Abstract
Background
The assisted reproductive techniques aimed to assist infertile couples have their own offspring carry significant risks of passing on molecular defects to next generations.
Results
Novel breakthroughs in gene and protein interactions have been achieved in the field of male infertility using genome-wide proteomics and transcriptomics technologies.
Conclusion
Male Infertility is a complex and multifactorial disorder.
Significance
This review provides a comprehensive, up-to-date evaluation of the multifactorial factors involved in male infertility. These factors need to be first assessed and understood before we can successfully treat male infertility.
Keywords: Male infertility, Spermatogenesis, Y chromosome, AZF region, Molecular genetics, Epigenetics
Introduction
Human male infertility is a major health problem that affects approximately 10 to 15 % of couples worldwide [1, 2]. Specifically, in the Western countries, 15 % of couples are diagnosed with male infertility [3]. In about 50 % of these cases, the underlying problem lies with the male, either solely or in combination with female factors [4]. Male infertility is a multifactorial syndrome encompassing a wide variety of disorders [5]. Anatomic defects underlying male infertility include varicocele, vesicular damage due to torsion, obstruction of testicular sperm passage and ejaculatory failures, genital tract infections, gametogenesis dysfunction, molecular genetics disorders, endocrine disturbances and immunologic problems [4, 6]. Additionally, factors such as life style, environment [7, 8] and smoking [9] have also been reported to affect gamete and embryo development. Infertile men with no past history and normal semen analyses are designated as ‘idiopathic infertile’. Several factors including oxidative stress induced by reactive oxygen species (ROS), sperm DNA damage and molecular genetic abnormalities are responsible for the symptoms of Idiopathic infertility [10–13]. The most common causes of male infertility are summarized in Table 1.
Table 1.
Distribution of the most common causes of male infertility
Genetic abnormalities leading to male infertility are responsible for 15 to 30 % of cases, with a prevalence of Y chromosome abnormalities [14, 15]. Whereas many other genetic abnormalities that cause male infertility are still unknown, it is likely that most cases of idiopathic infertility could be accounted for by underlying genetic causes [16]. Molecular defects and genetic alterations responsible for male infertility disrupt physiological processes including hormonal homeostasis, spermatogenesis and sperm quality [17, 18]. Chromosomal abnormalities, single gene point mutations including mutation of cystic fibrosis transmembrane receptor gene (CFTR), polygenic or multifactorial genetic defects including Y chromosome deletions or micro deletions, mitochondrial DNA (mtDNA) mutations, endocrine disorders of genetic origin are the most frequently reported genetic causes of male infertility (Table 2). In addition, mutations and polymorphisms of various genes involved in spermatogenesis and other aspects of male reproduction have also been found to be associated with male infertility [19].
Table 2.
Prevalence and phenotype of the most common genetic anomalies associated with male infertility
| Genetic abnormalities | Phenotype | Prevalence (%) | References |
|---|---|---|---|
| Chromosome aberrations | Azoospermia to normospermia | 2–10 % | [1] |
| Numerical disorders | |||
| Klinefelter’s syndrome | Azoospermia to severe oligospermia | 5–10 % azoospermia 2–5 % oligospermia | [1] |
| Other sex chromosomes | Azoospermia to normospermia | 0.1–0.2 % | [20] |
| Structural disorders | |||
| Robertsonian translocations | Azoospermia to severe oligospermia | 0.5–1 % | [20] |
| Reciprocal translocations | Azoospermia to severe oligospermia | 0.5–1 % | [32] |
| Y chromosome deletions and microdeletions | Azoospermia to severe oligospermia | 5–10 % | [226] |
| AZFa | Azoospermia to SCOS | 0.5–1 % | [34] |
| AZFb | Azoospermic to arrest of spermatogenesis | 0.5–1 % | [34] |
| AZFc | Azoospermia to severe oligospermia | 3–7 % | [20] |
| AZFb,c | SCOS to arrest of spermatogenesis | 0.5–1 % | [34] |
| Partial deletions of AZFc | Azoospermia to normozoospermia | 3–5 % | [34] |
| Genetic mutations | |||
| CFTR | Obstructive azoospermia | 4–5 % | [20] |
| AR | Azoospermia to oligospermia | 2–3 % | [82] |
| KAL-1 | Hypogonadism hypogonadotropic | 5 % | [18] |
| INSL3-LGR8 | Cryptorchidism | 4–5 % | [227] |
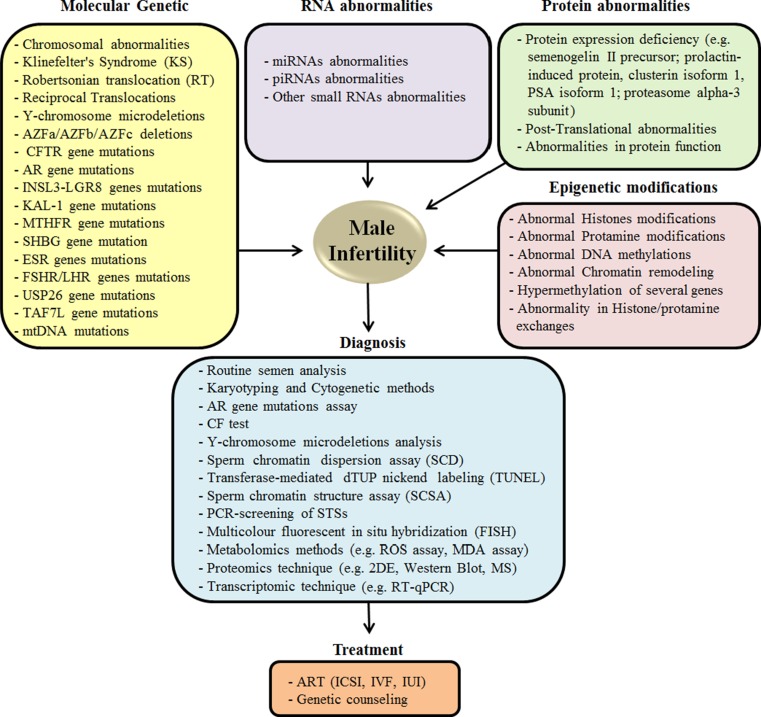
With recent advances in assisted-reproductive technologies such as, in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI), it is possible to offer reproductive hope to men once considered to be irreversibly sterile. However, in these instances, if genetic anomalies (Mendelian and chromosomal disorders) are the cause of their infertility, then there is an increased risk of transmitting the genetic defects to future generations. Also of importance to note is that it is becoming increasingly clear that abnormalities, both qualitative and quantitative, in semen analysis might be the ‘presenting symptom or phenotype’ of a variety of pathologies that can also affect non-reproductive organs (Fig. 1). In following sections, we will discuss important molecular and genetic causes of human male infertility and describe the state-of-the art technologies utilized to investigate this disorder, as well as highlight the increased need for genetic counseling and testing.
Fig. 1.
Common genetic, molecular and epigenetic factors contributing to the development, and the diagnostic and preventive measures to treat male infertility
Genetic causes of male infertility
Chromosomal abnormalities
Chromosomal defects are very common in infertile men, with a reported incidence of 2–15 %. The chromosomal aberrations involve the sex chromosomes in 3.8 % cases and the autosomes in 1.3 % of cases [20]. In addition to sex or autosomal chromosome abnormalities, numerical and structural defects of chromosomes also abound in infertile males. Numerical disorders result from changes in the normal number of chromosomes per cell. Chromosomal aneuploidy is a genetic condition in which an individual has either an abnormal number of one or more chromosomes, or has fragments of chromosomes lacking, or in excess. The severity of the phenotypic manifestations depends on which chromosomes are affected [21]. Structural disorders include deletions, duplications or translocations of chromosomes, as well as chromosomal rearrangements.
Klinefelter’s syndrome (KS)
Klinefelter’s syndrome (47, XXY) is the major sex chromosomal/numerical defect detected in newborn males (0.1–0.2 %) [22]. The prevalence of KS among infertile men is very high, up to 5 % in severe oligozoospermia and 10 % in azoospermia [23]. This karyotype arises spontaneously when paired X chromosomes fail to disjoin in the first or second phase of meiosis during oogenesis or spermatogenesis. Extra copies of genes on the X chromosome in these patients interfere with male sexual development and prevents the testes from functioning normally, consequently reducing the levels of testosterone that are responsible for the impaired spermatogenesis [4].
Although most patients with KS are infertile, there have been few patients, typically in mosaic cases (46, XY/47, XXY), who have conceived normally without assisted medical technology [24]. Within the testis of azoospermic men with KS, sperm can be retrieved in approximately 50 % of cases on testicular exploration from focal areas of spermatogenesis. In such cases, ICSI can be performed with an estimated success rate ranging from 30 to 50 % [4, 24]. In light of these findings, testicular sperm extraction and ICSI may be considered a potential option to attain pregnancy in males with azoospermia and KS patients [24]. Preimplantation genetic diagnosis using FISH techniques is another option that has shown promise in selecting normal embryos from KS men. In the future, it can be visualized that early sperm retrieval and sperm banking would be highly recommended for men with KS [25].
Chromosomal translocations
Robertsonian and reciprocal translocations are the most frequently observed structural chromosomal abnormalities in humans [26]. Although the prevalence rate of robertsonian translocation (RT) is 0.9 in infertile males, this figure is 9 times higher than in the general population [27, 28]. Reciprocal translocations are observed in 0.9 of 1000 newborns and involve exchange of two unrelated chromosomal segments [29]. They have been found in approximately 1 % of infertile men and commonly occur in azoospermic men compared to oligozoospermic males [16, 30]. Robertsonian translocations could potentially affect male fertility and pregnancy by altering the gene expression pattern of the spermatozoa at different stages [31, 32]. This is very evident in oligozoospermic (1.6 %) and azoospermic (0.09 %) men [30]. In all carriers, reciprocal translocations have impacted spermatogenesis, demonstrated by poor sperm quality or sperm aneuploidies, severe azoospermia and infertility. Although carriers of RT may exhibit a normal phenotype, spermatogenesis in male carriers of RT is variably affected, ranging from normal to oligoasthenospermia or azoospermia [17, 30]. It is thereby of paramount importance to analyze the chromosomal constitution in the sperm of RT carriers, in order to provide preimplantation genetic diagnosis and adaptive genetic counseling to persons undergoing IVF treatment.
Other sex chromosomal aneuploidies
In addition to these numerical chromosome abnormalities, men with karyotypes 47XYY, mosaic 45X/46XY, 45× and 46XX also occur but their fertility status is variable [1]. A 47, XYY male birth occurs in approximately 1:1000 live births. Although they are phenotypically normal, this karyotype can also result in infertility [16]. In patients with spermatogenic failure, maturation arrest or the Sertoli-cell-only syndrome (SCOS) with associated hormonal abnormalities and eventually oligospermia or azoospermia may be observed [21]. The mosaic karyotype 45×/46XY is found in infertile men with an incidence of 4 % and known to cause infertility [33]. All males with discordant sex chromosomal pattern are azoospermic due to absence of long arm of the Y chromosome containing the azoospermic factor (AZF) gene, which is necessary for normal spermatogenesis [34]. Translocation of SRY gene (testis-determining factor) from chromosome Y to chromosome X has also been reported [35]. In a study by Abusheikha et al., [36], they reported a male with normal male phenotype, in whom seminal analysis showed complete azoospermia. Moreover, polymerase chain reaction analysis of genomic DNA failed to detect the presence of the SRY gene. Men with azoospermia and karyotype 45, X have also mutations in AZF gene. A comprehensive investigation detailing the frequency, etiology and diagnosis of these rare syndromes is thereby necessary for these patients to receive gentle management and counseling through a cooperative interdisciplinary approach [36].
Y chromosome and male infertility
Extensive research has been performed on the association of Y chromosome deletions with male infertility, since it contains several genes that are critical for spermatogenesis and the development of male gonads. The prevalence of Y chromosome deletions is estimated to be ~1:2000 to 1:3000 males. It represents the second most common genetic cause of spermatogenetic failure in infertile males after Klinefelter’s syndrome [18, 37]. The incidence of Y micro deletions in infertile men varies from 1 to 55 % [38, 39].
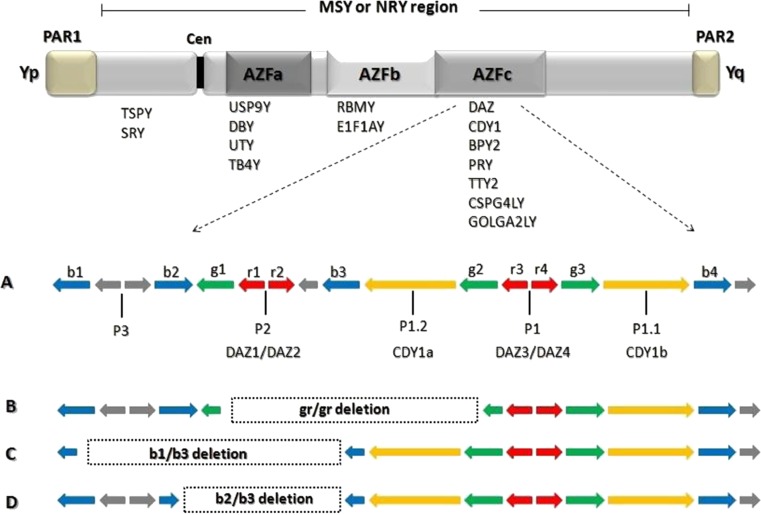
The Y chromosome locus is divided into three regions, the proximal, middle, and distal Yq11 and is designated as AZFa, AZFb and AZFc (Fig. 2), after the azoospermia factor AZF [40, 41]. Further, a fourth AZF region that overlaps with AZFb and AZFc regions is termed as AZFd [42]. These regions contain important genes, notable for their role in germ cell development. Furthermore, these regions are of greater significance since studies have revealed that gene deletions in these regions have been shown to be pathogenically involved in male infertility, specifically, azoospermia or severe oligozoospermia [43].
Fig. 2.
A schematic picture of Y chromosome displaying AZF regions (AZFa-c) and associated genes. Enlarged portion of AZFc region illustrates the discussed microdeletions. (a) Normal AZFc region, (b) gr/gr deletion, (c) b1/b3 deletion, and (d) b2/b3 deletion
Increased awareness has created increased demand for Yq deletion screening. Y micro deletion screening is strongly recommended for infertile men with severe sperm defect of unknown origin (ranging from azoospermia to oligozoospermia). Although, ART techniques such as ICSI, testicular sperm extraction (TESE) and IVF now facilitate infertile men associated with Y-chromosome micro deletion to achieve pregnancies [44], there is concern that such pregnancies may produce male offspring with similar micro deletions and consequent infertility [45]. In the following sections, we will discuss the different AZF regions of the Y chromosome and the functional significance of the genes they harbor, in detail.
The AZFa region
The AZFa locus is located at the proximal portion of long arm of the Y chromosome (Fig. 2) and spans about 1.1 Mb [46]. This region includes 4 genes including USP9Y (Ubiquitin-specific protease 9, Y chromosome), DBY (dead box on the Y), UTY (ubiquitous TPR motif on the Y) and TB4Y (thymosin B4-isoform). Deletion of the candidate genes, USP9Y and DBY are considered to be responsible for infertility. USP9Y is present as a single copy gene that is expressed in a wide variety of tissues and functions as an ubiquitin hydrolase [47]. DBY encodes an RNA helicase, is ubiquitously expressed and is the major gene located in AZFa region. While deletions or small mutations in USP9Y seem to be associated with severe hypo-spermatogenesis only, patients with deletions of DBY may present with a combination of the two: sertoli cell-only syndrome (SCOS), a condition characterized by the presence of complete Sertoli cells in the testes but a lack of spermatozoa in the ejaculate and severe hypo-spermatogenesis [17, 48]. On the contrary, patients with deletions of both USP9Y and DBY seem to invariably demonstrate azoospermia with a testicular histology of SCOS [17, 49]. Deletion of the USP9Y gene can additionally cause azoospermia, oligozoospermia, or oligoasthenozoospermia [15, 50].
The AZFb (P5/proximal-P1) region
The AZFb locus spans about 3.2 Mb of the Y chromosome long arm (Fig. 2) and includes two important genes, EIF1AY (translation initiation factor 1 A, Y isoform) and RBMY (RNA binding motif on Y) critical for male infertility. EIF1AY encodes a ubiquitously expressed translation initiation factor. The other main gene in the AZFb region is RBMY. Approximately 15–30 copies of these genes are dispersed in the short and long arm of the Y chromosome [51]. RBMY1 encodes a testis-specific RNA binding protein/splicing factor that is expressed in the nuclei of spermatogonia, spermatocytes, and round spermatids [52]. Deletions of the AZFb region include at least one functional copy of the RBMY1 gene. A family of PRY genes, essential in the regulation of apoptosis is also found in the AZFb region [53]. Whereas small AZFb deletions have been related to variable testicular phenotypes [54], larger AZFb deletions and those that include genes PRY1/PRY2 seem to cause complete meiotic arrest [34]. Recently, studies have revealed that in cases where all the genes in the AZFb region excluding RBMY and PRY are deleted, patients present with hypo spermatogenesis [20]. However, if both the RBMY and PRY genes are removed, spermatogenesis is arrested completely, indicating that RBMY and PRY are the major genes involved in fertility [17, 34].
