Abstract
Activated T regulatory cells (Treg) express latent TGF-β1 on their cell surface bound to GARP. Although integrins have been implicated in mediating the release of active TGF-β1 from the complex of latent TGF-β1 and latent TGF-β1 binding protein, their role in processing latent TGF-β1 from the latent TGF-β1/GARP complex is unclear. Mouse CD4+Foxp3+ Treg, but not CD4+Foxp3− T cells, expressed integrin β8 (Itgb8) as detected by qRT-PCR. Itgb8 expression was a marker of thymically-derived (t)Treg, as it could not be detected on Foxp3+Helios− Tregs or on Foxp3+ T cells induced in vitro. Tregs from Itgb8 conditional knockouts exhibited normal suppressor function in vitro and in vivo in a model of colitis, but failed to provide TGF-β1 to drive Th17 or iTreg differentiation in vitro. In addition, Itgb8 knockout Tregs expressed higher levels of latent TGF-β1 on their cell surface consistent with defective processing. Thus, integrin αvβ8 is a marker of tTregs and functions in a cell intrinsic manner in mediating the processing of latent TGF-β1 from the latent TGF-β1/GARP complex on the surface of tTregs.
Activated Foxp3+ T regulatory cells (Tregs) express the latent TGF-β1 binding protein GARP/LRRC32 that is required for expression of latent TGF-β1 on the surface of human and mouse Tregs. In an in vitro culture system in which activated Tregs are used as source of TGF-β1 to drive iTreg or Th17 differentiation, GARP is required for efficient production of biologically active TGF-β1(1, 2). Previous studies have indicated, among other mechanisms, that the αv-integrins, αvβ6 and αvβ8, bind the RGD site in latent TGF-β1 and facilitate the release of biologically active TGF-β from the complex of latent TGF-β1 and latent TGF-β1 binding protein (LTBP, the large latent complex). Activation mediated by αvβ6 requires the interaction of its cytoplasmic domain with the actin cytoskeleton, which appears to drive the shear forces needed to activate TGF-β1 from large latent complex (2). In contrast, αvβ8 has a short cytoplasmic tail that does not interact with the actin cytoskeleton. Some studies have demonstrated αvβ8-dependent processing of latent TGF-β1 from the large latent complex requires the co-expression of matrix metalloproteases (MMP), specifically MMP-14 (or MT1-MMP)(3). It is unknown, however, if this is the only MMP which can function in αvβ8-mediated activation of TGF-β1.
Previous studies have focused on processing of latent TGF-β1 from the large latent complex by integrin αvβ8 on dendritic cells. Conditional deletion of Itgb8 from leukocytes or only from DCs resulted in severe inflammatory bowel disease and age-related autoimmunity (4). Deletion of Itgb8 on dendritic cells also resulted in an inability to drive endogenous Th17 differentiation in the gut and in a failure to generate highly pathogenic Th17 cells during EAE resulting in markedly milder symptoms (5). The role of integrins in processing latent TGF-β1 from the latent TGF-β1/GARP complex is less clear. αvβ6 and to a lesser extent αvβ8 were shown to activate latent TGF-β1 from 293 cells transfected with αv integrins, pro-TGF-β1, and GARP(6). However, interpretation of these studies was complicated by the fact that TGF-β1 was equally activated in the absence of GARP, probably due the presence of endogenous LTBP. Nevertheless, it appeared that the ability of αvβ6 to activate TGF-β1 from the latent TGF-β/GARP complex was real as it was preserved in the presence of the ECR3E fragment of LTBP, which inhibits endogenous LTBP, but not GARP. It remains unclear whether similar mechanisms are present in Tregs or platelets that physiologically express GARP.
The purpose of the present study was to examine how biologically active TGF-β1 is released from the GARP/latent TGF-β1 complex by mouse Treg. Here, we demonstrate that integrin αvβ8 is a marker of mouse thymic-derived Tregs (tTregs) and functions in a cell intrinsic manner to release active TGF-β1.
Materials and Methods
Mice
C57BL/6 were obtained from the NCI Mouse Repository (Frederick, MD). Foxp3-GFP, C57BL/6-Rag1−/−, and OVA-specific TCR transgenic OT-II (CD45.1, Rag1−/−) mice were obtained by NIAID and were maintained by Taconic Farms (Germantown, NY) under contract by NIAID. Itgb8Fl/Fl mice, which have been previously described (7), were obtained from the Mutant Mouse Regional Resource Center (MMRRC stock number 014108-UCD). These mice contain loxP sites within 3′ and 5′ introns of exon 4 of the Itgb8 gene. Lrrc32Fl/Fl (GARP), Tgfb1Fl/Fl, Ikzf2Fl/Fl (Helios) mice have been previously described (1, 8, 9). Floxed mice were crossed to CD4-CRE mice (Taconic). Helios-GFP reporter mice were developed by Taconic Artemis and will be described in detail at a later date. Helios-GFP reporter mice were crossed to Foxp3-mRFP mice obtained from Jackson (Bar Harbor, Maine). OVA-specific TCR-transgenic OT-II mice were obtained from Taconic Farms and bred to Foxp3-GFP mice to generate OT-II Foxp3-GFP mice, as previously described (10). All animal protocols used in this study were approved by the NIAID Animal Care and Use Committee.
