Abstract
Purpose
The present study is a case–control analysis of a SNP (rs28368082) in exon 7 of the SPO11 gene and its possible association with male infertility in three provinces of Iran. We also searched for genetic differences among populations.
Methods
Using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) analysis, we genotyped 113 infertile men and 50 fertile controls. Then, samples consisting SNP, as determined by PCR-RFLP, were genotyped by sequencing. The differences in genotype distributions between cases and fertile controls were examined using Chi-squared analysis. The genetic difference between individuals with mutated nucleotide was investigated by phylogenetic trees. Genetic difference among populations (provinces) was analyzed through ANOVA test, and homogeneity was investigated using STRUCTURE and K-means clustering analysis.
Results
According to the statistical analysis, the SNP was significantly associated with male infertility in all populations except oligozoospermic cases of the Center region. The phylogenetic trees showed partial genetic variation among the individuals, although ANOVA test showed no significant genetic difference between populations (provinces) for both azoospermic, and oligozoospermic cases. Eventually, we affirmed that individuals in the inclusive populations had genetic difference, but it was not statistically significant for dividing underlying populations to separate groups, so each population was homogenous.
Conclusion
Our study indicates that the mentioned polymorphism in SPO11 gene may be linked to the susceptibility of azoospermia and oligozoospermia male infertility in three provinces of Iran. Further studies are required to support obtained results. It finally should be noted that the possible association between a particular SNP and a specific disease completely depends on the underlying population.
Keywords: SNP, Male Infertility, SPO11 gene, Azoospermia, Oligozoospermia
Introduction
Infertility is a worldwide reproductive health problem, affecting men and women roughly equally [1]. While there is no universal definition of infertility, a couple is generally considered clinically infertile when pregnancy has not occurred after at least 12 months of regular unprotected sexual activity [2]. Infertility affects 10–15 % of couples worldwide, and male factors account for nearly half of all infertility cases [3].
Although modern diagnostic methods identified much more pathogenesis of infertility, unfortunately, approximately 50 % of infertility cases are still unexplained or idiopathic [4].
The genetic factors involved in male infertility are: chromosomal or monogenic disorders, mitochondrial DNA (mtDNA) mutations, Y chromosome deletions, multifactorial disorders, imprinting disorders, as well as endocrine disorders with genetic origin [5]. Studies have shown that genetic abnormalities account for approximately 5 % of infertility in males [6].
Impaired spermatogenesis is an important etiology of male infertility. Spermatogenesis is a complex physiological process, regulated by several genes. With the development of molecular biology, many infertility-related genes have been identified [7]. About 10 % genes in the genome are related to spermatogenesis [5]. Understanding the molecular mechanism of abnormal spermatogenesis and the genes involved is important in developing both diagnostic tools and treatment strategies for male infertility [8]. Meiosis is a specialized form of cell division, which is essential for sexually reproducing organisms to generate gametes [9].
S. cerevisiae
sporulation protein (Spo11) is an evolutionarily conserved topoisomerase-like protein that, in mammals, is functionally expressed in gonads of both male and female during meiosis. It is responsible for physiological DNA Double Strand Breaks (DSB) formation during the early meiotic prophase in spermatocytes and oocytes [10].
Human SPO11 was detected in several somatic tissues, with its gene localized to chromosome 20q13.2-q13.3. This gene contains 13 exons [11]. When the SPO11 gene was disrupted in mice by homologous recombination, there was a generalized arrest of spermatogenesis in spermatocytes before the pachytene stage, resulting in male infertility [12]. Zhang et al. (2011) reported that SNP (rs28368082) in the exon 7 of SPO11 gene is associated with idiopathic male infertility in Chinese’s population [13].
The present study was designed to investigate the association of an SNP (rs28368082) in the exon7 of SPO11 gene with azoospermia and oligozoospermia male infertility in three different populations (provinces) of Iran (South, Center and North provinces).
Materials and methods
Ethnical standards
The study was supported by Shahid Beheshti University in Iran. All participants were aged from 25 to 55 years. We obtained written informed consent from all participating individuals. Details that might disclose the identity of the subjects under study were omitted.
Subjects and sample collection
We recruited a total of 113 infertile Iranian patients, consisting of 58 men with azoospermia, and 55 men with oligozoospermia from Royan Institute of Tehran. Racial/ethnic backgrounds were important factors for recruitment. So we selected our subjects from three regions; including: South, Center and North provinces of Iran. Thirty-six infertile men were from North of Iran (19 azoospermia and 17 oligozoospermia); 37 infertile men were from South of Iran (19 azoospermia and 18 oligozoospermia), and 40 infertile men were from Central region of Iran (20 azoospermia and 20 oligozoospermia).
