Summary
Objectives
Non-structural protein 1 (NS1)-based tests may offer a larger window of opportunity for dengue diagnosis and could constitute a very useful diagnostic tool. The aim of this study was to establish the overall accuracy of NS1-based tests for diagnosing dengue infection.
Methods
A meta-analysis was conducted including 18 studies published up to October 1, 2012 identified using PubMed, ISI Web of Science, Google Scholar, and the Chinese National Knowledge Infrastructure (CNKI) database.
Results
For the single NS1-based tests – ELISA (Panbio Dengue Early ELISA Kit, Dengue NS1 Ag ELISA Kit, and Platelia Dengue NS1 Ag-ELISA Kit) and immunochromatography (Dengue NS1 Ag STRIP Kit and SD BIOLINE Dengue Duo Strip Kit) – the summarized sensitivities and specificities were 67% (95% confidence interval (CI) 59–74%) and 99% (95% CI 97–99%), and 71% (95% CI 61–79%) and 99% (95% CI 98–100%), respectively. The hierarchical summary receiver operating characteristics (HSROCs) were 0.92 and 0.96, respectively. For NS1 combined with an anti-dengue-specific IgM test, the summarized sensitivity, specificity, and HSROC were 83% (95% CI 68–92%), 86% (95% CI 79–91%), and 0.91, respectively. The accuracy for serotypes was 50.0–90.9% for DENV-1, 38.5–85.7% for DENV-2, 46.7–91.3% for DENV-3, and 21.7–87.0% for DENV-4.
Conclusions
These results support the use of single NS1-based tests; they have good diagnostic utility for confirming dengue and for distinguishing serotypes DENV-1 and 3 from DENV-2 and 4, while they can be used as a screening tool when combined with an IgM test [Au?2]. Moreover, the Dengue NS1 Ag STRIP Kit appears to be the best for confirming and serotyping dengue infection.
Keywords: Dengue, NS1, Diagnostic accuracy, Sensitivity, Specificity
Introduction
Dengue is a vector-borne disease caused by dengue virus (DENV), occurring throughout tropical and subtropical areas. It has become one of the most serious public health problems due to the increasing morbidity.1 The World Health Organization (WHO) 2009 guidelines identify three diagnostic tests as gold standards for dengue diagnosis: viral isolation and identification, nucleotide detection, and serological tests for IgM or IgG seroconversion.2 However, these have limitations, such as requests for acute infection (0–5 days post-onset) samples, the time required for viral isolation and identification (more than 1 week), the possibility of false-positive or false-negative final results, and the need for further serum samples to confirm serological tests [Au?3].3
An affordable, time-saving, and convenient diagnostic test for confirming dengue infection is thus urgently needed. It was recently reported that serum or plasma DENV non-structural protein 1 (NS1) can be detected in the peripheral blood from 9 to 18 days after illness onset.4–7 Thus, the detection of NS1 may offer a larger window of opportunity for dengue diagnosis. NS1-capture based detection methods have been evaluated comprehensively. The two main methods for detecting dengue virus infection are currently ELISA (Panbio Dengue Early ELISA Kit, Dengue NS1 Ag ELISA Kit, and Platelia Dengue NS1 Ag-ELISA Kit) and immunochromatography (Dengue NS1 Ag STRIP Kit and SD BIOLINE Dengue Duo Strip Kit). Many of these have shown results comparable to those of the gold standard detection methods (antibody detection, nucleotide detection, or viral identification) [Au?4].
We conducted a meta-analysis to comprehensively assess the performance of NS1-based detection in the evaluation of dengue. Through this analysis, we provide evidence of the adequate sensitivities and specificities of NS1-based tests for the diagnosis of dengue in a large population [Au?5].
Materials and methods
Data sources and searches
A search was performed to identify published articles reporting studies of dengue diagnosis methods, including virus isolation and identification, RNA detection, serological tests for IgM or IgG seroconversion, and the NS1-based capture method. NCBI PubMed, ISI Web of Science, Google Scholar, and the Chinese National Knowledge Infrastructure (CNKI) databases were searched for studies published prior to October 1, 2012, using the following search terms: dengue, NS1 or non-structure 1, diagnosis. No language limitation was applied.
Selection criteria
For inclusion, studies had to meet the following criteria: (1) patients or samples with dengue infection confirmed by one of the three standard methods (viral isolation and identification, RNA detection, or serological tests for IgM and/or IgG seroconversion); (2) patients or samples also investigated by NS1-based capture method combined or not with an IgM test; (3) report of the data necessary to calculate the true positive, false positive, true negative, and false negative diagnostic results of NS1 for dengue diagnosis; (4) the inclusion of at least 50 samples from participants and a control group respectively, for good reliability; (5) a Quality Assessment for Diagnostic Accuracy Studies (QUADAS)8 score of <7. Studies with an overlapping patient sample were excluded; only the study with the larger number of patients was included.