Other genes including SMCY, CDY2, XKRY, HSFY, LOC170324, CYorf14, CYorf15A/B, TTY5, TTY6, TTY9, TTY10, TTY13, TTY14, XKRY and RPS4Y2 have been mapped to this region, but their role in the spermatogenic process is not well elucidated yet [1, 54]. Recent evidence points to a role of CDY2 in male germ cell development, specifically in the histone to protamine transition [20, 55]. HSFY encodes for a heat shock factor type DNA binding domain and is predicted to bind to heat shock promoter elements, as a transcriptional activator [54]. Mahanta et al. [55] reported that the frequency of Y chromosome micro deletions in XKRY gene is 23.1 %. Although very little information exists for other AZFb genes, there is sufficient evidence to suggest that that five single copy genes (LOC170324, SMCY, EIF1AY, RPS4Y2, and GAPD-similar), and two duplicate genes (HSFY and LOC14007/140020) are deleted in some infertile patients [56].
The AZFc (b2/b4) region
The AZFc region spans about 3.5 Mb of the long arm of Y chromosome (distal Yq11) and contains testis-specific genes involved in male spermatogenesis (Fig. 2) [57]. Deletions of the AZFc locus occur more frequently (65–70 %) than AZFa or AZFb locus [15]. Deletions of the AZFc locus manifest in 10–20 % of infertile men [58] and specifically in cases of moderate oligozoospermia (0.7 %), severe oligozoospermia (4–14 %), or secretory azoospermia (11–18 %) [41]. In addition, AZFc deletions are associated with testicular histologies varying from hypo-spermatogenesis (HP) to MA or SCOS [59]. Recently, studies have demonstrated that although only the AZFa and AZFb regions are required to initiate spermatogenesis, without the AZFc region, spermatogenesis will not be completely normal [29].
The genomic structure of AZFc region is composed mainly of large repeats of sequence blocks termed “amplicons”, organized into palindromic structures that show high (>99.9 %) sequence identity between the arms [60]. Such structures may undergo frequent inversion and gene-conversion events [61], as well as duplications and deletions [62]. These small partial deletions or sub-deletions produce a wide array of phenotypes, ranging from normospermic to azoospermic, aided by their interaction with the environment and their genetic background [17]. The gr/gr, b1/b3, and g1/g3 (b2/b3) are the most frequent sub deletions in the AZFc region of the Y chromosome [63] (Fig. 2). It is distressing to note that these rearrangements or polymorphisms can be transmitted to their male progeny and may represent a risk factor for decreased testis function and male sub fertility. However, in a study by Hopps et al., they reported that the majority of men with AZFc deletion have sperm within the semen or testes available for use in IVF/ICSI [64].
The b1/b3 deletion removes the PRY genes and reduces the copy numbers of several other genes. However, the prevalence of the b1/b3 deletion in the human population is very low and its frequency varies, with only 18 deletions published to date [65]. Its effect on spermatogenesis is unknown [66]. The b2/b3 deletion removes 1.8 Mb of AZFc or DAZ3/DAZ4 genes and was detected only in men with spermatogenic failure [65]. The gr/gr deletion [67, 68] removes half of the AZFc region including two copies of the DAZ gene family (DAZ1/DAZ2), one copy of the CDY1 gene family (CDY1a) and one copy of the BPY2 gene [66]. The gr/gr deletion event occurs with a relatively high frequency (3.5 %) in different Y haplotypes among men with spermatogenic failure [66]. It varies from 2.1 to 12.5 % among all cases, and from 0 to 10.2 % among normozoospermic controls [69]. In a conflicting report by Eloualid et al., [65] they have reported that gr/gr deletions occur at similar frequencies of 5.83 and 6.25 % in patient and control populations respectively, suggesting that these deletions are not associated with spermatogenic failure.
Within the AZFc region, substantial evidence points to the importance of DAZ gene family in normal spermatogenesis. Deletion of DAZ accounts for 10 % of cases of men with spermatogenic defect [70, 71]. Several studies have identified a single nucleotide polymorphism of DAZL that confers susceptibility to defects in spermatogenesis [72]. The deletion of the DAZ1/DAZ2 doublet, as observed in a DAZ gene copy-specific deletion analysis, was considered responsible for severe oligozoospermia [73]. The importance of the DAZ1/DAZ2 doublets was also highlighted when deletion of DAZ1/DAZ2 was seen in incomplete MA and was responsible for the testicular phenotype of residual spermiogenesis. A very high incidence of AZFc deletions has been demonstrated in incomplete SCOS.
Another important gene harbored in the AZFc region is CDY1, which is involved in the replacement of histones during spermatogenesis [52]. Previously, it was revealed that loss of CDY1 variants (a/b) is significantly higher in infertile men than control men. But, in another conflicting study, no association between DAZ1/2, DAZ3/4, CDY1a, or CDY1b and spermatogenic failure was detected [74]. Screening for AZF micro deletions before undergoing ART is mandatory and very critical for azoospermic and severely oligozoospermic patients, since it is well established that their male offspring will inherit the Yq micro deletions and infertility as well [75]. Associations between other Y chromosome genes including SRY (Sex Determining Region) and TSPY (testis-specific protein Y-encoded) and male infertility have also been recently established. Deletion of the SRY gene occurs in 15 % of 46 XY sex reversed females [76]. Additionally, mutations in SRY gene have also been reported in 10–15 % of 46 XY males with gonadal dysgenesis [77]. Recent studies support the link between SRY alterations, gonadal dysgenesis and primary infertility [78]. Evidence to signify TSPY’s involvement in gonadoblastoma and prostate cancer has been reported [79].
Autosomal and X-linked genes mutations
In addition to sex chromosomal genes, many autosomal and X-linked genes are also necessary for normal sexual development, testis determination, testis descent, spermatogenesis and eventually fertility in men. Given the importance of these genetic regions, it is not surprising that mutations, translocations, deletions or inversions that alter these regions may result in severe infertility and azoospermia. The human X chromosome is enriched with testis-specific genes that may be crucial for spermatogenesis and male fertility. Mutations in autosomal CFTR, c-kit receptor (c-kitR), KAL-1, INSL3-RXFP2 and X-linked androgen receptor (AR) genes are most common and lead to infertility due to defects in germ cell proliferation or urogenital system. We will summarize the role of some autosomal and X linked genes in spermatogenesis and the effects of different mutations or polymorphisms on male infertility in the following sections.
The CFTR gene mutations
Cystic fibrosis (CF) is a heterogeneous genetic disease caused by mutations in CFTR gene, located on chromosome 7 (7Q31.2). There is a positive correlation between CF and congenital bilateral absence of the vas deferens (CBAVD), a form of male infertility that is a frequent cause of obstructive azoospermia. This suggests a direct association of CFTR mutations and poor sperm quality in healthy men with CF and CBAVD. More than 95 % of adult men with CF are infertile because of obstructive azoospermia. Additional mutations include the 5 T polymorphism within intron 8 in the CFTR gene that was found in 21 % of the subjects and is considered a mild mutation, specifically associated with male sterility [43, 80]. The 7 T polymorphism within intron 8 and the missense R117H mutation within exon 4 in the CFTR gene were also reported [4, 81]. Other common mutations including G542X, G551D, R553X, W1282X and N1303K occur with a frequency of 1–2 %. Recently, studies have reported that CFTR gene mutations are a relatively frequent cause of male infertility. Although techniques including IVF and ISCI have facilitated CBAVD patients to father children, the couples have an increased risk of bearing a child with CF. Thereby, it is imperative to advocate genetic testing and counseling for these couples [43]. Nowadays, PGD has been regarded as a useful tool to identify the presence of CFTR mutations in patients.
The androgen receptor (AR) gene mutations
Androgens (testosterone (T) and 5α-dihydrotestosterone (DHT)) are critical steroid hormones that determine the development and maintenance of spermatogenesis. Perturbations in their function can result in spermatogenic failure and male infertility. Mutations in AR gene, an X-linked genetic condition causes male infertility with a frequency of 1:60,000 of live deliveries. In infertile men, the AR mutations are believed to affect at least 2 % of patients [82]. Recent studies have shown that mutations or polymorphisms in AR gene and its expressed protein may also result in depressed spermatogenesis and idiopathic male infertility, without any abnormalities in male secondary sexual characteristics [83].
Mutations in TAD and LBD subunits of AR are probably most common. About 19 mutations in the TAD are reported to be associated with some form of androgen insensitivity, in which patients have a decreased sperm count and a high percentage of abnormal sperm [82]. A critical region known as CAG-nucleotide repeats exists in TAD exon and contains an average of 21 ± 2 repeats. It is polymorphic and reported to be either increased or decreased in infertile men. CAG-nucleotide repeats are also reported in TAD exon but examination of its polymorphism has produced limited evidence. Data from some studies have established that AR missense substitutions contribute about 2 % and CAG repeat expansion (>28 repeats) may contribute up to 35 % of male infertility cases [82]. A preliminary research indicated that the mean AR gene CAG repeat length was significantly larger in azoospermic subjects than in control fertile men [84]. Mutations in TAD range from point mutations, insertions or deletions, or altered CAG repeats of AR gene. Mutations in AR LBD have also been reported. Moreover, genetic screening of infertile males has also revealed several loci in the AR LBD that are associated with reduced testicular volume and severe oligospermia [83]. It is important to recommend and offer screening for AR mutations and appropriate pre-conception counseling before testicular sperm aspiration for ICSI in azoospermic men, at risk of having AR mutations [85].
The INSL3-LGR8 genes mutations
Insulin-like 3 growth gene (INSL3), also known as relaxin-like factor (RLF) or Leydig insulin-like protein (LEY I-L), localized on chromosome 19 and its receptor LGR8 (leucine-rich-repeat-containing G protein-coupled receptor 8), localized on chromosome 13 are both functionally involved in the molecular mechanism of testis descent from abdomen to scrotum during fetal development [1]. Cryptorchidism is one of the most frequent (2 % of male births) congenital abnormalities in humans and causes non-obstructive azoospermia in 20 % of cases [4]. It can result in male infertility due to impaired spermatogenesis. Reduced sperm concentration, increased FSH and reduced inhibin B plasma levels that correlate with reduced testicular volume in adult life have also been reported, even after treatment by orchidopexy [4]. Recent data have reported that mutations in INSL3 gene or LGR8/GREAT occur in approximately 5 % of men with cryptorchidism [86]. However, there is conflicting evidence to believe that these frequent pathologies could possibly be caused by mutations in other genes as well [1].
The Kallmann 1 (KAL-1) gene mutations
Kallmann syndrome (Ks) is an X-linked recessive disease that is characterized by isolated hypogonadotropic hypogonadism (HH) and infertility. It is a rare genetic condition that accrues in approximately 1 in 10,000–60,000 live births (1:10000 in men and 1:50000 in women). Most patients have gonadotropin-releasing hormone (GnRH) deficiency. Clinically, it is characterized by low serum concentrations of testosterone (T), low levels of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) with normal prolactin levels, as well as delayed puberty, small testes, incomplete sexual maturation, lack of secondary sexual features, erectile dysfunction and absence of ejaculation, anemia and osteoporosis and sometimes even cryptorchidism [87, 88]. Furthermore, testicular pathology displays a wide array of findings from SCSO to focal areas of complete spermatogenesis. Male infertility in these cases is treatable with gonadotropins (LH and FSH) replacement over 12 to 18 months, which induces sperm in the ejaculate in 80 % of men.
Although in about 75–80 % of Ks patients, the causative gene is unknown, mutations in several genes including KAL1, FGFR1 (Fibroblast growth factor receptor 1), PROKR2 (Prokineticin receptor 2), PROK2 (Prokineticin 2), CHD7 (Chromodomain helicase DNA binding protein) and FGF8 (Fibroblast growth factor 8) that are predominantly inherited in an autosomal dominant manner, are known to be associated with Ks [89]. Although several genes mutations are responsible in Ks, the discovery of the causes is clinically relevant because it can enable clinicians to better understand the development of the neuroendocrine axis.
The MTHFR (methylenetetrahydrofolate reductase) gene mutations
The enzyme 5-methylenetetrahydrofolate reductase (MTHFR), encoded by MTHFR gene, is a key enzyme in folate metabolism. Many studies have shown that folate deficiency is common in infertile men and the related hyperhomocysteinemia is a risk factor for spermatogenesis deficiency and male infertility [89]. The MTHFR enzyme deficiency and hypo-methylation may inhibit gene expression, cause absence of germinal cells and spermatogenesis arrest [90]. Recent studies have reported that mutations in MTHFR gene have a negative effect on male infertility. Among the significant mutations studied, C677T (alanine for valine) is most common in infertile men with MTHFR deficiency [91, 92] and causes a decrease in the activity of MTHFR enzyme causing dysregulation of folic acid metabolism, hyperhomocysteinemia, errors in the methylation of genomic DNA and subsequent implications in the expression of spermatogenesis genes induced by under methylation. Singh et al., [93] reported that the homozygous (C/C) A1298C polymorphism of the MTHFR gene was also present at a statistically high significance in idiopathic azoospermic infertile men. The findings suggest that C677T and 1298CC homozygosity are an additional risk for molecular genetics of male infertility. Infertile patients with hyperhomocysteinemia should be advised to screen for C677T mutation and simultaneously recommended a folate-rich diet or folate supplements.
The sex hormone–binding globulin (SHBG) gene mutation
The human sex hormone–binding globulin (SHBG) that is expressed in testicular germ cells contributes to the concentration of androgens in the testis by binding or increasing androgen bioavailability [46]. Recently, the SHBG gene located on chromosome 17 (17p13.1), has also been studied for a possible role in spermatogenesis and male infertility. The SHBG expression potentially influences SHBG levels in human sperm and consequently androgen bioavailability. In addition, the fact that SHBG accumulates between the outer acrosomal membrane and the sperm plasma membrane emphasizes the functional significance of SHBG expression in male reproduction [47].
In a study by Lazaros et al., [94], they investigated the possible role of SHBG (TAAAA) n polymorphism on male factor fertility. An association was found between SHBG (TAAAA) n polymorphism and sperm concentration. Higher SHBG levels were associated with higher androgen availability and thus increased levels of spermatogenesis. In particular, they found that short AR alleles (with <22 CAG repeats) were associated with lower sperm motility, while long AR alleles were associated with higher sperm motility. However, no effect of the AR (CAG)n polymorphism on sperm concentration was observed in accordance with previous studies. These results support that these genes might contribute to sperm’s basic parameters. Furthermore, this could provide an explanation of how SHBG collaborates with AR to influence sperm quality, by affecting intra-testicular androgen levels.
The estrogen receptors (ESR) genes mutations
The role of estrogens (ES) in regulation of male reproductive functions and testicular development is widely accepted [95]; however the molecular mechanism of their physiological role in spermatogenesis is not clearly defined. The ES signaling is known to be mediated by at least two functional isoforms of estrogen receptors (ER) known as ERα and ERβ that are encoded by two different genes on the long arm of chromosomes six (6q25) and 14 (14q23-24), respectively [22]. In human testis, ERα and ERβ were also found in ejaculated spermatozoa, early meiotic spermatocytes and elongated spermatids-independent ERα signaling is important for concentrating epididymal sperm, whereas estrogen-mediated ERβ signaling is essential for germ cell progression or viability [96]. The ER genes thereby represent a logical target for mutational analysis in the infertile male. Although some work evidenced an association between estrogen insufficiency and abnormal spermatogenesis, little attention has been paid to the role of ER gene mutations or polymorphisms in male infertility.
In the recent past, a possible link between ERα polymorphisms and male infertility has been reported in Greek and Japanese populations [97, 98]. In knockout models, removing the ERα (αERKO) or the aromatase genes (ArKO) showed deficiency in the process of spermatogenesis and subsequently, reduced epididymal sperm count, sperm motility and fertilizing ability [99]. Recently, the genetic screening of the ERα gene locus has revealed the existence of several polymorphic sites including the PvuII (T397C) and XbaI restriction fragment length polymorphisms (RFLPs) in intron I and the (TA)n variable number of tandem repeats (VNTR) within the promoter region of the gene [21]. In a study by Guarducci et al., [100] they investigated the (TA)n repeat polymorphism situated in the promoter region of the ERα gene in a large group of infertile and normospermic men. Although the (TA)n polymorphism failed to show a significant association with male infertility, they found a significant effect of this polymorphism on sperm count. They suggested that specific allelic combinations of the ERα that confer a stronger estrogen effect may negatively influence human spermatogenesis.