IBD Experiments
IBD experiments were performed similarly to those previously described (11). Briefly, 4×105 naive WT CD4+ T-cells (CD4+CD25− CD45RBhi) were transferred into Rag1−/− mice in the presence or absence of 2×105 (CD4+CD25hiCD45RBlow) Tregs from WT or Itgb8 conditional knockout mice. Weights were monitored two times weekly for up to 11 weeks.
Cell isolation and flow cytometry
For purification of dendritic cells, mouse spleens were fragmented and digested for 30 min at 37°C in the presence of liberase blendzyme II (Roche) and DNase (2 μg/ml) (Roche) in complete medium (modified RPMI 1640 supplemented by 10% FBS HyClone, 50 μM 2-ME (Sigma-Aldrich), 1% sodium pyruvate, 1% nonessential amino acids, 1% HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. They were labeled with anti-CD11c beads and purified on the AutoMACS Cell Separator (Miltenyi Biotec). T-cells from pooled lymph nodes and/or spleens were isolated using CD4-beads.
CD4+Foxp3+ Treg and Foxp3− conventional T-cells (Tconv) were sorted from the pooled lymph nodes of Foxp3-GFP mice. CD4+CD25+ Treg and CD4+CD25− Tconv were also sorted. All cell sorting was performed on FACSAria flow cytometers (BD Biosciences). Single cell suspensions were stained using the following antibodies according to the manufacturer’s protocol: Anti-mouse CD45.1 (A20), CD45.2 (104), CD4 (RM4-5), Foxp3 (FLK-16s), GARP (YGIC86), and IL-17A (eBio17B7). Purified mouse anti-mouse LAP clone TW7-16B4 was generously provided by Howard Weiner (Harvard Medical School, Boston, MA). The LAP antibody was labeled with SureLight-APC at Columbia Biosciences (Columbia, MD). For staining of Foxp3, cells were fixed and permeabilized using the Foxp3 fixation/permeabilization staining kit (eBiosciences). For staining of cytokines, cells were fixed and permeabilized using the Cytofix/Cytoperm kit from BD Biosciences.
Cytokines and other reagents
Recombinant mouse IL-6 was purchased from Biolegend (San Diego, CA). The MMP inhibitor GM6001 and the GM6001 control were both purchased from Santa Cruz Biotechnology (Dallas, TX).
In vitro suppression assay
In vitro suppression assays were performed as previously described(12).
In vitro T cell differentiation
To induce Th17 or iTreg differentiation using activated Tregs as a source of biologically active TGF-β1 as previously described (1), sorted Treg were activated and expanded using plate-bound anti-CD3 (1μg/well, 24-well plate) with IL-2 (100U/ml) for 2–3 days, followed by overnight culture with IL-2 alone. Cells were generally greater than 90–95% Foxp3+ after expansion. Tregs were washed and mixed 1:1 with naive CD45.1+ OT-II cells (RAG1−/−) in the presence of recombinant mouse IL-6 (10ng/ml) and stimulated with splenic dendritic cells and soluble anti-CD3 for 4 days. Similar experiments were performed using exogenous IL-2 (100U/ml) and activated Tregs to drive iTreg differentiation. Th17 differentiated T-cells were stimulated with the Cell Stimulation Cocktail and Protein Transport Inhibitors (eBioscience).
In other experiments, iTregs were differentiated to analyze the expression of Itgb8. Naive CD4+ T-cells (CD44lowCD62Lhi) from Foxp3-GFP mice were cultured with plate-bound anti-CD3 or dendritic cells and soluble anti-CD3. Naive CD4+ cells were also sorted from OT-II Foxp3-GFP mice and cultured with splenic dendritic cells and cognate peptide. In all conditions, cells were cultured in the presence of recombinant TGF-β1 and IL-2. After 4 days of culture, CD4+GFP(Foxp3+) cells were sorted and immediately subject to RNA isolation.
mRNA Isolation, cDNA production and Real-Time PCR
Tregs and conventional CD4+ T-cells were sorted and immediately subjected to RNA extraction or stimulated overnight with IL-2 and plate-bound anti-CD3 then subjected to RNA extraction using TRIzol reagent. The contaminating DNA was then removed by DNase I treatment. The SuperScript II First-Strand Synthesis Supermix for qRT-PCR (Invitrogen Life Technologies) was used to generate cDNA. Real-time PCR was conducted with the ABI Prism7900HT, using the Kapa Probe Fast Universal qPCR Kit and TaqMan Probes for GAPDH, ITGB8, and Foxp3.
