treff NR, Fedick A, Tao X, Devkota B, Taylor D, Scott RT., Jr Evaluation of targeted next-generation sequencing-based pre-implantation genetic diagnosis of monogenic disease. Fertil Steril. 2013;99:1377–1384. doi: 10.1016/j.fertnstert.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 9.Walne AJ, Vulliamy T, Kirwan M, Plagnol V, Dokal I. Constitutional mutations in RTEL1 cause severe Dyskeratosis Congenita. Am J Hum Genet. 2013;92(3):448–453. doi: 10.1016/j.ajhg.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballew BJ, Yeager M, Jacobs K, Giri N, Boland J, Burdett L, et al. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum Genet. 2013;132(4):473–480. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Treff NR, Tao X, Ferry KM, Su J, Taylor D, Scott RT., Jr Development and validation of an accurate quantitative real-time polymerase chain reaction-based assay for human blastocyst comprehensive chromosomal aneuploidy screening. Fertil Steril. 2012;97(4):819–824. doi: 10.1016/j.fertnstert.2012.01.115. [DOI] [PubMed] [Google Scholar]
- 12.Verlinsky Y, Cohen J, Munne S, Gianaroli L, Simpson JL, Ferraretti AP, et al. Over a decade of experience with pre-implantation genetic diagnosis: a multicenter report. Fertil Steril. 2004;82(2):292–294. doi: 10.1016/j.fertnstert.2003.09.082. [DOI] [PubMed] [Google Scholar]
- 13.Fedick A, Su J, Jalas C, Northrop L, Devkota B, Ekstein J, et al. High-throughput carrier screening using TaqMan allelic discrimination. PLoS One. 2013;8(3):1–9. doi: 10.1371/journal.pone.0059722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong ATG, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 15.Handyside AH, Harton GL, Mariani B, Thornhill AR, Affara NA, Shaw MA, et al. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. 2010;47(10):651–658. doi: 10.1136/jmg.2009.069971. [DOI] [PubMed] [Google Scholar]
- 16.Renwick P, Trussler J, Lashwood A, Braude P, Ogilvie CM. Preimplantation genetic haplotyping: 127 diagnostic cycles demonstrating a robust, efficient alternative to direct mutation testing on single cells. Reproductive biomedicine online; 2010. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 283 kb)