Abstract
Cryptococcus neoformans capsule structure modifications after prolonged in vitro growth or in vivo passaging have been reported previously. However, nothing is known about the dynamics of these modifications or about their environmental specificities. In this study, capsule structure modifications after mouse passaging and prolonged in vitro culturing were analyzed by flow cytometry using the glucuronoxylomannan-specific monoclonal antibody E1. The capsule structures of strains recovered after 0, 1, 8, and 35 days were compared by using the level of E1-specific epitope expression and its cell-to-cell heterogeneity within a given cell population. In vitro, according to these parameters, the diversity of the strains was higher on day 35 than it was initially, suggesting the absence of selection during in vitro culturing. In contrast, the diversity of the strains recovered from the brain tended to decrease over time, suggesting that selection of more adapted strains had occurred. The strains recovered on day 35 from the spleen and the lungs had different phenotypes than the strains isolated from the brain of the same mouse on the same day, thus strongly suggesting that there is organ specificity for C. neoformans strain selection. Fingerprinting of the strains recovered in vitro and in vivo over time confirmed that genotypes evolved very differently in vitro and in vivo, depending on the environment. Overall, our results suggest that organ-specific selection can occur during cryptococcosis.
Cryptococcus neoformans is a fungus that causes life-threatening infections of the central nervous system in immunocompromised individuals. Different phenotypic features, such as laccase expression (28), the ability to grow at 37°C, and especially the presence of a polysaccharide capsule (1), have been associated with yeast virulence. The main constituent of the capsule is glucuronoxylomannan (GXM), which is a large polymer of α-1,3 mannose with glucuronic acid, xylose, and O-acetyl branching (5). The quantities and positions of the O-acetyl and xylose residues determine the serospecificity of the strains, even though some differences exist between isolates within each serotype (5). Cherniak et al. also reported structural changes of the capsule structure during the course of human infections (4). Strain microevolution in vitro and in vivo, resulting in colony morphogenesis and changes in the capsule thickness and structure, has also been described (14, 15). The mechanism responsible for modifications in the capsule structure is not yet known, and only a few genes involved in capsule biosynthesis have been characterized to date (1). Moreover, although some transduction pathways have been shown to transduce signals regulating capsule size, the pathways involved in capsule structure regulation have yet to be identified (17). Members of our laboratory have previously shown that the capsule structure can influence the virulence of the strains since capsular xylose residues are necessary for C. neoformans virulence and de-O-acetylated capsule-producing strains are more virulent than control strains in a murine model of cryptococcosis (18, 24). Spontaneous and genetically controlled modifications of the capsule structure have been shown to influence different parameters of the cryptococcosis physiopathology (i.e., complement binding, cytokine production, phagocytosis, and GXM clearance) (20, 21, 26, 29).
Previous genetic analyses of the relationships between serial isolates from individual patients revealed that C. neoformans undergoes microevolution, thereby suggesting a mechanism responsible for the genotypic and phenotypic diversity in environmental and clinical isolates (2, 13). We also recently obtained different hybridization profiles for strains isolated from the same patient (16).
For this study, we investigated the dynamics of these spontaneous capsule structure modifications in a mouse model of cryptococcosis and in vitro. We first used the CENTEL probe, which has been proven to be useful for the characterization of isolates recovered from different body sites of the same patient over time (16). We then monitored capsule structure changes during the course of infection by flow cytometry using a capsule-specific monoclonal antibody (MAb). We show here that strain evolution is specific to the environment, and our data support the hypothesis of the organ-dependent selection of strains during the course of infection.
MATERIALS AND METHODS
Clinical isolates.
Seven C. neoformans var. grubii isolates were recovered from bronchoalveolar lavage fluid (C28 and C44), cerebrospinal fluid (C47), blood (C24), urine (C45 and C46) and bone marrow biopsy (C22) samples from an AIDS patient with cryptococcosis. The case was sent to the French National Reference Center for Mycoses during a multicentric prospective study (Etude Crypto A/D, Direction Générale de la Santé no. 970089). Isolates C44 and C47 were obtained the day treatment was started, and isolates C46, C45, C22, C24, and C28 were recovered 1, 6, 12, 15, and 29 days later, respectively. All of these isolates from patient P6 were already characterized in a previous study as having different CENTEL hybridization patterns (16).
Isolate C45 was arbitrarily chosen for a study of dynamics and was therefore streaked on Sabouraud agar and incubated for 48 h at 30°C. Five individual colonies were recovered. One of them was designated C45a and was stored at −80°C in 40% glycerol until further analysis, as were all of the strains that were subsequently isolated during the study (see below). These strains were assigned a code constructed as follows: C, B, S, or L to indicate strains recovered, respectively, from an in vitro culture or from the brain, spleen, or lungs of infected mice; the day of isolation followed by a period; and a number from 1 to 5. For example, strain C.8.3 was the third isolate recovered on the 8th day of in vitro culturing.
Animals.
BALB/c male mice (6 weeks old) and outbred male OF1 mice [Ico:OF1 (I.O.P.S. caw)] were obtained from Charles River Laboratory (St. Aubin-lès-Elbeuf, France). They were maintained in our animal facility and had access to food and water ad libitum.
