Abstract
Salmonella is a zoonotic pathogen with globally distributed serovars as well as serovars predominantly found in certain regions; for example, serovar Weltevreden is rarely isolated in the U.S., but is common in Thailand. Relative to our understanding of Salmonella diversity, our understanding of the global diversity of Salmonella phages is limited. We hypothesized that the serovar diversity in a given environment and farming system will affect the Salmonella phage diversity associated with animal hosts. We thus isolated and characterized Salmonella phages from 15 small-scale dairy farms in Thailand and compared the host ranges of the 62 Salmonella phage isolates obtained with host range diversity for 129 phage isolates obtained from dairy farms in the U.S. The 62 phage isolates from Thailand represented genome sizes ranging from 40 to 200 kb and showed lysis of 6 to 25 of the 26 host strains tested (mean number of strain lysed = 19). By comparison, phage isolates previously obtained in a survey of 15 U.S. dairy farms showed a narrow host range (lysis of 1 to 17; mean number of strains lysed = 4); principal coordinate analysis also confirmed U.S. and Thai phages had distinct host lysis profiles. Our data indicate that dairy farms that differ in management practices and are located on different continents can yield phage isolates that differ in their host ranges, providing an avenue for isolation of phages with desirable host range characteristics for commercial applications. Farming systems characterized by coexistence of different animals may facilitate presence of Salmonella phages with wide host ranges.
Keywords: Salmonella phage, phages from dairy farms, phage host range
1. Introduction
Salmonella is a common zoonotic pathogen that contaminates animals and animal-derived food-products. In Thailand, food animals are recognized sources of non-typhoidal Salmonella (Hendriksen et al., 2009). For example, Salmonella is commonly isolated from poultry, swine and cattle, which are important farming commodities in Thailand (Padungtod and Kaneene, 2006). One study in Thailand found three percent of Salmonella prevalence in dairy cattle (Padungtod and Kaneene, 2006). Another study reported 27 different serovars detected in clinically healthy dairy cattle (Chuanchuen et al., 2010). In the United States, dairy cattle have been typically reported as reservoir of Salmonella serovars (Cummings et al., 2010).
The Salmonella genus has >2,600 serovars (Guibourdenche et al. 2010). While some serovars (e.g., Typhimurium, Enteritidis) are globally distributed (Galanis et al., 2006); other serovars are regionally distributed. Importantly, some of the most common serovars in Thailand are different from the most common serovars in the U.S. Salmonella serovars Enteritidis, Typhimurium, Newport, Javiana, and 4,5,12:i:- are the five most common Salmonella serovars in the U.S. (CDC, 2011), whereas serovars Weltevreden, Enteritidis, Stanley, Corvallis, and Rissen are the most common Salmonella serovars in Thailand (Padungtod and Kaneene, 2006). Conversely to the knowledge of the global distribution of Salmonella serovars; the knowledge of the distribution of bacteriophages infecting the genus Salmonella is quite limited.
Salmonella phages have been isolated from different environments, including dairy farms, swine lagoons, poultry facilities, and sewage (McLaughlin et al. 2008; Andreatti Filho et al. 2007). In addition, with the advances in sequencing technologies, a number of Salmonella phages, from different geographical origins, have been fully sequenced (Moreno Switt et al. 2013b). Genomic characterizations have shown that some closely related phages are globally distributed; as phages that represent the same genus have been isolated from different continents (Moreno Switt et al. 2013b). Salmonella phages with different spectra of host range have been previously reported; for example, phage FelixO1 has a wide host range (Whichard et al., 2010) and FSL SP-031 has a narrow host range (Moreno Switt et al., 2013b). Among the factors that could drive phage host range is the density of suitable hosts (Guyader and Burch, 2008). While high host density appears to facilitate phage specialization; low host density, on the other hand, appears to facilitate generalist phages with wide host range (Guyader and Burch, 2008).
The aim of this study was to improve our understanding on the global Salmonella phage diversity; here we investigated the host range diversity on Salmonella phages, using as model dairy farms in two countries that have different farming practices; the U.S. and Thailand. Host ranges of the recently reported U.S. phages were compared with host range of newly isolated phages from Thai farms in this study.
2. Materials and Methods
2.1. Sample collection
Fifteen small-scale dairy farms in the Sai Yok district, Kanchanaburi province in Thailand were visited from February to December, 2010. In each farm, one freshly secreted fecal sample from the farm floor was collected and maintained on ice until processed in the laboratory. All the 15 farms sampled had small number (10 to 120 cows/farm) of Holstein Friesian cattle. The cows were fed with corns, beans, and molasses sugar and they obtained their roughage from hay and Napier grass. All farms were free-range, other free-range animals (e.g., hens and ducks) and penned animals (e.g., pigs and horses) were also present in some of the dairy farms sampled.