On the other hand, the ERβ knockout mice were fertile, but showed prostate and bladder hyperplasia [101]. Aschim et al., [102] reported that the RsaI polymorphism in the ERβ gene appears to be associated with infertility, showing a three times higher frequency of the heterozygous RsaI AG-genotype in the infertile group compared with controls. In contrast, in another study by Khattri et al., [103], they found no significant correlation in the incidence of the known SNPs or novel mutations between infertile and fertile men. Moreover, infertile men having ERβ mutations had normal reproductive tract and serum hormone levels. They postulated that the SNPs and mutations in ERβ gene are not a common cause of spermatogenesis failure in Indian men, although mutations specifically found in infertile men can affect transcription, translation or have synergic effect with other variants in causing infertility [104].
The FSH/LH receptors (FSHR/LHR) genes mutations
The follicle stimulating hormone (FSH) and luteinizing hormone (LH) regulate spermatogenesis. Although mutations in the LH or FSH receptor gene are rarely seen, a number of SNPs in the LHR gene and LH gene are associated with variable pathologies of the male gonad [21]. The SNPs give rise to different FSHR and LHR haplotypes that modify the action of FSH and LH. Mutations in the LHR gene lead to hypogonadal phenotypes (Leydig cell hypoplasia) because of a delay in fetal and pubertal male development. Mutational screening of the FSHR gene revealed several SNPs of the FSHR gene, 1 in the promoter region at nucleotide position-29 (G to A exchange) and 2 in exon 10 including positions 680 and 307 [105]. Furthermore, variable suppression of spermatogenesis and fertility was diagnosed in men homozygous for the inactivating A189V mutation in exon 7 of the FSHR gene [1]. Conflicting studies reported a different allelic frequency in azoospermic with respect to normozoospermic men in one study [106] while other studies revealed no differences in the distribution of FSHR polymorphisms between normal and infertile men. Additionally, a FSHR SNP has been discovered that may affect the activity of the gene [21]. Balkan et al., [105] investigated the genetic screening for SNPs at nucleotide position 29 in the core promoter region and codon 680 in exon 10 of the FSHR gene and the effect of the serum levels of FSH on male infertility in Southeast Turkey. They suggested that the FSHR haplotype does not demonstrate any association with different serum FSH levels. These different results suggest that an in-depth analysis of role of LHR and FSHR genes in male infertility is warranted.
The USP26 gene mutations
The ubiquitin-specific protease-26 (USP26) gene, located on human X chromosome (Xq26.2), is expressed throughout the testes in the preliminary stages of spermatogenesis. USP26 gene has emerged as an attractive novel candidate in the study of male infertility. Recent data have indicated increased number of mutations and SNPs in the USP26 gene in men with severe male factor infertility. Zhang et al., [107] investigated the incidence of SNPs in USP26 gene and its involvement in idiopathic male infertility in China. They showed three variations including a compound mutation (364insACA; 460G > A) and a 1044 T > A substitution in azoospermia, oligozoospermia and asthenozoospermia patients. All three variations led to changes in the coding amino acids. In a subsequent study, Zhang et al., [108] concluded that 22 % of infertile patients exhibited changes in the USP26 gene (364insACA and 460G > A (19.5 %) and 1044 T > A substitution (2.4 %)). Lee et al., [109] examined the association between some USP26 alleles and haplotypes with spermatogenic defect in Taiwanese and Chinese populations. In that study, they demonstrated that the allele frequencies of five SNPs (370-371insACA, 494 T > C, 576G > A, ss6202791C > T, 1737G > A) were significantly higher in infertile patients than control subjects. Although some USP26 alleles and haplotypes were found in the populations, the haplotype TACCGA was the most common (28 %) in patients with spermatogenic defect. Other novel variants such as 393A > T, 468 T > G, 520 T > G, 565 T > G, 2144A > T, 2182A > T, 2195 T > C, 2204 T > G, 2239A > T, 2247A > C, 2250A > G and 2271 T > C were also linked to impaired spermatogenesis. Recently, Ribarski et al., [110], reported high frequency (4.7 %) of 1090C > T mutations in infertile patients. Stouffs et al., [111] reported the presence of some mutations in USP26 gene such as 370-371insACA, 494 T > C and 1423C > T in patients (7.2 %) with SCOS.
All this data is underpins the importance of USP26 gene in male reproduction and spermatogenesis. Although these studies suggested that a specific genetic cluster might be associated with testicular dysfunction, further studies based on a larger group of patients with diversified ethnic backgrounds will be valuable to reinforce the role of USP26 gene in male infertility.
The TAF7L gene mutations
TAF7L (transcription initiation factor TFIID subunit7-like) is a member of TAFs that has an important role in transcription process. In humans, the TAF7L gene is an X-linked and single-copy testis-specific gene that may be essential for maintenance of spermatogenesis and a possible contributor to male infertility [112]. Recently, a number of publications have reported the importance of various TAFs genes such as TAF7L and TAF4b in mouse spermatogenesis. A study by Falender et al., [113] showed that TAF4b (a gonad-specific subunit of TFIID) is required for the maintenance of spermatogenesis in the mouse. In a knockout model study by Cheng et al., [112], they discovered that disruption of TAF7L gene in mice results in reduced sperm count and motility. They proposed that its counterpart, the human TAF7L gene may also play a similarly important role in spermatogenesis. Because of the hemizygous state of the X chromosome in men, mutations in TAF7L could potentially cause oligozoospermia in humans. On the other hand, Akinloye et al., [114], reported that TAF7L gene mutations are rare and are not a common cause of spermatogenic failure in normogonadotrophic nonobstructive azoospermia patients. Although the TAF7L gene is highly polymorphic, most of them were not significantly associated with gonadal dysfunction. In addition, they discovered that SNPs at exon 13 may be a risk factor for an infertile phenotype if combined with additional polymorphisms or mutations. These findings warrant additional research to elucidate the role of TAF7L gene as a molecular cause of male infertility.
The mitochondrial DNA (mtDNA) and male infertility
Significant strides in research have been made on the role of the mitochondria and its genome on male infertility [115]. Beside nuclear chromosome deficiencies that have profound effects on male fertility, there is increasing evidence that mitochondrial DNA (mtDNA) anomalies in sperm may lead to male infertility. Mitochondria play a critical role in energy metabolism by containing oxidative phosphorylation elements and enzymes (OXPHOS) [116]. There are 70–80 mitochondria in the midpiece of human mature spermatozoa that serve as a large source of energy (ATP) for the flagellum to move around in the early stages of fertilization [117]. As the mtDNA genes encode several polypeptides particularly for the OXPHOS, mtDNA damages cause deficiency in ATP production [118, 119]. The mitochondrial machinery plays a key role in the energy production and maintenance of spermatozoa motility and is proposed to be one of the major determinants of male fertility [120].
Substantial evidence has associated deficiencies of human sperm mitochondria with sperm dysfunction and poor sperm quality [121, 122]. Asthenozoospermia or oligoasthenozoospermia has been reported previously in patients suffering from typical mtDNA diseases [123, 124]. Other studies have shown a correlation between the quality of the sperm and the functionality of the respiratory chain in sperm mitochondria [125]. A direct and positive correlation between sperm motility and mitochondrial enzymatic activities has been demonstrated suggesting that motility largely depends on energy production by mitochondria [120]. In an interesting study by Carra et al., [120], they investigated sperm mtDNA from idiopathic oligoasthenozoospermic patients, with different sperm motility and sperm concentrations, by testing motile and non-motile sperm of the same individual. Their results suggested that only motile sperm have organelles functional in oxygen consumption, unequivocally demonstrating that motility depends on the mitochondrial activity. It has been recently demonstrated that in some asthenozoospermic patients the sperm midpiece was significantly shorter than in normal subjects [126]. This finding could be probably linked to defects in the energy producing machinery that is required to drive motility. Some studies have revealed that spermatogenesis is associated with a drastic reduction in mtDNA content, to about a tenth of its initial value, occurring mainly during spermiogenesis [127]. This result reinforces the idea that asthenozoospermia patients have defects in the sperm tail formation or energy production for sperm motility. The change of sperm mtDNA copy number is a very important event throughout spermatogenesis, fertilization and embryogenesis. However, in one study by May-Panloup et al., [122], they reported that the mtDNA with deletions was significantly higher (up to 28 times) in sperm samples of poor quality than in normal sperm samples.
Recent reports have shown that mtDNA point mutations or multiple deletions, mtDNA single nucleotide polymorphisms (SNPs) and mtDNA haplogroups can greatly influence sperm quality and are associated with asthenozoospermia or oligoasthenozoospermia [128–138]. Among the mitochondrial deletions observed, the deletion of 4977 bp was the most prevalent and abundant one. Kao et al., [132], for the first time reported that a 4977 bp mtDNA deletion is associated with diminished fertility and motility of human sperm and that the highest frequency of occurrence of the deletion coincided with reduced sperm motility. Further studies have analyzed sperm mtDNA deletions by long-range PCR techniques and found two other novel types of 7345 and 7599 bp large-scale deletions in the mtDNA of spermatozoa with poor motility [125]. Although these three large-scale deletions were found to occur either alone or in different combinations, the 4977 bp deletion is the most frequently (affecting 40 % of patients with mitochondrial myopathy) occurring deletion in spermatozoa with poor motility [125]. A higher percentage of multiple deletions, 4977-bp, 7599-bp and 7491-bp of mtDNA in spermatozoa of patients with poor sperm quality have also been recently reported [136]. Other studies have confirmed the persistence of multiple deletions in normozoospermic samples [139]. The ATPase6, ATPase8 and COII genes deletions have been reported previously in mtDNA of 49 asthenozoospermia patients [135]. However, there was a greater accumulation of multiple deletions present when the sperm quality was poor, with oligoasthenoteratozoospermic men harboring far more mtDNA deletions.
Recent studies have shown that mutations in mitochondrial genes such as COX II, ND1, ATPase6 and 8 disrupt the ATP production and effect spermatogenesis and sperm motility. In a study by Guney et al., [119], they investigated the relationship between the mtDNA genes (ATPase6, Cytb and ND1) and male infertility. They found 13 point mutations in the ATPase6 gene, of which three were novel and 8 caused amino acid changes. In addition, they examined for the first time the association of Cytb gene mutations with infertility and detected 20 point mutations in the Cytb gene, all of which were statistically non-significant. About 8 mutations in ND1 gene were also observed in their study. Holyoake et al., [134] showed that point mutation in the ATPase6 gene at 9055 was commonly seen associated with poor sperm quality. This substitution involves a G to A transition within the ATPase6 gene, changing an alanine to threonine. In their study, about 10.7 % of men with poor semen quality were shown to have the G9055A substitution, but only 1.3 % of men with normal fertility had the same substitution. Furthermore, a T8821C point mutation has been found in the ATPase6 gene of severely oligospermic men with immature spermatids. In another study, the T9098C transition was detected only in infertile cases, and the finding of this mutation was statistically significant when compared to controls [138, 140]. Thangaraj et al., [141] also established that mitochondrial mutations in sperms can cause low sperm motility that is not related to infertility. They observed 10 SNPs in the ATPase6 gene in the sperm DNA of an oligoasthenoteratozoospermic man. They found that these mutations were associated with low motility sperm but otherwise did not impair fertility. In another study by Spiropoulos et al., [142] they carried out semen analysis on a male harbouring the A3243G mtDNA mutation and showed that high levels of mutant mtDNA strongly correlated with low sperm motility. Folgero et al., [131] also previously noted reduced sperm motility in a patient with mtDNA disease due to the A3243G mtDNA mutation. Two specific point mutations in the region of mtDNA genome from COX I (complex IV) and ND 5 (complex I), at nt 9055 and nt 11719 are associated with poor semen quality parameters in 11 and 12 % of cases respectively. T26248G-transversion mutation in exon7 of the putative methyltransferase Nsun7 gene has been recently reported to lead to low sperm motility [143].
Nuclear gene defects may also result in mitochondrial disorders by predisposing the cell to multiple mtDNA deletions. Recently, the nuclear encoded proteins including mitochondrial-specific DNA polymerase gamma (POLG) [144] and glutathione S-transferase M1 have been reported to be involved in male infertility [145]. POLG is responsible for replication and repair of the mtDNA. Mutations in the gene for the catalytic subunit of POLG have been shown to be a frequent cause of mitochondrial disorders and sperm dysfunction [146]. Hence, mitochondrial respiratory chain function depends on the coordinated gene expression of both the mitochondrial and nuclear genomes and a mutation in either genome may lead to defective respiratory function of mitochondrion.
Mitochondria is a major intracellular source of reactive oxygen species (ROS). It is believed that mtDNA mutation may impair electron transport chain resulting in enhanced production of mitochondria ROS due to incomplete reduction of oxygen. In addition to mitochondrial inner membrane potential, ROS induces mtDNA strand breaks and large-scale deletions. The large-scale deletions result in complete removal or truncation of some structural and tRNA genes of mtDNA and results in male infertility [147].
Epigenetic modifications and male infertility
The recent advent of new technologies has spurred research on the role of epigenetics in spermatogenesis and male infertility. Epigenetics refer to process of gene regulation devoid of changes in DNA sequence and comprise of DNA methylation, posttranslational histone modifications or chromatin remodeling [148]. Recently, hyper methylation of promoters of several genes including MTHFR, PAX8, NTF3, SFN, HRAS, JHM2DA, IGF2, H19, RASGRF1, GTL2, PLAG1, D1RAS3, MEST, KCNQ1, LIT1 and SNRPN have been reported to be associated with poor semen parameters or male infertility [149].
DNA methylation is one of the best-characterized epigenetic processes and involves the addition of a methyl group to the 5′ position of the cytosine nucleotide, typically occurring in a CpG dinucleotide. This reaction is catalyzed by a group of proteins termed DNA (cytosine-5) methyltransferases (DNMTs) [150]. CpG islands have been found near promoters and have been documented to play an important role in regulating gene expression [149]. It has been reported that hyper methylation of DNA in CpG islands is associated with gene inactivation, while hypo methylation being associated with gene activation [151]. Improper DNA methylation of various genes has been implicated in abnormal semen parameters, as well as several instances of male infertility. Houshdaran et al., [152] demonstrated for the first time, that poor sperm concentration, motility and morphology were associated with broad DNA hyper methylation across a number of loci including PLAG1, DIRAS3, MEST, PAX8, NTF3, SFN and HRAS. They suggested that the underlying mechanism for these epigenetic changes may be improper erasure of DNA methylation during epigenetic reprogramming of the male germ line. In another study by Khazamipour et al., [153], 53 % of men with nonobstructive azoospermia revealed hypermethylation of the MTHFR promoter, while none of the men with obstructive azoospermia exhibited hypermethylation of this gene promoter, suggesting that MTHFR hypermethylation is a specific epigenetic aberration that may specifically contribute to certain types of male infertility. In addition, Wu et al., [154] in a study reported that hypermethylation of MTHFR gene promoter in sperm was associated with idiopathic male infertility. Abnormal DNA methylation of H19 and MEST imprinted genes has also been shown to be associated with oligozoospermia [140], suggesting that spermatogenesis may be particularly vulnerable to changes in the methyl pool brought about by deficiency in MTHFR enzyme. Hammoud et al., [155] examined CpG methylation patterns in infertile (oligozoospermic and abnormal protamine) and fertile donors at seven imprinted loci including LIT1, MEST, SNRPN, PLAGL1, PEG3, H19, and IGF2. At six of the seven imprinted genes, the overall DNA methylation patterns were significantly altered in both infertile patient populations. They demonstrated a link between abnormal spermatogenesis and abnormal methylation of genes. Contradictory reports demonstrated high methylation at the H19/IGF2 ICR1 locus and low methylation at the MEST locus in normozoospermia in one study, while another study revealed low methylation of the H19/IGF2 ICR1 locus and high methylation of the MEST locus in association with low sperm concentrations [156]. Similarly, Boissonnas et al., [157] found that many patients with teratozoospermia and oligoasthenoteratozoospermia exhibited hypo methylation at variable CpG islands at the H19 locus. However, recent studies have shown that hypermethylation at MEST was more strongly linked with poor sperm quality than hypo methylation at H19/IGF2 ICR1 [148]. They suggested that sperm from infertile patients, especially those with oligospermia, may carry a higher risk of transmitting incorrect primary imprints to their offspring, highlighting the need for more research into epigenetic changes, when considering ART.
Post-translational histone modifications, another component of epigenetic process is essential during many cellular processes including mitosis and spermatogenesis. The compactness of DNA is determined by histone proteins that are bound to DNA [148]. Histone methylation, one of the most prevalent modifications, is catalyzed by histone methyltransferase enzyme (HMTases) and is generally associated with gene silencing. Histone acetylation, modulated by both histone acetyl transferases (HATs) and histone deacetylases (HDACs), is associated with increased levels of transcription. HATs activate gene expression, while HDACs inhibit gene expression. Recent research has identified a critical role for the JHMD2A (Jumonji C-terminal containing histone demethylase 2A) histone demethylase in male infertility and spermatogenesis [158, 159]. Using knockout mice as models, this study identified a critical role for JHMD2A in the regulation and expression of two genes, protamine 1 (Prm1) and transition nuclear protein 1 (Tnp1) involved in the condensation and proper packaging of chromatin in the male sperm.