Results
Integrin αvβ8 is a marker of Foxp3+ Treg cells
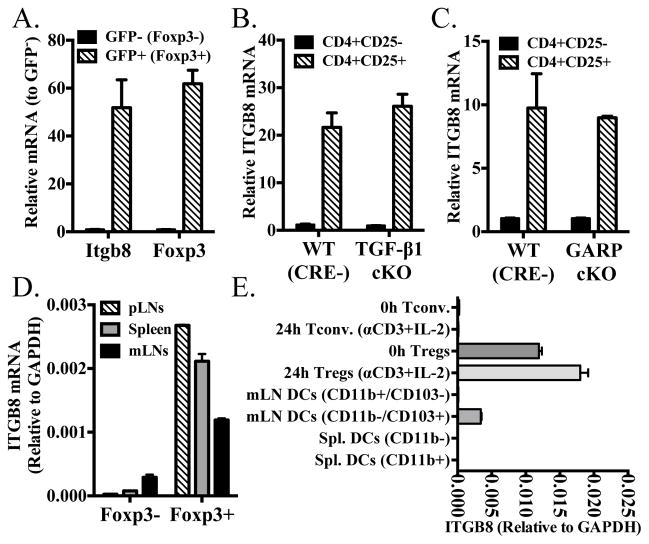
Previous studies from our group and others have demonstrated that activated Foxp3+ Treg express latent TGF-β1 on their cell surface bound to the tethering molecule, GARP (1, 13, 14). While αvβ8 on dendritic cells has been shown to mediate the release of active TGF-β1 from the large latent complex, we have previously shown that even in the absence of dendritic cells, Tregs are able to release active TGF-β1, primarily from the latent TGF-β1/GARP complex (1). We therefore decided to examine the potential expression and function of αvβ6 and αvβ8 by Treg cells. Sorted CD4+Foxp3+ expressed substantially higher levels of Itgb8 than CD4+ Foxp3− conventional T-cells, by qRT-PCR (Fig. 1A). Neither cell type expressed Itgb6 (data not shown). Unfortunately, we could not determine the expression of β8 on a single cell basis, as no antibody is currently available for staining and FACS analysis. The expression of Itgb8 was not dependent upon the expression of TGF-β1 or GARP (Fig. 1B, C). However, both cell populations expressed integrin αv (CD51) either freshly isolated or after activation with anti-CD3 and IL-2 (Fig. S1). CD51, however, is well known to be able to pair with multiple integrin β-chains in addition to β8 including β1, β3, β5, and β6 (15).
FIGURE 1.
Integrin αvβ8 is expressed by Foxp3+ Tregs. (A) Sorted CD4+Foxp3+(GFP+) and CD4+Foxp3− (GFP−) were measured for Itgb8 and Foxp3 message as measured by qRT-PCR. Data is expressed relative to CD4+Foxp3− cells. (B and C) Sorted CD4+CD25+ and CD4+CD25− from CD4 conditional knockouts of Tgfb1 and Lrrc32 and their CRE-littermate controls were measured for Itgb8 message. Data is expressed relative to wild-type CD4+CD25− cells. (D) Sorted CD4+Foxp3+(GFP+) and CD4+Foxp3-(GFP-) from spleen, peripheral lymph nodes, or mesenteric lymph nodes were measured for Itgb8 message as measured by qRT-PCR. (E) Sorted CD4+Foxp3(GFP) − and CD4+Foxp3(GFP)+ (fresh or after overnight stimulation with plate-bound anti-CD3+ IL-2), CD11b+CD103- and CD11b−CD103+ DCs from the mesenteric LNs, and CD11b+ or CD11b− DCs were measured for Itgb8 message. DCs were first gated on Thy1.2-CD19-CD11c+I-Ab-hi cells.
Itgb8 expression on Tregs was not dependent upon the lymphoid tissue from which they were isolated (Fig. 1D) and was only modestly increased upon activation (Fig. 1E). Itgb8 has been shown to be expressed in CD103+ dendritic cells in the mesenteric lymph nodes or gut associated lymphoid tissues (16, 17). We found that the level of expression of Itgb8 was higher in Tregs than in CD103+ dendritic cells (relative to GAPDH). As previously described (17), the level of Itgb8 was substantially higher in CD103+ dendritic cells from the mesenteric lymph nodes than in CD103− dendritic cells or dendritic cell populations from the spleen (Fig. 1E).
Integrin β8 is expressed primarily by Helios+ Foxp3+ Treg cells
Since we have previously shown that iTregs express the latent TGF-β1/GARP complex, it was of interest to determine if iTreg also express Itgb8 (1). iTregs were generated from naive CD4+Foxp3− T-cells from Foxp3-GFP or OT-II Foxp3-GFP mice. iTregs were generated using plate-bound anti-CD3, splenic dendritic cells with soluble anti-CD3, or splenic dendritic cells with OVA peptide (OT-II Cells) cultured for 4 days with IL-2 and TGF-β1, then sorted for Foxp3+ (GFP+) cells. Tregs and Foxp3− cells stimulated with plate-bound anti-CD3+IL-2 were cultured in parallel and sorted for GFP+ or GFP− cells, respectively. iTregs generated with plate-bound anti-CD3 did not express substantial levels of Itgb8, nor did those generated using dendritic cells with peptide. Tregs cultured in parallel maintained their expression while cultured Foxp3− cells did not show substantial expression. Interestingly, iTregs generated using splenic dendritic cells and soluble anti-CD3 did show some expression of Itgb8, while those generated using more physiologically relevant conditions with peptide did not. Their level of expression was still lower than cultured Tregs (Fig. 2A). As we have previously proposed (9) that Foxp3+Helios+ and Foxp3+Helios− Tregs may represent tTreg and peripherally derived Tregs (pTreg), respectively, the relative expression of Itgb8 was determined for each of these populations. CD4+Foxp3+Helios+, CD4+Foxp3+Helios−, and CD4+Foxp3−Helios− cells were isolated from a Helios(GFP)/Foxp3(RFP) double-reporter mouse. Only the CD4+Foxp3+Helios+ cells expressed substantial levels of Itgb8 (Fig. 2B). Furthermore, this difference in expression was maintained even after activation and expansion for 4 days in culture (Fig. 2C). Helios did not control the expression of Itgb8, as Tregs sorted from Ikzf2(Helios)Fl/Fl CD4-CRE conditional knockout mice maintained their expression of Itgb8 (Fig. 2D).