2.2. Salmonella isolation
Fecal sample (10 g) was mixed with 90 ml of buffered peptone water (BPW) (Becton, Dickinson, Sparks, MD). After 24 h at 35°C, 0.1 ml and 1 ml aliquots of the primary enrichment were transferred to 10 ml of Rappaport-Vassiliadis (RV) broth and 10 ml of tetrathionate (TT) broth, respectively. After incubation at 42°C for 24 h, loopful aliquots (10 μl) from RV and TT broths were streaked onto xylose lysine desoxycholate (XLD) agar. Plates were incubated at 35°C for 24 h. On XLD plate, typical Salmonella colonies appear pink with or without black centers. Selected pink colonies were subcultured and further used in PCR confirmation with Salmonella-specific primers targeting invA gene (Rahn et al., 1992).
2.3. Salmonella phage isolation, purification and propagation
Bacteriophage isolation, purification and propagation for samples collected on Thai farms were conducted as previously described (Moreno Switt et al., 2013a). Briefly, phage isolates were obtained using a direct isolation approach along with isolation after enrichment. Five Salmonella strains were selected as hosts for phage isolation (Table 1); representing serovars commonly reported in Thailand (i.e., Dublin, Typhimurium, Enteritidis, Newport and Weltevreden) (Hendriksen et al., 2009; Padungtod and Kaneene, 2006).
Table 1. Salmonella strains used for phage isolation and host range characterization.
| Strain FSL | Serovar | % (no.) of Thai phages that infect each strain2 | % (no.) of U.S. phages that infect each strain3 |
|---|---|---|---|
| S5-368 | Dublin1 | 57 (35) | 36 (46) |
| S5-370 | Typhimurium1 | 87 (54) | 26 (34) |
| S5-371 | Enteritidis1 | 77 (45) | 16 (21) |
| S5-548 | Newport1 | 90 (56) | 52 (67) |
| R8-798 | Weltevreden1 | 92 (57) | 6 (8) |
| A4-525 | Anatum | 73 (45) | 11 (14) |
| A4-737 | Typhimurium | 95 (59) | 36 (46) |
| R8-242 | Cerro | 77 (48) | 37 (48) |
| S5-431 | Kentucky | 58 (36) | 19 (24) |
| A4-793 | Mbandaka | 45 (28) | 18 (23) |
| R8-092 | Corvalis | 84 (52) | 10 (13) |
| R8-376 | Oranienburg | 53 (33) | 13 (17) |
| S5-369 | Saintpaul | 94 (58) | 20 (26) |
| S5-373 | Braenderup | 82 (51) | 3 (4) |
| S5-390 | 4,5,12:i:- | 71 (44) | 38 (49) |
| S5-406 | Javiana | 79 (49) | 21 (27) |
| S5-454 | Panama | 90 (56) | 15 (19) |
| S5-455 | Heidelberg | 53 (33) | 15 (19) |
| S5-464 | Stanley | 84 (52) | 16 (21) |
| S5-474 | Montevideo | 52 (32) | 16 (20) |
| S5-506 | Infantis | 50 (31) | 17 (22) |
| S5-515 | Newport | 97 (60) | 44 (57) |
| S5-917 | Muenster | 77 (48) | 5 (6) |
| S5-961 | Virchow | 81 (50) | 6 (8) |
| A1-125 | E. coli | 18 (11) | 18 (23) |
Strain used for phage isolation in Thailand; for isolation after enrichment, a cocktail of all five isolates mixed in equal ratios was used.
Data represent a total of 62 phage isolates from Thailand.
Data represent a total of 129 phage isolates from the U.S.; host range for 108 of these 129 phage isolates were previously reported by Moreno Switt et al., 2013a.
2.4. Genome size characterization
Genome size was characterized for 25 Thai phage isolates, which were selected to represent at least one phage isolate per farm. In farms where more than one phage isolate was characterized, phages representing different lysis profiles (LP) were selected (Suppl. Table 1). Nucleic acids were isolated using phenol-chloroform, dissolved in TE buffer (10 mM Tris, 1mM EDTA; pH8.0), and quantified (OD260 values) as previously described (Moreno Switt et al. 2013a). Pulsed-Field Gel Electrophoresis (PFGE) was used to determine the genome size of these phage isolates using the procedure described by Moreno Switt et al., 2013a.
2.5. Host range determination
Host range for each Thai phage isolate was determined with the same bacterial host panel, procedure, and data analysis as previously used to characterize the host range of Salmonella phages isolated in the U.S. (Moreno Switt et al., 2013a). The host panel includes 25 strains of Salmonella (representing 23 serovars) and one strain of E. coli (Table 1).