The control group included 50 fertile men who had at least one child without assisted reproductive technologies and had normal semen parameters, and all the cases had the normal karyotype. Controls were collected randomly from the South, Center, and North provinces of Iran.
The semen analysis for sperm concentration, motility and morphology was performed following the World Health Organization criteria [14] and serum levels of FSH, LH and testosterone were determined.
DNA isolation, SNP selection and genotyping (molecular analysis)
Genomic DNA was extracted from peripheral blood using a standard Salting Out procedure. Identification of the C5679T (rs28368082) polymorphic variant in the exon 7 of SPO11 gene (codon 211), Arginine change into Tryptophan, was performed by Polymerase Chain Reaction-Restriction Fragment Length polymorphism (PCR-RFLP) technique with the primers which were designed based on the published sequence of the human SPO11 gene.
Oligonucleotide primers were designed using Oligo5.0 software. PCR of the genomic DNA was conducted using the following primer pairs: 5′-TAACAGAGGAAGAAGTCTCTGATG-3′ (forward) and 5′-TTGTAAATCCTCTTA CCAATCACCA-3′ (reverse). These primers generated a 687 bp fragment. PCR amplification was carried out in a total volume of 25 μl containing approximately 50 ng of the genomic DNA, 0.4 μl of 40 mM dNTP-Mix, 0.8 μl of each primer at a final concentration of 2 μM, 0.3 μl of 100 mM MgCl2, 0.5 μl of 5u/μl Taq polymerase and 2 μl of 10× buffer.
The PCR reaction profile was: predenaturation at 95 °C for 4 min followed by denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s and extension at 72 °C for 90 s for 40 cycles, with a final extra extension at 72 °C for 10 min. PCR cycling was performed using the Applied Biosystems 2720 Thermal Cycler. Then the products were electrophoresed on a 1.5 % agarose gel, stained with ethidium bromide and visualized using ultraviolet illumination.
Restriction maps of the SPO11 gene sequences were acquired by the Webcutter 2.0 software then EcoRI enzyme was selected. PCR amplicons were digested with EcoRI enzyme. 5 μl of PCR product was digested with 0.7 μl of the selected enzyme [Bioron, Germany], incubated at 37 °C for 16 h.
Then the products of digestion were detected on 2 % agarose gel with ethidium bromide. Finally, mutated samples, obtained from PCR-RFLP, were sequenced with forward primer in Macrogen Company in Korea.
Statistical analysis
The differences in allele and genotype frequencies of C5679T polymorphism in the SPO11 gene between cases and controls were determined by standard Chi squared (χ2), the Odds Ratio (OR) tests. All P-values were based upon two-tailed tests and Odds Ratios (ORs), with 95 % confidence intervals (CIs). A P-value <0.05 was regarded as statistically significant. Both of Chi-squared and Odds Ratios (ORs) tests were done by SPSS Version 19.0.
To investigate the genetic difference between individuals with mutated nucleotide, we used phylogenetic trees such as Neighbor-Joining (NJ), Maximum Likelihood, Maximum Parsimony and UPGMA. Significant genetic difference among populations (provinces) was calculated by Analysis of Molecular Variance (ANOVA) method, which performed by GenAlex6.4. ]15.[
To check the genetic continuity versus populations’ stratification two methods were used: First STRUCTURE analysis was done by STRUCTURE software (2012) [16], second K-Means clustering was carried out in GenoDive version 2.0 (2012). We used two summary statistics to present K-Means clustering: 1. psedo-F [17] and 2. Bayesian Information Criterion [18].
We performed Principal Coordinate Analysis (PCoA) analysis to investigate the genetic relation between individuals. Its operation can be thought of as revealing the internal structure of the data in a way that best explains the variance in the data [19].