FIGURE 2.
(A) Naive CD4+ T-cells (CD44lowCD62Lhi) from Foxp3-GFP mice were cultured with plate-bound anti-CD3 (iTreg PB-αCD3) or dendritic cells and soluble anti-CD3 (iTreg DCs + soluble-αCD3). Naive CD4+ cells were also sorted from OT-II Foxp3-GFP mice and cultured with splenic dendritic cells and cognate peptide (iTreg OT-II w/DCs + Peptide). In all conditions, cells were cultured in the presence of recombinant TGF-β1 and IL-2 for 4 days. For the same experiment, naive T-cells (Tconv.) or CD4+GFP+(Foxp3+) cells were cultured on plate-bound anti-CD3 with IL-2 for 4 days. (B) Sorted CD4+GFP-(Helios)-RFP-(Foxp3)-, CD4+GFP-RFP+ and CD4+GFP+RFP+ cells from double reporter mice were measured for Itgb8 message. (C) Sorted CD4+GFP(Helios)-RFP-(Foxp3)-, CD4+GFP-RFP+ and CD4+GFP+RFP+ cells from double reporter mice were stimulated with plate-bound anti-CD3+IL-2 for 3 days then rested overnight in IL-2. The cells were then measured for Itgb8 message by qRT-PCR. (D) Sorted CD4+CD25+ and CD4+CD25− from CD4 conditional knockouts of Ikzf2 and their CRE-littermate controls were measured for Itgb8 message. In (A), data is expressed relative to T-conventional cell population, in (B) data is expressed relative to Helios-Foxp3+ cells, in (C) data is expressed relative to expanded (CD4+Foxp3−) non-Treg population, and in (D) data is expressed relative to wild-type CD4+CD25− cells.
Itgb8 is required for clearance of TGF-β1 from the GARP/Latent-TGF-β1 complex
To investigate the function of Itgb8 on Tregs, we bred CD4-conditional knockouts of Itgb8, using the previously described Itgb8Fl/Fl mice (7). When bred with the CD4-CRE, the percentages of thymocyte subpopulations (Figure S2, upper panels), including the percentages of Foxp3+ cells within the CD4 single-positive population (Figure S2, lower panels), were identical in CRE+ and CRE littermates. The percentages of CD4+Foxp3− and CD4+Foxp3+ T cells in the periphery were also identical in CRE+ and CRE littermates (Figure S2, Right Panels).
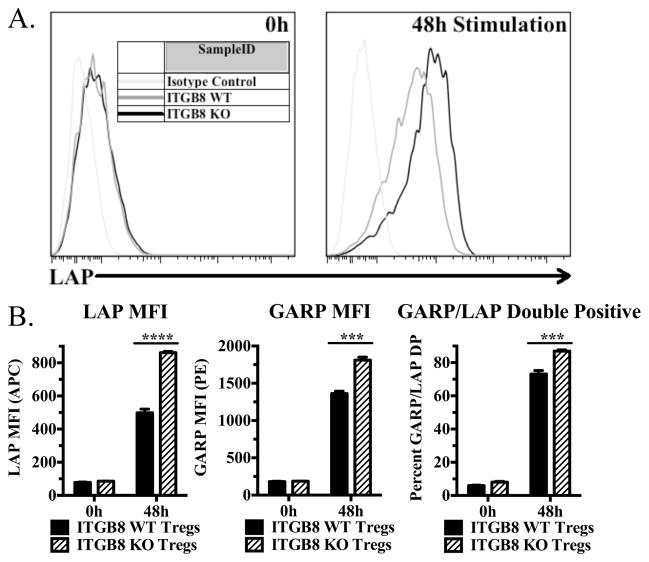
We hypothesized that if αvβ8 is involved in activation of TGF-β1 from the GARP/Latent TGF-β1 complex, that in the absence of Itgb8, latent-TGF-β1 would accumulate on the surface of Tregs. Freshly isolated Tregs from WT and Itgb8 cKO mice displayed similar low levels of cell surface latent-TGF-β1, while after activation, cKO Tregs displayed significantly higher levels of latent-TGF-β1 than activated WT Tregs (Fig. 3A, B). Because resting Tregs express higher level of GARP than latent TGF-β1, it is likely that the levels of latent TGF-β1 are probably limiting in forming the expression of the GARP/latent-TGF-β1 complex on the cell surface. Itgb8 cKO mice also expressed higher levels of GARP and GARP/LAP double positive cells (Fig. 3B).
FIGURE 3.
Activated Itgb8-deficient Tregs accumulate latent TGF-β1 on their surface. (A and B) Freshly isolated cells from pooled lymph nodes and enriched CD4+ cells from Itgb8 conditional knockout mice (CD4-CRE) or CRE- littermates were stimulated for 48 h with plate-bound anti-CD3 and IL-2, then stained for CD4, Foxp3, LAP (or IC), and GARP (or IC). (A) Representative histograms for LAP (latent TGF-β1) from CD4+Foxp3+ cells. (B) Graphical representations indicating LAP and GARP MFI on CD4+Foxp3+ cells as well as percentage of LAP and GARP double positive cells.