2.6. Comparison of host ranges between Salmonella phages isolated in Thailand and phages isolated in the U.S
We conducted a comparison of the host range profiles obtained from the Thai phages isolated in this study and from the previously characterized U.S. phages using the same panel of hosts (Moreno Switt et al., 2013a). To evaluate if host ranges can distinguish 62 Salmonella phages isolated in this study and 129 Salmonella phages isolated in the U.S. (108 of these phage isolates were previously reported, see Moreno Switt et al., 2013a), we conducted a Principal Coordinates Analysis (PCoA). Briefly, a distance matrix was calculated among host range phenotypes using the vegdist function with the Canberra method for presence of double-zeros correction in semi-quantitative data in the R package vegan. The resulting distance matrix was subjected to PCoA in the dudi.pco function of the R package ade4 (Gower 1966; Ramette 2007). The first 10 principal coordinate axes, explaining a sum of 72% of the total variation, were retained. The first versus the second sources of variation were plotted. Analysis of distance (ANODIS) was performed to test for significant variation between U.S. and Thai phages using an ANODIS model consisting of a single fixed effect for geographic source (McArdle and Anderson, 2001). Significance of the ANODIS model fit was evaluated by 999 Monte Carlo permutations of the objects in the Canberra distance matrix among geographic source levels.
3. Results
3.1. Salmonella phages were obtained from samples collected on 10 of the 15 Thai dairy farms included in this study
A total of 62 phage isolates were obtained from one sample collected from ten of the 15 visited farms (Table 2). All phage isolates were obtained from the enriched filtrate (using a Salmonella cocktail, see Table 1). The majority of the phage isolates were obtained on the host strain representing serovar Weltevreden (34%), followed by the strains representing serovars Newport (23%), Enteritidis (21%), Typhimurium (13%), and Dublin (10%) (see Table 2). Six out of 15 samples (i.e., from farms 1, 5, 9, 11, 12, and 13) were positive for Salmonella; these six samples yielded a total of 13 Salmonella isolates. Interestingly, while the six farms that were positive for Salmonella isolation were also positive for phage isolation (74.2% of the phage isolates were obtained from these six farms); phages were only obtained from samples collected on four of the nine farms negative for Salmonella (Table 2). Farms that were positive for Salmonella were significantly more likely to also yield Salmonella phages (p < 0.05). Further validation using a larger sample size is necessary though.
Table 2. Number of phage isolates obtained from 15 dairy farms in the Kanchanaburi province, Thailand.
| Farm ID | No. of cows | Salmonella1 positive samples | Number of phage isolates obtained on host strains representing serovar (%)2 | Total phage isolates | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Typhimurium | Enteritidis | Dublin | Weltevreden | Newport | ||||
| 1 | 15 | + | 1 | 2 | 3 | 2 | 0 | 8 |
| 2 | 8 | - | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 12 | - | 1 | 0 | 0 | 1 | 1 | 3 |
| 4 | 10 | - | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 70 | + | 1 | 2 | 2 | 0 | 3 | 8 |
| 6 | 24 | - | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 7 | - | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 16 | - | 0 | 3 | 0 | 1 | 2 | 6 |
| 9 | 140 | + | 1 | 1 | 0 | 14 | 2 | 18 |
| 10 | 40 | - | 3 | 0 | 0 | 0 | 2 | 5 |
| 11 | 15 | + | 0 | 1 | 0 | 0 | 1 | 2 |
| 12 | 32 | + | 1 | 2 | 0 | 1 | 2 | 6 |
| 13 | 20 | + | 0 | 1 | 1 | 1 | 1 | 4 |
| 14 | 8 | - | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 12 | - | 0 | 1 | 0 | 1 | 0 | 2 |
|
| ||||||||
| 8 (13) | 13 (21) | 6 (10) | 21 (34) | 14 (23) | 62 | |||
A total of 13 Salmonella isolates were isolated. According to Multi Locus Sequencing Typing (MLST), these isolates were initially predicted as Salmonella serovars Singapore (n=1), Weltevreden (n=1), Kentucky (n=4), Bergen (n=1), Mbandaka (n=1), Dublin (n=1), Typhimurium (n=2), Havana (n=1), and Virchow (n=1).
Percentage of phage isolates obtained using given serovars as hosts; a total of 62 phage isolates were isolated.
Genome size characterization of 25 phage isolates, representing at least one phage per farm, showed considerable diversity. Twelve phage isolates showed estimated genome sizes ranging from 40 to 200 kb (Suppl. Table 1). Six of these phage genomes showed two bands, one of approx. 55 kb and one of approx. 60 kb, similar pattern (two bands that are approx. 5 kb different) was also recently described for listeriaphage genomes using PFGE (Vongkamjan et al., 2012), further sequencing of listeriaphages that showed two bands on PFGE showed single phages with terminal redundant genomes (personal communication).
