Abstract
Complement factor H (CFH) protein is an inhibitor of the alternative pathway of complement (AP) both in the fluid phase and on the surface of host cells. Mouse and human complement factor H-related (CFHR) proteins also belong to the fH family of plasma glycoproteins. The main goal of the current study was to compare the presence of mRNA for two mCFHR proteins in spontaneously developing autoimmune diseases in mice such as dense deposit disease (DDD), diabetes mellitus (DM), basal laminar deposits (BLD), collagen antibody-induced arthrits (CAIA) and systemic lupus erythematosus (SLE). Here we report for the first time that the CFHR-C mRNA was universally absent in the liver from three strains of lupus-prone mice and in a diabetic-prone mouse strain. The mRNA levels (pg/ng) for CFH and CFHR-B in MRL-lpr/lpr, at 9 wks and 23 wks were 707.2 ± 44.4, 54.5 ± 5.75 and 729 ± 252.9, 74.04 ± 22.76 respectively. The mRNA levels for CFH and CFHR-B in NZB/NZW mice, at 9 wks and 54 wks were 579.9 ± 23.8, 58.8 ± 1.41 and 890.3 ± 135.2, 63.30 ± 9.2 respectively. CFHR-C protein was absent in the circulation of MRL-lpr/lpr and NZB/NZW mice before and after the development of lupus. Similarly, mRNA and protein for CFHR-C was universally absent in liver and other organs and in the circulation of NOD mice before and after the development of DM. In contrast, the mRNAs for CFH, CFHR-B and CFHR-C were universally present in the liver from mice with and without DDD, BLD and CAIA. The levels of mRNA for CFHR-B in mice with and without BLD were ~4 times higher than the mice with lupus. The complete absence of mRNA for CFHR-C in lupus and diabetic-prone strains indicates that polymorphic variation within the mouse CFHR family exists and raises the possibility that such variation contributes to lupus and diabetic phenotypes.
Keywords: Complement factor H related C, Dense deposit disease, diabetes mellitus, basal laminar deposits, collagen-induced arthritis, systemic lupus erythematosus
1. INTRODUCTION
Complement factor H (CFH) is a 155 kDa glycoprotein and consists of 20 short census repeat (SCR) domains. It is an essential inhibitor of the alternative pathway (AP) of complement both in the fluid phase and on the surface of host cells. It prevents the formation of AP C3 convertases and also acts as a cofactor for factor I (FI) in the cleavage of C3b (active) into iC3b (inactive). Mouse and human complement factor H-related (CFHR) proteins also belong to the fH family of plasma glycoproteins. Human CFH proteins consist of six CFHR proteins: CFHR1, CFHR2, CFHR3, CFHR4A, CFHR4B and CFHR5 (Estaller et al., 1991; Skerka and Zipfel, 2008). Human CFHR6 is a pseudogene and does not code for a mature protein (personal communication with Dr. Skerka). There is a high degree of sequence homology (36-100%) between human fH and human CFHR proteins in SCRs 18-20 indicating conserved function of this C terminal recognition region (Skerka et al., 2013; Zipfel et al., 2013). This is the region by which human CFHRs and CFH bind to the cell surface and recognize C3b and all five human CFHR proteins binds to C3b and to C3d (Goicoechea de Jorge et al., 2013; Heinen et al., 2009; Hellwage et al., 1999; McRae et al., 2005; Skerka et al., 2013; Tortajada et al., 2013). It has been shown that human CFHR1 protein binds to C3b component of the enzyme, C5 convertase and inhibits the cleavage of C5 into C5a and C5b (Heinen et al., 2009). In addition, CFHR1 also inhibits the formation of C5b-C9 (Membrane Attack Complex) (MAC)) (Heinen et al., 2009). Human CFHR2 protein inhibits the AP of the complement (Eberhardt, 2012). Human CFHR3 competes, like CFHR1, with fH for binding to C3b (Fritsche et al., 2010). The exact function of human CFHR4 is unknown, but complement activation by CFHR4A has been documented (Hebecker et al., 2010). CFHR4 also interacts with C-reactive protein (CRP) and is recruited to the surface of necrotic cells (Mihlan et al., 2009). Human CFHR5 has been shown to compete with CFH for binding to C3b and also to iC3b (Goicoechea de Jorge et al., 2013). It can also bind to CRP and then be recruited to sites of tissue damage to inactivate C3b (Park and Wright, 1996). In addition CFHR proteins due to copy number variations in the human cfHR gene cluster are associated with kidney disorders (Chen et al., 2014).
Although the functions of human CFHR proteins have been investigated extensively, the presence and function of mouse CFHR proteins are less well characterized and have not been demonstrated clearly in the pathogenesis of spontaneously occurring autoimmune diseases in mice. The first CFH-related transcripts and genomic sequences from 12 clones that were distinct from CFH in mouse liver were published more than two decades ago (Vik et al., 1990). Later on two CFHR proteins of approximately 38 and 100 kDa were identified first time in mouse plasma using CFH specific anti-serum and were named as complement FHR-B (CFHR-B) and FHR-C (CFHR-C) proteins, respectively (Hellwage et al., 2006). These two complement FHR proteins in mice were not the breakdown products of CFH, because these were also present in plasma and liver extracts from fH−/− mice. This study not only examined the expression of CFHR-B in various tissues by real-time polymerase chain reaction (RT-PCR), but also shows that mouse complement CFHR-B protein interacts with heparin and human C3b (Hellwage et al., 2006). Interestingly, differential expression levels of CFHR-B and fH in the liver and kidney were observed indicating their specific role in local complement activation. In general, fH deficiency in mice, which leads to absent cell surface and fluid phase complement regulation by fH, has been linked to hypocomplementemia and membranoproliferative glomerulonephritis (MPGN) (also termed Dense Deposit Disease [DDD]) (Pickering et al., 2002; Pickering et al., 2006),. Mice deficient in CFH develop spontaneous DDD characterized by C3 deposition along the glomerular basement membrane with subsequent development of glomerulonephritis (Pickering et al., 2002). However, specific deficiency of SCR16-20 of mouse CFH, leads to the development of spontaneous atypical hemolytic uremic syndrome (aHUS) (Goicoechea de Jorge et al., 2007). It has been shown in vitro that by replacing CFH in serum with a SCR16-20 deletion mutant (Pangburn, 2002) or by blocking CFH binding to cells by use of a recombinant competitive inhibitor containing short consensus repeats 19 and 20 (rfH19-20) of CFH, cell surface protection is impaired and AP activation and amplification is enhanced (Ferreira et al., JI 2006, blood 2009). The role of CFH in rheumatoid arthritis (RA) has been shown in a recent study using collagen antibody-induced arthritis (CAIA) in a mouse model of RA (Banda et al., 2013). The in vivo administration of the competitive fH inhibitor rfH19-20, but not of control rfH3-5, significantly worsened the arthritis in mice. AMD is the leading cause of blindness in older individuals. The generation of chimeric CFH transgenic mice using human CFH sequences for SCR6-8 flanked by the mouse sequence for SCR1-5 and SCR9-20, lead to the spontaneous AMD-like retinal disease (Ufret-Vincenty et al., 2010). An important role for CFH has been shown in the MRL-lpr/lpr mouse model of SLE (Bao et al., 2011). Renal disease was exacerbated in the absence of CFH in this strain.
Taken together, CFH is important in the pathogenesis of DDD, aHUS and AMD and influences tissue injury in rheumatoid arthritis and SLE. The roles of the FHR proteins remain unclear so here we investigated the presence of mRNAs for CFH as well as two mouse CFHR proteins B and C in spontaneous mouse models of autoimmune disease. We surprisingly found that the expression level of mRNA and protein for CFHR-C was missing not only in the liver, but also in the circulation, of MRL-lpr/lpr, LG/J and NZB/NZW lupus-prone and of NOD diabetic-prone mice. However, the mRNA for CFHR-C was present in the liver and also in other organs in mice with DDD, BLD and RA. This study regarding the complete absence of CFHR-C in lupus-prone and diabetic-prone mice compared with DDD, BLD, and RA suggests that it plays an important role in the pathogenesis of spontaneously occurring autoimmune diseases in mice.
2. Materials and Methods
2.1 Mice
Six to 44-week-old fH−/− and WT C57BL/6 mice were used for the DDD study. Our laboratory has maintained a colony of fH−/− C57BL/6 homozygous mice with the F10 progeny used for this study and older fH−/−mice develop DDD. DDD was renamed as membranoproliferative glomerulonephritis type II (MPGN-II) (Smith et al., 2011). For the current study, 6-week-old male fH−/− mice without DDD and 40-week-old male fH−/− mice with DDD were used.
Four to 20-week-old female non-obese diabetic (NOD) mice (Jackson Laboratories, Bar Harbor) were used for DM studies. NOD mice spontaneously develop clinical symptoms of diabetes at ~12 weeks of age. The clinical symptoms of diabetes before and after the development of DM in NOD mice were confirmed by measuring blood glucose levels using Ultima Blood Glucose Monitor (ReliON, Betoville, AR) and Abbot Glucose testing strips. Thus, glucose levels were used to differentiate between non-diabetic and diabetic mice.
Ninety-week-old Chimeric cfhTg/mCfh−/− mice were used for AMD studies. Dr. Ufret-Vincenty has maintained a colony of cfhTg/mCfh−/− mice. We obtained frozen organs such as liver and knee joints from cfhTg/mCfh−/− mice and also from age- and sex-matched littermate B6 control mice. Serum was also obtained from 90-week-old cfhTg/mCfh−/− and B6 mice. Serum was also obtained from 90-week-old cfhTg/mCfh−/− and B6 mice. These transgenic mice were chimeric (containing human fH SCR domains 6-8, flanked by murine 1-5 and 9-20) and spontaneously develop basal laminar deposits (BLD) under the retinal pigment epithelium by 12-months of age, which could be considered a model for early stages of AMD (Ufret-Vincenty et al., 2010). Mice cannot develop AMD because they do not have a macula (personal communication with Dr. Ufret-Vincenty). 50% of the AMD cases are associated with the domain 7 “H402 risk variant” of CFH and these chimeric mice are based on this model (Ufret-Vincenty et al., 2010). Liver from chimeric mice was also used as a negative control to confirm the absence of mouse CFH according to previously described methods and primers (Ufret-Vincenty et al., 2010).