Chromatin remodeling is a process in which ATP-dependent chromatin remodeling complexes (such as SWI/SNF, ISW1 and MI-2) use energy to alter the location and structure of nucleosomes [106]. The remodeling process activates or represses gene expression as required for proper development and maturation of gametes and proper meiosis [149]. It is a critical step in gamete development, and the compacted structure of the chromatin transmits vital information to the embryo to guide it through development. The lack of proper DNA packaging in sperm has been associated with infertility in mice [160]. During the chromatin re-packaging in spermatogenesis process, about 85 % of the histones are replaced by protamines [161]. In the initial stages of spermiogenesis, histones are hyper acetylated and undergo other modifications [162]. At the final stage of spermatogenesis, the nucleosomal structure is progressively disassembled, then replaced by transition proteins (TNPs) and finally by protamines [163]. The incorporation of protamines into sperm chromatin induces DNA compaction that is important for the formation of spermatozoa. However, it is known that protamines are phosphorylated before binding to DNA and that substantial dephosphorylation takes place concomitant with nucleoprotamine maturation. Indeed, mutation of the calmodulin-dependent protein kinase Camk4 that phosphorylates protamine 2, results in defective spermiogenesis and male infertility [164]. At fertilization, the mature sperm cells are packaged densely with protamines, whereas maternal genome is packaged with histones. Subsequently, upon fertilization, the highly compact nucleoprotamine structure must be unpacked and reorganized into a nucleosomal structure [165]. Recent studies have shown that some epigenetic errors in these processes are a possible cause of male infertility. Both protamines 1 and 2 are essential for sperm function, and the haploinsufficiency of either P1 or P2 results in a reduced amount of the respective protein, abnormalities in the structure of the chromatin, DNA damage and infertility [148]. Furthermore, it is known that the optimal protamine-1 to protamine-2 ratio (P1/P2 ratio) is critical and well regulated [166]. Indeed, the P1/P2 ratio in fertile men ranges from 0.8 to 1.2 and deviation from this ratio has been shown to lead to infertility [167]. A change in either direction of this ratio adversely affects semen quality, DNA integrity and fertility in men. Patients with abnormally depressed or elevated P1/P2 ratios are characterized by poor sperm concentration, motility and morphology as well as decreased fertilization capabilities. Impaired spermatogenesis has been reported to be associated with aberrant H4 acetylation [168]. A study by Sonnack et al., [169] showed that men exhibiting qualitative or quantitative infertility have significantly decreased levels of histone H4 acetylation associated with impaired spermatogenesis. Hyperacetylation of histone H4 is required in the transition from histones to protamines. This step decreases the affinity of the interaction between the sperm histones and DNA to allow the exchange for transition proteins to occur [149]. H4 hyperacetylation was also observed in infertile men exhibiting SCOS [150]. A significant difference in transcript expression of chromatin remodeling factors between normal spermatogenesis and round spermatid maturation arrest has been found in a previous study, suggesting impaired epigenetic information and aberrant transcription during sperm development. This could represent one possible reason for the developmental arrest of round spermatids [170].
Genetic imprinting, namely the expression of alleles in a sex-specific manner, is most often due to the differential methylation in the CpG islands in one of the alleles [148]. It determines which genes from the paternal and maternal genomes are expressed in the embryo and is critical for normal development [171]. Several loci are known to be imprinted. For instance, in humans, fetal spermatogonia seem to be mostly unmethylated at H19 differentially methylated regions, although spermatogonia in adult testis demonstrate significant methylation in this region [148]. Imprinted genes, including the paternally imprinted GTL2 and H19 loci, have been previously examined by Kobayashi et al., [171] in 97 infertile men. The importance of genomic imprinting during spermatogenesis has been reinforced by the association of decreased methylation of the paternal IGF2⁄H19 imprinting control region 1 (ICR1) and GTL2 imprints in spermatozoa of men with disturbed spermatogenesis. In another study by Poplinski et al., [156], fertile men had a high degree of IGF2⁄H19 ICR1 and a low degree of MEST methylation. Low sperm counts were clearly associated with IGF2⁄H19 ICR1 hypo methylation and, even stronger, with MEST hyper methylation. These results suggest that idiopathic male infertility is strongly associated with imprinting defects at IGF2⁄H19 ICR1 and MEST, with aberrant MEST methylation being a strong indicator for sperm quality. ART techniques such as ICSI and round spermatid injection (ROSI) may increase the incidence of imprinting disorders and adversely affect embryonic development by using immature spermatozoa that may not have established proper imprints or global methylation yet. Andrologists have voiced concern about concealing reproductive defects through ART that might have negative consequences at the epigenetic level.
Diversity of RNAs in sperm
The diverse RNA populations present in the spermatids are produced either throughout early spermatogenesis or during spermiogenesis and serve as a historical record of spermatogenesis. The highly condensed sperm nucleus is transcriptionally inert and contains mRNA, antisense RNAs, and miRNAs that have been transcribed prior to inactivation. Despite the inert status of the sperm RNAs, the delivery of sperm mRNAs to the oocyte at fertilization is speculated to be important in the early development of the zygote and embryo [172, 173].
Recent advances in technology have been instrumental in deciphering the role of sperm RNAs in development and disease. Transcriptomics involves profiling of gene expression using microarrays, RT-PCR and in situ hybridization (ISH) analysis. An in-depth spatial and temporal analysis of gene expression could be useful to determine the genes that are involved in each stage of the process [17]. Substantial evidence points to the presence of remarkably different RNA populations in mature sperm in fertile versus infertile men [139]. HSPA2 is correlated with sperm maturity, function and fertility, and its dysfunctional expression results in abnormal spermatogenesis. A study by Li et al., [174] demonstrated lower levels of HSPA2 expressed in sperm in oligoteratozoospermic men compared to normozoospermic controls. Yet another preliminary study [174] revealed significantly lower levels of BDNF and NGF receptor TrkA mRNAs in human sperm from oligoasthenozoospermic men compared to normal controls. Glycoprotein subunit 130 (GP130), upon binding to IL-6, is a known regulator of sperm motility. Low levels of GP130 mRNA and protein have been reported in asthenozoospermic men when compared to normozoospermic men [175]. Guo et al., [176] compared the levels of DDX4/VASA transcript, an RNA binding protein/helicase that is essential for germ cell development, between normozoospermic men and patients with oligozoospermia. VASA transcript and protein levels were significantly decreased in the sperm of oligozoospermic men, suggestive of the fact that VASA is potentially associated with pathogenesis in some selective subtypes of male infertility.
miRNAs in sperm
Awareness of the presence of myriad groups of RNAs and their elusive functions in spermatogenesis, fertility and early embryo development, have led to increased focus on small regulatory RNAs and their function in these processes. Small regulatory RNAs are non-coding RNAs that regulate gene expression at the transcriptional, post-transcriptional or through chromatin remodeling [177]. It has been established that human spermatozoa comprises of miRNAs (7 %) piRNAs (17 %) and repeat-associated small RNAs (65 %) [178].
miRNAs are single-stranded noncoding RNAs (20–25 nucleotides) that are mostly expressed in a tissue-specific fashion, and down regulate gene expression through their interaction with the 3′ untranslated region (UTR). Approximately one-third of human genes are regulated by miRNAs [179]. They are implicated in the regulation of many important biological pathways such as proliferation, apoptosis, and differentiation in various species, but a large number of them (about 68 %) show a tissue-specific expression [180]. They are expressed in various testicular cell populations as well as epididymis, indicating a critical role in the various steps of the highly organized process of spermatogenesis, sperm maturation and maintenance of male fertility. More than 200 miRNAs have also been identified in the human epididymis and this could suggest their potential involvement in regulating the physiological functions of sperm maturation and morphogenesis [181]. Members of the miR-888 cluster (miR-890, miR-891a, miR-891b, miR-892a and miR-892b) are significantly more abundant in the corpus/cauda regions of the epididymis, indicative of a prominent role of the miR-888 cluster in regulating physiological functions of the human epididymis. In another instance, expression of spag8 (sperm associated antigen 8) mRNA transcript encoding for a human sperm membrane protein (hSMP-1) negatively correlates with the expression of miR-892a in the epididymis.
Other recent reports have also highlighted the importance of miR-18 in the control of heat shock factor 2, a transcription factor that is required for normal spermatogenesis in mouse [182]. miR-34c and miRNA34b are important players in the later steps of spermatogenesis in germ cells [233] and differentiation of male germ cells [183]. The role of miR-122a that regulates Tnp2, a testis-specific gene involved in chromatin remodeling during spermatogenesis cannot be underscored [184, 185]. Additionally, miR-17–92 cluster functions in the regulation of apoptosis and E2F-1 transcription factor during normal spermatogenesis [186]. In a recent study, high levels of expression of specific miRNAs was observed in low fertility bulls, which could suggest that miRNAs might be responsible for down regulating gene expression and thereby, regulate proteins that have important roles in fertilization and early embryonic development [187]. Other studies revealed significant differences in the expression of miRNAs between immature and mature individuals, suggesting miRNAs have a role in regulating spermatogenesis [188, 189]. Special mention needs to be made of miR-322, miR-323, miR-372 and miR-373 that are presumed to be novel oncogenes that participate in the development of human testicular germ cell tumors [190].
Even deletions or mutations in miRNA processing machinery of Dicer1 and Drosha can cause impaired spermatogenesis and male infertility. Primary miRNAs molecules exist in intronic regions and are produced by RNA Pol II, then further processed through a complex process in nucleus (by the microprocessor machinery known as DROSHA) and subsequently transported to cytoplasm (by an endonuclease DICER1) [191, 192], leading to mature miRNAs production. DICER1and DROSHA are also reported to play crucial roles in the normal function of somatic nursing cells of the seminiferous epithelium and the process of normal spermatogenesis. Many studies have highlighted the critical roles of DICER1 and DROSHA in male infertility. For instance, knockout of Dicer1 in mice Sertoli cells or male germ cells leads to infertility due to impaired spermatogenesis, progressive testicular degeneration and both meiotic and spermiogenic defects [193]. Papaioannou et al., [194] also reported that ablation of Dicer1 in Sertoli cells leads to infertility due to the complete absence of functional spermatozoa and progressive testicular degeneration. Another study indicated that Dicer1 is unnecessary for spermatogonial stem cell renewal and mitotic proliferation, but is required for germ cell differentiation through the meiotic and haploid phases of spermatogenesis. Of relevance to the function of DICER1and DROSHA in miRNAs biogenesis and spermatogenesis, it has also been demonstrated that mutations or single nucleotide polymorphisms (SNP) in Dicer1 and Drosha is associated with male infertility. A mutation in Dicer1 or Drosha may cause global alterations of miRNA expression and result in the up-regulation of their target genes, which in turn could affect other interacting factors such as the miRNA 17–92 cluster, leading to apoptosis of spermatozoa and abnormal semen quality [195]. In a study by Qin et al., [196], they reported a significant association between Dicer1 and Drosha SNP with abnormal semen parameters and idiopathic male infertility. The genetic variants of rs12323635 (Dicer1), observed in Han-Chinese population were also associated with idiopathic male infertility. Additionally, men with the rs642321TT genotype (Drosha) may have a higher risk of oligozoospermia. The results showed that variants of Dicer1 and Drosha may modify the risk of abnormal semen parameters, and could result in male infertility. Due to their functional diversity and the increasing complexity of miRNAs, it is clear that we have just touched the tip of the iceberg and a lot more needs to be done to understand the diverse and crucial roles of miRNAs and other DICER-dependent RNA pathways in male germ cell biology.
Recent studies have also revealed the presence of miRNAs in serum, plasma, semen and other body fluids. The production of miRNAs in body fluids and their biogenesis patterns are closely correlated with various diseases [197, 198]. Human semen contains several species of miRNAs that can be readily detected by RT-qPCR and used as a diagnostic tool for semen stain identification. It is assumed that miRNAs may leak either passively or actively from apoptotic or broken sperm cells, germ cells or from cells of accessory glands into seminal plasma [198]. The binding of miRNAs with complex organic molecules and the RNA-macromolecular complex render them more or less resistant to RNase degradation and they are stable in seminal plasma. Additionally, fertile and infertile patients have some specific miRNAs in their semen that can be used as potential biomarkers to distinguish between them. Since reproductive disorders-associated miRNAs can be identified in seminal plasma, a non-invasive semen test could be developed to diagnose male infertility. Thus, it is reasonable to speculate that the concentrations of miRNAs in seminal plasma may provide some useful information about gene expression in the male reproductive system and may represent a new source of novel, minimally invasive biomarkers of male infertility.
Recent studies also reported a significant difference in the concentration of some specific miRNAs in seminal plasma of infertile men [198, 199]. For instance, Wang et al., [199], observed that 7 special miRNAs (miR-34c-5p, miR-122, miR-146b-5p, miR-181a, miR-374b, miR-509 –5p and miR-513a-5p) are markedly decreased in semen of azoospermia but increased in asthenozoospermia. They revealed a strong relationship between the seminal plasma miRNAs and male infertility, suggesting that the seminal plasma concentrations of these miRNAs can accurately distinguish infertile patients from fertile controls. These recent discoveries of differential expression of miRNAs in testis have generated a new approach, miRNAs profiling, to reveal functional information of miRNAs relevant to this disorder. Needless to say, miRNA research is an emerging and exciting field that holds promise for improvements in diagnosis and treatment of male infertility.
piRNAs and male infertility
Another newly identified class of small RNAs deemed to regulate spermatogenesis and male infertility, is that of piRNAs. They are the largest and most complex class of small non-coding RNAs that interact with piwi-family proteins and include MIWI, MIWI2, HIWI, PRG-1, PRG-2 and MILI [200]. Not only are they DICER-independent, they are also distinct from miRNAs in their length (24–30 nucleotides) and expression patterns. They are expressed particularly in pachytene spermatocytes and round spermatids during spermatogenesis, indicative of a role of these small RNAs in development and protection of male germ cells from invasive transposable elements [201]. The piRNAs repress retrotransposons and regulate at the post-transcriptional level. The roles of piRNAs in spermatogenesis are supported by the well-established functions of their partner, the Piwi proteins. Recent evidence indicates that the piwi subfamily proteins are essential for stem cell self-renewal, the development of male germ cells and spermatogenesis [202]. For instance, spermatogenesis is arrested at the pachytene spermatocyte stage in Mili-knockout mice [203]. In Miwi2-knockout mice, spermatogenesis stops with meiotic arrest at the leptotene stage [204]. Germ cells in Miwi-deficient mice testis undergo meiosis without elongated spermatids or mature spermatozoa production [205]. Furthermore, the small non-coding RNAs Nct1 and Nct2 have recently been suggested to be piRNA precursors and are expressed particularly in pachytene spermatocytes [206], which also points to a role of piRNAs in the regulation of the meiotic stage in spermatogenesis. Although, Nct1 and Nct2-deficient mice display a decrease in a small cluster of piRNAs located on chromosome 2, it does not affect mouse spermatogenesis or fertility, suggesting that those piRNAs located on chromosome 2 are necessary to maintain transposon silencing [202]. Thus, it is likely that piRNAs are potentially involved in regulating the processes of meiosis and post-meiosis of male germ cell development.
Power of proteomics in male infertility
The proteomics techniques allows for the measurement of protein levels and accurately determines the changes in all proteins expressed and translated from a single genome in a tissue or cell. Proteins are identified with two-dimensional electrophoresis (2-DE) and mass spectrometry (MS) techniques, and the data are used to create the profiles of proteins in the given sample [260]. Proteomic technology serves as a diagnostic tool to probe the pathology and idiopathic causes of male infertility, and the seminal plasma and sperm proteins identified can also be validated by Western blot [207]. In addition, comprehensive studies of the sperm or seminal proteome can be instrumental in the identification of differences in protein expression between normal and abnormal sperm populations, enabling the identification of potential novel biomarkers for diagnosis, prognosis and development of therapeutic options to infertile men.
During the spermatogenesis process and sperm maturation in the epididymis and post-ejaculatory capacitation in the female reproductive tract, post-translational modifications (PTM) are critical for the functionality of spermatozoa [208–210]. PTMs that are formed by glycosylation, methylation, or phosphorylation are able to change the functional properties of spermatozoa and seminal plasma proteins [211]. Thus, identification of differentially expressed proteins and their modification patterns between fertile and infertile patients could provide an opportunity to study candidate genes and other abnormalities in search for mutations.