3.2. Thai phage isolates were found to have wide host ranges including those that lysed all 25 Salmonella strains used in the host range panel
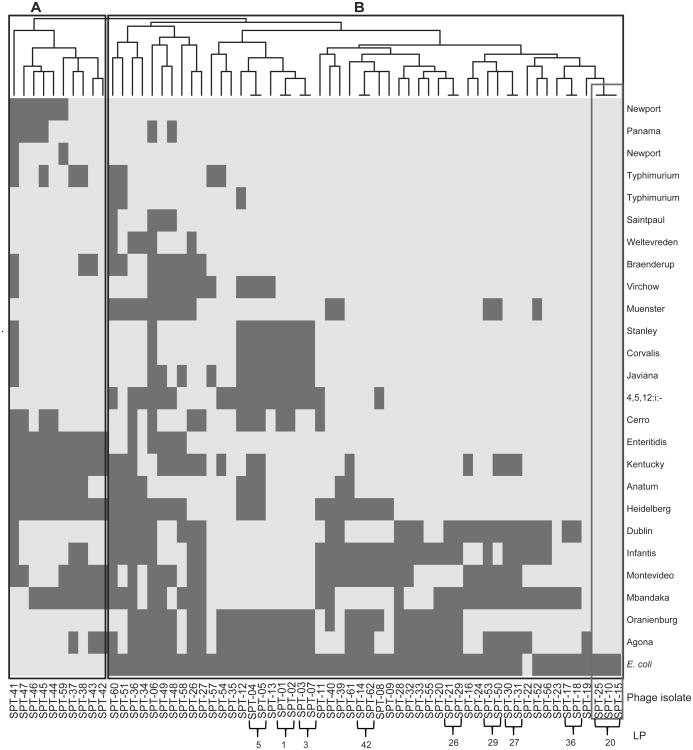
Host range characterization of all 62 phage isolates obtained from dairy farms in Thailand was performed with 25 Salmonella strains (representing 23 serovars) and one E. coli strain, the same host range panel was previously used to characterize 108 phage isolates obtained from dairy farms in the U.S. (Moreno Switt et al., 2013a). The 62 Thai phage isolates lysed between 6 and 25 of the Salmonella host strains; the E. coli strain was only lysed by 11 phages (18%, see Table 1 and Fig. 1). Five highly susceptible host strains representing Salmonella serovars Newport, Typhimurium, Saintpaul, Weltevreden, and Panama were lysed by > 90% of the Thai phage isolates (Table 1). Conversely, less susceptible strains representing serovars Agona, Mbandaka, and Infantis were only lysed by <50% of the phage isolates. According to the host strains that a given phage lysed, phage isolates were classified into 53 Lysis Profiles (LPs) (Suppl. Table 2). Whereas wide variability in LPs was observed, a number of these LPs differ by only one or two strains, for example, phage isolates with LP1 lysed 18 strains and phage isolates with LP3 lysed the same 18 strains, plus the strain representing S. Cerro (Fig. 1). Nine LPs were found in more than one phage, four of these LPs (i.e., LP20, LP26, LP27, and LP42) were found in phage isolates obtained from different farms (Fig. 1). The three phage isolates that were classified as LP20, were obtained from two farms (Farms 1 and 5), and represented phages that lysed all Salmonella strains tested. Interestingly, the two farms where phage isolates with LP20 were isolated were positive for Salmonella isolation (Table 2).
Figure 1. Heat map representation of lysis profiles of the 62 Thai phage isolates tested on 26 host strains.
The right vertical axis lists the host strains and the horizontal axis is labeled with the phage isolates and the lysis profiles (LP) found in more than one phage. Lysis is represented as light gray; no lysis is represented as dark gray. At the top of the figure is the clustering performed with the Ward's method of binary distance; two clusters of Thai phages were identified and labeled as “A” and “B”.
Phage isolates were also classified into (i) narrow (previously defined as lysis of < 4 host strains (Moreno Switt et al., 2013a)); (ii) wide (defined as lysis between 4 and 19 host strains); and (iii) very wide host range (defined here as lysis of > 20 host strains) (Fig. 2); cut-offs for the host ranges were arbitrary. Using these criteria, the 62 Thai phage isolates were classified as (i) wide host range (36 phage isolates) and (ii) very wide host range (26 phage isolates) (Fig. 2). Cluster analysis based on host range revealed two clusters (Fig. 1); one of these clusters (cluster A) contains 10 wide host range phage isolates, all obtained from Farm 9, which showed lysis on several Salmonella serovars and on the E. coli strain. Cluster B contains phage isolates with wide and very wide host range; in addition, in cluster B are the three phage isolates with LP20 (i.e., SPT-010, SPT-015, and SPT-025), which lysed all 25 Salmonella strains (Fig. 1). Phage isolate SPT-010 from Farm 1 with LP20 showed a genome size of 200 kb.