CAIA (a.k.a. RA) is a mouse model of human RA. Eight-week-old C57BL/6 WT male mice (Jackson Laboratories) were used for the RA studies described below. Additionally, 8-week to 54-week-old MRL-lpr/lpr (MRL/MpJ-Faslpr/J), NZB/NZW and LG/J mice were used for the current studies. All strains of lupus-prone mice i.e. MRL-lpr/lpr, and NZB/NZW females were directly purchased at the age of 8 weeks from the Jackson Laboratories. The MRL-lpr/lpr strain of mice started showing signs of lupus at around 18 weeks. However, the NZB/NZW strain of mice started showing the signs of lupus at around 44 weeks. We also obtained livers from 8-week-old LG/J female mice directly from Jackson Laboratories. MRL-lpr/lpr mice were originated in a colony from LG/J mice due to a spontaneous mutation. MRL-lpr/lpr, the most commonly studied mouse model of the SLE, develops an autoimmune disease that reflects pathologies of human SLE, including lymph node enlargement, increased IgG levels, anti-nuclear antibody production, proteinuria, and kidney failure caused by inflammation of the glomeruli (Perry et al., 2011). Urine was also collected before and after the development of disease and we examined the signs of lupus development in MRL-lpr/lpr and NZB/NZW by testing for proteinuria.
Age-matched and sex-matched C57BL/6 mice (Jackson Laboratories) were used as wild type (WT) controls for DDD, BLD and RA studies. All animals were kept in a barrier animal facility with a climate-controlled environment having 12-h light/dark cycles. Filter-top cages were used with 3 mice in each cage. During the course of this study, all experimental mice were fed breeder’s chow provided by the Center for Laboratory Animal Care, University of Colorado School of Medicine. All animal studies were approved by the University of Colorado IACUC.
2.2 Induction of collagen antibody-induced arthritis
We used CAIA, a mouse model of RA, due to the non-availability of a spontaneous mouse model of arthritis at the time these studies were conducted. CAIA was induced in WT C57BL/6 mice by using a cocktail of 4 mAb to bovine CII (Arthrogen-CIA, Chondrex) suspended in sterile Dulbecco’s PBS according of our published studies (Banda et al., 2013; Banda et al., 2006). All mice received i.p. injections of 4 mg/mouse of Arthrogen (a mixture of 5 clones) on day 0 and 50 μg/mouse of LPS from E. coli strain 0111B4 on day 3 to synchronize the development of arthritis. All mice started to develop arthritis at day 4 and were sacrificed at day 10, including 4 age-matched WT mice that were not treated with Arthrogen-CIA. Severity of clinical disease (CDA) in WT mice was examined according to our published studies (Banda et al., 2013; Banda et al., 2006). At day 10, all mice with and without RA, were sacrificed and above-mentioned tissues were removed and snap frozen prior to the extraction of RNA.
2.3 Surgical removal and processing of various organs for RNA extraction
Various organs including brain, liver, lung, heart, spleen, kidney, knee joint and skin from mice before and after the development of DDD, DM, BLD, CAIA and SLE were surgically removed after sacrificing mice via anesthesia followed by cervical dislocation. Fat and muscle were carefully removed from each organ. All organs were rinsed three times in 1xPBS to remove blood and/or debris and then snap frozen in liquid nitrogen. Total RNA was extracted from all organs using RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s suggested protocol.
2.4 Expression levels of mRNAs for CFH, CFHR-B and CFHR-C
The main focus of current studies was on liver. Therefore, RNA was extracted using liver from fH−/−, MRL-lpr/lpr (MRL/MpJ-Faslpr/J), NZB/NZW, LG/J, cfhTg/mCfh−/− and WT C57BL/6 mice. RNA was also extracted from various other tissues including brain, lung, heart, spleen, kidney, knee joint and skin. All RNA samples were pretreated with DNAase. cDNA was synthesized from all RNA samples and also analyzed for 18srRNA. All qRT-PCR data were analyzed using 18srRNA cDNA based standard curve. All tissue samples were run in duplicate from each mouse. All qRT-PCR reactions were run in a 96-well plate using a 7900HT- Real Time PCR System (Applied Biosystems). RNA samples for CFH, CFHR-B and CFHR-C were analyzed according to published methods by qRT-PCR using 40 cycles (Schmittgen and Livak, 2008). A Standard curves were made using liver from WT C57BL/6 mice with no disease and were used as a positive control to calculate the mRNAs for CFH, CFHR-B and CFHR-C. Gene-specific primers and probes for qRT-PCR for fH, CFHR-B and CFHR-C were designed by using exon # 3, exons # 4/5 and exon # 11, respectively. Specifically, exon # 11 from the CFHR-C nucleotide data base sequence (Accession numbers: Variant 1: NM_001029977; Variant 2: NM_001160303 and Variant 3: NM_001160304) was used to design primers for CFHR-C, because it has three variants and all share identical nucleotide sequences in this region. Moreover, there was no homology in nucleotide sequences between CFHR-B and CFHR-C in exon # 11 and this strategy was used to design primers so that the mRNA for both can be detected precisely. The reason we have selected primers from a single exon for CFHR-C because all three variants happened to be present in this exon. All RNA samples were pre-treated with DNAase to avoid contamination from genomic DNA and also during cDNA preparation an internal control without RT enzyme was used. No amplification was seen in samples without RT compared with RT. The liver and knee joints from cfhTg/mCfh−/− chimeric mice contains mRNA for CFH when primers from exon #3 were used. The sequences of primers and probes used to determine mRNAs for CFH, CFHRB and for CFHR-C levels have been listed in table 1.
Table 1.
Primers and probes for quantitative RT-PCR used to determine the levels of mouse CFH, CFHR-B and CFHR-C mRNAs
| Gene name: CFH* | |
| Forward Primer (5′-3′) | GTGGGCATCCCGGAGAC |
| Reverse Primer (5′-3′) | CACCAAACTCAAATTGAGATCCAA |
| Probe (5′-3′) | CACCCTTTGGGTCCTTTAGGCTGGC |
| Product size | 69 bp |
|
| |
| Gene name: CFHR-B | |
| Forward Primer (5′-3′) | AAAATGGTCAAGACACAATGACATG |
| Reverse Primer (5′-3′) | TTGTTGAGTTGATACGGATGCAT |
| Probe (5′-3′) | AGAATGGCTGGTCCC |
| Product size | 78 bp |
|
| |
| Gene name: CFHR-C | |
| Forward Primer (5′-3′) | TCCCAGCCCCCTACAATAGA |
| Reverse Primer (5′-3′) | TCATGACTGCTGAACTCAATGGA |
| Probe (5′-3′) | ACCCAGAACATCTGAAAA |
| Product size | 89 bp |
primers for mouse CFH have been designed using exon #3 except for cfhTg/mCfh−/− mice. The designs for CFHR-B and CFHR-B primers and probes have been explained in the Methods section. Primers and probes were purchased from Life Technologies, Grand Island, NY
2.5 Western blot analysis of serum for detecting CFH and CFHR-C
To examine for the presence of CFH and CFHR-C proteins, sera from WT, fH−/−, NOD, cfhTg/mCfh−/− and MRL-lpr/lpr mice were electrophoresed on 8% Tris-glycine gel running buffer (Life Technologies) under reducing conditions with 1xTris-glycine buffer. Serum samples were diluted 1:240 in 1x SDS buffer. After transfer, the PVDF membrane was blocked with 5% milk in 1x PBS/0.5% Tween 20 solution for 2 h. To detect CFH and CFHR-C simultaneously the blots were then incubated for 24 h at 4° C with goat anti-human CFH polyclonal Ab (dilution 1:1000) (Quidel), followed by a rabbit anti-goat HRP-conjugated secondary antibody (dilution 1:2000) (Santa Cruz Biotechnology). The blots were washed three times for 10 min each in 1x PBS/0.5% Tween 20 solution. The blots were then developed for 3 min by using a 1:1 mixture of SuperSignal West Pico chemiluminescent substrate (Thermo Scientific). It was noticed that the band of CFHR-C was too close to a non-specific band when using 10% NuPAGE Bis-Tris gel. Thus, to clearly separate the band of CFHR-C we used 8% Tris-glycine gel. Nonetheless, most of the anti-CFH antibodies bind to non-specific bands. In spite of multiple attempts we failed to find an antibody that specifically recognizes the mouse CFHR-B protein (expected MW of ~40 kDa). We cannot rule out the possibility that the anti-CFH antibody used in this study also recognizes the CFHR-B band. A sharp band of 155 kDa shows the presence of CFH and a sharp band of 98-100 kDa shows the presence of CFHR-C. The band of CFHR-C was normally less intense than the CFH band and it indicated that less amount of this protein was naturally present in the circulation (not shown). For western blot sera from WT and fH−/− mice have been used as a positive and negative controls respectively.
2.6 Statistical analyses
All p - values were calculated using Student’s t test with the GraphPad PrismR 4 statistical program. The PCR data were first analyzed with the Kolmogorov-Smirnov test for normal distribution. The qRT-PCR data in histograms are shown as the mean + SEM, with p < 0.05 considered significant using an unpaired, two-tailed t-test.