Determination of microevolution events in C. neoformans. (i) In vivo procedure.
After being precultured on Sabouraud dextrose agar, strain C45a was transferred as a cell mass into yeast nitrogen base medium (Difco Laboratories, Inc., Detroit, Mich.) supplemented with 2% glucose and then incubated for 18 h at 30°C. The overnight culture was washed twice with sterile physiological saline, yeast cells were counted on a hemacytometer, and their number was adjusted to the appropriate concentration. Mice were infected by intravenous injection via the lateral tail vein (five mice infected with 105 yeast cells per mouse). The number of viable cells was verified by colony counts (CFU) on Sabouraud agar-filled petri dishes and five colonies (day 0 strains) were arbitrarily selected for analysis. For each time point after a yeast challenge (days 1, 8, and 35), one mouse was sacrificed by chloroform inhalation. The brain, lungs, and spleen were removed and homogenized, and the homogenates were plated onto Sabouraud agar-filled petri dishes. Five colonies were then arbitrarily selected from each of the organ cultures.
(ii) In vitro procedure.
After being precultured on Sabouraud dextrose agar, a loopful of strain C45a was inoculated into 50 ml of yeast-peptone-dextrose (YPD) liquid medium (10 g of yeast extract, 20 g of Bacto Peptone, and 20 g of glucose per liter) and incubated at 30°C with agitation at 150 rpm. After 1, 8, and 35 days of culture (with renewal of the medium every 3 days), one aliquot of the culture was sampled, plated onto Sabouraud agar, and allowed to grow for 48 h at 30°C. Five colonies were recovered at each time point.
RFLP analysis.
For restriction fragment length polymorphism (RFLP) analysis, genomic DNAs were prepared as previously described (33). All of the clones recovered in vivo from infected mice (n = 15 per organ) and in vitro (n = 15) as well as the initial strain C45a and the five clones recovered from the inoculum were typed by Southern blotting with the CENTEL probe (16) labeled with digoxigenin-11-dUTP by use of a DIG-High Prime kit (Boehringer Mannheim, Mannheim, Germany). The CENTEL probe was labeled by using the whole pCnTel-1 plasmid as a substrate. The pCnTel-1 plasmid contains the URA5 gene and C. neoformans telomeric repeats (10). The genomic DNAs of the 36 strains were digested with AccI. The resulting fragments were then separated by electrophoresis through a 0.8% agarose gel and transferred onto positively charged nylon membranes (Boehringer Mannheim). After an overnight hybridization at 65°C and stringent washes, bands were detected after exposure to a chemiluminescent substrate according to the manufacturer's instructions.
Virulence studies.
To evaluate the relative virulence of the strains, we inoculated 104 cryptococci intravenously into groups of seven mice per strain after preparation of the inoculum as described above. The animals were observed daily and deaths were recorded.
Growth rate and capsule measurements.
For each strain, a growth curve at 37°C on YPD medium was drawn and doubling times were calculated. To determine the capsule thickness, we incubated cells overnight at 30°C in induction medium [1.7 g of yeast nutrient base without amino acids and without ammonium sulfate (Difco Laboratories) supplemented with 1.5 g of asparagine and 20 g of glucose per liter of buffer containing 12 mM NaHCO3 and 35 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 7.1] (18). Yeast cells were then suspended in India ink, and the distance from the cell wall to the outer border of the capsule was measured for 20 cells with a magnification of ×100 by using a grid with a resolution of 0.12 μm. The mean thickness (± standard deviation [SD]) was determined for each strain.
Antifungal drug susceptibility testing.
Fluconazole (Pfizer Central Research, Sandwich, United Kingdom), itraconazole (Janssen Pharmaceuticals, Beerse, Belgium), and amphotericin B (Squibb, Princeton, N.J.) powders were used. Final drug concentrations ranged from 0.125 to 64 μg/ml for fluconazole and 0.015 to 8 μg/ml for itraconazole and amphotericin B. MICs were determined by a broth microdilution technique performed according to a protocol from the National Committee for Clinical Laboratory Standards (25). The MIC was considered to be the lowest drug concentration that inhibited growth at least 80% compared to the control well for the azole drugs and 100% for amphotericin B. Candida kefyr 706 was used as the reference strain. MICs were considered to be significantly different when they differed by ±2 dilutions.
Laccase production.
Whole-cell laccase assays were performed according to a modified version of a previously described method (34). Briefly, freshly harvested cultured cells were grown on minimal asparagine medium (1 g of asparagine, 0.25 g of MgSO4, and 3 g of sodium phosphate per liter [pH 6.5]) supplemented with 20 g of glucose per liter for 16 h at 30°C. Cells were harvested, washed once in minimal asparagine medium without glucose, and resuspended in the same medium for 24 h at 30°C. Cells were then recovered, washed once with water, resuspended in 5 ml of phosphate-buffered saline (PBS; 10 mM phosphate buffer, 150 mM NaCl, pH 6.5), and diluted to 107 yeast cells/ml. Aliquots (1 ml) were mixed with 10 mM epinephrine (Sigma Chemical Co., St. Louis, Mo.) and incubated for 30 min at room temperature. Supernatants were recovered and optical densities were read at 475 nm by a Pharmacia Biotech (Cambridge, United Kingdom) Ultrospec 2000 spectrophotometer. One unit of laccase activity was defined as 0.001 absorbance units after 30 min of incubation. The results are expressed in units of laccase activity per 107 cells.