Figure 2. Host ranges of Thai and U.S. phage isolates.
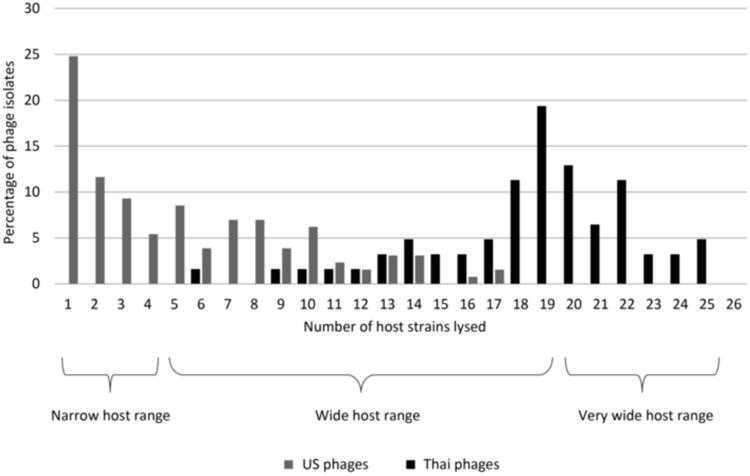
Percentage of phage isolates is labeled on the y-axis and number of host strains being lysed is on the x-axis. In black are the Thai phage isolates and in gray are the U.S. phage isolates. Phage isolates were arbitrarily classified as having narrow (lysed from 1-4 host strains), wide (lysed from 5-19 host strains) and very wide host range (lysed from 20-25 host strains).
3.3.Comparison of host range profiles characterized on 23 Salmonella serovars distinguished Thai and U.S. phages
Comparison of the host range profiles, using the same panel of 26 strains, of (i) 129 Salmonella phage isolates obtained from dairy farms in the U.S. (Moreno Switt et al., 2013a) and (ii) 62 phage isolates of this study reveals major differences in the host range phenotypes. While phages in the U.S. were mostly characterized as narrow host range phages (51.1% of phages lysed between 1-4 strains), Thai phages showed, as described above, wide and very wide host ranges (61.3% of phages lysed between 19 and 25 strains) (Fig. 2). Strains representing Salmonella serovars Newport and Typhimurium were susceptible to Thai (e.g., 90% of the phage isolates lysed strain FSL S5-548) and U.S. phages (e.g., 52% of phage isolates lysed strain FSL S5-548) (Table 1). However, susceptibility of the 23 Salmonella serovars tested showed important differences; for example, S. Weltevreden was lysed by 92% of Thai phages and only by 6 % of U.S. phages (Table 1). Cluster analysis of the host range phenotypes of U.S. and Thai phages showed two clusters, A and B. Cluster A contains cluster A1, representing a cluster of U.S. phages, and cluster A2, representing a cluster of Thai phages. Cluster B represents the other Thai phage cluster (Suppl. Fig. 2). Exceptions were five U.S. phage isolates (i.e., two in cluster A2 and three in cluster B) that clustered with Thai phages and one Thai phage isolate (SPT-006) clustered with U.S. phages. Whereas all five U.S. phages that clustered with Thai phages were classified to have a wide host range (infected >14 hosts), the Thai phage that clustered with U.S. phages is the phage that, among Thai phages, showed the narrower host range (only lysed 6 hosts) (Suppl. Table 2).
Noncanonical principal coordinates analysis (PCoA) was applied to determine whether U.S. and Thai phages could be distinguished by their host ranges. PCoA demonstrated that the largest source of variation in host ranges distinguished U.S. and Thai phages (first principal coordinate axis [PCoA1: 6.8%]) (Fig. 3). The second principal coordinate axis partially explained the host ranges within the U.S. [PCoA2: 4.6%]), which exhibited greater variation than their Thai counterparts. ANODIS analysis confirmed the existence of a significant difference between the U.S. and Thai phage host ranges (R2=0.118, p < 0.001). Importantly, the variation of host ranges within U.S. phage isolates was much greater than the variation observed in Thai phage isolates.
Figure 3. Principal Coordinates Analysis (PcoA) of the host range phenotypes.