3. Results
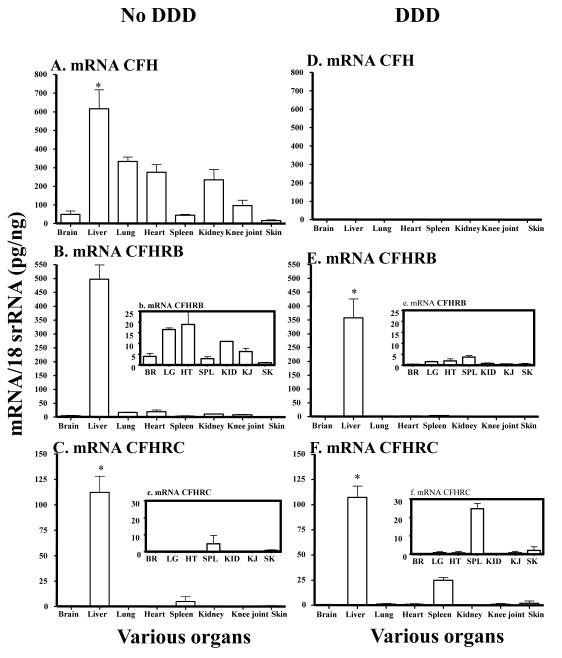
3.1 mRNA levels in the liver and various organs of mice with and without DDD
To determine the expression of mRNAs for CFH, CFHR-B and CFHR-C in liver and various organs from aged fH−/− with DDD and WT (fH+/+) mice with no DDD, total RNA was analyzed by qRT-PCR (Fig. 1). The mRNA levels for CFH, CFHR-B and C have been expressed in pg/ng 18S rRNA throughout this study. The mRNAs for CFH and CFHR-B were present in all tissues of WT mice with no DDD (Fig.1A, 1B). The levels of mRNAs for CFHR-B and CFHR-C in liver of mice with no DDD were 613.1 ± 36.1 and 183.2 ± 25.9 respectively (Fig.1B, 1C). The mRNA expression for CFH was absent in mice with DDD, as expected, not only in the liver but also in the brain, lung, heart, spleen, kidney, knee joint and skin (Fig.1D). The levels of mRNAs for CFHR-B and CFHR-C in liver of mice with DDD were 141.4 ± 13.8 and 107.1 ± 11.2 respectively (Fig.1E, 1F). The levels of mRNA for CFHR-B and CFHR-C in mice with DDD in liver were significantly (P < 0.0001) lower than the WT mice with DDD. The levels of mRNA for CFHR-C in spleen of mice with DDD vs. in the spleen of mice with no DDD were 24.1 ± 2.83 and 4.74 ± 10.60 and these differences were significant (P < 0.011) (Fig. 1C, 1F). In contrast, no major differences were seen in kidney (Fig.1C, 1F). Overall, mRNAs for CFHR-B and CFHR-C were present in the liver and also in other organs in mice with and without DDD.
FIGURE 1.
mRNA expression level analysis of CFH, CFHR-B and CFHR-C in various organs in mice with and without DDD. Aged fH−/− mice, but not fH+/+, were susceptible to DDD. Total RNA was extracted from brain, liver, lung, heart, spleen, kidney, knee joint and skin from fH−/− and fH+/+ with and without DDD. All RNA samples were analyzed in duplicate by qRT-PCR. The mRNA for CFHR-B and CFHR-C were not only universally present, but were present at very high levels in the liver of mice with and without DDD vs. brain, lung, heart, spleen, kidney, knee joint and skin. A. CFH mRNA in various organs of fH+/+ mice with no DDD. B. CFH mRNA in various organs of fH−/− mice with DDD and no CFH was present as expected. b. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-B in brain, lung, heart, spleen, kidney, knee joint and skin. C. CFHR-B mRNA in various organs of fH+/+ mice with no DDD. The mRNA was detectable in all organs. c. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-B only in SPL and SK. D. The CFH mRNA in various organs of fH−/− mice with DDD was absent, as expected. E. CFHR-B mRNA in various organs of fH−/− mice with DDD. e. A small histogram embedded in the main histogram shows that it was also detectable in LG, HT, SPL, KID, KJ and SK. F. CFHR-C mRNA in various organs of fH−/− mice with DDD. f. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-C in LG, HT, SPL, KJ and SK. All data presented in these graphs, as Mean ± SEM, were from fH+/+ mice with no DDD (n = 5) and fH−/− mice with DDD (n =5) and these experiments were repeated three times. BR = brain, LG = lung, HT = heart, SPL = spleen, KID = kidney, KJ = knee joint and SK = skin. *p-value < 0.05 compared to liver
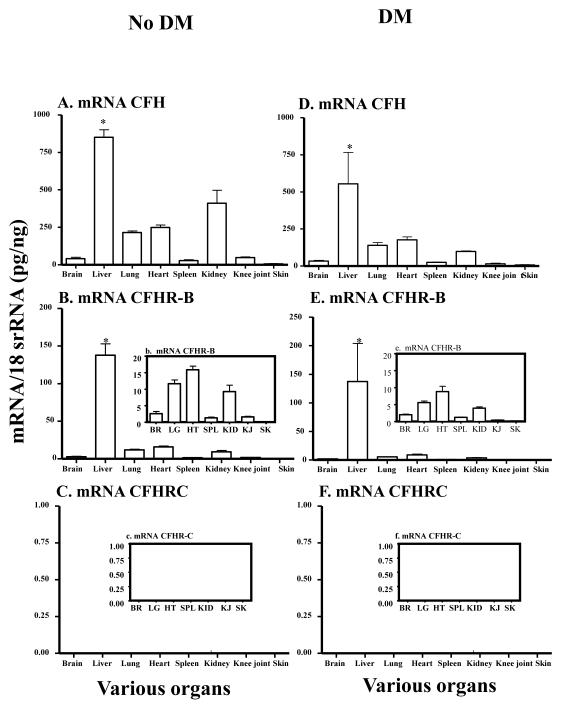
3.2 mRNA levels in the liver and various organs of mice with and without DM
Total RNA was analyzed by qRT-PCR, as mentioned above, to determine the expression of mRNAs for CFH, CFHR-B and CFHR-C in liver and various organs from female NOD mice with and without DM (Fig. 2). The glucose levels before and after the development of DM were 114.2 + 6.46 and >500 respectively. The mRNA for CFH was present not only in the liver of NOD mice with and without DM but it was also present in various organs such as brain, lung, heart, spleen, kidney, knee joint and skin (Fig. 2A, 2D). The mRNAs for CFHR-B were present in all tissues of NOD mice with and without DM (Fig. 2B, 2E). The levels of mRNAs for CFH, CFH-B and CFHR-C in liver of mice with no DM were 851.0 ± 49.9, 137.9 ± 15.0 and 0 ± 0 respectively (Fig. 2A, 2B, 2C). In liver, CFH and CFHR-B were significantly (P < 0.05) higher than brain, lung, heart, spleen, kidney, knee joint and skin in mice with and without DM (Fig. 2A, 2B, 2D, 2E). The levels of mRNAs for CFH, CFHB and CFHR-C in liver of mice with DM were 554.5 ± 212.24, 137.4 ± 66.6 and 0 ± 0 respectively (Fig. 2D, 2E, 2F). Thus, mRNA for CFHR-C was universally absent not only in the liver, but also in all other organs of the NOD mouse with and without DM (Fig. 2C, 2F). No significant differences were seen in the mRNAs for CFH and CFHRB in the liver of mice with and without diabetes (Fig. 2A, 2D). However, the differences in the mRNAs levels were significant (p < 0.034) for only CFH but not for CFHRB in kidney of mice with and without DM. The presence of mRNAs for CFH and CFHR-B in liver of mice with and without DM and the universal absence of CFHR-C in liver of diabetic NOD-prone mice and also in various organs of these mice indicated that it has important relevance in the pathogenesis of diabetes.
FIGURE 2.
mRNA expression levels of CFH, CFHR-B and CFHR-C analysis of various organs of NOD mice without and with diabetes. Eighteen-week-old, but not 4-week-old NOD mice were susceptible to the DM. Total RNA was extracted from brain, liver, lung, heart, spleen, kidney, knee joint and skin from NOD mice with and without DM. All RNA samples were analyzed in duplicate by qRT-PCR. The mRNA for CFH and CFHR-B were universally present in liver, brain, lung, heart, spleen, kidney, knee joint and skin of mice with and without DM. However, CFHR-C mRNA was universally absent in all organs. A. CFH mRNA levels in various organs of NOD mice with no DM. B. CFHR-B mRNA level in various organs of NOD mice with no DM. b. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-B in BR, LG, HT, SPL, KID, and KJ. C. No detection of CFHR-C mRNA in various organs including liver of NOD mice with no DM. c. A small histogram embedded in the main histogram showing universal absence of mRNA for CFHR-C in BR, LG, HT, SPL, KID, KJ and SK in mice without DM. D. CFH mRNA levels in various organs of NOD mice with DM was detectable in all organs. E. CFHR-B mRNA levels in various organs of NOD mice with DM. e. A small histogram embedded in the main histogram showing presence of detectable levels of mRNAs for CFHR-B in BR, LG, HT, SPL, KID, KJ and SK in mice with DM. F. No detection of CFHR-C mRNA in various organs including liver of NOD mice with DM. f. A small histogram embedded in the main histogram showing complete absence of mRNAs for CFHRC in BR, LG, HT, SPL, KID, KJ and SK in mice with DM. All data presented in these graphs, as Mean ± SEM, were from NOD mice with no DM (n = 5) and NOD mice with DM (n = 5). The q-RTPCR was repeated two times. BR = brain, LG = lung, HT = heart, SPL = spleen, KID = kidney, KJ = knee joint and SK = skin. *p-value < 0.05 compared to liver
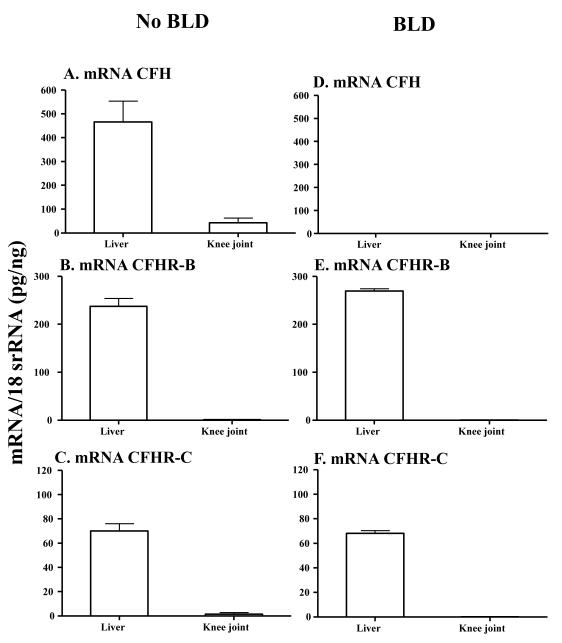
3.3 mRNA levels in the liver and knee joints of mice with and without BLD
The mRNA levels were determined only from the liver and knee joints of CfhTg/mCfh−/− mice with BLD and B6 mice with no BLD (Fig. 3). The mRNA levels from brain, lung, heart, spleen, kidney and skin were not determined because of the non-availability of enough CfhTg/mCfh−/− mice at the time these studies were conducted. The levels of mRNAs for CFH, CFHR-B and CFHR-C in liver of mice with no BLD were 465.8 ± 1.7, 246.0 ± 15.9 and 70.1 ± 5.9 respectively (Fig. 3A, 3B, 3C). The mRNA for CFH was completely absent in the liver of mice with BLD (Fig.3A), but the mRNA levels for CFHR-B and CFHR-C were 270.5 ± 2.3 and 68.1 ± 2.3 respectively (Fig. 3E, 3F). CFH was absent in the knee joints of mice with BLD (Fig. 3D) but the levels for CFHR-B and CFHR-C were 0.40 ± 0.06 and 0.11 ± 0.08 respectively (Fig. 3E, 3F). Nonetheless, liver was positive for the human/mouse fH chimeric transgene as confirmed by using specific primers and probes (data not shown) (Ufret-Vincenty et al., 2010). There was no statistically significant difference in the mRNA levels for CFHR-B (p < 0.15) (Fig.3B, 3E) and CFHR-C (p < 0.75) in the liver of mice with and without BLD (Fig. 3E, F). The mRNA for CFHR-B and CFHR-C were present but very low in the knee joints of mice with and without BLD (Fig. 3E, F). No statistically significant differences were seen in the levels of mRNA for CFHR-B (p < 0.18) and CFHR-C (p < 0.38) in the knee joints of mice with and without BLD. In these studies, age-matched and sex-matched littermate non-chimeric WT C57BL/6 mice without BLD were used. These results showed that CFHR-B and CFHR-C mRNAs were present in the liver but very low knee joint of mice with and without BLD.