Flow cytometry.
Clinical in vivo- and in vitro-passaged strains were analyzed by fluorescence-activated cell sorting (FACS) with an EPICS XL-4C instrument (Beckman Coulter, Miami, Fla.). Cells were grown for 48 h in liquid Sabouraud medium at 30°C and subcultured overnight in induction medium. They were washed twice with sterile PBS and diluted to 107 yeast cells per ml. The cells were then incubated for 30 min at room temperature with the GXM-specific MAb E1 (8) (hybridoma supernatant diluted 1:100) and a 1:100 dilution of fluorescein isothiocyanate-labeled goat anti-mouse immunoglobulin G (Sigma). Next, the cells were washed once with PBS, fixed in 2% paraformaldehyde (Sigma), and stored at 4°C in the dark until they were used for FACS analysis. Cells incubated without antibodies or with only the fluorescein isothiocyanate conjugate were considered negative controls. A total of 10,000 cells were analyzed for each strain. Histograms of fluorescence distribution were generated by plotting the numbers of cells (y axis) versus fluorescence intensities (log scale; x axis) with System II software (Beckman Coulter). Two parameters were considered: first, the median fluorescence intensity (MdF), which reflects the overall level of expression of the epitope specifically recognized by E1, and second, the coefficient of variation of fluorescence (CVF). which reflects the cell-to-cell heterogeneity in a given cell population. The CVF was calculated as follows: CVF = (SD × 100)/mean fluorescence intensity. The values obtained from a typical experiment are reported. The interstrain variability was assessed by calculating the SD for both the MdF and the CVF at each sampling time. Experiments were performed twice independently, and the results were reproducible.
Statistical analysis.
Statistical analyses were performed with GraphPad Prism, version 3.03, for Windows (GraphPad Software, San Diego, Calif.). Epitope expression and cell-to-cell heterogeneity were compared by the Kruskal-Wallis test and by Dunn's posthoc test for multiple comparisons. Mortality rates were compared by the log rank test. Statistical significance was defined as P levels of <0.05.
RESULTS
Genotypic and phenotypic studies on clinical isolates recovered from a single patient.
We investigated whether the genotypic differences previously noted for seven clinical isolates from the same patient (16) were associated with any particular phenotypic changes. Indeed, in a murine model of cryptococcosis, the isolates exhibited different degrees of virulence (P < 0.02 by the log rank test). The median survival time for the most virulent isolate (C24) was 28 days, whereas it was 45 days for the least virulent isolate (C47) (Fig. 1). Isolate C45 had an intermediate level of virulence. These results were confirmed in two independent experiments using inbred (BALB/c) or outbred (OF1) mice (data not shown).
FIG. 1.
Clinical isolate virulence assays. The survival of mice infected with 104 viable yeast cells from one of seven clinical isolates recovered from the same patient are shown.
We then focused on the characterization of phenotypes which have typically been associated with cryptococcal virulence in other studies (3). As shown in Table 1, only small differences were seen among isolates regarding their laccase activities or fluconazole, itraconazole, and amphotericin B MICs. All of the isolates but one produced dry colonies with similar growth rates. We also noted slight differences in the capsule thickness from one isolate to another, with a median capsule thickness of 0.20 μm.
TABLE 1.
Phenotypic characteristics of clinical isolates
| Isolate | Laccase activity (U/107 cells)b | Doubling time (h) | Capsule thickness (μm)b | MIC (μg/ml) of drug for isolatea
|
Capsule structure
|
|||
|---|---|---|---|---|---|---|---|---|
| ITZ | FCZ | AmB | Fluorescence intensity (MdF) | Heterogeneity of the cell population (CVF) | ||||
| C22 | 199 ± 36 | 4.4 | 0.22 ± 0.07 | 1 | 4 | 1 | 1.5 | 244 |
| C24 | 173 ± 9 | 4.3 | 0.23 ± 0.08 | 0.25 | 2 | 1 | 1.8 | 238 |
| C28 | 178 ± 2 | 4.0 | 0.18 ± 0.07 | 0.25 | 2 | 0.5 | 2.1 | 233 |
| C44 | 181 ± 9 | 3.8 | 0.20 ± 0.07 | 0.5 | 4 | 0.5 | 1.9 | 224 |
| C45 | 224 ± 33 | 3.6 | 0.16 ± 0.08 | 0.5 | 4 | 0.5 | 2.3 | 250 |
| C46 | 188 ± 29 | 3.9 | 0.17 ± 0.07 | 0.5 | 4 | 1 | 2.5 | 231 |
| C47 | 182 ± 21 | 3.7 | 0.20 ± 0.06 | 0.5 | 4 | 0.5 | 4.6 | 236 |
ITZ, itraconazole; FCZ, fluconazole; AmB, amphotericin B.
Data are means ± SD.