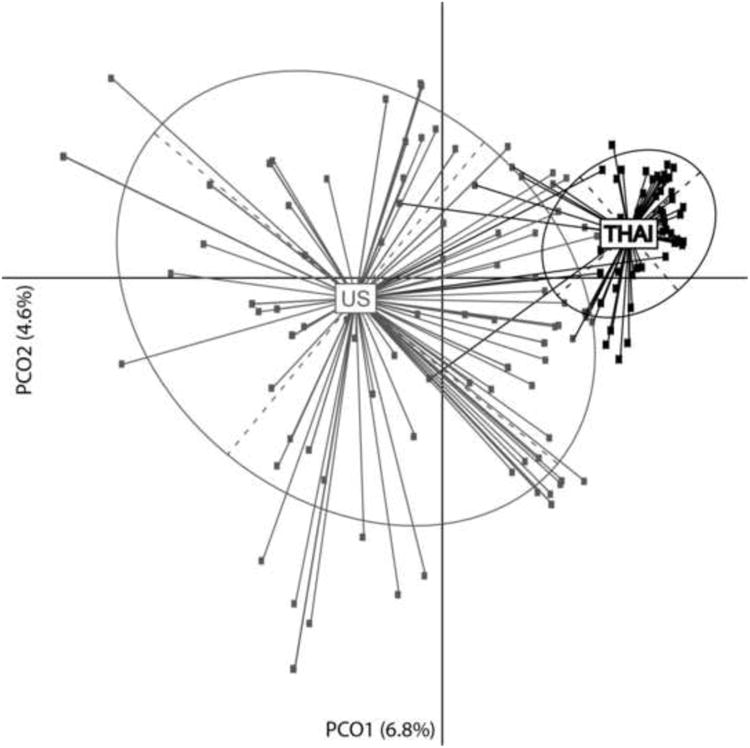
In gray are the U.S. phage isolates, in black are the Thai phage isolates. X-axis is the first source of variation that distinguished U.S. and Thai phages and Y-axis is the second source of variation that shows the diversity within U.S phage isolates. The two components explained 6.8% and 4.6% of the variance, respectively. Four Thai phage isolates exhibited host ranges similar to those observed in the U.S. phage isolates and five U.S. phage isolates exhibited host ranges similar to those observed in the Thai phage isolates.
4. Discussion
4.1. Salmonella phages from Thai dairy farm show considerably wider host ranges than phage isolates from U.S. dairy farms
The characterized host ranges of Salmonella phage isolates obtained from two different countries showed considerable differences. While most of the Thai phage isolates showed wide and very wide host range (mean number of strain lysed = 19); most of the U.S. phage isolates showed narrow host range (mean number of strain lysed = 4). Here we found three Thai phages that lysed all 25 Salmonella strains tested representing 23 serovars. Previously, wide host range Salmonella phages have been well characterized (e.g., phages FelixO1 and PVP-SE1 (Santos et al., 2011; Whichard et al., 2010)). FelixO1 is described as one of the phages with the widest host range, when > 600 strains were tested, FelixO1 lysed > 98% of the strains (Welkos et al., 1974). PVP-SE1 was isolated from wastewater plants in Europe; this phage was found to lyse multiple Salmonella serovars (Santos et al., 2011). Salmonella phages with narrow host range have been reported as well, including phages specific to a single serovar (e.g., phage FSL SP-031 only lyses S. Cerro (Moreno Switt et al., 2013b)). One of the factors that drive the phage host range is the abundance of a suitable host, it has been reported that phages isolated from environments with high host density tend to have a narrow host range, or are specific to the host that is in abundance (Guyader and Burch, 2008). Concordant with these findings, a previous per-farm analysis on the host range characterization of U.S. phages showed that phages specialized to lyse a single serovar were isolated from farms where that given serovar was in high prevalence (10-50%) (Moreno Switt et al., 2013a). The most crucial differences between the dairy farms in these two countries are the farming practices. While in Thailand the farms were all small-scale and free-range farms (15 cows/farm on average), in the U.S. the sampled farms were all intensive and confined farm systems (1,323 cows per farm on average). In addition, free-range hens were typically found in the dairy farms in Thailand (Koonawootrittriron, 2010). We thus hypothesize here that agricultural practices may have a role in the diversity of Salmonella phages in Thailand and the U.S. Whereas confined-intensive farms may facilitate high density of a single serovar, which have been reported for dairy farms in New York (Cummings et al. 2010); free-range cows and presence of other animals, may facilitate low density of Salmonella, and co-presence of Salmonella serovars associated with different animal hosts (e.g., S. Enteritidis is associated with poultry) (Gantois et al. 2009). Consequently, high density of a single serovar might drive toward phages with narrow host range; on the contrary, low density of multiple serovars might drive toward phages with wide host range. Our hypothesis could be further tested by phage isolation and host range characterization from multispecies/free-range and single species/confined farms.
Dairy farm environments are rich in phage abundance and diversity, as previously described for U.S. phages and also found in this study for Thai phages (Moreno Switt et al., 2013a). Phage isolation using samples from different continents, representing different production systems, can further enhance the ability to isolate phages with desired host ranges. These phages could further be used as biocontrol agents and in diagnostic applications.