Itgb8 is not required for Treg-mediated suppression in vitro or in vivo in a transfer model of colitis
We next determined if Treg expression of Itgb8 is required for suppression of T effector cell proliferation in vitro. Using the traditional T-cell suppression assay, Tregs from both Itgb8 cKO and WT floxed littermates equally suppressed proliferation with increasing ratios of Treg to effector T-cells (Fig. 4A). This was not unexpected, however, as it has previously been shown by our group and others that neither TGF-β1 nor GARP expression is required for in vitro suppression(1, 8). Next, we determined if Itgb8 plays a role in Treg-mediated suppression in the transfer model of colitis. Transfer of wild type CD4+CD25−CD45RBhi T cells resulted in significant weight loss in the recipients; both wild type and cKO Tregs were equally capable of protecting from disease (Fig. 4B). Importantly, 75 days post-transfer, both wild type and cKO Tregs equally maintained Foxp3+ cells in the mesenteric lymph nodes (Fig. 4C). This result is consistent with a previous report (18), which demonstrated that TGF-β1 was required for suppression of IBD in this model, but that the source of the TGF-β1 was not the Treg.
FIGURE 4.
Tregs from Itgb8 conditional knockout mice suppress normally in vitro and in vivo. (A) T-cell suppression assays using sorted CD4+CD25− cells from WT mice in the presence or absence of increasing ratios of Itgb8 WT and KO CD4+CD25+ cells, as previously described(12). (B) WT CD4+CD25−CD45RBhi T cells were transferred into Rag1−/− mice in the presence or absence of CD4+CD25hiCD45RBlow Tregs from WT or Itgb8 conditional knockout mice and monitored for weight loss for up to 11 weeks. Indicated are the average weight changes from 5 mice in each group. (C) At the end of the experiment, mesenteric lymph nodes were stained for CD4 and Foxp3. Left panels are representative lymph node samples. Right panels indicate the percentage of CD4+ cells that were Foxp3+ in individual mice.
ITGB8 expression by Tregs is required for the bioavailability of active TGF-β1 from Tregs
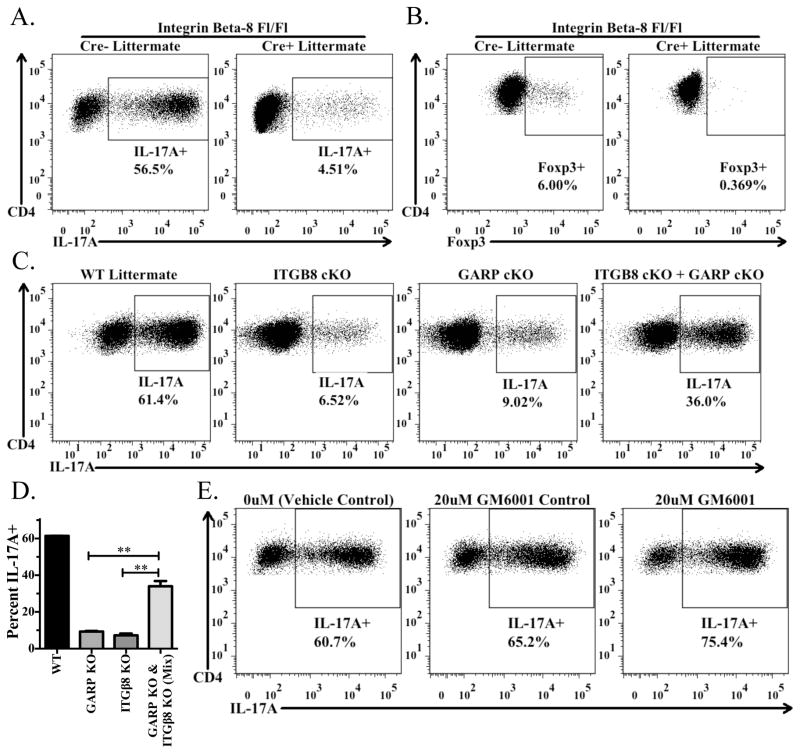
In order to evaluate the availability of biologically active TGF-β1 released from wild type and Itgb8 cKO Tregs, we used an in vitro culture system in which activated Tregs are cultured with naive T-cells under activating conditions (splenic DCs with soluble anti-CD3) in the presence of IL-6 or IL-2 to drive Th17 or iTreg differentiation, respectively. We have previously shown that TGF-β1 derived from the TGF-β1/GARP complex plays a major role as the source of TGF-β1 required for differentiation of Th17 cells or iTreg (1). Wild type, but not Itgb8 cKO, Tregs, promoted the differentiation of Th17 or iTreg as measured by IL-17A and Foxp3 expression, respectively (Fig. 5A, B). The same result was observed when the Tregs were only activated overnight prior to being cultured with naive cells (data not shown). To determine if the GARP/latent-TGF-β1 complex and αvβ8 needed to be expressed on the same cells or if αvβ8 could result in release of biologically active TGF-β1 from the GARP/latent TGF-β1 complex on a different cell, we cultured Tregs from GARP (Lrrc32) cKO or Itgb8 cKO mice separately or together with naive T cells in the Th17 differentiation assay. Neither GARP nor Itgb8 cKO cells alone could efficiently drive Th17 differentiation, however when mixed they were able to substantially restore Th17 differentiation, indicating that αvβ8 expressed on one cell could result in the activation of TGF-β1 from the GARP/latent-TGF-β1 complex on a distinct cell (Fig. 5C, D). Previous studies indicated that integrin αvβ8 expressed by human epithelial cell lines required the co-expression of a matrix metalloproteinase, in particular MMP-14(3)), in order to activate latent TGF-β1 from the large latent complex. The contribution of MMP-14 was demonstrated by the use of the MMP inhibitor GM6001. Thus far, we have not observed any effect of this inhibitor over a wide dose range on Treg-mediated Th17 induction up to 20μM, as compared to the vehicle control or GM6001-control (Fig. 5E). It is important to note, however, that it is unknown which MMPs are expressed by Tregs and whether αvβ8-mediated activation of latent TGF-β1 activation from the GARP/latent TGF-β1 complex is dependent on MMPs.