We also analyzed the structure of the capsule by using the anti-GXM MAb E1 (8). A preliminary observation of isolate C45 under an epifluorescence microscope showed that even though most of the cells were positive for E1, the fluorescence intensity was considerably variable from one cell to another (Fig. 2). In order to characterize the capsule structures of the different isolates, we considered two parameters which estimated the fluorescence intensity and the cell-to-cell heterogeneity (see Materials and Methods). The capsule structures of the clinical isolates differed slightly from one isolate to another. However, no obvious relationships were observed between the phenotype or genotype, the day of sampling, or the organ from which the clinical isolate was recovered.
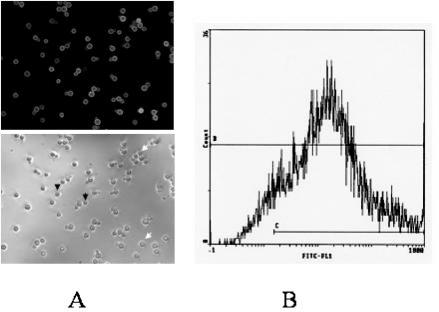
FIG. 2.
Heterogeneity of the capsule structure within a clonal cell population. (A) Cells from strain C45a were grown on induction medium and labeled with the anti-GXM MAb E1. The arrows indicate strongly labeled cells (white arrows) and negative cells (black arrows). (B) FACS analysis of this cell population.
Genotypic variability in mice and during in vitro culturing.
The dynamics of microevolution events in vivo were studied in a murine model of cryptococcosis after the recovery at different times (days 1, 8, and 35) of five clones from each infected organ (brain, spleen, and lungs). We characterized all of the strains by using the CENTEL probe. Of the five strains studied from the inoculum subculture (day 0), four had the original C45a profile (I) and one had a new profile (II) (Fig. 3). For the mouse-passaged strains, pattern differences were detected early during the course of the infection (as early as day 1 for one strain each isolated from the spleen and lungs and on day 8 for two strains isolated from the brain) (Fig. 3). All of the strains recovered from the brain on day 1 had profile I. However, on day 35, none of the strains had the initial profile. In the lungs and spleen, two or three patterns, including the original pattern I, were observed over time (Fig. 3).
FIG. 3.
(A) Evolution of CENTEL hybridization patterns of strains isolated from the brain over time. Stars indicate profile changes compared to the original strain C45a (profile I). Roman numerals refer to the different profiles. (B) Schematic representation of the evolution of CENTEL hybridization patterns of strains isolated from continuous culture (▴) or from different organs (▪). B, brain; S, spleen; L, lungs. Roman numerals refer to the different profiles. Only two strains isolated from the lungs on day 8 gave interpretable profiles, and they are not included in this figure.
We then investigated the behavior of strain C45a in terms of genotypic variability after in vitro growth for 1, 8, and 35 days. Changes were detected in one of five strains after 1 and 8 days of culture, and no changes were observed among the five clones recovered after 35 days of culture (Fig. 3).
Phenotypic variability in mice and during in vitro culturing.
Since the dynamics of microevolution of the clones recovered from the brain and after in vitro culturing clearly differed, the phenotypes of the corresponding strains were assessed by using all of the parameters that were evaluated for the clinical isolates, except for laccase activity, which was not informative and not reproducible in our hands. The phenotypic characteristics of the strains recovered from brains are reported in Table 2. All strains but one produced colonies with a dry morphology. The differences seen in the capsule thickness and drug MICs had no obvious relationship with the day of sampling.
TABLE 2.
Phenotypic characteristics of strains recovered from the brainc
| Strain | Doubling time (h) | Capsule thickness (μm)b | MIC of drug for straina
|
Capsule structure
|
|||
|---|---|---|---|---|---|---|---|
| ITZ | FCZ | AmB | Fluorescence intensity (MdF) | Heterogeneity of cell population (CVF) | |||
| B.1.1 | 5.0 | 0.17 ± 0.05 | 0.5 | 8 | 1 | 1.2 | 202 |
| B.1.2 | 4.2 | 0.20 ± 0.10 | 0.5 | 8 | 0.5 | 31.7 | 203 |
| B.1.3 | 4.1 | 0.16 ± 0.06 | 0.25 | 8 | 0.5 | 79.6 | 144 |
| B.1.4 | 3.9 | 0.16 ± 0.05 | 0.5 | 4 | 0.5 | 9.7 | 227 |
| B.1.5 | 3.7 | 0.23 ± 0.09 | 0.125 | 4 | 0.5 | 70.2 | 163 |
| B.8.1 | 4.0 | 0.20 ± 0.08 | 0.5 | 8 | 1 | 34.4 | 200 |
| B.8.2 | 4.1 | 0.17 ± 0.05 | 0.25 | 8 | 0.5 | 86.4 | 143 |
| B.8.3 | 4.1 | 0.20 ± 0.06 | 0.125 | 8 | 0.5 | 13.3 | 247 |
| B.8.4 | 3.7 | 0.17 ± 0.06 | 0.25 | 8 | 0.5 | 17.4 | 209 |
| B.8.5 | 4.2 | 0.19 ± 0.05 | 0.06 | 4 | 0.5 | 43.7 | 162 |
| B.35.1 | 6.1 | 0.17 ± 0.04 | 0.03 | 4 | 0.5 | 5.4 | 250 |
| B.35.2 | 6.6 | 0.16 ± 0.05 | 0.03 | 2 | 0.5 | 8.7 | 235 |
| B.35.3 | 7.1 | 0.16 ± 0.06 | 0.03 | 4 | 0.5 | 7.0 | 232 |
| B.35.4 | 3.9 | 0.23 ± 0.06 | 0.125 | 8 | 0.5 | 14.7 | 224 |
| B.35.5 | 19.6 | 0.17 ± 0.05 | 0.015 | 2 | 0.25 | 8.0 | 249 |
ITZ, itraconazole; FCZ, fluconazole; AmB, amphotericin B.