Proteomic profiling studies revealed that human sperm and seminal plasma contain a large number of proteins such as heat shock protein (HSP), fibronectin, semenogelins, and laminin, as well as enzymes such as serine protease, prostate-specific antigen (PSA), prostatic specific acid phosphatase (PSAP), and creatine kinase [212]. Additionally, antimicrobial proteins including lactoferrin, protease inhibitors such as R-1-antitrypsin, and transport proteins including albumin are also abundant in the semen [213]. The large number of identified proteins indicates the complex composition and function of human sperm. Wang et al., [199] identified 4675 human sperm proteins, of which 227 were testis-specific proteins. Approximately 403 different proteins have also been identified from the isolated sperm nuclei, the most prominent among them being the histones. Majority of sperm proteins have been functionally categorized as those involved in sperm movement and structural organization (34 %), energy and metabolism (27 %), stress response, protein folding and protein turnover (22 %), signaling and transport (8 %) and antioxidant activity (6 %) [214]. The others are essential in the capacitation of the spermatozoa, modulation of the immune responses in the uterus, formation of the tubal sperm reservoir and finally in both the sperm-zona pellucida (ZP) interaction and sperm and egg fusion.
Differential profiling studies between fertile and infertile patients have demonstrated that the identification of differentially expressed proteins could help elucidate mutations in candidate genes as well as identify biomarkers that can, in turn, help clinicians determine certain peptides or metabolites that may be linked to male infertility [215]. For instance, Sharma et al., [212] have reported that there is significant difference in protein expression between men with normal and abnormal sperm. In a study detailing oxidative stress, Sharma et al., also demonstrated that the increased expression of several proteins especially histone cluster 1H2ba (HIST1H2BA), mitochondrial malate dehydrogenase precursor (MDH2), transglutaminase-4 (TGM4), glutathione peroxidase-4 isoform A precursor (GPX4), glutamine synthetase (GLUL), heat shock proteins (HSP9 0B1 and HSPA5) in the ROS+ patients, suggests that these proteins can serve as potential biomarkers of oxidative stress [216]. Thacker et al., [217], found that four unique proteins including semenogelin II precursor, prolactin-induced protein, clusterin isoform 1, and PSA isoform 1 preproprotein were predominantly present in the semen of healthy men. In another study based on 2D SDS–PAGE followed by MALDI MS/MS analysis, Siva et al., [214] confirmed significant changes in intensity of proteasome alpha-3 subunit in asthenozoospermic samples when compared with normozoospermic controls. Significant positive correlation was found between proteasome alpha-3 subunit levels and rapid, linear progressive motility of the spermatozoa. In a preliminary study, Martinez-Heredia et al., [218] identified 17 proteins, particularly cytoskeletal actin-B, annexin-A5, cytochrome C oxidase-6B, histone H2A, prolactin-inducible protein and precursor, calcium binding protein-S100A9, clusterin precursor, dihydrolipoamide dehydrogenase precursor, fumarate hydratase precursor, heat shock protein-HSPA2, inositol-1 mono phosphatase, 3-mercapto-pyruvate sulfur transferase, dienoyl-CoA isomerase precursor and proteasome subunit-PSMB3, in asthenozoospermic sperm samples when compared with the normozoospermic individuals. Similarly, Zhao et al., [219], found 10 proteins to be differentially expressed in these samples when compared with the normozoospermic individuals. Li and Zhang [211] reported that HSP70-2, PLC, GPX4, β-tubulin and GAPDHS are associated with sperm motility, and thereby male fertility. They showed these proteins are potential markers for the mechanisms of infertility or radiotherapy, because their expression pattern is changed in these cases. β-tubulin is essential for sperm axoneme migration, flagellar movement and regulation. GAPDHS is also related to sperm energy supply. Sperm proteome studies on asthenozoospermia samples have also revealed that some component of the proteasome complex is differentially expressed, suggesting an importance of the proteasome complex in sperm motility [220]. These results suggest the validity of the proteomic approach to identify proteins with altered amounts in the human semen and sperm cells of idiopathic infertile patients. Therefore, proteomics studies will allow for a better understanding of PTMs and offer the opportunity to identify proteins that are differentially expressed in abnormal semen samples and potentially involved in idiopathic male infertility.
Diagnosis of male infertility
To date, the diagnosis of male infertility has been primarily based on physical exams, post-ejaculatory urine sample, blood tests and traditional semen analysis, with a strong emphasis on the assessment of semen volume and sperm concentration, motility and morphology. But mounting evidence suggests that routine evaluation of semen quality is insufficient to reflect the overall quality of sperm [213]. Large-scale molecular genetics analyses have indicated that ~3000 genes are involved in male reproduction [173]. Detailed mechanistic studies of these genes have aided in the development of diagnostic tests to test male infertility and therapeutic strategies to overcome infertility, particularly in men undergoing the ICSI procedure. To name a few, testicular mapping, genetic analysis at the chromosomal level (karyotyping), androgen receptor gene mutations, CF test and Y chromosome micro deletion analysis are being used routinely in diagnosis of male factor infertility [15]. In addition, some molecular cytogenetic methods such as comet assay, sperm-chromatin-dispersion assay (SCD), transferase-mediated dUTP nicked labeling (TUNEL), or sperm chromatin structure assay (SCSA) have been developed for the detection of DNA damage [4]. Although, the pre-implantation genetic diagnosis (PGD) technology is not a routine method in the evaluation of male infertility, it could potentially prevent the transmission of genetic anomalies to the offspring [4]. These tests can help identify DNA fragmentation, chromosomal defects, or the possibility of genetic diseases that can be passed on to children. Currently, Y chromosome micro deletions are analyzed by PCR-screening of STSs from different regions of the AZF on the long arm of the Y, as well as by using DNA probes of the genes (Fig. 1). In the recent years, it has become possible to detect the incidence of chromosomal abnormalities using a variety of high-powered polymerase chain reaction (PCR) techniques and multi-colour fluorescent in situ hybridization (FISH) analysis [4]. Before patients can be subjected to ICSI, FISH-sperm studies are necessary to understand whether there is a genetic cause for male infertility. If genetic abnormalities are suspected in either partner, counseling is recommended. Recently, further tests, such as ROS determination and chromatin fragmentation, have also been proposed for research purposes [115]. In light of recent proteome and transcriptome profiling analyses, researchers strongly advocate for additional new markers of fertility or infertility that could include sperm and seminal plasma proteins, complex population of RNAs including miRNAs and other small noncoding RNAs that are retained in mature sperm.
Conclusions
It appears that, in the recent decades, the quality of human sperm, especially, sperm counts are decreasing across the world. About 75 % of men who are infertile suffer from oligospermia or from asthenozoospermia. Decreasing sperm counts are attributed to the deleterious effects of many factors including environmental contamination to molecular genetics factors that affect sperm development and function. Spermatogenesis is a complex process that is regulated by many genes; however the molecular mechanisms involved are beginning to be unraveled. The male infertile state is produced by chromosomal aneuploidies and rearrangements, deletions of gene families present in AZF regions and other parts of the Y chromosome. Furthermore, there are an increasing number of autosomic and X-linked genes identified in humans that have critical roles in spermatogenesis and sperm functions. Apart from nuclear DNA defects, mtDNA mutations and deletions also contribute to the pathogenesis of male infertility. Although the majority of children conceived through ART (such as ICSI) seem normal, these artificial procedures may transmit these molecular genetic abnormalities to the offspring who manifest with higher risk of infertility, congenital abnormalities and morbidity. Consequently, it is necessary to evaluate male infertility utilizing a multidisciplinary approach involving genetics, epigenetics and at molecular levels to develop appropriate screens for abnormal phenotypes and to discover more effective solutions for an infertile couple’s problems. In addition, the development of techniques to test for genetic abnormalities or unfavorable polymorphisms before performing ART is critical. Technological progresses in molecular genetics and accumulation of data from animal models and from the human genome project represent an excellent background for a large-scale research in the field of genetics of male infertility. With the availability of modern technologies such as proteomics and transcriptomics we are now able to identify hundreds and thousands of proteins and RNAs, as biomarkers, in complex biological samples such as seminal plasma. Moreover, epigenetic studies offer an important window to understanding the role of cellular and molecular interactions with the genome in causing spermatogenesis deficiency and male infertility. This rapid burgeoning of research will better prepare clinicians to diagnose and treat infertile patients and take knowledgeable decisions and actions about the use of ART as well.
Footnotes
Eisa Tahmasbpour and Dheepa Balasubramanian have contributed equally
Capsule This review reports the genetic, molecular and phenotypic factors that greatly facilitate the understanding and treatment of a multi-factorial disorder, Male infertility.
References
- 1.Vogt PH. Molecular genetic of human male infertility: from genes to new therapeutic perspectives. Curr Pharm Des. 2004;10:1–29. doi: 10.2174/1381612043453261. [DOI] [PubMed] [Google Scholar]
- 2.Ji G, Long Y, Zhou Y, Huang C, Gu A, Wang X. Common variants in mismatch repair genes associated with increased risk of sperm DNA damage and male infertility. BMC Med. 2012;10:1–10. doi: 10.1186/1741-7015-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Kretser DM. Male infertility. Lancet. 1997;349:787–90. doi: 10.1016/s0140-6736(96)08341-9. [DOI] [PubMed] [Google Scholar]
- 4.Dada R, Thilagavathi J, Venkatesh S, Esteves SC, Agarwal A. Genetic testing in male infertility. Open Reprod Sci J. 2011;3:42–56. [Google Scholar]
- 5.Varghese AC, du Plessis SS, Agarwal A. Male gamete survival at stake: causes and solutions. Reprod Bio Med Online. 2008;17:866–880. doi: 10.1016/s1472-6483(10)60416-6. [DOI] [PubMed] [Google Scholar]
- 6.Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2008;4:41–9. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- 7.Benoff S, Jacob A, Hurley IR. Male infertility and environmental exposure to lead and cadmium. Hum Reprod. 2000;6:107–121. doi: 10.1093/humupd/6.2.107. [DOI] [PubMed] [Google Scholar]
- 8.Sharpe RM. Lifestyle and environmental contribution to male infertility. Br Med Bull. 2000;56630–642. [DOI] [PubMed]
- 9.Hosseinzadeh Colagar A, Jorsaraee GA, Tahmasbpour ME. Cigarette smoking and the risk of male infertility. Pakistan Biol Sci. 2007;10(21):3870–3874. doi: 10.3923/pjbs.2007.3870.3874. [DOI] [PubMed] [Google Scholar]
- 10.Hosseinzadeh Colagar A, Tahmasbpour ME. Ascorbic acid levels in seminal plasma of fertile and idiopathic infertile men: determination and its relationship to sperm quality. J Clin Biochem Nutr. 2009;45:144–149. doi: 10.3164/jcbn.08-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosseinzadeh Colagar A, Tahmasbpour Marzony E, Chaichi MJ. Zinc levels in seminal plasma are associated with sperm quality in fertile and infertile men. Nutr Res. 2009;29:82–88. doi: 10.1016/j.nutres.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal A, Allamaneni SSR. Oxidants and antioxidants in human fertility. Middle East Fertil Society J. 2004;9:187–197. [Google Scholar]
- 13.Hosseinzadeh Colagar A, Pouamir M, Tahmasbpour Marzony E, Jorsaraee GA. Relationship between seminal malondialdehyde and sperm parameters quality in the fertile and infertile men. Brazilian Archive Biol Tech. 2009;52(6):1387–1392. [Google Scholar]
- 14.Krausz C, Forti G, Mcelreavey K. The Y chromosome and male fertility and infertility. Int J Androl. 2003;26:70–75. doi: 10.1046/j.1365-2605.2003.00402.x. [DOI] [PubMed] [Google Scholar]
- 15.Krausz C, Degl’Innocenti S, Nuti F, Morelli A, Felici F, Sansone M, Varriale G, Forti G. Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility. Hum Mol Genet. 2006;15:2673–81. doi: 10.1093/hmg/ddl198. [DOI] [PubMed] [Google Scholar]
- 16.Walsh TJ, Pera RR, Turek PJ. The genetics of male infertility. Semin Reprod Med. 2009;27:124–136. doi: 10.1055/s-0029-1202301. [DOI] [PubMed] [Google Scholar]
- 17.O'Flynn O’Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: A review. Fertil Steril. 2010;93:1–12. doi: 10.1016/j.fertnstert.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 18.Asero P, La Vignera S, Lanzafame F. Genetic aspects of male infertility. J Androl Sci. 2010;17:1–16. [Google Scholar]
- 19.Tüttelmann F, Simoni M. Current Recommendations for Genetic Testing in Male Infertility. European Urological Review, 2008; 88-92.