Results
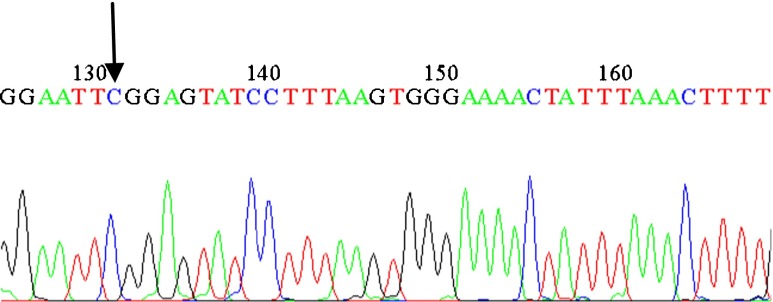
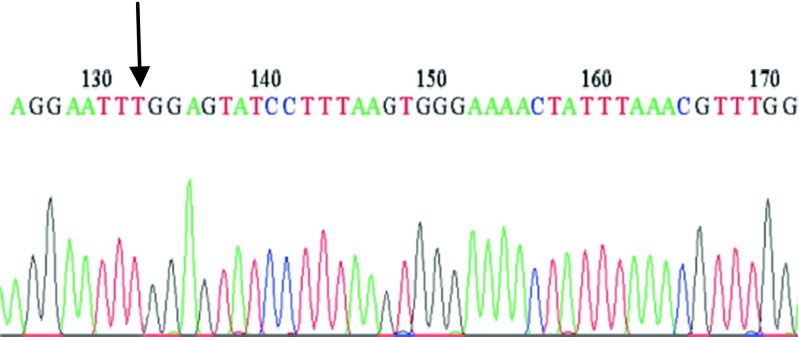
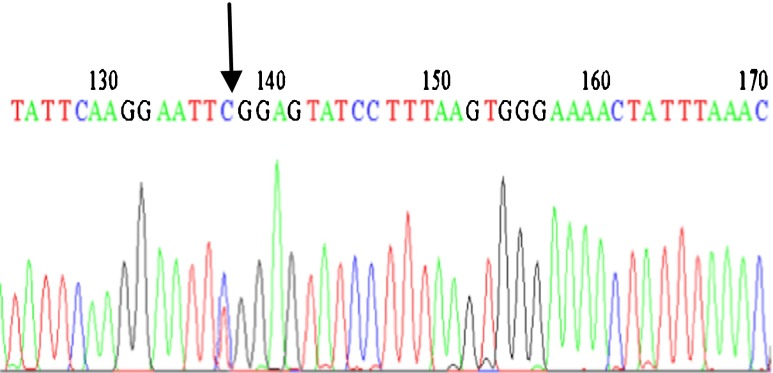
In the present study, we analyzed C5679T polymorphism in SPO11 gene in 58 azoospermia, 55 oligozoospermia and 50 fertile men as the control group, from North, Center and South provinces of Iran using the PCR-RFLP method. Three different patterns were appeared: the wild-type homozygote (CC), heterozygote (CT) and homozygote (TT). The uncleaved fragment, homozygous for T (TT), was 687 bp; whereas the cleaved fragment, including homozygous for C (CC) and heterozygote (CT) showed two bands (533 and 154 bp) and three bands (687, 533 and 154 bp) respectively, because the substitution disrupts an EcoRI recognition site which digests the 687 bp into 533 and 154 bp fragments. Finally, samples consisting one band and three bands, as determined by PCR-RFLP, were genotyped by sequencing. The results of mutated samples, and one wild-type sample are displayed (Figs. 1, 2, and 3).
Fig. 1.
Sequence profile from a wild type
Fig. 2.
Sequence profile from an infertile man carrying the SNP (Homozygous)
Fig. 3.
Sequence profile from an infertile man carrying the SNP (Heterozygous)
Statistical analysis
The genotype and allele frequencies of SNP (rs28368082) for all of 163 samples are presented in Tables 1 and 2 respectively. We used Chi-squared test to compare genotype distribution between infertile subjects and fertile controls in each population, and we performed an Odds Ratio (OR) test to indicate the strength of association between the SNP and studied populations. The results of Chi-squared and Odds Ratio (OR) tests are presented in Table 3.
Table 1.
Genotype frequencies of SNP (rs28368082) in the SPO11 gene for azoospermic and oligozoospermic patients and controls
| Genotype | Frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Azoo. North N = 19 | Azoo. Center N = 20 | Azoo. South N = 19 | Azoo. Total N = 58 | Oligo. North N = 17 | Oligo. Center N = 20 | Oligo. South N = 18 | Oligo. Total N = 55 | Controls N = 50 | |
| CC | 16(84.21 %) | 18(90 %) | 17(89.47 %) | 51(87.93 %) | 14(82.35 %) | 19(95 %) | 16(88.89 %) | 49(89.09 %) | 49(98 %) |
| CT | 2 (10.53 %) | 2(10 %) | 2(10.53 %) | 6(10.34 %) | 3(17.65 %) | 1(5 %) | 2(11.11 %) | 6(10.91 %) | 1(2 %) |
| TT | 1(5.26 %) | 0(0 %) | 0(0 %) | 1(1.72 %) | 0(0 %) | 0(0 %) | 0(0 %) | 0(0 %) | 0(0 %) |
Azoo azoospermia, Oligo oligozoospermia
Table 2.