FIGURE 5.
Treg-mediated Th17 differentiation requires activation of latent TGF-β1 via integrin αvβ8. (A) Naive CD45.1+ OT-II cells from Rag1−/− mice were activated by soluble anti-CD3 in the presence of IL-6, splenic DCs, and preactivated CD4+CD25+ T cells from Itgb8 conditional knockout mice (CD4-CRE) or CRE-littermates. Cells were reactivated with Cell Stimulation Cocktail and Protein Transport Inhibitors and then stained for CD4, CD45.1, CD45.2, and IL-17A. The percentage of IL-17A+ cells derived from the CD4+CD45.1+CD45.2−cells in the culture are shown in each panel. (B) Same as in (A), except in the presence of IL-2 (100U/ml). Culture was stained for CD4, CD45.1, CD45.2, and intracellularly for Foxp3. (C and D) Same culture set up as in (A), except using preactivated CD4+CD25+ T cells from Itgb8 or Lrrc32 conditional knockout mice or CRE- littermates. In the right panel, half of the CD4+CD25+ T cells were from Itgb8 cKO mice and half were from Lrrc32 cKO mice. (D) Bar graph indicates the average (± SD) of duplicates within the experiment from (C). (E) Same as in (A), except using sorted CD4+Foxp3+ cells from Foxp3-GFP reporter mice and plate-bound anti-CD3 to activate the cells in the culture. Cells were cultured in the presence of a vehicle control (DMSO), the MMP-inhibitor GM6001, or the GM6001 control and measured for IL-17A as in (A).
Helios −/lowTregs are less efficient at producing biologically active TGF-β1
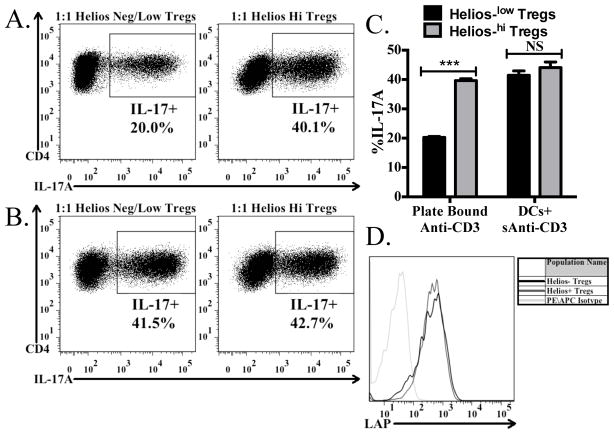
Since we found that Helios+ Tregs expressed substantially higher levels of Itgb8 (Fig. 2B), we wished to assess the ability of the Helios+ versus Helios−/low Tregs to drive Th17 differentiation in the same experimental setup as used in Fig. 5. When using plate-bound anti-CD3 to activate the cells in the culture, Helios+ Tregs were more efficient at driving Th17 differentiation (Fig. 6A, C) than Helios−/low Tregs. This result is consistent with the higher level of expression of Itgb8 on the Helios+ cells. It is not due to an inability of the Helios−/low cells to express latent TGF-β1 on their surface, as total Tregs activated in vitro, when gated on the Helios−/low or Helios+ subsets both have similar levels of latent TGF-β1 on their surface (Fig. 6D). It also remains possible that Helios−/low Tregs can process the latent-TGF-β1 by an avβ8-independent mechanism. Surprisingly, when the co-cultures were activated using soluble anti-CD3 with splenic dendritic cells, which both the Helios+ and Helios−/low Tregs were equally capable of driving Th17 differentiation (Fig. 6B, C). As splenic dendritic cells were unable to restore biologically active TGF-β1 production from Itgb8 KO Tregs (Fig. 5A), it is unlikely that low levels of Itgb8 on the dendritic cells contribute to TGF-β1 activation, but instead it is possible that the dendritic cells act as a platform to force a close proximity between the Tregs and naive T-cells, thus resulting in a decreased threshold for TGF-β1 concentrations. As result, lower levels of biologically active TGF-β1 produced by Helios−/low (Fig. 6A, C) are sufficient to drive a full response in the presence of dendritic cells (Fig. 6B, C).
FIGURE 6.
Helios low/- cells are less efficient at producing biologically active TGF-β1. Naive CD45.1+ OT-II cells from Rag1 −/− mice were activated by plate-bound (A) or soluble ani-CD3+ splenic DCs in the presence of IL-6 and preactivated Helios+ or Helios-/low cells from Helios (GFP)/Foxp3 (RFP) double reporter mice. After 4 days, cells were reactivated with Cell Stimulation Cocktail and Protein Transport Inhibitors and then stained for CD4, CD45.1, CD45.2, and IL-17A. The percentage of IL-17A+ cells derived from the CD4+CD45.1+CD45.2− cells in the culture are shown in each panel. (C) Bar graph indicates the average (± SD) of duplicates within the experiment from (A) and (B). (D) Tregs were activated for 48 hours, then stained for CD4, Foxp3, Helios, and LAP. Histograms shown represent the expression of LAP on CD4+Foxp3+Helioshi cells and CD4+Foxp3+Helios−/low cells, as compared to the isotype control.