- 20.Ferlin A, Raicu F, Gatta V, Zuccarello D, Palka G, Foresta C. Male infertility: role of genetic background. Reprod Biomed Online. 2007;14:734–45. doi: 10.1016/s1472-6483(10)60677-3. [DOI] [PubMed] [Google Scholar]
- 21.Maduro MR, Lamb DJ. Understanding the new genetics of male infertility. J Urol. 2002;168:2197–2205. doi: 10.1016/S0022-5347(05)64355-8. [DOI] [PubMed] [Google Scholar]
- 22.Ferlin A, Arredi B, Foresta C. Genetic causes of male infertility. Reprod Toxicol. 2006;22:133–141. doi: 10.1016/j.reprotox.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 23.Schiff JD, Palermo GD, Veeck LL, Goldstein M, Rosenwaks Z, Schlegel PN. Success of testicular sperm extraction and intracytoplasmic sperm injection in men with Klinefelter syndrome. J Clin Endocrinol Metab. 2005;90(11):6263–7. doi: 10.1210/jc.2004-2322. [DOI] [PubMed] [Google Scholar]
- 24.Visootsak J, Graham JM., Jr Klinefelter syndrome and other sex chromosomal aneuploidies. Orphanet J of Rare Diseases. 2006;1:42. doi: 10.1186/1750-1172-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichioka K, Utsunomiya N, Kohei N, Ueda N, Inoue K, Terai A. Adult onset of declining spermatogenesis in a man with nonmosaic Klinefelter's syndrome. Fertil Steril, 2006; 85(5): 1511 e1-2. [DOI] [PubMed]
- 26.Therman E, Susman M. Human Chromosome: Structure, Behaviour and Effects. New York: Springer-Verlag; 1993. [Google Scholar]
- 27.De Braekeleer M, Dao TN. Cytogenetic studies in male infertility: a review. Hum Reprod. 1991;6:245–50. [PubMed] [Google Scholar]
- 28.Ananthapur V, Avvari S, Tella S, Nallari P, Akka J. A Robertsonian Translocation rob (14;15) (q10:q10) in a patient with recurrent abortions: A case report. J Reprod Infertil. 2010;11(3):197–200. [PMC free article] [PubMed] [Google Scholar]
- 29.Georgiou I, Syrrou M, Pardalidis N, Karakitsios K, Mantzavinos T, Giotitsas ND, Dimitriadis F, Saito M, Miyagawa I, Tzoumis P, Sylakos A, Kanakas N, Moustakareas T, Baltogiannis D, Touloupides S, Giannakis D, Fatouros M, Sofikitis N. Genetic and epigenetic risks of intracytoplasmic sperm injection method. Asian J Androl. 2006;8:643–73. doi: 10.1111/j.1745-7262.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- 30.Keymolen K, Van Berkel K, Vorsselmans A, Staessen C, Liebaers L. Pregnancy Outcome in Carriers of Robertsonian Translocations. Am J Med Genet Part A. 2011;155:2381–2385. doi: 10.1002/ajmg.a.33941. [DOI] [PubMed] [Google Scholar]
- 31.Gunel M, Cavkaytar S, Ceylaner G, Batioglu S. Azoospermia and cryptorchidism in a male with a de novo reciprocal t(Y;16) translocation. Genet Couns. 2008;19:277–280. [PubMed] [Google Scholar]
- 32.Scriven PN, Flinter FA, Braude PR, Ogivie CM. Robertsonian translocations—reproductive risks and indications for preimplantation gen-etic diagnosis. Hum Reprod. 2001;16:2267–2273. doi: 10.1093/humrep/16.11.2267. [DOI] [PubMed] [Google Scholar]
- 33.Papadimas J, Goulis DG, Giannouli C, Papanicolaou A, Tarlatzis B, Bontis JN. Ambiguous genitalia, 45, X/46, XY mosaic karyotype, and Y chromosome microdeletions in a 17-year-old man. Fertil Steril. 2001;76:1261–1263. doi: 10.1016/s0015-0282(01)02877-1. [DOI] [PubMed] [Google Scholar]
- 34.Vogt PH. Human chromosome deletions in Yq11, AZF candidate genes and male infertility: history and update. Mol Hum Reprod. 1998;4:739–44. doi: 10.1093/molehr/4.8.739. [DOI] [PubMed] [Google Scholar]
- 35.Van der Auwera B, Van Roy N, De Paepe A, Hawkins JR, Liebaers I, Castedo S, Dumon J, Speleman F. Molecular cytogenetic analysis of XX males using Y-specific DNA sequences, including SRY. Hum Genet. 1992;89:23–28. doi: 10.1007/BF00207036. [DOI] [PubMed] [Google Scholar]
- 36.Abusheikha N, Lass A, Brisden P. XX males without SRY gene and with infertility. Hum Reprod. 2001;16:717–718. doi: 10.1093/humrep/16.4.717. [DOI] [PubMed] [Google Scholar]
- 37.Vogt PH, Fernandes S. Polymorphic DAZ gene family in poly-morphic structure of AZFc locus: artwork or functional for human sperma-togenesis? Acta Pathol Microbiol Immunol Scand. 2003;111:115–127. doi: 10.1034/j.1600-0463.2003.11101161.x. [DOI] [PubMed] [Google Scholar]
- 38.Foresta C, Ferlin A, Garolla A, Moro E, Pistorello M, Barbaux S, Rossato M. High frequency of well-defined Y-chromosome deletions in idiopathic Sertoli cell-only syndrome. Hum Reprod. 1998;13:302–307. doi: 10.1093/humrep/13.2.302. [DOI] [PubMed] [Google Scholar]
- 39.van der Ven K, Montag M, Peschka B, Leygraaf J, Schwanitz G, Haidl G, Krebs D, van der Ven H. Combined cytogenetic and Y chromosome microdeletion screening in males undergoing intracytoplasmic sperm injection. Mol Hum Reprod. 1997;3:699–704. doi: 10.1093/molehr/3.8.699. [DOI] [PubMed] [Google Scholar]
- 40.Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Kohn FM, Schill WB, Farah S, Ramos C, Hartmann M, Hartschuh W, Meschede D, Behre HM, Castel A, Nieschlag E, Weidner W, Gröne HJ, Jung A, Engel V, Haidl G. Human Y chromo-some azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1991;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 41.Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr Rev. 2001;22:226–39. doi: 10.1210/edrv.22.2.0425. [DOI] [PubMed] [Google Scholar]
- 42.Muslumanoglu MH, Turgut M, Cilingir O, Can C, Ozyurek Y, Artan S. Role of the AZFd locus in spermatogenesis. Fertil Steril. 2005;84:519–22. doi: 10.1016/j.fertnstert.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 43.Sertic J, Cvitkovic P, Myers A, Saiki RK, Rukavina AS. Genetic Markers of Male Infertility: Y Chromosome Microdeletions and Cystic Fibrosis Transmembrane Conductance Gene Mutations. Croat Med J. 2001;42(4):416–420. [PubMed] [Google Scholar]
- 44.Silber SJ, Alagappan R, Brown LG, Page DC. Y chromosome deletions in azoospermic and severely oligozoospermic men undergoing intracytoplasmic sperm injection after testicular sperm extraction. Hum Re-prod. 1998;13:3332–7. doi: 10.1093/humrep/13.12.3332. [DOI] [PubMed] [Google Scholar]
- 45.Fujisawa M, Shirakawa T, Kanzaki M, Okada H, Arakawa S, Kamidono S. Y-chromosome microdeletion and phenotype in cytogenetically normal men with idiopathic azoospermia. Fertil Steril. 2001;76:491–495. doi: 10.1016/s0015-0282(01)01955-0. [DOI] [PubMed] [Google Scholar]
- 46.Selva DM, Hammond GL. Human sex hormone-binding globulin is expressed in testicular germ cells and not in Sertoli cells. Horm Metab Res. 2006;38:230–235. doi: 10.1055/s-2006-925336. [DOI] [PubMed] [Google Scholar]
- 47.Selva DM, Hogeveen KN, Seguchi K, Tekpetey F, Hammond GL. A human sex hormone-binding globulin isoform accumulates in the acrosome during spermatogenesis. J Biol Chem. 2002;277:45291–45298. doi: 10.1074/jbc.M205903200. [DOI] [PubMed] [Google Scholar]
- 48.Foresta C, Ferlin A, Moro E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum Mol Genet. 2000;9:1161–1169. doi: 10.1093/hmg/9.8.1161. [DOI] [PubMed] [Google Scholar]
- 49.Vogt PH. Azoospermia factor (AZF) in Yq11: towards a molecular understanding of its function for human male fertility and spermatogenesis. Reprod Biomed Online. 2005;10:81–93. doi: 10.1016/s1472-6483(10)60807-3. [DOI] [PubMed] [Google Scholar]
- 50.Galan JJ, Guarducci E, Nuti F, Gonzalez A, Ruiz M, Ruiz A, Krausz C. Molecular analysis of estrogen receptor alpha gene AGATA haplotype and SNP12 in European populations: potential protective effect for cryptorchidism and lack of association with male infertility. Hum Reprod. 2007;22:444–9. doi: 10.1093/humrep/del391. [DOI] [PubMed] [Google Scholar]
- 51.Delbridge ML, Harry JL, Toder R, O'Neill RJ, Ma K, Chandley AC, Graves JA. A human candidate spermatogenesis gene, RBM1, is conserved and amplified on the marsupial Y chromosome. Nat Genet. 1997;15:131–136. doi: 10.1038/ng0297-131. [DOI] [PubMed] [Google Scholar]
- 52.Vogt PH. AZF deletions and Y chromosomal haplogroups: history and update based on sequence. Hum Reprod Update. 2005;11:319–336. doi: 10.1093/humupd/dmi017. [DOI] [PubMed] [Google Scholar]
- 53.Elliot DJ, Millar MR, Oghene K, Ross A, Kiesewetter F, Pryor J, McIntyre M, Hargreave TB, Saunders PTK, Vogt PH, Chandley AC, Cooke H. Expression of RBM in the nuclei of human germ cells is dependent on a critical region of the Y chromosome long arm. Proc Natl Acad Sci. 1997;94:3848–3853. doi: 10.1073/pnas.94.8.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lahn BT, Tang ZL, Zhou J, Barndt RJ, Parvinen M, Allis CD, Page DC. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci U S A. 2002;99:8707–8712. doi: 10.1073/pnas.082248899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahanta R, Gogoi A, Chaudhury PNB, Roy S, Bhattacharyya IK, Sharma P, Lahkar K. Microdeletions in the Y Chromosome of Infertile Males with Azoospermia, Oligozoospermia and Asthenozoospermia from Assam, India. World Applied Sciences Journal. 2012;17(10):1265–1270. [Google Scholar]
- 56.Vineeth VS, Malini SS. A Journey on Y Chromosomal Genes and Male Infertility. Int J Hum Genet. 2011;11(4):203–215. [Google Scholar]
- 57.de Carvalho CMB, Zuccherato LW, Fujisawa M, Shirakawa T, Ribeiro-dos-Santos AKC, Santos SEB, Pena SDJ, Santos FR. Study of AZFc partial deletion gr/gr in fertile and infertile Japanese males. J Hum Genet. 2006;51:794–799. doi: 10.1007/s10038-006-0024-2. [DOI] [PubMed] [Google Scholar]
- 58.Krausz C, Quintana-Murci L. McElreaveyK. Prognostic value of Y deletion analysis: what is the clinical prognostic value of Y chromosome microdeletion analysis? Hum Re-prod. 2000;15:1431–1434. doi: 10.1093/humrep/15.7.1431. [DOI] [PubMed] [Google Scholar]
- 59.Pinho MJ, Neves R, Costa P, Ferras C, Sousa M, Alves C, Almeida C, Fernandes S, Silva J, Ferras L, Barros A. Unique t(Y;1)(q12;q12) reciprocal translocation with loss of the heterochromatic region of chromosome 1 in a male with azoospermia due to meiotic arrest: a case report. Hum Reprod. 2005;20:689–696. doi: 10.1093/humrep/deh653. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J, Qiu SD, Li SB, Zhou DX, Tian H, Huo YW, Ge L, Zhang QY. Novel mutations in ubiquitin-specific protease 26 gene might cause spermatogenesis impairment and male infertility. Asian J Androl. 2007;9(6):809–814. doi: 10.1111/j.1745-7262.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 61.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 62.Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JW, Oates RD, Silber S, van der Veen F, Page DC, Rozen S. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 63.Lardone MC, Parodi DA, Ebensperger M, Penaloza P, Cornejo V, Valdevenito R, et al. AZFc partial deletions in Chilean men with severe sper-matogenic failure. Fertil Steril. 2007;88:1318–26. doi: 10.1016/j.fertnstert.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 64.Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, Schlegel PN. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod. 2003;18:1660–1665. doi: 10.1093/humrep/deg348. [DOI] [PubMed] [Google Scholar]
- 65.Eloualid A, Rhaissi H, Reguig A, Bounaceur S. El houate B, Abidi O, Charif M, Louanjli N, Chadli E, Barakat A, Bashamboo A, McElreavey K, Rouba H. Association of Spermatogenic Failure with the b2/b3 Partial AZFc Deletion. PLoS ONE. 2012;7(4):e34902. doi: 10.1371/journal.pone.0034902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Repping S, van Daalen SK, Korver CM, Brown LG, Marszalek JD, Gianotten J, Oates RD, Silber S, van der Veen F, Page DC, Rozen L. A family of human Y chromosomes has dispersed throughout northern eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics. 2004;83:1046–1052. doi: 10.1016/j.ygeno.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 67.de Llanos M, Ballesca JL, Gazquez C, Margarit E, Oliva R. High frequency of gr/gr chromosome Y deletions in consecutive oligospermic ICSI candidates. Hum Reprod. 2005;20:216–220. doi: 10.1093/humrep/deh582. [DOI] [PubMed] [Google Scholar]
- 68.Tuttelmann F, Rajpert-De Meyts E, Nieschlag E, Simoni M. Gene polymorphisms and male infertility- a meta-analysis and literature review. RBM Online. 2007;15:643–658. doi: 10.1016/s1472-6483(10)60531-7. [DOI] [PubMed] [Google Scholar]
- 69.Stouffs K, Lissens W, Tournaye H, Haentjens P. What about gr/gr deletions and male infertility? Systematic review and meta-analysis. Hum Reprod Update. 2011;17:197–209. doi: 10.1093/humupd/dmq046. [DOI] [PubMed] [Google Scholar]
- 70.Ferlin A, Moro E, Garolla A, Foresta C. Human male infertility and Y chromosome deletions: role of the AZF-candidate genes DAZ, RBM and DFFRY. Hum Reprod. 1999;14:1710–6. doi: 10.1093/humrep/14.7.1710. [DOI] [PubMed] [Google Scholar]
- 71.Kuo PL, Wang ST, Lin YM, Lin YH, Teng YN, Hsu CC. Expression profiles of the DAZ gene family in human testis with and without spermatogenic failure. Fertil Steril. 2004;81:1034–1040. doi: 10.1016/j.fertnstert.2003.08.047. [DOI] [PubMed] [Google Scholar]
- 72.Teng YN, Lin YM, Lin YH, Tsao SY, Hsu CC, Lin SJ, Tsai WC, Kuo PL. Association of a single-nucleotide polymorphism of the deleted-in-azoospermia-like gene with susceptibility to spermatogenic failure. J Clin Endocrinol Metab. 2002;87:5258–5264. doi: 10.1210/jc.2002-020016. [DOI] [PubMed] [Google Scholar]
- 73.Fernandes S, Huellen K. Gonc¸alves J, Dukal H, Zeisler J, De Meyts ER, Skakkebaek NE, Habermann B, Krause W, Sousa M et al. High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligo-zoospermia. Mol Hum Reprod. 2002;8:286–298. doi: 10.1093/molehr/8.3.286. [DOI] [PubMed] [Google Scholar]
- 74.Ravel C, Chantot-Bastaraud S, Houate BE, Rouba H, Legendre M, Lorencxo D, Mandelbaum J, Siffroi JP, McElreavey K. Y-chromosome AZFc structural architecture and relationship to male fertility. Fertil Steril. 2009;92:1924–33. doi: 10.1016/j.fertnstert.2008.08.135. [DOI] [PubMed] [Google Scholar]
- 75.Sadeghi-Nejad H, Farrokhi F. Genetics of azoospermia: current knowledge, clinical implications, and future directions. Part II. Y chromosome microdeletions. Urol J. 2007;4:192–206. [PubMed] [Google Scholar]
- 76.Knower KC, Kelly S, Harley VR. Turning on the male-SRY, SOX9 and sex determination in mammals. Cytogenet Genome Res. 2003;101:185–198. doi: 10.1159/000074336. [DOI] [PubMed] [Google Scholar]
- 77.de Carvalho CMB, Santos FR. Human Y-chromosome variation and male dysfunction. J of Mol and Genet Med. 2005;1(2):63–75. doi: 10.4172/1747-0862.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shahid M, Dhillion VS, Jain N, Hedau S, Diwakar S, Sachdeva P, et al. Two new novel point mutations localized upstream and downstream of the HMG box region of the SRY gene in three Indian 46, XY females with sex reversal and gonadal tumour formation. Mol Hum Reprod. 2004;10:521–526. doi: 10.1093/molehr/gah071. [DOI] [PubMed] [Google Scholar]
- 79.Dasari VK, Goharderakhshan RZ, Perinchery G, Li LC, Tanaka Y, Alonzo J, Dahiya R. Expression analysis of Y chromosome genes in human prostate cancer. Urol. 2001;165:1335–1341. [PubMed] [Google Scholar]
- 80.Eckardstein S, Cooper TG, Rutscha K, Meschede D, Horst J, Nieschlag E. Seminal plasma characteristics as indicators of cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations in men with obstructive azoospermia. Fertil Steril. 2000;73:1226–31. doi: 10.1016/s0015-0282(00)00516-1. [DOI] [PubMed] [Google Scholar]
- 81.Chillon M, Casals T, Mercier B, Bassas L, Lissens W, Silber S, Romey MC, Ruiz-Romero J, Verlingue C, Claustres M, Nunes V, Férec C, Estivill X. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. N Engl J Med. 1995;332(22):1475–80. doi: 10.1056/NEJM199506013322204. [DOI] [PubMed] [Google Scholar]
- 82.Hiort O, Holterhus PM, Horter T, Schulze W, Kremke B, Bals-Pratsch M, Sinnecker GH, Kruse K. Significance of mutations in the androgen receptor gene in males with idiopathic infertility. J Clin Endocrinol Metab. 2000;85(8):2810–5. doi: 10.1210/jcem.85.8.6713. [DOI] [PubMed] [Google Scholar]