Data are means ± SD.
Different phenotypes of strains recovered from the brain on day 1 (B.1.X), day 8 (B.8.X), and day 35 (B.35.X) were studied.
For the clinical isolates, MAb E1 binding to the capsule was examined by flow cytometry. The fluorescence intensity values for the 15 mouse-passaged strains recovered from brains differed, as did the cell-to-cell heterogeneity of the capsule structure within each cell population (Table 2). We observed that the expression of the epitope recognized by E1 was decreased on day 35 compared to that on day 1 or 8, with a concomitant increase in cell-to-cell heterogeneity (Fig. 4A). However, due to the high diversity of the strains recovered on days 1 and 8, these differences were not statistically significant even though this trend was confirmed with a smaller inoculum (P < 0.001) (Fig. 4). In contrast, the strains recovered from the spleen or the lungs of the same mouse on day 35 had a more homogeneous phenotype (Fig. 4A).
FIG. 4.
Capsule structure evolution as assessed by median fluorescence intensity (A) and heterogeneity of the cell population (CVF; see Materials and Methods) (B) after in vitro or in vivo passaging. Two in vivo independent experiments were done, one with a large inoculum (105 cells per mouse) and one with a small inoculum (104 cells per mouse).
We also analyzed the strain-to-strain diversity at each sampling time. As shown in Fig. 4B, the diversity of the strains recovered from brains on day 35 was comparable to that observed on day 0 and much lower than what was seen on days 1 and 8. Similarly, the levels of diversity among strains recovered on day 35 from the spleen and the lungs of the same mouse were low and comparable to the day 0 measurement. These data were reproduced in an independent experiment using a lower inoculum.
Table 3 summarizes the phenotypic characteristics of strains recovered from a continuous culture over time. Small differences were seen for in vitro susceptibilities to itraconazole, but not to amphotericin B or fluconazole. All colonies obtained on days 1, 8, and 35 exhibited the dry phenotype. Capsule thicknesses varied widely, with a median of 0.20 μm. The capsule structures were analyzed by flow cytometry (Fig. 4). The fluorescence intensity values tended to increase over time (P < 0.02), while the cell-to-cell heterogeneity declined, but not statistically significantly due to the high diversity of the strains recovered on day 35. For both parameters, and in contrast to the results obtained in vivo in the brain, the diversity of the strains was higher on day 35 than at the earlier sample times.
TABLE 3.
Phenotypic characteristics of strains recovered in vitroa
| Strain | Doubling time (h) | Capsule thickness (μm)c | MIC of drug for strainb
|
Capsule structure
|
|||
|---|---|---|---|---|---|---|---|
| ITZ | FCZ | AmB | Fluorescence intensity (MdF) | Heterogeneity of cell population (CVF) | |||
| C.0.1 | 4.1 | 0.12 ± 0.00 | ND | ND | ND | 7.2 | 209 |
| C.0.2 | 4.0 | 0.16 ± 0.05 | ND | ND | ND | 3.8 | 225 |
| C.0.3 | 3.7 | 0.13 ± 0.17 | ND | ND | ND | 8.7 | 194 |
| C.0.4 | 3.7 | 0.17 ± 0.06 | ND | ND | ND | 8.8 | 203 |
| C.0.5 | 3.8 | 0.17 ± 0.07 | ND | ND | ND | 12.5 | 215 |
| C.1.1 | 4.1 | 0.19 ± 0.06 | 0.125 | 8 | 0.5 | 15.5 | 202 |
| C.1.2 | 3.8 | 0.21 ± 0.04 | 0.125 | 8 | 0.5 | 21.5 | 192 |
| C.1.3 | 4.0 | 0.16 ± 0.06 | 0.125 | 8 | 0.5 | 23.3 | 172 |
| C.1.4 | 4.2 | 0.21 ± 0.07 | 0.125 | 8 | 0.5 | 16.7 | 198 |
| C.1.5 | 3.5 | 0.23 ± 0.06 | 0.06 | 4 | 0.5 | 25.8 | 178 |
| C.8.1 | 4.6 | 0.19 ± 0.06 | 0.125 | 8 | 0.5 | 20.4 | 182 |
| C.8.2 | 4.0 | 0.20 ± 0.06 | 0.125 | 8 | 0.5 | 23.4 | 190 |
| C.8.3 | 3.7 | 0.20 ± 0.06 | 0.125 | 4 | 0.5 | 22.8 | 183 |
| C.8.4 | 3.2 | 0.21 ± 0.05 | 0.125 | 4 | 0.5 | 21.8 | 184 |
| C.8.5 | 3.7 | 0.22 ± 0.06 | 0.125 | 4 | 0.5 | 30.2 | 156 |
| C.35.1 | 4.1 | 0.25 ± 0.10 | 0.250 | 8 | 0.5 | 86.4 | 100 |
| C.35.2 | 3.9 | 0.19 ± 0.07 | 0.125 | 4 | 0.5 | 10.7 | 220 |
| C.35.3 | 3.7 | 0.23 ± 0.06 | 0.125 | 4 | 0.5 | 58.9 | 144 |
| C.35.4 | 4.0 | 0.22 ± 0.06 | 0.06 | 4 | 0.5 | 25.9 | 148 |
| C.35.5 | 4.2 | 0.22 ± 0.06 | 0.06 | 8 | 0.5 | 14.1 | 211 |
Different phenotypes of the strains recovered from cultures after day 1 (C.1.X), day 8 day (C.8.X), and day 35 (C.35.X) were studied.