FIGURE 3.
mRNA expression levels of CFH, CFHR-B and CFHR-C analysis from liver and knee joint of CfhTg/mCfh−/− mice without and with BLD. Only liver and knee joint from 12-month-old CfhTg/mCfh−/− chimeric mice (with BLD) and age-matched littermate B6 mice (with no BLD) were analyzed. All RNA samples were analyzed in duplicate by qRT-PCR. A. CFH mRNA was present in liver and knee joints of B6 mice with no BLD using primers from exon #3. B. CFHR-B was present in liver and knee joints of B6 mice with no BLD. C. CFHR-C was present in liver and knee joints of B6 mice with no BLD. D. No mouse fH mRNA was detected in liver and knee joint of CfhTg/mCfh−/− chimeric mice with BLD using primers described previously (Ufret-Vincenty et al., 2010) but mRNA for CFH was present using primers from exon #3 (data not shown). E. CFHR-B mRNA in liver and knee joints of CfhTg/mCfh−/− mice with BLD. F. CFHR-C mRNA in liver and knee joint of CfhTg/mCfh−/− mice with BLD mice. All data presented in these graphs, as Mean ± SEM, were from CfhTg/mCfh−/− mice with BLD (n = 5) and B6 mice (n = 5) with no BLD. These experiments were repeated three times. *p-value < 0.05 compared to liver
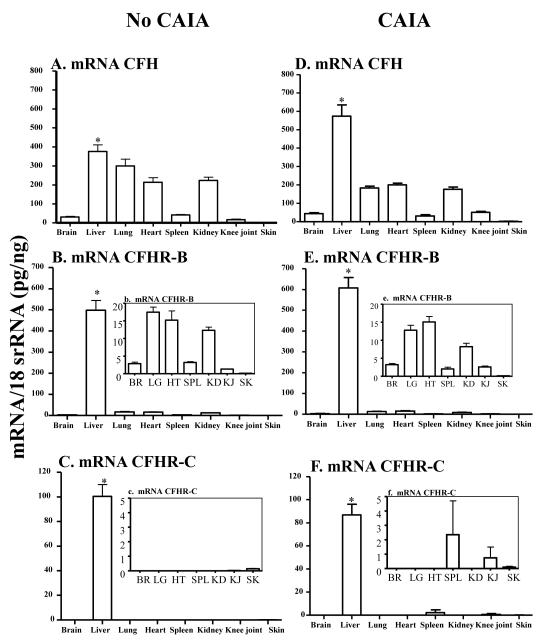
3.4 mRNA levels in the liver and various organs of mice with and without CAIA
The arthritis in WT mice was confirmed visually and Clinical disease scores were used to assess the severity of arthritis according to our published studies (Banda et al., 2013; Banda et al., 2006). All mice with CAIA were sacrificed at day 10. Age- and sex-matched WT mice with no arthritis were also sacrificed at the same time. The mRNA levels of CFH in the liver of mice with and without CAIA were 573.95 ± 61.37 and 375.63 ± 35.51, respectively (Fig. 4A, 4D). The increases in the mRNA levels of fH in the liver of mice with disease were significant (p <0.04). The levels of mRNAs for CFHR-B and CFHR-C in liver of mice with no CAIA were 498.42 ± 46.12 and 100.33 ± 9.60, respectively (Fig.4B, 4C). The levels of mRNAs for CFHR-B and CFHR-C in liver of mice with RA were 607.62 ± 50.10 and 86.81 ± 9.36, respectively (Fig.4E, 4F). Interestingly, there was an increase in the mRNA levels of CFHR-C in the spleen of mice with RA compared with mice with no CAIA (Fig. 4f, c).
FIGURE 4.
mRNA expression levels of CFH, CFHR-B and CFHR-C analysis of various organs in mice with and without CAIA. CAIA was induced in C57BL/6 mice as described in Materials and Methods. All mice with CAIA were sacrificed at day 10. Age-matched WT mice were also sacrificed in parallel at day 10 without the induction of CAIA. Total RNA was extracted from brain, liver, lung, heart, spleen, kidney, knee joint and skin from WT mice with and without arthritis. A. CFH mRNA various organs of mice with no CAIA. B. CFHR-B mRNA in various organs of mice with no CAIA. b. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-B in BR, LG, HT, SPL, KID, and KJ but not in SK. C. CFHR-C mRNA in various organs of mice with no CAIA. c. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-B only in SK. D. CFH mRNA in various organs of mice with CAIA. E. CFHR-B mRNA in various organs of mice with CAIA. e. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-B in BR, LG, HT, SPL, KID, KJ and SK. F. CFHR-C mRNA in various organs of mice with CAIA. f. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-B in SPL, KJ and SK. All RNA samples were analyzed in duplicate by qRT-PCR. All data presented in these graphs, as Mean ± SEM, were from WT mice with no CAIA (n = 4) and WT mice with CAIA (n = 5). BR = brain, LG = lung, HT = heart, SPL = spleen, KID = kidney, KJ = knee joint and SK = skin. *p-value < 0.05 compared to liver
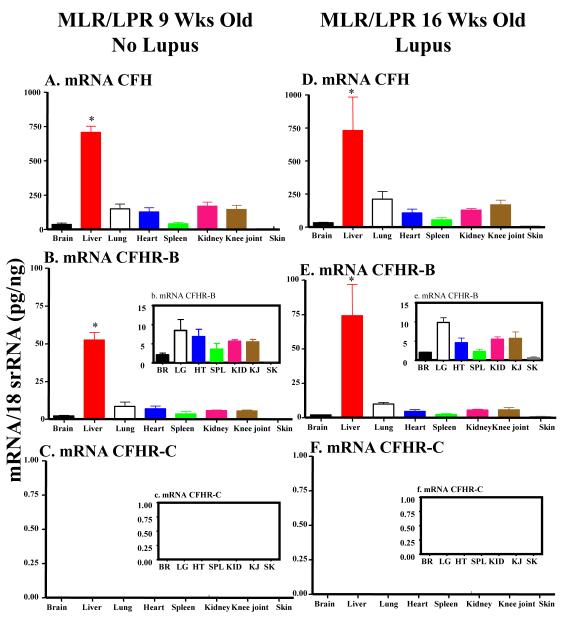
3.5 mRNA levels in the liver and various organs of mice with SLE
Three different strains of lupus-prone mice were used to determine the mRNA expression of CFH, CFHR-B and CFHR-C in the liver. The mRNA for CFH and CFHR-B in liver of MRL-lpr/lpr before the development of lupus were 707.2 ± 44.4 and 54.5 ± 5.75 respectively (Fig.5A, 5B). The mRNA for CFH and CFHR-B in liver of MRL-lpr/lpr after the development of lupus were 729.3 ± 252.9 and 74.0 ± 22.76 respectively (Fig. 5D, 5E). These differences in the mRNA levels, before and after the development of lupus for both CFH and CFHR-B, were not statistically significant (p < 0.91) and (p < 0.37), respectively. However, the mRNA for CFHRC was completely absent in the liver of these mice before and after the development of lupus (Fig. 5C, 5F). The CFHR-C mRNA was again absent in all other organs including brain, lung, heart, spleen, kidney, knee joint and skin before (Fig. 5C, 5c) and after the development of lupus (Fig. 5F, 5f).
FIGURE 5.
mRNA expression levels of CFH, CFHR-B and CFHR-C analysis of various organs in mice with and without SLE. At 23 weeks of age vs. 9 weeks MRL-lpr/lpr showed clinical signs of lupus. Total RNA was extracted from brain, liver, lung, heart, spleen, kidney, knee joint and skin from MRL-lpr/lpr with and without lupus. All RNA samples were analyzed in duplicate by qRT-PCR. The mRNA for CFHR-C was absent not only in the liver of mice with and without lupus, but it was absent universally in brain, lung, heart, spleen, kidney, knee joint and skin. A. fH mRNA in various organs of MRL-lpr/lpr mice with no lupus. B. CFHR-B mRNA in various organs of MRL-lpr/lpr mice with no lupus. The mRNA was present in all organs, but in skin it was very low. b. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-B in BR, LG, HT, SPL, KID, KJ and SK. C. Undetectable levels of CFHR-C mRNA in all organs of MRL-lpr/lpr mice with no lupus. D. CFH mRNA in various organs of MRL-lpr/lpr mice with lupus. It was detectable in all organs except in skin it was low. E. CFHRB mRNA in various organs of MRL-lpr/lpr mice with lupus. e. A small histogram embedded in the main histogram shows detectable mRNA levels of CFHR-B in BR, LG, HT, SPL, KID, KJ and SK. F. Undetectable levels of CFHR-C mRNA in various organs of FH mRNA in various organs of MRL-lpr/lpr mice with lupus. All data presented in these graphs, as Mean ± SEM, were from MRL-lpr/lpr mice with no lupus (n = 5) and MRL-lpr/lpr mice with lupus (n = 5). These experiments were repeated three times. BR = brain, LG = lung, HT = heart, SPL = spleen, KID = kidney, KJ = knee joint and SK = Skin. *p-value < 0.05 compared to liver.