- 83.Yong EL, Tut TG, Ghadessy FJ, Prins G, Ratnam SS. Partial androgen insensitivity and correlations with the predicted three dimensional structure of the androgen receptor ligand-binding domain. Mol Cell Endocrinol. 1998;137:41–50. doi: 10.1016/s0303-7207(97)00229-3. [DOI] [PubMed] [Google Scholar]
- 84.Mifsud A, Sim CK, Boettger-Tong H, Moreira S, Lamb DJ, Lipshultz LI, Young EL. Trinucleotide (CAG) repeat polymorphisms in the androgen receptor gene: molecular markers of risk for male infertility. Fertil Steril. 2001;75:275–81. doi: 10.1016/s0015-0282(00)01693-9. [DOI] [PubMed] [Google Scholar]
- 85.Mengual L, Oriola J, Ascaso C, Ballesca JL, Oliva R. An Increased CAG Repeat Length in the Androgen Receptor Gene in Azoospermic ICSI Candidates. J Androl. 2003;24:279–284. doi: 10.1002/j.1939-4640.2003.tb02673.x. [DOI] [PubMed] [Google Scholar]
- 86.Cram DS, Song B, McLachlan RI, Trounson AO. CAG trinucleotide repeats in the androgen receptor gene of infertile men exhibit stable inheritance in female offspring conceived after ICSI. Mol Hum Reprod. 2000;6(9):861–6. doi: 10.1093/molehr/6.9.861. [DOI] [PubMed] [Google Scholar]
- 87.Foresta C, Ferlin A. Role of INSL3 and LGR8 in cryptorchidism and testicular functions. Reprod BioMed Online. 2004;9:294–298. doi: 10.1016/s1472-6483(10)62144-x. [DOI] [PubMed] [Google Scholar]
- 88.Mokosch A, Bernecker C, Willenberg HS, Neumann NJ. Kallmann syndrome. Hautarzt. 2011;62:728–30. doi: 10.1007/s00105-011-2237-3. [DOI] [PubMed] [Google Scholar]
- 89.Hwang SH, Lee SM, Seo EJ, Choi KU, Park HJ, Park NC, Choi J, Lee EY. A case of male infertility with a reciprocal translocation t(X;14)(p11.4;p12) Korean J Lab Med. 2007;27(2):139–42. doi: 10.3343/kjlm.2007.27.2.139. [DOI] [PubMed] [Google Scholar]
- 90.Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 91.Kelly TL, Neaga OR, Schwahn BC, Rozen R, Trazler JM. Infertility in 5, 10 methylenetetrahydrofolate reductase (MTHFR)-deficient male mice is partially alleviated by lifetime dietary betaine supplementation. Biol Reprod. 2005;72:667–77. doi: 10.1095/biolreprod.104.035238. [DOI] [PubMed] [Google Scholar]
- 92.Singh AR, Vrtel R, Vodicka R, Dhaifalah I, Konvalinka D, Janikova M, Santavy J. Y Chromosome and Male Infertility. Int J Hum Genet. 2005;5(4):225–235. [Google Scholar]
- 93.Singh K, Singh SK, Raman R. MTHFR A1298C polymorphism and idiopathic male infertility. J Grad Med. 2010;56:267–269. doi: 10.4103/0022-3859.70935. [DOI] [PubMed] [Google Scholar]
- 94.Lazaros L, Xita N, Kaponis A, Zikopoulos K, Sofikitis N, Georgiou I. Evidence for association of sex hormone-binding globulin and androgen receptor genes with semen quality. Andrologia. 2008;40:186–91. doi: 10.1111/j.1439-0272.2008.00835.x. [DOI] [PubMed] [Google Scholar]
- 95.Rochira V, Granata AR, Madeo B, Zirilli L, Rossi G, Carani C. Estrogens in males: what have we learned in the last 10 years?Asian. J Androl. 2005;7:3–20. doi: 10.1111/j.1745-7262.2005.00018.x. [DOI] [PubMed] [Google Scholar]
- 96.Sinkevicius KW, Laine M, Lotan TL, Woloszyn K, Richburg JH, Greene GL. Estrogen-dependent and independent estrogen receptor-alpha signaling separately regulate male fertility. Endocrinology. 2009;150:2898–2905. doi: 10.1210/en.2008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ferlin A, Tessari A, Ganz F, Marchina E, Barlati S, Garolla A, Engl B, Foresta C. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet. 2005;42:209–213. doi: 10.1136/jmg.2004.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Suzuki Y, Sasagawa I, Itoh K. AshidaJ, Muroya K, Ogata T. Estrogen receptor alpha gene polymorphismis associated with idiopathic azoospermia. Fertil Steril. 2002;78:1341–1343. doi: 10.1016/s0015-0282(02)04267-x. [DOI] [PubMed] [Google Scholar]
- 99.Robertson KM, O’Donnell L, Jones ME, Meachem SJ, Boon WC, Fisher CR, Graves KH, McLachlan RI, Simpson ER. Impairment of sperma-togenesis in mice lacking a functional aromatase (cyp 19) gene. Proc Natl Acad Sci. 1999;96:7986–7991. doi: 10.1073/pnas.96.14.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guarducci E, Nuti F, Becherini L, Rotondi M, Balercia G, Forti G, Krausz C. Estrogen receptor α promoter polymorphism: stronger estrogen action is coupled with lower sperm count. Hum Reprod. 2006;21:994–1001. doi: 10.1093/humrep/dei439. [DOI] [PubMed] [Google Scholar]
- 101.Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, Sar M, Korach KS, Gustafsson JA, Smithies O. Generation and reproductive phenotypes of mice lacking estrogen receptor beta.Proc Natl Acad Sci, 1998; 95:15677. [DOI] [PMC free article] [PubMed]
- 102.Aschim EL, Giwercman A, Stahl O, Eberhard J, Cwikiel M, Nordenskjold A, Haugen TB, Grotmol T, Giwercman YL. The RsaI polymorphism in the estrogen receptor-beta gene is associated with male infertility. J Clin Endocrinol Metab. 2005;90:5343. doi: 10.1210/jc.2005-0263. [DOI] [PubMed] [Google Scholar]
- 103.Khattri A, Pandey RK, Gupta NJ, Chakravarty B, Deendayal M, Singh L, Thangaraj K. CA repeat and RsaI polymorphisms in ERbeta gene are not associated with infertility in Indian men. Int J Androl. 2007;32:81–87. doi: 10.1111/j.1365-2605.2007.00821.x. [DOI] [PubMed] [Google Scholar]
- 104.Khattri A, Pandey RK, Gupta NJ, Chakravarty B, Deenadayal M, Singh L, Thangaraj K. Estrogen receptor β gene mutations in Indian infertile men. Mol Hum Reprod. 2009;15:513–520. doi: 10.1093/molehr/gap044. [DOI] [PubMed] [Google Scholar]
- 105.Balkan M, Gedik A, Akkoc H, Ay O, Erdal ME, Isi H, Budak T. FSHR Single Nucleotide Polymorphism Frequencies in Proven Fathers and Infertile Men in Southeast Turkey. J Biomed Biotechnol. 2010;2010:1–5. doi: 10.1155/2010/640318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zamudio NM, Chong S, O’Bryan MK. Epigenetic regulation in male germ cells. Reproduction. 2008;136:131–146. doi: 10.1530/REP-07-0576. [DOI] [PubMed] [Google Scholar]
- 107.Zhang F, Lu C, Li Z, Xie P, Xia Y, Zhu X, Wu B, Cai X, Wang X, Qian J, Wang X, Jin L. Partial deletions are associated with an increased risk of complete deletion in AZFc: a new insight into the role of partial AZFc deletions in male infertility. J Med Genet. 2007;44:437–444. doi: 10.1136/jmg.2007.049056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang J, Shao XG, Shi YB, Yan L, Wang L, Tian H, Qiu SD. Polymorphism of Usp26 correlates with idiopathic male infertility. Zhonghua Nan Ke Xue. 2012;18(2):105–8. [PubMed] [Google Scholar]
- 109.Lee IW, Kuan LC, Lin CH, Pan HA, Hsu CC, Tsai YC, Kuo PL, Teng YN. Association of USP26 haplotypes in men in Taiwan, China with severe spermatogenic defect. Asian J Androl. 2008;10:896–904. doi: 10.1111/j.1745-7262.2008.00439.x. [DOI] [PubMed] [Google Scholar]
- 110.Ribarski I, Lehavi O, Yogev L, Hauser R, Bar-Shira Maymon B, Botchan A, Paz G, Yavetz H, Kleiman SE. USP26 gene variations in fertile and infertile men. Hum Reprod. 2009;24:477–484. doi: 10.1093/humrep/den374. [DOI] [PubMed] [Google Scholar]
- 111.Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. Possible role of USP26 in patients with severely impaired spermatogenesis. Eur J Hum Genet. 2005;13:336–40. doi: 10.1038/sj.ejhg.5201335. [DOI] [PubMed] [Google Scholar]
- 112.Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, Davidson I, Wang PJ. Abnormal Sperm in Mice Lacking the Taf7l Gene. Molecular and Cellular Biology, 2007; 2582-2589. [DOI] [PMC free article] [PubMed]
- 113.Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Akinloye O, Gromoll J, Callies C, Nieschlag E, Simoni M. Mutation analysis of the X-chromosome linked, testis-specific TAF7L gene in spermatogenic failure. Andrologia. 2007;39:190–195. doi: 10.1111/j.1439-0272.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- 115.Kumar DP, Sangeetha N. Mitochondrial DNA mutations and male infertility. Indian J Hum Genet. 2009;15(3):93–97. doi: 10.4103/0971-6866.60183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Alcivar AA, Hake LE, Millette CF, Trasler JM, Hecht NB. Mitochondrial gene expression in male germ cells of the mouse. Dev Biol. 1989;135:263–271. doi: 10.1016/0012-1606(89)90178-4. [DOI] [PubMed] [Google Scholar]
- 118.Solano A, Playan A, Lopez-Perez MJ, Montoya J. Genetic diseases of human mitochondrial DNA. Salud Publica Mex. 2001;43:1–11. [PubMed] [Google Scholar]
- 119.Guney AI, Javadova D, Kırac D, Ulucan K, Koc G, Ergec D, Tavukcu H, Tarcan T. Detection of Y chromosome microdeletions and mitochondrial DNA mutations in male infertility patients. Genet Mol Res. 2012;11:1039–1048. doi: 10.4238/2012.April.27.2. [DOI] [PubMed] [Google Scholar]
- 120.Carra E, Sangiorgi D, Gattuccio F, Rinaldi AM. Male infertility and mitochondrial DNA. Biochem Biophys Res Commun. 2004;322:333–339. doi: 10.1016/j.bbrc.2004.07.112. [DOI] [PubMed] [Google Scholar]
- 121.St John JC, Jokhi RP, Barratt CL. Men with oligoasthenoteratozoospermia harbour higher numbers of multiple mitochondrial DNA deletions in their spermatozoa, but individual deletions are not indicative of overall etiology. Mol Hum Reprod. 2001;7:103–111. doi: 10.1093/molehr/7.1.103. [DOI] [PubMed] [Google Scholar]
- 122.May-Panloup P, Chretien MF, Savagner F, Vasseur C, Jean M, Malthiery Y, Reynier P. Increased sperm mitochondrial DNA content in male infertility. Hum Reprod. 2003;18:550–556. doi: 10.1093/humrep/deg096. [DOI] [PubMed] [Google Scholar]
- 123.Folgero T, Bertheussen K, Lindal S, Torbergsen T, Oian P. Mitochondrial disease and reduced sperm motility. Hum Reprod. 1993;8:1863–1868. doi: 10.1093/oxfordjournals.humrep.a137950. [DOI] [PubMed] [Google Scholar]
- 124.Lestienne P, Reynier P, Chretien MF, Penisson-Besnier I, Malthièry Y, Rohmer V. Oligoasthenospermia associated with multiple mitochondrial DNA rearrangements. Mol Hum Reprod. 1997;3:811–814. doi: 10.1093/molehr/3.9.811. [DOI] [PubMed] [Google Scholar]
- 125.Hoshi K, Kasai T, Ogawa K, Mizuno K, Nagai S, Uchida Y, Ohta S, Fujie M, Suzuki K, Hirata S. Relationship between sperm mitochondrial membrane potential, sperm motility, and fertility potential. Asian J Androl. 2002;4:97–103. [PubMed] [Google Scholar]
- 126.Kao SH, Chao HT, Wei YH. Multiple deletions of mitochondrial DNA are associated with the decline of motility and fertility of human spermatozoa. Mol Hum Reprod. 1998;4:657–666. doi: 10.1093/molehr/4.7.657. [DOI] [PubMed] [Google Scholar]
- 127.Mundy AJ, Ryder TA, Edmonds DK. Asthenozoospermia and the human sperm mid-piece. Hum Reprod. 1995;10:116–119. doi: 10.1093/humrep/10.1.116. [DOI] [PubMed] [Google Scholar]
- 128.Hecht NB, Liem H, Kleene KC, Distel RJ, Ho SM. Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev Biol. 1984;102:452–461. doi: 10.1016/0012-1606(84)90210-0. [DOI] [PubMed] [Google Scholar]
- 129.Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 130.Kumar V, Hassan MI, Tomar AK, Kashav T, Nautiyal J, Singh S, Yadav S. Proteomic analysis of heparin-binding proteins fromhuman seminal plasma: a step towards identification of molecular markers of male fertility. J Biosci. 2009;34:899–908. doi: 10.1007/s12038-009-0104-5. [DOI] [PubMed] [Google Scholar]
- 131.Folgero T, Torbergsen T, Oian P. The 3243 MELAS mutation in a pedigree with MERRF. Eur Neurol. 1995;35:168–171. doi: 10.1159/000117115. [DOI] [PubMed] [Google Scholar]
- 132.Kao SH, Chao HT, Wei YH. Mitochondrial deoxyribonucleic acid 4977-bp deletion is associated with diminished fertility and motility of human sperm. Biol Reprod. 1995;52:729–736. doi: 10.1095/biolreprod52.4.729. [DOI] [PubMed] [Google Scholar]
- 133.Sampson MJ, Decker WK, Beaudet AL, Ruitenbeek W, Armstrong D, Hicks MJ, Craigen WJ. Immotile sperm and infertility in mice lacking mitochondrial voltage-dependent anion channel type 3. J Biol Chem. 2001;276:39206–12. doi: 10.1074/jbc.M104724200. [DOI] [PubMed] [Google Scholar]
- 134.Holyoake AJ, Sin IL, Benny PS, Sin FY. Association of a novel human mtDNA ATPase6 mutation with immature sperm cells. Andrologia. 1999;31:339–345. doi: 10.1046/j.1439-0272.1999.00150.x. [DOI] [PubMed] [Google Scholar]
- 135.O’Connell M, McClure N, Lewis SE. A comparison of mitochondrial and nuclear DNA status in testicular sperm from fertile men and those with obstructive azoospermia. Hum Reprod. 2002;17:1571–1577. doi: 10.1093/humrep/17.6.1571. [DOI] [PubMed] [Google Scholar]
- 136.Hosseinzadeh Colagar A, Karimi F. large scale deletions of the mitochondrial DNA in an asthenoterato and oligoasthenoterato-spermic men. Mitochondrial DNA. 2013 doi: 10.3109/19401736.2013.796512. [DOI] [PubMed] [Google Scholar]
- 137.St John JC, Jokhi RP, Barratt CLR. The impact of mitochondrial genetics on male infertility. Int J Androl. 2005;28:65–73. doi: 10.1111/j.1365-2605.2005.00515.x. [DOI] [PubMed] [Google Scholar]
- 138.Kumar R, Bhat A, Bamezai RN, Shamsi MB, Kumar R, Gupta NP, Ammini AC, Aron M, Sharma RK, Dada R. Necessity of nuclear and mitochondrial genome analysis prior to ART/ICSI. Indian J Biochem Biophys. 2007;44:437–42. [PubMed] [Google Scholar]
- 139.Lambard S, Galeraud-Denis I, Martin G, Levy R, Chocat A, Carreau S. Analysis and significance of mRNA in human ejaculated sperm from normozoospermic donors: relationship to sperm motility and capacitation. Mol Hum Reprod. 2004;10:535–41. doi: 10.1093/molehr/gah064. [DOI] [PubMed] [Google Scholar]
- 140.Kumar R, Venkatesh S, Kumar M, Tanwar M, Shasmsi MB, Kumar R, Gupta NP, Sharma RK, Talwar P, Dada R. Oxidative stress and sperm mitochondrial DNA mutation in idiopathic oligoasthenozoospermic men. Indian J Biochem Biophys. 2009;46:172–177. [PubMed] [Google Scholar]
- 141.Thangaraj K, Joshi MB, Reddy AG, Rasalkar AA, Singh L. Sperm mitochondrial mutations as a cause of low sperm motility. J Androl. 2003;24:388–392. doi: 10.1002/j.1939-4640.2003.tb02687.x. [DOI] [PubMed] [Google Scholar]
- 142.Spiropoulos J, Turnbull DM, Chinnery PF. Can mitochondrial DNA mutations cause sperm dysfunction? Mol Hum Reprod. 2002;8:719–721. doi: 10.1093/molehr/8.8.719. [DOI] [PubMed] [Google Scholar]
- 143.Khosronezhad N, Hosseinzadeh Colagar A, Jorsarayi SGA. T26248G-transversion mutation in exon7 of the putative methyltransferase Nsun7 gene causes a change in protein folding associated with reduced sperm motility in asthenospermia men. Reprod Fertil and Dev. 2013 doi: 10.1071/RD13371. [DOI] [PubMed] [Google Scholar]
- 144.Rovio AT, Marchington DR, Donat S, Schuppe HC, Abel J, Fritsche E, Elliott DJ, Laippala P, Ahola AL, McNay D, Harrison RF, Hughes B, Barrett T, Bailey DM, Mehmet D, Jequier AM, Hargreave TB, Kao SH, Cummins JM, Barton DE, Cooke HJ, Wei YH, Wichmann L, Poulton J, Jacobs HT. Mutations at the mitochondrial DNA polymerase (POLG) locus associated with male infertility. Nature Genet. 2001;29:261–262. doi: 10.1038/ng759. [DOI] [PubMed] [Google Scholar]
- 145.Chen SS, Chang LS, Chen HW, Wei YH. Polymorphisms of glutathione S-transferase M1 and male infertility in Taiwanese patients with varicocele. Hum Reprod. 2002;17:718–725. doi: 10.1093/humrep/17.3.718. [DOI] [PubMed] [Google Scholar]
- 146.St John JC, Sakkas D, Barrat CLR. A role for mitochondrial DNA in sperm survival. J Androl. 2000;21:189–199. [PubMed] [Google Scholar]
- 147.Shamsi MB, Kumar R, Bhatt A, Bamezai RNK, Kumar R, Gupta NP, Das TK, Dada R. Mitochondrial DNA Mutations in etiopathogenesis of male infertility. Indian J Urol. 2008;24(2):150–154. doi: 10.4103/0970-1591.40606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rajender S, Avery K, Agarwal A. Epigenetics, spermatogenesis and male infertility. Mutat Res. 2011;727:62–71. doi: 10.1016/j.mrrev.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 149.Rajender S, Agarwal A. Aberrant epigenetic modifications in male infertility. Open Reprod Sci J. 2011;3:57–64. [Google Scholar]
- 150.Dada R, Kumar M, Jesudasan R, Fernández LJ, Gosalvez J, Agarwal A. Epigenetics and its role in male infertility. J Assist Reprod Genet. 2012 doi: 10.1007/s10815-012-9715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Biermann K, Steger K. Epigenetics in male germ cells. J Androl. 2007;28:466–480. doi: 10.2164/jandrol.106.002048. [DOI] [PubMed] [Google Scholar]