Discussion
TGF-β1 is critical to the maintenance of immune homeostasis as deletion of TGF-β1 results in a generalized inflammatory syndrome and autoimmunity. T and B lymphocytes play a critical role in disease development in TGF-β1−/− mice as inflammation does not develop in TGF-β1−/− mice on a SCID background(19). Almost every cell type expresses TGF-β1 receptors, but TGF-β1 is always produced in an inactive form associated with LAP that prevents its binding to its receptor. Activation of TGF-β1 is closely regulated so that its effects can be mediated in an appropriate environment. A number of mechanisms have been proposed for the activation of TGF-β1 including plasmin, matrix metalloproteases, lysosomal proteases, and thrombospondin. Recent studies have demonstrated that physiologically, integrins are the key activations of TGF-β1. The strongest evidence in favor of this is that mice with a single point mutation in the RGD sequence of LAP that cannot bind to integrins phenotypically copy mice with a global deficiency of TGF-β1 (20).
While 6 of the 24 integrins can bind latent TGF-β1 via the RGD sequence in LAP, only αvβ3, αvβ5, αvβ6, and αvβ8 have been demonstrated to liberate active TGF-β1. A number of studies have shown that αvβ6 and αvβ8 are the key activators of TGF-β1 in vivo. Integrin αvβ6 is primarily expressed in epithelial cells and β6−/− mice develop a lung and skin inflammation and pulmonary emphysema (21). The cytoplasmic domain of αvβ6 connects to the actin cytoskeleton and αvβ6-mediated TGF-β1 activation is controlled by cell contraction (2). In contrast, αvβ8 is widely expressed by many different cell types including neurons, astrocytes, airway epithelial cell, fibroblasts, dendritic cells, and T cells. The cytoplasmic domain of αvβ8 does not connect to the actin cytoskeleton. It appears that the role of this integrin is to present latent TGF-β1 to a membrane bound protease resulting in release of active TGF-β1 (3). It is not clear whether αvβ8 utilizes different mechanisms to activate TGF-β 1 in different cell types.
While most cell types are capable of secreting latent TGF-β1 bound to LTBP as the large latent complex, Tregs and platelets express a unique TGF-β1 binding protein, GARP that targets latent TGF-β1 to the cell surface (1, 13, 14). Within the immune system, αvβ8 expression on leukocytes, particularly CD103+ dendritic cells, appears to be critical in generating active TGF-β1 from the large latent complex (4). In this report, we have demonstrated that Itgb8 is selectively expressed on tTreg and mediates the release of active TGF-β1 from the latent TGF-β1/GARP complex. In contrast to the latent TGF-β1/GARP complex whose expression is markedly upregulated during the course of T cell activation, expression of Itgb8 on Treg was constitutive and only modestly upregulated during activation.
Thus far, mice with selective deficiency of GARP on T-cells cells show no evidence of spontaneous autoimmune disease or inflammation in any organ. Similarly, mice with a deletion of β8 in CD4+ T cells did not have a phenotype (4). In contrast, mice lacking αv or β8 only on dendritic cells develop systemic autoimmunity and colitis and may also have a defect in generating Treg cells (4, 16). In addition, mice with a myeloid cell specific β8 deficiency exhibit a reduction in Th17 cells and are protected from EAE (5). Importantly, the same myeloid cell must both present the antigen and express the integrin. It is unclear why the expression of αvβ8 on cells on dendritic cells is so critical for the maintenance of immune homeostasis, while loss of αvβ8 expression on Treg does not lead to a defect in immune regulation, at least in the steady state. One possibility is that the number of dendritic cells that express αvβ8 is much higher than the number of αvβ8 expressing Treg and that they are much more broadly distributed in the body. However, this is clearly not the case as αvβ8 expression is highly restricted to intestinal CD103+ DC and αvβ8 expression is not detected on splenic DC (16). The most likely explanation for the different phenotypes of β8−/− DC and T cells is that dendritic cells are constantly exposed to the large latent complex at sites where they access it in the extracellular matrix. Furthermore, in the gut, potentiation of Th17 induction may be modulated by the intestinal microflora and Treg induction can be enhanced by the production of retinoic acid by the CD103+ DC. In contrast, while Treg express relatively high levels of αvβ8 (Fig. 1), Treg only express the latent TGF-β1/GARP complex when activated and the conditions leading to expression of this complex in vivo have yet to be defined.
It remains unclear why Tregs should express a second pathway for the delivery and activation of TGF-β1 on their cell surface. We have demonstrated that Treg-derived active TGF-β1 can play a role both in the differentiation of iTreg and Th17 cells. Tregs may primarily exert their effects via recognition of self-antigens during the process of priming and differentiation of naive T cells on the surface of antigen-presenting dendritic cells. It is likely that not all dendritic cells express Itgb8 and that Treg cell intrinsic expression of Itgb8 may be required to facilitate activation of Treg-expressed latent TGF-β1 during the priming/differentiation of naive T cells. Depending on the make up of the inflammatory environment (IL-2 versus IL-6), Treg-derived active TGF-β1 would then promote infectious tolerance (existing Tregs driving de novo Treg differentiation (22)) by the generation of iTreg or promote the induction of potentially regulatory or pathogenic Th17 cells.