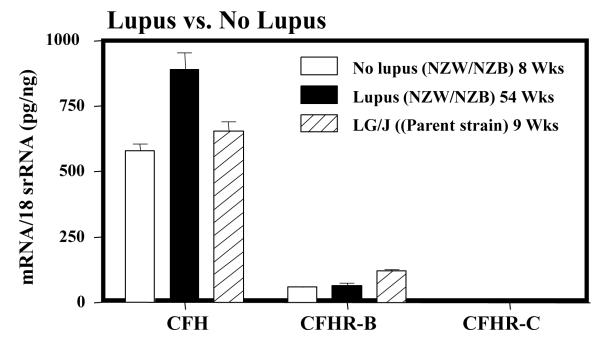
Next we examined the mRNA for CFH, mCFHR-B and CFHR-C in NZB/NZW mice, which is a different strain of lupus-prone mice with similar disease phenotype (Fig. 6). The mRNA for CFH and CFHR-B in liver of NZB/NZW before the development of lupus were 579.9 ± 23.8 and 58.87 ± 1.41, respectively (Fig. 6). The mRNA for CFH and CFHR-B in liver of NZB/NZW after the development of lupus were 890.3 ± 135.2 and 63.3 ± 9.2, respectively (Fig. 6). These differences in the mRNA levels, before the development of lupus in NZB/NZW mice for both CFH and CFHR-B, were barely statistically significant (p < 0.05). However, again the mRNA for CFHR-C was completely absent in the liver of these mice before and after the development of lupus (Fig. 6).
FIGURE 6.
Analysis of the mRNA expression levels of CFH, CFHR-B and CFHR-C in various organs of NZB/NZW mice, with and without SLE, and in LG/J mice without SLE. At ~54 weeks of age NZB/NZW showed clinical signs of lupus. Total RNA was extracted from liver of NZB/NZW with (54 weeks) and without lupus (9 weeks). Liver was also obtained from LG/J mice without lupus (age 9 weeks). All RNA samples were analyzed in duplicate by qRT-PCR. The mRNA for CFH and CFHR-B were present in liver of NZB/NZW and LG/J mice with and without lupus. The mRNA for CFHR-C was totally absent in liver of both NZB/NZW and LG/J mice regardless of disease.
Finally, we examined for the mRNA levels in the liver from LG/J mice with no lupus. LG/J is the mouse line from which MRL-lpr/lpr lupus-prone mice was originated that also develop lupus, but slower than the MRL-lpr/lpr and NZB/NZW mice. We found that the mRNAs for CFH and CFHR-B in liver of these mice before the development of lupus were 654.6 ± 33.4 and 119.6 ± 3.8, respectively (Fig. 6). Surprisingly, the mRNA for CFHR-C was again totally absent in the liver of LG/J mice (Fig. 6). The complete universal absence of CFHRC mRNA in liver of all three strains of lupus-prone mice and also in various organs of MRL-lpr/lpr mice, despite the presence of mRNAs for CFH and CFHR-B in liver, indicate that CFHRC is might have an important relevance to lupus pathogenesis.
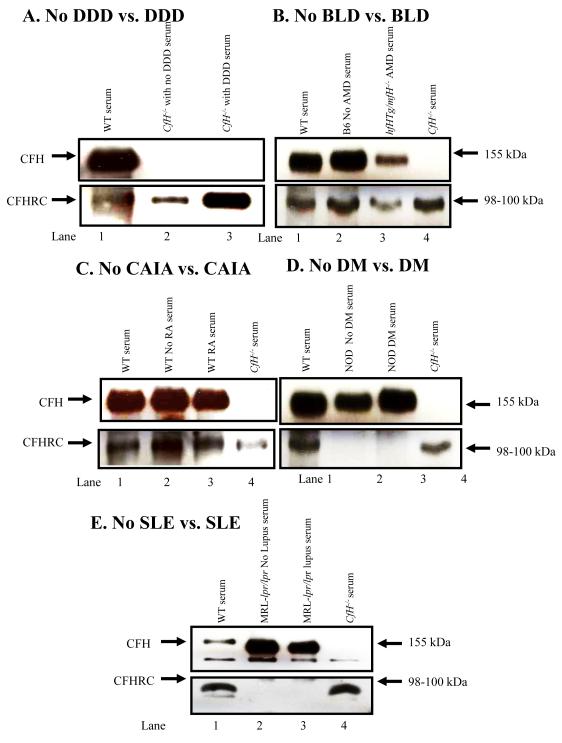
3.6 Absence of CFHR-C protein in the serum from mouse with and without DM and SLE
When analyzed by western blot, the serum from mice with DDD, BLD, and CAIA all showed the presence of CFHRC (Fig. 7A, 7B, 7C). We also examined sera from NOD mice before (4 weeks) and after (20 weeks) the development of DM (Fig. 7D). To confirm the absence of CFHR-C protein in circulation, serum from MRL-lpr/lpr mouse was also examined by western blot analysis before (9 weeks) and after (23 weeks) the development of lupus (Fig. 7E). We used a specific antibody as mentioned in methods that recognized both CFH and CFHR-C simultaneously. A band of CFH at 155 kDa was present using sera from WT and MRL-lpr/lpr mice (Fig. 7E, lanes 1, 2 & 3). It was absent in the serum from fH−/− mouse as expected (Fig. 7E, lane 4). A band of ~98-100 kDa of CFHR-C was present only in the sera from WT and fH−/− mice (Fig. 7, lanes 1 & 4) and it was completely absent in the sera from MRL-lpr/lpr mice before and after the development disease (Fig. 7, lanes 2 & 3). Qualitatively, by Western blot analysis, the protein expression of CFHR-C in the sera from WT and fH−/− mice was low and these results are consistent with the low mRNA levels of this protein in liver as well as in various tissues from WT and fH−/− mice (Fig. 7). The complete absence of CFHR-C protein in sera from MRL-lpr/lpr mice (Fig.7E, lanes 2 & 3) was consistent with the universal absence of mRNA for CFHR-C in lupus-prone mice (Fig. 5 C & F; Fig. 6). A band of CFHR-C was also absent in the sera from NOD mice with and without DM (Fig. 7D, lanes 2 & 3). We did not carry out a western blot analysis for CFHR-B, because specific antibody to detect it was not available, even though we screened over four anti-CFH antibodies for this purpose. We did not analyze cell lysates from various organs to detect intracellular CFHR-C protein, because the anti-CFH antibody we used for western blot analysis failed to recognize a clean band instead multiple non-specific bands in the SDS-PAGE gels (not shown).
FIGURE 7.
Western blot analyses showing the presence of CFHR-C in mouse serum with and without DDD, BLD and CAIA and also showing the universal absence of mice with and without DM and SLE. Sera from WT C57BL/6 and fH−/− mice were used as a positive and a negative control for CFH and CFHR-C, respectively. Mouse serum was analyzed by using 8% Trisglycine SDS-PAGE under reducing conditions. Immuno-blotting was done using a primary anti-CFH and a secondary HRP-conjugated antibody. A. Serum was analyzed from WT (fH+/+) and fH−/− mice with no DDD and with DDD. Top panel (lanes 1, 2 & 3): Serum from WT (fH+/+) mice, 6-week-old fH−/− mice with no DDD and 40-week-old fH−/− mice with DDD. No CFH is present in fH−/− mice with DDD, as expected. Bottom panel (lanes 1, 2 & 3): Serum from WT (fH+/+) mice with no DDD and fH−/− mice with DDD. A band of ~98-100 kDa of CFHR-C was present in serum from WT, and in fH−/− with no DDD and with DDD, respectively. B. Sera from B6 mice with no BLD and from CfhTg/mCfh−/− chimeric mice with BLD were analyzed. Top panel (lanes 1, 2 & 3): A band of CFH was present in serum from WT, with no BLD and with AMD mouse. Bottom panel (Lanes 1, 2, 3 & 4): CFHR-C was present in serum from WT, B6, CfhTg/mCfh−/− and fH−/− mouse. C. Sera from WT C57BL/6 mice with no CAIA, at day 0, and WT C57BL/6 mice with CAIA, at day 10, were analyzed. Top panel (lanes 1, 2 & 3): A band of CFH was present in the sera from WT, with no CAIA and with CAIA mice. Lane 4. Absence of CFH in serum from fH−/− mice, as expected. Bottom panel (Lanes 1, 4): A band of CFHR-C was present in sera from WT, with no arthritis, with arthritis and fH−/− mice. D. Sera from NOD mice with no DM, and NOD mice with DM were analyzed. Top panel (lanes 1, 2 & 3): A band of fH ~155 kDa was present in serum from WT, no DM and DM mice. Lane 4. Absence of CFH in serum from fH−/− mouse as a negative control. Bottom panel (Lanes 1, 4): A band of ~98-100 kDa of CFHR-C was present in serum from WT and fH−/− mice. Lanes 2, 3. CFHR-C was absent in serum from mice with no DM or with DM. E. Sera from MRL-lpr/lpr without disease and MRL-lpr/lpr with disease mice were analyzed. Top panel (lanes 1, 2 & 3): A band of CFH was present in sera from WT, MRL-lpr/lpr (12 Wks) and MRL-lpr/lpr (23 Wks) mouse. Lane 4. Absence of CFH in the sera from fH−/− mice as expected. Bottom panel (lanes 1): A band of CFHR-C was present in sera from WT mouse. Occasionally, twin bands were also seen at ~96-100 kDa and these may be variants of CFHR-C. Lanes 2 & 3. No CFHR-C protein was present in sera from MRL-lpr/lpr mice with and without lupus, respectively. Lane 4. CFHR-C protein was present in sera from fH−/− mice even in the absence of CFH. Western blots using each serum were repeated at least three times and results were highly reproducible.
4. Discussion
Although the functions of human CFHR proteins have been investigated extensively, their biological role remains uncertain. FHR1, FHR2 and FHR5 may act as competitive inhibitors of CFH, a process termed CFH de-regulation (Goicoechea de Jorge et al., 2007). The characterization of mouse CFHR proteins is incomplete. The main goal of this study was to define if levels (through measurement of mRNA and where possible, protein quantification in plasma) of CFH, CFHR-B and CFHR-C differed amongst strains with different pathologies. Most of the complement proteins are synthesized in the liver so we assessed expression in liver tissue in addition to other organs.