ITZ, itraconazole; FCZ, fluconazole; AmB, amphotericin B. ND, not determined.
Data are means ± SD.
DISCUSSION
The authors of previous studies reported a marked genetic diversity for C. neoformans strains as well as the occurrence of microevolution events during the course of yeast infections. We previously described a wide variety of RFLP profiles (which show microevolution in vivo) for nine clinical isolates that were serially recovered from one C. neoformans-infected patient by use of the new CENTEL probe (16). In this study, we investigated the dynamics of microevolution during prolonged in vitro culturing and in a murine model of cryptococcosis and the phenotypes associated with these genotypic modifications. Indeed, serial isolates differed in their levels of virulence in mice, as assessed by the different mean survival times obtained. This finding is in agreement with the results of Fries and Casadevall (12), who demonstrated that parameters such as virulence in mice, growth rate, and capsule thickness varied among strains and from one serial clinical isolate to another. However, in contrast with previous studies (2, 19, 31) that noted variations in fluconazole and amphotericin B susceptibilities among recurrent isolates, we found no significant differences in antifungal susceptibilities. Similarly, the growth rates for all of our isolates were comparable, with only minor variations in the capsule thickness.
Changes in the GXM structure in the C. neoformans capsule have been observed by nuclear magnetic resonance with isolates from patients with recurrent cryptococcosis (4). For our study, we analyzed the capsule structure by flow cytometry using the anti-GXM MAb E1, therefore enabling the characterization of more isolates than by biochemical methods. Moreover, Cleare et al. reported that a C. neoformans population could have heterogeneous and reproducible patterns when analyzed by flow cytometry (6). Thus, the use of this technique allowed us not only to compare the expression of one epitope among the strains, but also to examine the cell-to-cell heterogeneity of the capsule structure within a clonal population. Our results showed that the degree of heterogeneity and the level of epitope expression recognized by MAb E1 were reproducible in independent experiments, thereby clearly determining marked phenotypes of the different strains. Phenotypic heterogeneity has been described previously for Saccharomyces cerevisiae and other microorganisms (for a review, see reference 32). In S. cerevisiae, some of the key parameters that control this heterogeneity can be cell cycle progression, cell aging, mitochondrial activity, epigenetic regulation, and potentially stochastic variation. In C. neoformans, antigenic heterogeneity seems to be a general phenomenon and is not specific to a serotype or to the anti-GXM antibody used (G. Janbon, unpublished observation). However, the mechanism that drives this cell-specific phenotype is currently unknown. Moreover, assessing whether the modifications of the capsule structure can explain the differences in virulence noted among the serial clinical isolates is impossible with this experimental design. Similarly, there were no obvious relationships between a specific CENTEL pattern and a particular phenotype or virulence level of the clinical isolates. The modifications of the CENTEL pattern observed in this study are only indicative of the microevolution of the isolates.
Using CENTEL genotyping and the flow cytometry approach to analyze the capsule structure, we were able to detect the microevolution of a strain in a murine model and to look at its phenotypic consequences. Genotypic changes occurred very early in this model, as one strain each from the spleen and the lungs 1 day after inoculation had already modified its CENTEL patterns. Moreover, the number of strains with modified hybridization patterns among those recovered from the infected organs increased over time until day 35 in vivo, whereas in vitro all of the recovered strains retained the initial hybridization pattern. Thus, four of five strains isolated from the brains and three of five strains isolated from the lungs were identical to each other but different from the original strain. These results suggest that microevolution events are not unique to the infection process, but rather that the mouse microenvironment might select for more adapted strains over time (22, 23). Moreover, the brain is a very peculiar organ and is quite isolated from the blood circulation by the brain-blood barrier. This could explain why strains with a modified CENTEL pattern accumulated in this organ. The same strains were also studied with the CNRE-1 probe (30), but we observed very rare modifications of the hybridization pattern (data not shown). This result confirms that the CENTEL probe is a more adapted tool for the study of strain microevolution than the CNRE-1 probe (16). Finally, it is also probable that the CENTEL probe does not reveal all of the strain microevolution steps. The strains recovered from the other organs might have undergone some microevolution steps that are not visible by CENTEL analysis.