- 152.Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS ONE. 2007;2:e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Khazamipour N, Noruzinia M, Fatehmanesh P, Keyhanee M, Pujol P. MTHFR promoter hypermethylation in testicular biopsies of patients with non-obstructive azoospermia: the role of epigenetics in male infertility. Hum Reprod. 2009;24:2361–2364. doi: 10.1093/humrep/dep194. [DOI] [PubMed] [Google Scholar]
- 154.Wu W, Shen O, Qin Y, Niu X, Lu C, Xia Y, Song L, Wang S, Wang X. Idiopathic male infertility is strongly associated with aberrant promoter methylation of methylenetetrahydrofolate reductase (MTHFR) PLoS ONE. 2010;5:e13884. doi: 10.1371/journal.pone.0013884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–33. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 156.Poplinski A, Tuttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–649. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 157.Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, Gut I, Vaiman D, Jouannet P, Tost J, Jammes H. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18:73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, Grootegoed JA. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 160.Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- 161.Oliva R. Protamines and male infertility. Hum Reprod Update. 2006;12:417–35. doi: 10.1093/humupd/dml009. [DOI] [PubMed] [Google Scholar]
- 162.Oliva R, Bazett-Jones D, Mezquita C, Dixon GH. Factors affecting nucleosome disassemble by protamines ‘in vitro’. Histone hyperacetylation and chromatin structure, time dependence, and the size of the sperm nuclear proteins. J Biol Chem. 1987;262:17016–17025. [PubMed] [Google Scholar]
- 163.Oliva R, Dixon GH. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog Nucleic Acid Res Mol Biol. 1991;40:25–94. doi: 10.1016/s0079-6603(08)60839-9. [DOI] [PubMed] [Google Scholar]
- 164.Wu JY, Means AR. Ca2+/calmodulin-dependent protein kinase IV is expressed in spermatids and targeted to chromatin and the nuclear matrix. J Biol Chem. 2000;275:7994–9. doi: 10.1074/jbc.275.11.7994. [DOI] [PubMed] [Google Scholar]
- 165.Oliva R, Mezquita C. Histone H4 hyperacetylation and rapid turnover of acetyl groups in transcriptionally inactive rooster testis spermatids. Nucleic Acids Res. 1982;10:8049–8059. doi: 10.1093/nar/10.24.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Torregrosa N, Dominguez-Fandos D, Camejo MI, Shirley CR, Meistrich ML, Ballesca JL, Oliva R. Protamine 2 precursors, protamine 1/protamine 2 ratio, DNA integrity and other sperm parameters in infertile patients. Hum Reprod. 2008;21:2084–2089. doi: 10.1093/humrep/del114. [DOI] [PubMed] [Google Scholar]
- 167.Aoki VW, Liu L, Carrell DT. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod. 2005;20:1298–1306. doi: 10.1093/humrep/deh798. [DOI] [PubMed] [Google Scholar]
- 168.Faure AK, Pivot-Pajot C, Kerjean A, Hazzouri M, Pelletier R, Peoc’h M, Sele B, Khochbin S, Rousseaux S. Misregulation of histone acetylation in Sertoli cell-only syndrome and testicular cancer. Mol Hum Reprod. 2003;9:757–763. doi: 10.1093/molehr/gag101. [DOI] [PubMed] [Google Scholar]
- 169.Sonnack V, Failing K, Bergmann M, Steger K. Expression of hyperacetylated histone H4 during normal and impaired human spermatogenesis. Andrologia. 2002;34:384–390. doi: 10.1046/j.1439-0272.2002.00524.x. [DOI] [PubMed] [Google Scholar]
- 170.Steilmann C, Cavalcanti MCO, Bartkuhn M, Pons-Kuhnemann J, Schuppe H, Weidner W, Steger K, Paradowska A. The interaction of modified histones with the bromodomain testis-specific (BRDT) gene and its mRNA level in sperm of fertile donors and subfertile men. Reproduction. 2010;140:435–443. doi: 10.1530/REP-10-0139. [DOI] [PubMed] [Google Scholar]
- 171.Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–51. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 172.Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- 173.Li C, Zhou X. Gene transcripts in spermatozoa: Markers of male infertility. Clin Chim Acta. 2012;413:1035–1038. doi: 10.1016/j.cca.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 174.Li C, Zheng L, Wang C, Zhou X. Absence of nerve growth factor and comparison of tyrosine kinase receptor A levels in mature spermatozoa from oligoasthenozoospermic, asthenozoospermic and fertile men. Clin Chim Acta. 2010;411:1482–6. doi: 10.1016/j.cca.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 175.Cai ZM, Gui YT. GuoX, Zhang LB, Wang H, Yu J. Low expression of glycoprotein subunit 130 in ejaculated spermatozoa from asthenozoospermic men. J Androl. 2006;27:645–52. doi: 10.2164/jandrol.106.000562. [DOI] [PubMed] [Google Scholar]
- 176.Guo X, Gui YT, Tang AF, Lu LH, Gao X, Cai ZM. Differential expression of VASA gene in ejaculated spermatozoa from normozoospermic men and patients with oligozoospermia. Asian J Androl. 2007;9:339–44. doi: 10.1111/j.1745-7262.2007.00253.x. [DOI] [PubMed] [Google Scholar]
- 177.Papaioanou MD, Nef S. MicroRNAs in the testis: building up male fertility. J Andeol. 2010;31:26–33. doi: 10.2164/jandrol.109.008128. [DOI] [PubMed] [Google Scholar]
- 178.Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, Diamond MP. A survey of small RNAs in human sperm. Hum Reprod. 2011;26:3401–3412. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Esquela-Kerscher A, Slack FJ. Oncomirs- microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 180.Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard M, Durand P, Samarut J, Pain B, Rouault J. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA. 2010;16:720–731. doi: 10.1261/rna.1963810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Belleannee C, Calvo E, Thimon V, Cyr DG, Legare C, Garneau L, Sullivan R. Role of MicroRNAs in Controlling Gene Expression in Different Segments of the Human Epididymis. PLoS ONE. 2012;7:1–13. doi: 10.1371/journal.pone.0034996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bjork JK, Sandqvist A, Elsing AN, Kotaja N, Sistonen L. miR-18, a member of Oncomir-1, targets heat shock transcription factor 2 in spermatogenesis. Development. 2010;137:3177–3184. doi: 10.1242/dev.050955. [DOI] [PubMed] [Google Scholar]
- 183.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A. B entwich I, E inav U, Gilad S, Hurban P, Karov Y, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yu ZR, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. 2005;73:427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 185.Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transi-tion protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. 2005;73:427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 186.Novotny GW, Sonne SB, Nielsen JE, Jonstrup SP, Hansen MA, Skakkebaek NE, Rajpert-De Meyts E, Kjems J, Leffers H. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell Death and Differentiation, 2007; 14879–882. [DOI] [PubMed]
- 187.Govindaraju A, Uzun A, Robertson L, Atli MO, Kaya A, Topper E, Crate EA, Padbury J, Perkins A, Memili E. Dynamics of microRNAs in bull spermatozoa. Reprod Biol Endocrinol. 2012;10(82):1–10. doi: 10.1186/1477-7827-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yan N, Lu Y, Sun H, Tao D, Zhang S, Liu W, Ma Y. A microarray for microRNA profiling in mouse testis tissues. Reproduction. 2007;134:73–79. doi: 10.1530/REP-07-0056. [DOI] [PubMed] [Google Scholar]
- 189.Lian C, Sun B, Niu S, Yang R, Liu B, Lu C, Meng J, Qiu Z, Zhang L, Zhao Z. A comparative profile of the microRNA transcriptome in immature and mature porcine testes using Solexa deep sequencing. FEBS J. 2012;279:964–975. doi: 10.1111/j.1742-4658.2012.08480.x. [DOI] [PubMed] [Google Scholar]
- 190.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J. D rost J, G riekspoor A et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Adv Exp Med Biol. 2007;604:17–46. doi: 10.1007/978-0-387-69116-9_2. [DOI] [PubMed] [Google Scholar]
- 191.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 192.Sun W, Julie Li YS, Huang HD, Shyy JY, Chien S. MicroRNA: a master regulator of cellular processes for bioengi-neering systems. Annu Rev Biomed Eng. 2010;12:1–27. doi: 10.1146/annurev-bioeng-070909-105314. [DOI] [PubMed] [Google Scholar]
- 193.Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O’Carroll D, Das PP, Tarakhovsky A, Miska EA, Surani MA. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One, 2008; 3:e1738. [DOI] [PMC free article] [PubMed]
- 194.Papaioannou MD, Pitetti JL, Ro S, Park C, Aubry F, Schaad O, Vejnar CE, Kuhne F, Descombes P, Zdobnov EM, McManus MT, Guillou F, Harfe BD, Yan W. Sertoli cell DICER1 is essential for spermatogenesis in mice. Dev Biol. 2009;326:250–259. doi: 10.1016/j.ydbio.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Romero Y, Meikar O, Papaioannou MD, Conne B, Grey C, Weier M, Pralong F, Massy BD, Kaessmann H, Vassalli JD, Kotaja N, Nef S. Dicer1 Depletion in Male Germ Cells Leads to Infertility Due to Cumulative Meiotic and Spermiogenic Defects. PLoS ONE. 2011;6:e25241. doi: 10.1371/journal.pone.0025241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Qin Y, Xia Y, Wu W, Han X, Lu C, Ji G, Chen D, Wang H, Song L, Wang S, Wang X. Genetic variants in microRNA biogenesis pathway genes are associated with semen quality in a Han-Chinese population. Reprod BioMed Online. 2012;24:454–461. doi: 10.1016/j.rbmo.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 197.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–9. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 198.Huang S, Li H, Ding X, Xiong C. Presence and characterization of cell-free seminal RNA in healthy individuals: implications for noninvasive disease diagnosis and gene expression studies of the male reproductive system. Clin Chem. 2009;55:1967–76. doi: 10.1373/clinchem.2009.131128. [DOI] [PubMed] [Google Scholar]
- 199.Wang C, Yang C, Chen X, Yao B, Yang C, Zhu C, Li L, Wang J, Li X, Shao Y, Liu Y, Ji J, Zhang J, Zen K, Zhang CY, Zhang C. Altered Profile of Seminal Plasma MicroRNAs in the Molecular Diagnosis of Male Infertility. Clin Chem. 2011;57:1722–1731. doi: 10.1373/clinchem.2011.169714. [DOI] [PubMed] [Google Scholar]
- 200.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 201.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development, 135: 3–9. [DOI] [PubMed]
- 202.He Z, Kokkinaki M, Pant D, Gallicano GI, Dym M. Small RNA molecules in the regulation of spermatogenesis. Reproduction. 2009;137:901–911. doi: 10.1530/REP-08-0494. [DOI] [PubMed] [Google Scholar]
- 203.Kuramochi-Miyagawa S. K imura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 204.Carmell MA, Girard A. van de K ant HJ, Bourchis D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2006;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 205.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 206.Xu M, You Y, Hunsicker P, Hori T, Small C, Griswold MD, Hecht NB. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol Reprod. 2008;79(1):51–7. doi: 10.1095/biolreprod.108.068072. [DOI] [PubMed] [Google Scholar]
- 207.du Plessis SS, Kashou AH, Benjamin DJ, Yadav SP, Agarwal A. Proteomics: a subcellular look at spermatozoa. Reprod Biol Endocrinol. 2011;9(36):1–12. doi: 10.1186/1477-7827-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Aitken RJ, Baker MA. The role of proteomics in understanding sperm cell biology. Int J Androl. 2008;31:295–302. doi: 10.1111/j.1365-2605.2007.00851.x. [DOI] [PubMed] [Google Scholar]
- 209.Caron C, Govin J, Rousseaux S, Khochbin S. How to pack the genome for a safe trip. Prog Mol Subcell Biol. 2005;38:65–89. doi: 10.1007/3-540-27310-7_3. [DOI] [PubMed] [Google Scholar]
- 210.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 211.Li HY, Zhang H. Proteome analysis for profiling infertility markers in male mouse sperm after carbon ion radiation. Toxicol. 2013;306(5):85–92. doi: 10.1016/j.tox.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 212.Sharma R, Agarwal A, Mohanty G, Jesudasan R, Gopalan B, Willard B, Yadav SP, Sabanegh E. Functional proteomic analysis of seminal plasma proteins in men with various semen parameters. Reprod Biol Endocrinol. 2013;11:11–38. doi: 10.1186/1477-7827-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Milardi D, Grande G, Vincenzoni F, Messana I, Pontecorvi A, De Marinis L, Castagnola M, Marana R. Proteomic approach in the identification of fertility pattern in seminal plasma of fertile men. Fertil Steril. 2012;97:67–73. doi: 10.1016/j.fertnstert.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 214.Siva AB, Kameshwari DB, Singh V, Pavani K, Sundaram CS, Rangaraj N, Deenadayal M, Shivaji S. Proteomics-based study on asthenozoospermia: differential expression of proteasome alpha complex. Mol Hum Reprod. 2010;16:452–462. doi: 10.1093/molehr/gaq009. [DOI] [PubMed] [Google Scholar]
- 215.Kashou AH, Benjamin DJ, Agarwal A, du Plessis SS. The Advent of Sperm Proteomics has Arrived. Open Reprod Sci J. 2011;3:92–97. [Google Scholar]
- 216.Sharma A, Agarwal A, Mohanty G, Hamada AJ, Gopalan B, Willard B, Yadav S, du Plessis S. Proteomic analysis of human spermatozoa proteins with oxidative stress. Reproductive Biology and Endocrinology. 2013;11(48):1–18. doi: 10.1186/1477-7827-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Thacker S, Yadav SP, Sharma RK, Kashou A, Willard B, Zhang D, Agarwal A. Evaluation of Sperm Proteins in Infertile Men: A Proteomic Approach. Fertil Steril. 2011;95:2745–2748. doi: 10.1016/j.fertnstert.2011.03.112. [DOI] [PubMed] [Google Scholar]
- 218.Martinez-Heredia J, de Mateo S, Vidal-Taboada JM, Ballesca JL, Oliva R. Identification of proteomic differences in asthenozoospermic sperm samples. Hum Reprod. 2008;23:783–791. doi: 10.1093/humrep/den024. [DOI] [PubMed] [Google Scholar]
- 219.Zhao L, Burkin HR, Shi X, Li L, Reim K, Miller DJ. Complexin I is required for mammalian sperm acrosomal exocytosis. Dev Biol. 2007;309:236–244. doi: 10.1016/j.ydbio.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.de Mateo S, Martinez-Heredia J, Estanyol JM, Dominguez-Fandos D, Vidal-Taboada JM, Ballesca JL, Oliva R. Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics. 2007;7:4264–4277. doi: 10.1002/pmic.200700521. [DOI] [PubMed] [Google Scholar]
- 221.Agarwal A. Relationship between oxidative stress, varicocele and infertility: a meta-analysis. Reprod BioMed Online. 2006;12:630–633. doi: 10.1016/s1472-6483(10)61190-x. [DOI] [PubMed] [Google Scholar]
- 222.Vaidya D. The hormonal assessment of the infertile male. In: A Publication of the Hope Infertility Clinic. 2006; Available at: education. vsnl.com/hic/hic.html.
- 223.Askienazy-Elbhar M. Male genital tract infection: the point of view of the bacteriologist. Gynecol Obstet Fertil. 2005;33(9):691–7. doi: 10.1016/j.gyobfe.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 224.Peterson CM. Human reproduction: clinical, pathologic and pharmacologic correlations. In: Human Reproduction. 2006; Available at: library.med.utah.edu/kw/human_reprod/seminars/seminar2B.html.
- 225.Collins JA, Burrows EA, Yeo J, YoungLai EV. Frequency and predictive value of antisperm antibodies among infertile couples. Hum Reprod. 1993;8:592–8. doi: 10.1093/oxfordjournals.humrep.a138102. [DOI] [PubMed] [Google Scholar]
- 226.Ambasudhan R, Singh K, Agarwal JK, Singh SK, Khanna A, Sah RK, Singh I, Raman R. Idiopathic cases of male infertility from a region in India show low incidence of Y-chromosome microdeletions. J Biosci. 2003;28:605–612. doi: 10.1007/BF02703336. [DOI] [PubMed] [Google Scholar]
- 227.Hwang SK, Makita Y, Kurahashi H, Cho YW, Hirose S. Autosomal dominant nocturnal frontal lobe epilepsy: a genotypic comparative study of Japanese and Korean families carrying the CHRNA4 Ser284Leu mutation. J Hum Genet. 2011;56:609–612. doi: 10.1038/jhg.2011.69. [DOI] [PubMed] [Google Scholar]