Our unexpected finding was the complete absence of CFHR-C in NOD, LG/J, MRL-lpr/lpr and NZB/NZW strains. CFHR-C was detectable in C57BL/6 and CFH-deficient animals inter-crossed onto the C57BL/6 genetic background. This suggested that there could have been recombination within the murine FHR locus that has resulted in a copy number variation in which there is deletion of the CFHR-C gene and that this allele is present in homozygosity in NOD, LG/J, MRL-lpr/lpr and NZB/NZW strains. Deletion of the human FHR genes, FHR3 and FHR1, is a common polymorphism resulting in an allele in which there is complete absence of the genes for these two CFHRs (Malik et al., 2012). Notably the FH-deficient mouse strain (Pickering et al., 2002) was generated using gene-targeting techniques in 129/Sv embryonic stem cells so the original 129/Sv targeted locus contains an intact CFHR-C gene both before and after targeted mutation of the CFH gene.
Here, the data show an important and surprising finding that CFHR-C was universally absent in three different strains of lupus-prone and in one strain of diabetic-prone mice. We provided evidence by comparing the mRNA levels in liver and in other vital organs of mice with and without lupus. CFHR-C protein was also absent in the circulation of MRL-lpr/lpr mice and NOD mice, with and without lupus and diabetes, respectively. However, the mRNA for CFHR-C was present in the liver and other vital organs of mice with and without DDD, BLD and CAIA. CFHR-C protein was also present in the circulation of mice with and without DDD, BLD and CAIA. In mouse models of all autoimmune diseases tested in this study the mRNA for fH in brain, liver, lung, heart, spleen, kidney, knee joints and skin was consistently more than the mRNA for CFHR-B and the latter was more than the mRNA for CFHR-C. We found a significant increase in the levels of mRNA for CFHR-C in the spleen of mice with DDD and CAIA compared with the mRNA levels in the spleen of mice with no DDD and CAIA, which indicated its pro-inflammatory role. CFHR-B mRNA was 4-fold higher in mice with BLD vs. mice with lupus. Finally, the liver of mice with DDD, BLD, DM, CAIA and SLE had the highest, and skin had the lowest, levels of CFH, CFHR-B and CFHR-C mRNA.
The molecular weight of mouse CFHR-C protein was ~98-100 kDa and thus consistent with that found in a previous study (Hellwage et al., 2006). The sequence of CFHR-C and its variants has also been published previously (Hellwage et al., 2006). Our study indicates that genetic deficiency of CFHR-C in lupus-prone mice might play a critical role in the pathogenesis of lupus. The mRNA for CFHR-C was also universally absent in the diabetic-prone NOD mice before and after the development of diabetes. Older MRL-lpr/lpr and NOD mice develop lupus and diabetes, respectively. We thought that the complete absence of CFHR-C in mice susceptible to two different autoimmune diseases that occur spontaneously might be a coincidence, but we found that the strains of lupus and diabetic-prone mice from the Jackson database were different. MRL-lpr/lpr and LG/J mice belong to group 1 of the mouse family tree, while NOD and NZB/NZW mice belong to group 2 and group 3, respectively (Petkov et al., 2004). LG/J mice, which do not have CFHR-C mRNA in liver, are actually the parent strain of MRL-lpr/lpr mice. These results indicate that CFHR-C might be a “common risk factor” for two different autoimmune diseases that develop spontaneously in MRL-lpr/lpr, NZB/NZW, LG/J and NOD mice at different ages due to specific mutations. We do not know the exact function of CFHR-C and it could be a pro-inflammatory protein. Our study also indicated that CFH and CFHR-B proteins could not substitute for CFHR-C, because the mRNA for both of these proteins was present in the liver and also in various organs of lupus- and diabetic-prone mice. We have not examined the presence or absence of complement regulatory proteins, because it was not the focus of current study.
There are three known variants of CFHR-C in mouse and Fhrc gene codes for all three variants (Hellwage et al., 2006; Vik et al., 1990). In order to show the specificity and sensitivity of the qRT-PCR, the primers and probes to examine the mRNA levels of all variants of CFHR-C have been carefully designed so as to include all three variants, but to exclude mouse CFHR-B. Furthermore, the specificity of primers and probes has also been confirmed by using RNA from various organs from fH−/− mice. Liver samples from fH−/− mice contain the mRNAs for CFHR-B and CFHR-C, but completely lack the mRNA for fH as expected since we used primers that targeted CFH exon 3 and this has been deleted in the FH-deficient mouse. Mouse CFH exon 3 mRNA was present in CfhTg/mCfh−/− chimeric mice (data not shown), which develop BLD spontaneously due to the expression of a CFH chimeric protein. We confirmed by using different primer sets that these transgenic mice have exon 3 RNA because the transgene present which contain mouse exon 3 as only SCR 6 7 8 are human. The reason fH protein is also detected in the serum from CfhTg/mCfh−/− mice in western blot analysis (Fig. 7C) is because these mice express a human/mouse transgenic CFH and the anti-CFH antibody used for the western blots cross-reacts with both mouse and human CFH.
C3 glomerulonephritis (C3GN) is subtype of C3 glomerulopathy that results from diverse abnormalities of the AP of complement (Sethi et al., 2012). In this study, in addition to the presence of H402 and V62 alleles of CFH, other abnormalities such as CFH autoantibodies, mutations in CFH, FI, CFHR genes as well as copy number variations in the human cFHR gene cluster were reported (Sethi et al., 2012). A mutation in hCFHR5 has also been associated with C3 glomerulonephritis (C3GN) in Greek Cypriots (Gale and Maxwell, 2013). In addition in two related patients diagnosed for C3G with DDD pattern, a chromosomal deletion of a 25 kbp fragment links the CFHR2 and CFHR5 genes and results in expression of a hybrid CFHR2-CFHR5 protein that in patient plasma deregulates the fluid phase C3 convertase (Chen et al., 2014). One study has linked variants of genes encoding fH and five CFH-related proteins (CFHR1-CFHR5) within the chromosome 1q32 locus linked to SLE (Zhao et al., 2011). We hypothesized that there must be a sequence homology among human CFHR1-CFHR5 and mouse CFHR-C, because the mRNA as well as the protein was universally absent in lupus-prone mice. Therefore, we examined the sequence similarities between hCFHR5 and mouse CFHR-C variants using available bioinformatics information. The sequence homology between human CFHR5 and all three variants of mouse CFHR-C was modest at ~47%. Other human CFHR proteins, such as human CFHR2 and human CFHR3, have an identical sequence homology with mouse CFHR-C and this sequence homology was ~45% to 47% respectively. Furthermore, the sequence similarity among all three variants of mouse CFHR-C with all three variants of human CFHR4 ranged from 33% to 47.7%. SLE has been associated with a single nucleotide polymorphism (SNPs) in CFH-CFHRs regions in various ethnic groups (Zhao et al., 2011). This study has shown that human CFHR3-CFHR1 deletion led to absence of human CFHR1 and human CFHR3 proteins and it was the causal variant for high risk of SLE within the SLE-associated block. Interestingly, CFHR1-CFHR3 deletions in humans has also been associated with AMD (Hageman et al., 2005), the auto immune form, DEAP-HUS of atypical hemolytic uremic syndrome (aHUS) (Pickering et al., 2007; Zipfel et al., 2010) and MPGN II (Abrera-Abeleda et al., 2006; Pickering et al., 2007). Complete absence of fH in mouse plasma has been reported to cause DDD (Appel et al., 2005). Bioinformatics data related to the moderate sequence homologies between human CFHR proteins and mouse CFHR-C show that the later may be a “common moderate risk factor” in autoimmune diseases that develop spontaneously, but specific mutations are necessary for certain autoimmune disease phenotypes.
We found that the mRNA for mouse CFHR-B was significantly lower in the liver of DDD-prone fH−/− mice compared with the liver of DDD-resistant mice (Fig. 1B, 1E). In contrast, the mRNA for CFHR-C was significantly higher in spleen of DDD-prone mice compared with the spleen of DDD- resistant mice (Fig. 1C, 1F). The liver and spleen differ anatomically as well as functionally, but these vital organs might play differential roles in locally regulating the CFHR proteins (Fig. 1E, 1F). It has previously been reported that fH−/− and fB−/− mice develop DDD because of the lack of CFH while also maintaining a very low level of C3 in the circulation (Pickering et al., 2002). We have reported that fH−/− mice are resistant to the development of RA and these mice maintain very low levels of C3 in their circulation (Banda et al., 2013). Interestingly, our studies show that CFHR-C protein was still present in the circulation of fH−/− mice at a level almost equivalent to that of the WT mice. The presence of CFHR-C in DDD-prone fH−/− mice separates these mice from lupus-prone, MRL-lpr/lpr and DM-prone, NOD mice. The mRNA of CFHR-C was also present in lung, heart, spleen, and knee joint of fH−/− mice with DDD, but absent in brain indicating the unique nature of this protein as it may not cross blood-brain barrier in certain autoimmune diseases. In contrast, the mRNA and protein for CFHR-C was present in the sera from DDD-prone, fH−/−, AMD-prone, CfhTg/mCfh−/−, and RA-susceptible WT mice.
One main reason for the complete absence of CFHR-C in lupus- and diabetic-prone mice could be that spontaneous lupus and DM develops in strains of mice that are different from those that develop DDD, BLD, and CAIA. DDD, BLD and CAIA develop in mice on C57BL/6 background, which are deficient in CFH, chimeric for CFH, or have normal levels of CFH, respectively. Therefore, the complete absence of CFHR-C mRNA and protein in lupus- and DM-prone mice could be due to two reasons. First, the presence or absence of CFHR-C may be strain-specific and this is the most likely explanation for these disease-specific differences. Second, DDD and BLD develop in older CFH-deficient or CFH-chimeric mice, respectively, on a C57BL/6 background, even in the absence of genetic mutations, while lupus and DM develops spontaneously due to specific genetic mutations in aged MRL and NOD mice, respectively. The lower level of mRNA for fH only in the kidney but not in the liver of mice with DM showed that there is an organ-specific dysregulation of the AP in diabetic prone mice. Furthermore no differences were seen in the mRNA of CFHRB in the kidney and liver of mice with and without DM confirm the hypothesis that various CFHR proteins play a distinct and an organ-specific role in regulating the AP in various autoimmune diseases. Although CFHR-C is universally absent in both MRL-lpr/lpr and NOD mice, the outcome of the autoimmune diseases are different due to specific genetic mutations in lupus-prone mice. Therefore, we conclude that the complete absence of CFHR-C in lupus- and DM-prone mice may be a “common risk factor” that may play a vital role in the pathogenesis of these two spontaneously occurring autoimmune diseases in mice. At present we do not know whether complete absence of CFHR-C promotes or protects mice from developing lupus and DM. It has been reported that treatment of pre-diabetic NOD mice (i.v. injection) with heat-killed Mycobacterium bovis (bacillus Calmette-Guerin (BCG)) protects them from DM, but leads to the development of SLE-like disease (Baxter et al., 1994b). This study led to the possibility that genetic susceptibility for DM and SLE might be conferred by single collection of genes in NOD mice, because BCG treatment of normal non-autoimmune C57BL/6 mice did not cause the development of SLE (Baxter et al., 1994a). To further explore the role of CFHR-C in the pathogenesis of DDD studies are in progress to cross-breed fH−/− mice with NOD mice to generate fH−/−/NOD mice. This double KO mouse will be deficient not only in CFH, but also in CFHR-C. We hypothesize that fH−/−/NOD will develop rapid and severe DDD vs. fH−/− alone due to the lack of CFHR-C. Similarly, loss of fH in fH−/−/MRL-lpr mice has been shown to accelerate the development of lupus nephritis (Bao et al., 2011). This study also reported high mortality rate in fH−/−/MRL-lpr mice due to renal failure. Nonetheless, this study did not explore the role of CFHR-C. Thus, it remains possible that the high mortality rate observed due to renal failure in the fH−/−/MRL-lpr mice is due to the probable absence of CFHRC in these mice.