According to a previous report, mouse passaging of C. neoformans strains results in alterations of yeast membrane sterol contents and amphotericin B susceptibility (7). In addition, it has been shown that a laboratory C. neoformans strain can undergo microevolution in vitro, resulting in subtypes that differ in capsule thickness, growth, virulence, morphology, and GXM structure (11, 14). In this study, we confirmed that certain phenotypes, such as growth rate, antifungal susceptibility, and capsule thickness, can vary after infection in mice or growth in a synthetic medium.
Most importantly, monitoring of the capsule structure phenotypes during infection revealed an apparent selection of strains over time. Cleare et al. (6) already noted modifications of capsular epitopes after murine passaging of strains, but they did not attempt to establish the dynamics of this phenotypic evolution. Our results showed notable differences between the evolution of the fluorescence patterns of strains recovered from the brains of infected mice and those recovered from continuous culturing in vitro over time. Strains recovered on days 1 and 8 after infection were quite homogeneous, in contrast to day 35, when broad cell-to-cell capsule structure heterogeneity was observed. More striking was the in vivo evolution of the interstrain variability over time, which increased early in the course of the infection and then decreased to the value observed at day 0. This observation contrasted strongly with the in vitro results in which the interstrain variability was higher on day 35 than on day 0. These data suggest that selection of the more adapted strains occurs in vivo, while in vitro the absence of selection pressure and the regular appearance of random variant phenotypes might explain the increased diversity of the population over time. In addition, we found that the expression of the epitope recognized by E1, and thus the capsule structure, differed at day 35 when we compared strains from the brain and strains from the spleen and lungs. These results suggest that strain selection may be dependent on the organ that is seeded. A possible relationship between the characteristics of the isolates and body localization has already been evoked (9). In addition, Rivera and colleagues (27) observed that differences in capsule thickness were organ related. It is obvious that the environmental pressure in the brain differs from that in the spleen and lungs. Lortholary et al. previously reported the organ-dependent evolution of fungal burden and inflammation and emphasized the particularity of the brain (23). Our results are consistent with the idea that the host environment invokes pressure (via nutrients, pH, and host effector cells) on C. neoformans strains, selecting for subtypes that are better adapted to survive and escape host defenses.
In conclusion, strains recovered from mice at different times after C. neoformans inoculation and from different organs presented genotypic and phenotypic modifications. These modifications could have been different under other experimental conditions or with other animals and should thus not be seen as definitive phenotypic characteristics of isolates recovered from infected organs or in vitro cultures. Nevertheless, our experiments show that a number of C. neoformans subtypes exist in mice infected with a pure strain after a few days postinfection. These subtypes may be the result of host- and organ-specific selective pressure.
Acknowledgments
We are grateful to B. Wickes (University of Texas Health Science Center at San Antonio, San Antonio) for providing plasmid pCnTel-1. We thank L. Improvisi (Institut Pasteur) for her contribution during the animal studies and M. T. Nugeyre (Institut Pasteur) for her assistance in the flow cytometry experiments. We also thank J. Jacobson for editorial assistance and Nancy Lee (University of British Columbia) for her critical reading of the manuscript.
Financial support for this work was provided by SIDACTION and the Pasteur Institute (Contrat de Recherche Clinique).
Editor: T. R. Kozel
REFERENCES
- 1.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt, M. E., C. Hutwagner, L. A. Klug, W. S. Baughman, D. Rimland, E. A. Graviss, R. J. Hamill, C. Thomas, P. G. Pappas, A. L. Reingold, R. W. Pinner, and the Cryptococcal Disease Active Surveillance Group. 1996. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 34:912-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan, K. L., and J. W. Murphy. 1998. What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 4:71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherniak, R., L. C. Morris, T. Belay, E. D. Spitzer, and A. Casadevall. 1995. Variation in the structure of glucuronoxylomannan in isolates from patients with recurrent cryptococcal meningitis. Infect. Immun. 63:1899-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherniak, R., H. Valafar, L. C. Morris, and F. Valafar. 1998. Cryptococcus neoformans chemotyping by quantitative analysis 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab. Immunol. 5:146-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleare, W., R. Cherniak, and A. Casadevall. 1999. In vitro and in vivo stability of Cryptococcus neoformans glucuronoxylomannan epitope that elicits protective antibodies. Infect. Immun. 67:3096-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Currie, B., H. Sanati, A. F. Ibrahim, J. E. Edwards, Jr., A. Casadevall, and M. A. Ghannoum. 1995. Sterol compositions and susceptibilities to amphotericin B of environmental Cryptococcus neoformans isolates are changed by murine passage. Antimicrob. Agents Chemother. 