4.2. Trade of agricultural products from different production systems and/or continents may contribute to dispersal of different phages
In this study we found that U.S. and Thai phages showed different phenotypic characteristics. International trade of food and agricultural products could facilitate the dispersal of different phage populations. Dissemination of Salmonella through the international food trade have been documented and it is a concern for food safety authorities in the U.S. (Buzby, 2003). International dissemination of Salmonella phages have not been reported, despite the well-known fact that phages play important roles in the evolution of foodborne zoonotic pathogens, by horizontal gene transfer and lysogenic conversion (Brüssow et al.,2004; Casjens, 2003). Dispersal of distinct phages could facilitate horizontal gene transfer across a wide range of Salmonella serovars. If wide host range phage isolates characterized in this study are transducers, further experimental validations are needed to identify undesirable traits in transduced Salmonella strains.
To determine if U.S. and Thai phages are in fact different phages (e.g., different genera), genomic characterizations are necessary. Current data show that whereas some phages appear to be widely distributed through the globe (PhageDB.org); other phages appear to have a limited distribution (Held and Whitaker, 2009). Importantly, if the differences in the phenotypes between U.S. and Thai phages correspond with genomic differences, distinct phages in different geographical regions could be used to track sources, particularly in the case of outbreaks associated with food imported from multiple locations.
5. Conclusion
An improved understanding of Salmonella phage diversity will likely provide a better insight into the roles of phages in Salmonella ecology and diversity. This study provides initial insights that distinct dairy farm managements could drive toward narrow or wide host range phage populations. While the presence of wide host range phages could facilitate horizontal gene transfer; these wide host range phages, isolated here, can also be used for biocontrol and for diagnostic applications.
Supplementary Material
Highlights.
Host ranges of Salmonella phages isolated from dairy farms in Thailand and in the US were compared.
In comparison to US phage isolates, Thai phages showed much wider host ranges.
Different farm management practices and locations could yield phages with different host ranges.
Acknowledgments
We thank Felix d'Herelle Collection, Laval University, Quebec, Canada for providing the control phage FelixO1. We thank Yumin Xu for her support with some experiments. Support for Sarach Wongsuntornpoj was provided by the Overseas Research Scholarship from Distinction Program, Faculty of Science, Mahidol University. Supports for this project were provided by NIH/NIAID Food and Waterborne Integrated Research Network/Zoonosis Research Unit Project # ZC009-09 and by New Researcher Scholarship of CSTS/NSTDA (Thailand) Project # SCH-NR-2010-14-02. The funding sources had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarestrup FM, Hendriksen RS, Lockett J, Gay K, Teates K, McDermott PF, White DG, Hasman H, Sørensen G, Bangtrakulnonth A, Pornreongwong S, Pulsrikarn C, Angulo FJ, Gerner-Smidt P. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg Infect Dis. 2007;13:726–31. doi: 10.3201/eid1305.061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens EM, Ackermann HW, Anany H, Blasdel B, Connerton IF, Goulding D, Griffiths MW, Hooton SP, Kutter EM, Kropinski AM, Lee JH, Maes M, Pickard D, Ryu S, Sepehrizadeh Z, Shahrbabak SS, Toribio AL, Lavigne R. A suggested new bacteriophage genus: “Viunalikevirus”. Arch Virol. 2012;157:2035–46. doi: 10.1007/s00705-012-1360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatti Filho RL, Higgins JP, Higgins SE, Gaona G, Wolfenden AD, Tellez G, Hargis BM. Ability of bacteriophages isolated from different sources to reduce Salmonella enterica serovar enteritidis in vitro and in vivo. Poult Sci. 2007;86:1904–9. doi: 10.1093/ps/86.9.1904. [DOI] [PubMed] [Google Scholar]
- Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- Centers for Diseases Control and Prevention. National Emteric Disease Surveillance. Salmonella annual Report. 2011 Avilable at: http://www.cdc.gov/ncezid/dfwed/PDFs/salmonella-annual-report-2011-508c.pdf.