Further studies are needed in lupus- and DM-prone mice to verify that in vivo administration of recombinant CFHR-C or of plasmid coding for CFHR-C could significantly attenuate the severity of these diseases in terms of less proteinuria, lupus nephritis, double stranded DNA auto-antibodies, and glucose levels. Furthermore, the 47% homology between all three CFHR-C variants and the huCFHR5 gene indicate that CFHR-C might be playing a role in murine lupus, because a mutation in huCFHR5 that leads to a reduced amount of the protein has been linked with C3 glomerlopathy in a Greek Cypriot family (Gale et al., 2010).
To the best of our knowledge, this is the first qRT-PCR study comparing simultaneously the mRNA expression levels of mouse CFHR proteins in various spontaneously-occurring autoimmune diseases in mice and also showing the complete absence of CFHR-C mRNA as well as protein in the circulation of mice with SLE and DM. These studies will contribute to developing templates for needed gene therapy approaches to treat lupus and DM. Interestingly, serum from NOD mice is already being used as a negative control in C5 complement ELISA assays because these mice have a natural deficiency of C5 and are resistant to arthritis (Banda et al., 2010; Baxter and Cooke, 1993; Johansson et al., 2001). Thus, sera from lupus- and DM-prone mice can be used as controls for future studies involving CFHR proteins. CFHR proteins of mice are significantly different in overall sequence, while in some domains within fH they are homologous to those described in human. Thus, similar studies to those described herein, but with human tissue from patients with DDD, DM, BLD, SLE and RA, are warranted.
Conclusions
The mRNA of mouse CFHR-C was universally absent in brain, liver, lung, heart, spleen, kidney, knee joint and skin of mice with and without lupus and DM. CFHR-C protein was also absent in the circulation of lupus- and DM-prone mice. In contrast the mRNAs for CFH and CFHR-B were universally present in lupus- and DM-prone mice. The complete absence of CFHR-C in lupus- and DM-prone mice was disease-specific and also mouse strain-specific. CFH, CFHR-B and CFHR-C mRNAs were universally present in all organs of mice with and without DDD, BLD and CAIA. The exact function of CFHR-C is unknown, but the increase in the levels of mRNA in the spleen of mice with DDD and CAIA, compared with mice with no DDD and CAIA, suggest that it could be a pro-inflammatory protein in the pathogenesis of these two diseases. Furthermore, the presence of differential levels of the mRNAs for CFHR vs. CFH proteins in mice with various autoimmune diseases adds credence to the hypothesis that CFHR proteins have regulatory and deregulatory roles and that their roles are disease-specific.
Highlights.
CFHR-C was universally absent in the circulation of lupus- and diabetic- prone mice.
CFHR-C was present in the circulation of DDD, BLD and CAIA susceptible mice.
Increase in the mRNA of CFHR-C in spleen of mice with DDD and CAIA indicated its pro-inflammatory role.
CFHR-B mRNA was 4 fold higher in mice with BLD vs. mice with lupus.
Liver has the highest and skin has the lowest levels of CFH, CFHR-B and CFHR-C mRNA.
Acknowledgements
The authors thank: Dr. V. Michael Holers, University of Colorado Anschutz Medical Campus for in-depth discussion and valuable input related to these studies. Dr. Rafael L. Ufret-Vincenty, UT Southwestern Medical Center, Dallas, Texas for providing tissues samples from CfhTg/mCfh−/−chimeric mice with and without BLD and also going through this manuscript critically. Ms. Umarani Pugazhenthi, PCR Core University of Colorado Anschutz Medical Campus, for performing quantitative RT-PCR on samples used in this study; Dr. Lee, Barbara Davis Center, University of Colorado School of Medicine, for helping to identify diabetic NOD mice vs. non-diabetic NOD mice using glucose levels and Dr. Subhajyoti De, University of Colorado School of Medicine, Associate Member, University of Colorado Cancer Center for analyzing and providing critical bioinformatics information regarding sequence homology among various human and mouse complement factor H-related proteins and its variants. CS has received funding for these studies from the Deutsche Forschungsgemeinschaft (DFG SK-46). CS has also received funding from the European Community’s Seventh Framework Programme under grant agreement no 2012-305608 “European Consortium for High-Throughput Research in Rare Kidney Diseases (EURenOmics)”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- CP
classical pathway
- AP
alternative pathway
- LP
lectin pathway
- MAC or C5b-9
membrane attack complex
- CFH
complement factor H
- CFHR
complement factor H related proteins
- mouse CFHR-C
mouse complement factor H related C
- mouse CFHR-B
mouse complement factor H related B
- human CFHR-1
human complement factor H related-1
- human CFHR-2
human complement factor H related -2
- human CFHR-3
human complement factor H related-3
- hCFHR-4
human complement factor H related-4
- human CFHR-5
human complement factor H related-5
- DDD
dense deposit disease
- BLD
basal laminar deposits
- AMD
macular degeneration
- DM
diabetes mellitus
- SLE
systemic lupus erythematotsus
- CAIA
collagen antibody-induced arthritis
- CII
type II collagen
- DAS
disease activity score
- RA
rheumatoid arthritis
Footnotes
Disclosure All authors have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrera Abeleda M. A., Nishimura C, Smith JL, Sethi S, McRae JL, Murphy BF, Silvestri G, Skerka C, Jozsi M, Zipfel PF, Hageman GS, Smith RJ. Variations in the complement regulatory genes factor H (CFH) and factor H related 5 (CFHR5) are associated with membranoproliferative glomerulonephritis type II (dense deposit disease) J Med Genet. 2006;43:582–9. doi: 10.1136/jmg.2005.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel GB, Cook HT, Hageman G, Jennette JC, Kashgarian M, Kirschfink M, Lambris JD, Lanning L, Lutz HU, Meri S, Rose NR, Salant DJ, Sethi S, Smith RJ, Smoyer W, Tully HF, Tully SP, Walker P, Welsh M, Wurzner R, Zipfel PF. Membranoproliferative glomerulonephritis type II (dense deposit disease): an update. J Am Soc Nephrol. 2005;16:1392–403. doi: 10.1681/ASN.2005010078. [DOI] [PubMed] [Google Scholar]
- Banda NK, Levitt B, Wood AK, Takahashi K, Stahl GL, Holers VM, Arend WP. Complement activation pathways in murine immune complex-induced arthritis and in C3a and C5a generation in vitro. Clin Exp Immunol. 2010;159:100–8. doi: 10.1111/j.1365-2249.2009.04035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banda NK, Mehta G, Ferreira VP, Cortes C, Pickering MC, Pangburn MK, Arend WP, Holers VM. Essential role of surface-bound complement factor H in controlling immune complex-induced arthritis. J Immunol. 2013;190:3560–9. doi: 10.4049/jimmunol.1203271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banda NK, Thurman JM, Kraus D, Wood A, Carroll MC, Arend WP, Holers VM. Alternative complement pathway activation is essential for inflammation and joint destruction in the passive transfer model of collagen-induced arthritis. J Immunol. 2006;177:1904–12. doi: 10.4049/jimmunol.177.3.1904. [DOI] [PubMed] [Google Scholar]
- Bao L, Haas M, Quigg RJ. Complement factor H deficiency accelerates development of lupus nephritis. J Am Soc Nephrol. 2011;22:285–95. doi: 10.1681/ASN.2010060647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter AG, Cooke A. Complement lytic activity has no role in the pathogenesis of autoimmune diabetes in NOD mice. Diabetes. 1993;42:1574–8. doi: 10.2337/diab.42.11.1574. [DOI] [PubMed] [Google Scholar]
- Baxter AG, Healey D, Cooke A. Mycobacteria precipitate autoimmune rheumatic disease in NOD mice via an adjuvant-like activity. Scandinavian journal of immunology. 1994a;39:602–6. doi: 10.1111/j.1365-3083.1994.tb03419.x. [DOI] [PubMed] [Google Scholar]
- Baxter AG, Horsfall AC, Healey D, Ozegbe P, Day S, Williams DG, Cooke A. Mycobacteria precipitate an SLE-like syndrome in diabetes-prone NOD mice. Immunology. 1994b;83:227–31. [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Wiesener M, Eberhardt HU, Hartmann A, Uzonyi B, Kirschfink M, Amann K, Buettner M, Goodship T, Hugo C, Skerka C, Zipfel PF. Complement factor H-related hybrid protein deregulates complement in dense deposit disease. The Journal of clinical investigation. 2014;124:145–55. doi: 10.1172/JCI71866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt HU, Skerka C, Zipfel PF, Hallstrom T, Hartmann A, Chen Q. C3-glomerulopathy associated human factor H-related proteins 2 (CFHR2) and 5 (CFHR5) regulate complement C3b and TCC. Immunobiology. 2012;217:1143. [Google Scholar]
- Estaller C, Koistinen V, Schwaeble W, Dierich MP, Weiss EH. Cloning of the 1.4-kb mRNA species of human complement factor H reveals a novel member of the short consensus repeat family related to the carboxy terminal of the classical 150-kDa molecule. J Immunol. 1991;146:3190–6. [PubMed] [Google Scholar]
- Fritsche LG, Lauer N, Hartmann A, Stippa S, Keilhauer CN, Oppermann M, Pandey MK, Kohl J, Zipfel PF, Weber BH, Skerka C. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD) Hum Mol Genet. 2010;19:4694–704. doi: 10.1093/hmg/ddq399. [DOI] [PubMed] [Google Scholar]