39:1934-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dromer, F., J. Salamero, A. Contrepois, C. Carbon, and P. Yeni. 1987. Production, characterization, and antibody specificity of a mouse antibody reactive with Cryptococcus neoformans capsular polysaccharide. Infect. Immun. 55:742-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dromer, F., A. Varma, O. Ronin, S. Mathoulin, and B. Dupont. 1994. Molecular typing of Cryptococcus neoformans serotype D clinical isolates. J. Clin. Microbiol. 32:2364-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edman, J. C. 1992. Isolation of telomere-like sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol. Cell. Biol. 12:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzot, S. P., J. Mukherjee, R. Cherniak, L. Chen, J. S. Hamdan, and A. Casadevall. 1998. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect. Immun. 66:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fries, B., and A. Casadevall. 1998. Serial isolates of Cryptococcus neoformans from patients with AIDS differ in virulence for mice. J. Infect. Dis. 178:1761-1766. [DOI] [PubMed] [Google Scholar]
- 13.Fries, B. C., F. Chen, B. P. Currie, and A. Casadevall. 1996. Karyotype instability in Cryptococcus neoformans infection. J. Clin. Microbiol. 34:1531-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fries, B. C., D. L. Goldman, and A. Casadevall. 2002. Phenotypic switching in Cryptococcus neoformans. Microbes Infect. 4:1345-1352. [DOI] [PubMed] [Google Scholar]
- 15.Fries, B. C., C. P. Taborda, E. Serfass, and A. Casadevall. 2001. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J. Clin. Investig. 108:1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Hermoso, D., F. Dromer, S. Mathoulin-Pellissier, and G. Janbon. 2001. Are two Cryptococcus neoformans strains epidemiologically linked? J. Clin. Microbiol. 39:1402-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hull, C. M., and J. Heitman. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557-615. [DOI] [PubMed] [Google Scholar]
- 18.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42:453-469. [DOI] [PubMed] [Google Scholar]
- 19.Klepser, M. E., and M. A. Pfaller. 1998. Variation in electrophoretic karyotype and antifungal susceptibility of clinical isolates of Cryptococcus neoformans at a university-affiliated teaching hospital from 1987 to 1994. J. Clin. Microbiol. 36:3653-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozel, T. R., S. M. Levitz, F. Dromer, M. A. Gates, P. Thorkildson, and G. Janbon. 2003. Antigenic and biological characteristics of mutant strains of Cryptococcus neoformans lacking capsular O-acetylation or xylosyl side chains. Infect. Immun. 71:2868-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipovsky, M. M., G. Gekker, S. Hu, L. C. Ehrlich, I. M. Hoepelman, and P. K. Peterson. 1998. Cryptococcal glucuronoxylomannan induces interleukin-8 (IL-8) production by human microglia but inhibits neutrophil migration toward IL-8. J. Infect. Dis. 177:260-263. [DOI] [PubMed] [Google Scholar]
- 22.Lortholary, O., L. Improvisi, M. Nicolas, F. Provost, B. Dupont, and F. Dromer. 1999. Fungemia during cryptococcosis sheds some light on pathophysiology. Med. Mycol. 37:169-174. [PubMed] [Google Scholar]
- 23.Lortholary, O., L. Improvisi, N. Rayhane, F. Gray, C. Fitting, J. M. Cavaillon, and F. Dromer. 1999. Cytokine profiles of AIDS patients are similar to those of mice with disseminated Cryptococcus neoformans infection. Infect. Immun. 67:6314-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyrand, F., B. Klaproth, U. Himmelreich, F. Dromer, and G. Janbon. 2002. Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol. Microbiol. 45:837-849. [DOI] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Pietrella, D., B. L. Fries, P. Lupo, F. Bistoni, A. Casadevall, and A. Vecchiarelli. 2003. Phenotypic switching of Cryptococcus neoformans can influence the outcome of the human immune response. Cell. Microbiol. 5:513-522. [DOI] [PubMed] [Google Scholar]
- 27.Rivera, J., M. Feldmesser, M. Cammer, and A. Casadevall. 1998. Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect. Immun. 66:5027-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salas, S. D., J. E. Bennett, K. J. Kwon-Chung, J. R. Perfect, and P. R. Williamson. 1996. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J. Exp. Med. 184:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Small, J. M., and T. G. Mitchell. 1989. Strain variation in antiphagocytic activity of capsular polysaccharides from Cryptococcus neoformans serotype A. Infect. Immun. 57:3751-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spitzer, E. D., and S. G. Spitzer. 1992. Use of dispersed repetitive DNA element to distinguish clinical isolates of Cryptococcus neoformans. J. Clin. Microbiol. 30:1094-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan, D., K. Haynes, G. Moran, D. Shanley, and D. Coleman. 1996. Persistence, replacement, and microevolution of Cryptococcus neoformans strains in recurrent meningitis in AIDS patients. J. Clin. Microbiol. 34:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Summer, E. R., and S. V. Avery. 2002. Phenotypic heterogeneity: differential stress resistance among individual cells of the yeast Saccharomyces cerevisiae. Microbiology 148:345-351. [DOI] [PubMed] [Google Scholar]
- 33.Varma, A., and K. J. Kwon-Chung. 1991. Rapid method to extract DNA from Cryptococcus neoformans. J. Clin. Microbiol. 29:810-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue, C., L. M. Cavallo, J. A. Alspaugh, P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. 1999. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics 153:1601-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]