Supplementary Material
Acknowledgments
We would like to thank Julie Edwards and Carol Henry from the NIAID Flow Cytometry Section for sorting our cells.
Abbreviations used in this article
- Itgb8
Integrin β8
- GARP
Glycoprotein A Repetitions Predominant
- LAP
Latency Associated Peptide
- Treg
Regulatory T-cell
- LTBP
latent TGF-β binding proteins
Footnotes
Supported by funds from the Intramural Program of the National Institute of Allergy and Infectious Diseases
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Edwards JP, Fujii H, Zhou AX, Creemers J, Unutmaz D, Shevach EM. Regulation of the expression of GARP/latent TGF-beta1 complexes on mouse T cells and their role in regulatory T cell and Th17 differentiation. J Immunol. 2013;190:5506–5515. doi: 10.4049/jimmunol.1300199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahamed J, Burg N, Yoshinaga K, Janczak CA, Rifkin DB, Coller BS. In vitro and in vivo evidence for shear-induced activation of latent transforming growth factor-beta1. Blood. 2008;112:3650–3660. doi: 10.1182/blood-2008-04-151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H, Sheppard D, Broaddus VC, Nishimura SL. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travis MA, Reizis B, Melton AC, Masteller E, Tang Q, Proctor JM, Wang Y, Bernstein X, Huang X, Reichardt LF, Bluestone JA, Sheppard D. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449:361–365. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melton AC, Bailey-Bucktrout SL, Travis MA, Fife BT, Bluestone JA, Sheppard D. Expression of alphavbeta8 integrin on dendritic cells regulates Th17 cell development and experimental autoimmune encephalomyelitis in mice. J Clin Invest. 2010;120:4436–4444. doi: 10.1172/JCI43786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R, Zhu J, Dong X, Shi M, Lu C, Springer TA. GARP regulates the bioavailability and activation of TGFbeta. Mol Biol Cell. 2012;23:1129–1139. doi: 10.1091/mbc.E11-12-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proctor JM, Zang K, Wang D, Wang R, Reichardt LF. Vascular development of the brain requires beta8 integrin expression in the neuroepithelium. J Neurosci. 2005;25:9940–9948. doi: 10.1523/JNEUROSCI.3467-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Q, Kim YC, Laurence A, Punkosdy GA, Shevach EM. IL-2 controls the stability of Foxp3 expression in TGF-beta-induced Foxp3+ T cells in vivo. J Immunol. 2011;186:6329–6337. doi: 10.4049/jimmunol.1100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thornton AM, Shevach EM. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J Exp Med. 1998;188:287–296. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proc Natl Acad Sci U S A. 2009;106:13445–13450. doi: 10.1073/pnas.0901944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R, Kozhaya L, Mercer F, Khaitan A, Fujii H, Unutmaz D. Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells. Proc Natl Acad Sci U S A. 2009;106:13439–13444. doi: 10.1073/pnas.0901965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Worthington JJ, Czajkowska BI, Melton AC, Travis MA. Intestinal dendritic cells specialize to activate transforming growth factor-beta and induce Foxp3+ regulatory T cells via integrin alphavbeta8. Gastroenterology. 2011;141:1802–1812. doi: 10.1053/j.gastro.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paidassi H, Acharya M, Zhang A, Mukhopadhyay S, Kwon M, Chow C, Stuart LM, Savill J, Lacy-Hulbert A. Preferential expression of integrin alphavbeta8 promotes generation of regulatory T cells by mouse CD103+ dendritic cells. Gastroenterology. 2011;141:1813–1820. doi: 10.1053/j.gastro.2011.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahlen L, Read S, Gorelik L, Hurst SD, Coffman RL, Flavell RA, Powrie F. T cells that cannot respond to TGF-beta escape control by CD4(+)CD25(+) regulatory T cells. J Exp Med. 2005;201:737–746. doi: 10.1084/jem.20040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diebold RJ, Eis MJ, Yin M, Ormsby I, Boivin GP, Darrow BJ, Saffitz JE, Doetschman T. Early-onset multifocal inflammation in the transforming growth factor beta 1-null mouse is lymphocyte mediated. Proc Natl Acad Sci U S A. 1995;92:12215–12219. doi: 10.1073/pnas.92.26.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Mu Z, Dabovic B, Jurukovski V, Yu D, Sung J, Xiong X, Munger JS. Absence of integrin-mediated TGFbeta1 activation in vivo recapitulates the phenotype of TGFbeta1-null mice. J Cell Biol. 2007;176:787–793. doi: 10.1083/jcb.200611044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris DG, Huang X, Kaminski N, Wang Y, Shapiro SD, Dolganov G, Glick A, Sheppard D. Loss of integrin alpha(v)beta6-mediated TGF-beta activation causes Mmp12-dependent emphysema. Nature. 2003;422:169–173. doi: 10.1038/nature01413. [DOI] [PubMed] [Google Scholar]
- 22.Andersson J, Tran DQ, Pesu M, Davidson TS, Ramsey H, O'Shea JJ, Shevach EM. CD4+ FoxP3+ regulatory T cells confer infectious tolerance in a TGF-beta-dependent manner. J Exp Med. 2008;205:1975–1981. doi: 10.1084/jem.20080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.