Allele frequencies of SNP (rs28368082) in the SPO11 gene for azoospermic and oligozoospermic patients and controls
| Allele | Frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Azoo. North N = 19 | Azoo. Center N = 20 | Azoo. South N = 19 | Azoo. Total N = 58 | Oligo. North N = 17 | Oligo. Center N = 20 | Oligo. South N = 18 | Oligo. Total N = 55 | Controls N = 50 | |
| C | 89.47 % | 95 % | 94.74 % | 93.1 % | 91.18 % | 97.5 % | 94.44 % | 94.55 % | 99 % |
| T | 10.53 % | 5 % | 5.26 % | 6.9 % | 8.82 % | 2.5 % | 5.56 % | 5.45 % | 1 % |
Table 3.
The results of chi-squared and Odds Ratio (OR) tests
| Parameters | Cases | |||||||
|---|---|---|---|---|---|---|---|---|
| Azoo. North N = 19 | Azoo. Center N = 20 | Azoo. South N = 19 | Azoo. Total N = 58 | Oligo. North N = 17 | Oligo. Center N = 20 | Oligo. South N = 18 | Oligo. Total N = 55 | |
| χ 2 | 11.966 | 5.674 | 6.664 | 7.680 | 14.22 | 1.332 | 6.664 | 6.664 |
| P- value | 0.001a | 0.017a | 0.010a | 0.006a | 0.00a | 0.248b | 0.010a | 0.010a |
| df | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Odds Ratio | 9.33 | 5.444 | 6.056 | 6.682 | 10.756 | 2.579 | 6.056 | 6.056 |
aStatistically significant
bNot significant
According to the Table 3, the SNP was significantly associated with male infertility in all populations except oligozoospermic cases of the Center region.
The genetic difference among individuals with mutated nucleotide was determined by phylogenetic trees such as Neighbor-Joining (NJ) ،Maximum Likelihood، Maximum Parsimony and UPGMA, all of them produced similar results. Therefore, only NJ trees are discussed here. According to the NJ trees, in azoospermic cases with mutated nucleotide, we obtained two main groups, while in oligozoospermic cases, the patients were placed in three groups. The NJ trees showed partial genetic variation among the studied individuals with mutated nucleotide.
Genetic difference among populations was analyzed by ANOVA test. Azoospermic cases with mutated nucleotide were classified in four different groups while, oligozoospermic cases with mutated nucleotide were classified in two different groups.
The ANOVA test showed no significant genetic difference between provinces for both azoospermic (Fst = −0.026, P-value = 0.5), and oligozoospermic cases (Fst = −0.025 and P-value = 0.39). Therefore, both groups differ in very minor genetic differences as revealed in NJ tree. Genetic continuity versus populations’ stratification was tested using STRUCTURE and K-means clustering analysis.
According to the STRUCTURE results for azoospermic and oligozoospermic men, there were two and three separated sub-groups respectively, but the differences were not statistically significant. The STRUCTURE results for both groups confirmed the NJ tree results. Here we proved that genetic diversity was not significant and the STRUCTURE results showing no separation for both groups.
Then we used K-means clustering analysis. Concerning the results in azoospermic cases; best clustering according to pseudo-F was k = 2 and according to Bayesian Information Criterions was k = 5. Based on the NJ tree k = 2 was better. Considering the results in oligozoospermic cases; best clustering according to both of pseudo-F and Bayesian Information Criterion was k = 4. Finally, we checked PCoA test to estimate the best relation between individuals with mutated nucleotide.