- Gale DP, de Jorge EG, Cook HT, Martinez-Barricarte R, Hadjisavvas A, McLean AG, Pusey CD, Pierides A, Kyriacou K, Athanasiou Y, Voskarides K, Deltas C, Palmer A, Fremeaux Bacchi V., de Cordoba SR, Maxwell PH, Pickering MC. Identification of a mutation in complement factor H-related protein 5 in patients of Cypriot origin with glomerulonephritis. Lancet. 2010;376:794–801. doi: 10.1016/S0140-6736(10)60670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale DP, Maxwell PH. C3 glomerulonephritis and CFHR5 nephropathy. Nephrol Dial Transplant. 2013;28:282–8. doi: 10.1093/ndt/gfs441. [DOI] [PubMed] [Google Scholar]
- Goicoechea de Jorge E, Caesar JJ, Malik TH, Patel M, Colledge M, Johnson S, Hakobyan S, Morgan BP, Harris CL, Pickering MC, Lea SM. Dimerization of complement factor H-related proteins modulates complement activation in vivo. Proc Natl Acad Sci U S A. 2013;110:4685–90. doi: 10.1073/pnas.1219260110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea de Jorge E, Harris CL, Esparza-Gordillo J, Carreras L, Arranz EA, Garrido CA, Lopez-Trascasa M, Sanchez-Corral P, Morgan BP, Rodriguez de Cordoba S. Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc Natl Acad Sci U S A. 2007;104:240–5. doi: 10.1073/pnas.0603420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–32. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebecker M, Okemefuna AI, Perkins SJ, Mihlan M, Huber-Langy M, Jozsi M. Molecular basis of C-reactive protein binding and modulation of complement activation by factor H-related protein 4. Mol Immunol. 2010;47:1347–55. doi: 10.1016/j.molimm.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Heinen S, Hartmann A, Lauer N, Wiehl U, Dahse HM, Schirmer S, Gropp K, Enghardt T, Wallich R, Halbich S, Mihlan M, Schlotzer-Schrehardt U, Zipfel PF, Skerka C. Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood. 2009;114:2439–47. doi: 10.1182/blood-2009-02-205641. [DOI] [PubMed] [Google Scholar]
- Hellwage J, Eberle F, Babuke T, Seeberger H, Richter H, Kunert A, Hartl A, Zipfel PF, Jokiranta TS, Jozsi M. Two factor H-related proteins from the mouse: expression analysis and functional characterization. Immunogenetics. 2006;58:883–93. doi: 10.1007/s00251-006-0153-y. [DOI] [PubMed] [Google Scholar]
- Hellwage J, Jokiranta TS, Koistinen V, Vaarala O, Meri S, Zipfel PF. Functional properties of complement factor H-related proteins FHR-3 and FHR-4: binding to the C3d region of C3b and differential regulation by heparin. FEBS Lett. 1999;462:345–52. doi: 10.1016/s0014-5793(99)01554-9. [DOI] [PubMed] [Google Scholar]
- Johansson AC, Sundler M, Kjellen P, Johannesson M, Cook A, Lindqvist AK, Nakken B, Bolstad AI, Jonsson R, Alarcon-Riquelme M, Holmdahl R. Genetic control of collagen-induced arthritis in a cross with NOD and C57BL/10 mice is dependent on gene regions encoding complement factor 5 and FcgammaRIIb and is not associated with loci controlling diabetes. European journal of immunology. 2001;31:1847–56. doi: 10.1002/1521-4141(200106)31:6<1847::aid-immu1847>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Malik TH, Lavin PJ, Goicoechea de Jorge E, Vernon KA, Rose KL, Patel MP, de Leeuw M, Neary JJ, Conlon PJ, Winn MP, Pickering MC. A hybrid CFHR3-1 gene causes familial C3 glomerulopathy. Journal of the American Society of Nephrology : JASN. 2012;23:1155–60. doi: 10.1681/ASN.2012020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae JL, Duthy TG, Griggs KM, Ormsby RJ, Cowan PJ, Cromer BA, McKinstry WJ, Parker MW, Murphy BF, Gordon DL. Human factor H-related protein 5 has cofactor activity, inhibits C3 convertase activity, binds heparin and C-reactive protein, and associates with lipoprotein. Journal of Immunology. 2005;174:6250–6. doi: 10.4049/jimmunol.174.10.6250. [DOI] [PubMed] [Google Scholar]
- Mihlan M, Hebecker M, Dahse HM, Halbich S, Huber-Lang M, Dahse R, Zipfel PF, Jozsi M. Human complement factor H-related protein 4 binds and recruits native pentameric C-reactive protein to necrotic cells. Mol Immunol. 2009;46:335–44. doi: 10.1016/j.molimm.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Pangburn MK. Localization of the host recognition functions of complement factor H at the carboxyl-terminal: implications for hemolytic uremic syndrome. J Immunol. 2002;169:4702–6. doi: 10.4049/jimmunol.169.9.4702. [DOI] [PubMed] [Google Scholar]
- Park CT, Wright SD. Plasma lipopolysaccharide-binding protein is found associated with a particle containing apolipoprotein A-I, phospholipid, and factor H-related proteins. J Biol Chem. 1996;271:18054–60. doi: 10.1074/jbc.271.30.18054. [DOI] [PubMed] [Google Scholar]
- Perry D, Sang A, Yin Y, Zheng YY, Morel L. Murine models of systemic lupus erythematosus. J Biomed Biotechnol. 2011;2011:271694. doi: 10.1155/2011/271694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome research. 2004;14:1806–11. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MC, Cook HT, Warren J, Bygrave AE, Moss J, Walport MJ, Botto M. Uncontrolled C3 activation causes membranoproliferative glomerulonephritis in mice deficient in complement factor H. Nat Genet. 2002;31:424–8. doi: 10.1038/ng912. [DOI] [PubMed] [Google Scholar]
- Pickering MC, de Jorge EG, Martinez-Barricarte R, Recalde S, Garcia-Layana A, Rose KL, Moss J, Walport MJ, Cook HT, de Cordoba SR, Botto M. Spontaneous hemolytic uremic syndrome triggered by complement factor H lacking surface recognition domains. J Exp Med. 2007;204:1249–56. doi: 10.1084/jem.20070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MC, Warren J, Rose KL, Carlucci F, Wang Y, Walport MJ, Cook HT, Botto M. Prevention of C5 activation ameliorates spontaneous and experimental glomerulonephritis in factor H-deficient mice. Proc Natl Acad Sci U S A. 2006;103:9649–54. doi: 10.1073/pnas.0601094103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sethi S, Fervenza FC, Zhang Y, Zand L, Vrana JA, Nasr SH, Theis JD, Dogan A, Smith RJ. C3 glomerulonephritis: clinicopathological findings, complement abnormalities, glomerular proteomic profile, treatment, and follow-up. Kidney international. 2012;82:465–73. doi: 10.1038/ki.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerka C, Chen Q, Fremeaux-Bacchi V, Roumenina LT. Complement factor H related proteins (CFHRs) Mol Immunol. 2013;56:170–80. doi: 10.1016/j.molimm.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Skerka C, Zipfel PF. Complement factor H related proteins in immune diseases. Vaccine. 2008;26(Suppl 8):I9–14. doi: 10.1016/j.vaccine.2008.11.021. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Harris CL, Pickering MC. Dense deposit disease. Molecular Immunology. 2011;48:1604–10. doi: 10.1016/j.molimm.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortajada A, Yebenes H, Abarrategui-Garrido C, Anter J, Garcia-Fernandez JM, Martinez-Barricarte R, Alba-Dominguez M, Malik TH, Bedoya R, Cabrera Perez R, Lopez Trascasa M, Pickering MC, Harris CL, Sanchez-Corral P, Llorca O, Rodriguez de Cordoba S. C3 glomerulopathy-associated CFHR1 mutation alters FHR oligomerization and complement regulation. J Clin Invest. 2013;123:2434–46. doi: 10.1172/JCI68280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufret-Vincenty RL, Aredo B, Liu X, McMahon A, Chen PW, Sun H, Niederkorn JY, Kedzierski W. Transgenic mice expressing variants of complement factor H develop AMD-like retinal findings. Invest Ophthalmol Vis Sci. 2010;51:5878–87. doi: 10.1167/iovs.09-4457. [DOI] [PubMed] [Google Scholar]
- Vik DP, Munoz-Canoves P, Kozono H, Martin LG, Tack BF, Chaplin DD. Identification and sequence analysis of four complement factor H-related transcripts in mouse liver. J Biol Chem. 1990;265:3193–201. [PubMed] [Google Scholar]
- Zhao J, Wu H, Khosravi M, Cui H, Qian X, Kelly JA, Kaufman KM, Langefeld CD, Williams AH, Comeau ME, Ziegler JT, Marion MC, Adler A, Glenn SB, Alarcon-Riquelme ME, Pons-Estel BA, Harley JB, Bae SC, Bang SY, Cho SK, Jacob CO, Vyse TJ, Niewold TB, Gaffney PM, Moser KL, Kimberly RP, Edberg JC, Brown EE, Alarcon GS, Petri MA, Ramsey-Goldman R, Vila LM, Reveille JD, James JA, Gilkeson GS, Kamen DL, Freedman BI, Anaya JM, Merrill JT, Criswell LA, Scofield RH, Stevens AM, Guthridge JM, Chang DM, Song YW, Park JA, Lee EY, Boackle SA, Grossman JM, Hahn BH, Goodship TH, Cantor RM, Yu CY, Shen N, Tsao BP. Association of genetic variants in complement factor H and factor H-related genes with systemic lupus erythematosus susceptibility. PLoS Genet. 2011;7:e1002079. doi: 10.1371/journal.pgen.1002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel PF, Hallstrom T, Riesbeck K. Human complement control and complement evasion by pathogenic microbes--tipping the balance. Molecular Immunology. 2013;56:152–60. doi: 10.1016/j.molimm.2013.05.222. [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Mache C, Muller D, Licht C, Wigger M, Skerka C. DEAP-HUS: deficiency of CFHR plasma proteins and autoantibody-positive form of hemolytic uremic syndrome. Pediatric nephrology. 2010;25:2009–19. doi: 10.1007/s00467-010-1446-9. [DOI] [PubMed] [Google Scholar]