- Chuanchuen R, Ajariyakhajorn K, Koowatananukul C, Wannaprasat W, Khemtong S, Samngamnim S. Antimicrobial resistance and virulence genes in Salmonella enterica isolates from dairy cows. Foodborne Pathog Dis. 2010;7:63–9. doi: 10.1089/fpd.2009.0341. [DOI] [PubMed] [Google Scholar]
- Cummings KJ, Warnick LD, Elton M, Gröhn YT, McDonough PL, Siler JD. The effect of clinical outbreaks of salmonellosis on the prevalence of fecal Salmonella shedding among dairy cattle in New York. Foodborne Pathog Dis. 2010;7:815–23. doi: 10.1089/fpd.2009.0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanis E, Lo Fo Wong DMA, Patrick ME, Binsztein N, Cieslik A, Chalermchikit T, Aidara-Kane A, Ellis A, Angulo FJ, Wegener HC. Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg Infect Dis. 2006;12:381–8. doi: 10.3201/eid1203.050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Gast R, Humphrey TJ, Van Immerseel F. Mechanisms of egg contamination by Salmonella enteritidis. FEMS Microbiol Rev. 2009;33:718–38. doi: 10.1111/j.1574-6976.2008.00161.x. [DOI] [PubMed] [Google Scholar]
- Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–338. [Google Scholar]
- Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PAD, Weill FX. Supplement 2003-2007 (No 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol. 2010;161:26–9. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Guyader S, Burch CL. Optimal foraging predicts the ecology but not the evolution of host specialization in bacteriophages. PLoS One. 2008;3:e1946. doi: 10.1371/journal.pone.0001946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–66. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- Hendriksen RS, Bangtrakulnonth A, Pulsrikarn C, Pornruangwong S, Noppornphan G, Emborg HD, Aarestrup FM. Risk factors and epidemiology of the ten most common Salmonella serovars from patients in Thailand: 2002-2007. Foodborne Pathog Dis. 2009;6:1009–19. doi: 10.1089/fpd.2008.0245. [DOI] [PubMed] [Google Scholar]
- Hoelzer K, Cummings KJ, Wright EM, Rodriguez-Rivera LD, Roof SE, Moreno Switt AI, Dumas N, Root T, Schoonmaker-Bopp DJ, Grohn YT, Siler JD, Warnick LD, Hancock DD, Davis MA, Wiedmann M. Salmonella Cerro isolated over the past twenty years from various sources in the US represent a single predominant pulsed-field gel electrophoresis type. Vet Microbiol. 2011;150:389–93. doi: 10.1016/j.vetmic.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzby Jean C. International Trade and Food Safety: Economic Theory and Case Studies. Agricultural Economic Report No (AER-828) 2003 Nov;:145. 2013. Available at: http://www.ers.usda.gov/publications/aer-agricultural-economic-report/aer828.aspx#.UvJZNbSj8kI.
- Koonawootrittriron S, Mauricio AE. Challenges and oportunities for improvement in dairy production and genetic progress in Thailand. 2010 Available at: http://www.animal.ufl.edu/elzo/Presentations/2010/14AAAP/Presentation-SKK-MAE_20100819-Final.pdf.
- McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- McLaughlin MR, Balaa MF, Sims J, King R. Isolation of Salmonella bacteriophages from swine effluent lagoons. J Environ Qual. 2006;35:522–8. doi: 10.2134/jeq2005.0080. [DOI] [PubMed] [Google Scholar]
- Moreno Switt AI, den Bakker HC, Vongkamjan K, Warnick LD, Cumming KJ, Wiedmann M. Salmonella bacteriophage diversity reflects host diversity on dairy farms. Food Microbiol. 2013;36:275–85. doi: 10.1016/j.fm.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Switt AI, Orsi RH, den Bakker HC, Vongkamjan K, Altier C, Wiedmann M. Genomic characterization provides new insight into Salmonella phage diversity. BMC Genomics. 2013;14:481. doi: 10.1186/1471-2164-14-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Mycobacteriophage database. Phage isolation location by GPS coordinates. Available at: http://phagesdb.org/GPSmap/
- Padungtod P, Kaneene JB. Salmonella in food animals and humans in northern Thailand. Int J Food Microbiol. 2006;108:346–54. doi: 10.1016/j.ijfoodmicro.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galán JE, Ginocchio C, Curtiss R, Gyles CL. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–9. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62:142–60. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SB, Kropinski AM, Ceyssens PJ, Ackermann HW, Villegas A, Lavigne R, Krylov VN, Carvalho CM, Ferreira EC, Azeredo J. Genomic and proteomic characterization of the broad host range Salmonella phage PVP-SE1 - The creation of a new phage genus. J Virol. 2011;85:11265–73. doi: 10.1128/JVI.01769-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongkamjan K, Moreno Switt AI, den Bakker HC, Fortes ED, Wiedmann M. Silage collected from dairy farms harbors an abundance of listeriaphages with considerable host range and genome size diversity. Appl Environ Microbiol. 2012;78:8666–75. doi: 10.1128/AEM.01859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welkos S, Schreiber M, Baer H. Identification of Salmonella with the O-1 bacteriophage. Appl Microbiol. 1974;28:618–22. doi: 10.1128/am.28.4.618-622.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whichard JM, Weigt LA, Borris DJ, Li LL, Zhang Q, Kapur V, Pierson FW, Lingohr EJ, She YM, Kropinski AM, Sriranganathan N. Complete genomic sequence of bacteriophage felix o1. Viruses. 2010;2:710–30. doi: 10.3390/v2030710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.