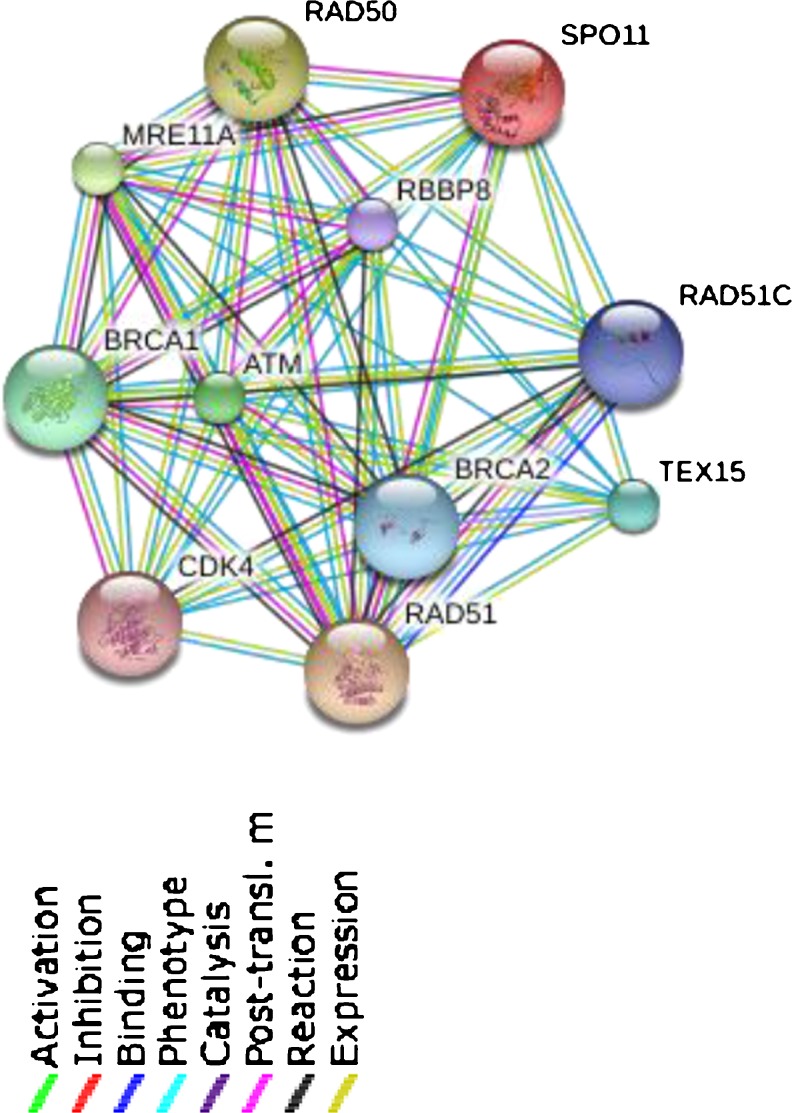
Finally, we presented the network of SPO11 protein in Fig. 4, which adapted from data presented in SWISS-Prot database [20]. It displays the other partners of SPO11 protein; according to the Fig. 4, these proteins are probably other candidates for investigating male infertility, but we need more experimental tests to prove such roles Table 4. Predicted functional partners of SPO11 protein are shown in Table 4.
Fig. 4.
The network of SPO11 protein (Homo sapiens). This network shows proteins, which interact with SPO11 protein [20]
Table 4.
Predicted functional partners of SPO11 protein [20]

Discussion
Genetic polymorphisms may increase susceptibility to some forms of male infertility [7]. Recent reports showed that genetic disorders affecting spermatogenesis might be responsible for many cases of idiopathic male infertility [21]. In this study, we investigated an SNP (rs28368082) in the exon 7 of SPO11gene and its association with male infertility in azoospermic and oligozoospermic groups. We carried out our study based on an ethnic background.
The results revealed this SNP was associated with male infertility in all of populations except oligozoospermic cases of the Center region; it might be resulted from ethnic differences among populations. Inconsistent results might be attributed to differences in ethnic background and geographic variations. Our study indicates that the mentioned polymorphism in SPO11 gene may be linked to the susceptibility of azoospermia and oligozoospermia in three provinces of Iran.
In this study, the number of cases and controls was not large enough, regarding that current research is based on an ethnic background so the results can be premature and further studies are required to support obtained results. With respect to the development of molecular biology techniques, the role of SPO11 gene in the male infertility will be cleared.
We also provided data, which shows SPO11 network from SWISS-Prot database [20]; all of them may be candidate genes for male infertility, investigating all of them can be useful.
It finally should be noted that the relation between a particular SNP with the disease completely depends on the population. Perhaps an SNP in a specific population of patients has a strong association while it has no such association in other populations at all.
Acknowledgments
We acknowledge Shahid Beheshti University for supporting this study. Special thanks to Dr. Mohseni for helping in performing the study. We thank all the clinicians (Mrs. Mokhtari and et al.) who provided samples for this study from the Molecular laboratory unit of the Royan Institute of Tehran. We also thank patients and volunteers for their participation in the study.
Footnotes
Capsule
A case–control study was designed to investigate the possible association between an SNP in SPO11 gene and azoospermia, oligozoospermia male infertility in three provinces of Iran, according to our data the SNP was associated with male infertility in all populations except oligozoospermic cases of the Center region.
References
- 1.Zheng K, Yang F, Wang PJ. Regulation of male fertility by X-linked genes. J Androl. 2010;31(1):79–85. doi: 10.2164/jandrol.109.008193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowe PJ, Comhaire FH, Hargreave TB, Mahmoud AMA. WHO manual for the standardized investigation, diagnosis and management of the infertile male. Cambridge: Cambridge University Press; 2000.
- 3.Irvine DS. Epidemiology and aetiology of male infertility. Hum Reprod. 1998;1(13 Suppl):33–44. doi: 10.1093/humrep/13.suppl_1.33. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Qiu SD, Li SB, Zhou DX, Tian H, Huo YW, et al. Novel mutations in ubiquitin-specific protease 26 gene might cause spermatogenesis impairment and male infertility. Asian J Androl. 2007;9:809–14. doi: 10.1111/j.1745-7262.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 5.Shamsi MB, Kumar K, Dada R. Genetic and epigenetic factors: role in male infertility. Indian J Urol. 2011;27(1):110–20. doi: 10.4103/0970-1591.78436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferlin A, Raicu F, Gatta V, Zuccarello D, Palka G, Foresta C. Male infertility: role of genetic background. Reprod Biomed Online. 2007;14:734–45. doi: 10.1016/S1472-6483(10)60677-3. [DOI] [PubMed] [Google Scholar]
- 7.O’Flynn O’Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: a review. Fertil Steril. 2010;93:1–12. doi: 10.1016/j.fertnstert.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 8.Wilson GR, Sim ML, Brody KM, Taylor JM, McLachlan RI, O’Bryan MK, et al. Molecular analysis of the PArkin co-regulated gene and association with male infertility. Fertil Steril. 2010;93:2262–8. doi: 10.1016/j.fertnstert.2009.01.079. [DOI] [PubMed] [Google Scholar]
- 9.Prieler S, Penkner A, Borde V, Klein F. The control of Spo11’s interaction with meiotic recombination hotspots. Genes& Development. 2005;19:255–69. doi: 10.1101/gad.321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking SPO11. Mol Cell. 2000;6:989–98. doi: 10.1016/S1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 11.Romanienko PJ, Camerini-Otero RD. Cloning, characterization and localization of mouse and human SPO11. Genomics. 1999;61:156–69. doi: 10.1006/geno.1999.5955. [DOI] [PubMed] [Google Scholar]
- 12.Carrell DT, De Jonge C, Lamb DJ. The genetics of male infertility: a field of study whose time is now. Arch Androl. 2006;52:269–74. doi: 10.1080/01485010500503603. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Zhou D, Wang H, Tian Z. An association study of SPO11 gene single nucleotide polymorphisms with idiopathic male infertility in Chinese Han population. J Assist Reprod Genet. 2011;28:731–6. doi: 10.1007/s10815-011-9571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO laboratory manual for the examination of human semen and semen-cervical mucus interaction. 4. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- 15.Peakall R, Smouse PE. GenAlex6: genetic analysis in excel population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–95. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard JK, Stephens M, Donnelly P. Inference of populations structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caliński T, Harabasz J. A dendrite method for cluster analysis. Commun Stat. 1974;3:1–27. doi: 10.1080/03610928308827180. [DOI] [Google Scholar]
- 18.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–4. doi: 10.1214/aos/1176344136. [DOI] [Google Scholar]
- 19.Podani J. Introduction to the exploration of multivariate data. English translation. Leide, Backhuyes Publisher. 2000; p. 407.
- 20.www.uniprot.org/uniprot/Q9Y5K1.stringdb/newstring_cgi/show_network_section.pl/identifier=9606.ENSP00000360310
- 21.Nishimune Y, Tanaka H. Infertility caused by polymorphisms or mutations in spermatogenesis-specific genes. J Androl. 2006;27:326–34. doi: 10.2164/jandrol.05162. [DOI] [PubMed] [Google Scholar]