In order to evaluate the accuracy of serotyping of dengue, the studies that reported dengue serotyping were also retrieved. These studies had to meet the following criteria: (1) the inclusion of information on the serotyping of dengue by NS1-based captured test, and also accompanied by an RT-PCR test; (2) no fewer than 100 participants.
Study selection and data extraction
The eligible studies were assessed independently by two of the authors and were verified reciprocally, with disagreements resolved in consultation with a third investigator. The data were extracted independently by two investigators. For each study, the following information was abstracted: author, study publication year, country, study design, study population, number of patients or samples, the standard methods used, and the NS1-based capture method used. True positive, false positive, true negative, and false negative results were extracted, allowing the calculation of sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for each reported test threshold. For the accuracy of serotyping analysis, the percentage, author, publication year, and total number of participants were extracted.
Quality assessment
The quality of the studies included in the meta-analysis was assessed using the QUADAS questionnaire. The QUADAS tool has 11 items that assess study design-related issues and the validity of the results of the study. Each item may be scored `yes' if reported (1 point), `no' if not reported, or `unclear' if there is no adequate information in the article to make an accurate judgment (0 points) [Au?6]. If the QUADAS score was <6 points, the study was classified as low methodological quality and was excluded.
Data synthesis and statistical analysis
The summarized sensitivity, specificity, and diagnostic odds ratio (OR) with corresponding 95% confidence interval (CI) were used to examine the accuracy of NS1 tests for dengue. The diagnostic OR represents how much larger the odds of dengue infection is for those with positive test results than the odds of dengue infection for those with negative test results. The diagnostic OR is calculated as follows: diagnostic OR = positive likelihood ratio/negative likelihood ratio. A hierarchical summarized receiver operating characteristic (HSROC) curve9 was also plotted to graphically present the results. The heterogeneity among studies was evaluated by computing Higgins' I2 and Q tests using the generic inverse variance method of meta-analysis of diagnostic OR. An I2 value of >50% or a p-value of <0.05 was considered substantial heterogeneity. If heterogeneity existed between primary studies, the random effect method was used for pooled analyses.
In addition, a meta-regression technique was used according to the following pre-defined characteristics, to explore source of heterogeneity in the studies: different kits, study design, publication year, sample size, and QUADAS score. A p-value of <0.05 was considered to be representative of statistical significance. The Deeks funnel plot asymmetry test was used to investigate publication bias.10
Pre-test probabilities of 25%, 50%, and 75% vs. corresponding post-test probabilities were evaluated. This was followed by a `positive' or `negative' NS1 result based on the summarized sensitivity and specificity using Fagan plot analysis, which showed the relationship between the prior probability specified, the likelihood ratio, and the posterior test probability (a ruler for interpreting diagnostic test results). Analyses were performed using STATA version 11.0 (Stata Corp., College Station, TX, USA) and the orders of Metandi11 and Midas.12
The percentage of correct serotype identification using NS1-based capture methods was calculated. The accuracy for each serotype was also calculated. The graphs were drawn using GraphPad Prism 5 and Photoshop software.
Results
Search results
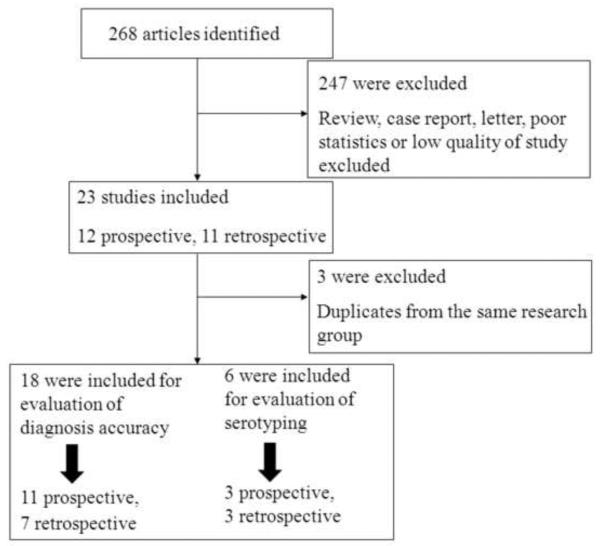
On the basis of the described search strategies, a total of 268 articles were retrieved. After eliminating the duplicates (n = 11) and the studies not related to the topic (n = 213), 44 potentially relevant studies were identified for further evaluation. A further 14 studies were excluded as it was not possible to extract the statistics, 10 were excluded for small sample sizes, and two were excluded for low quality. Thus, 18 studies in total met the inclusion criteria. A flow chart of the study selection process is shown in Figure 1. For the evaluation of accuracy of serotyping, another two studies were included.13,14
Figure 1.
Flow diagram of selection and disposition of studies.
Characteristics of the studies included in the analysis
A total of 12 313 patients and samples were included. Six commercially available NS1-based capture tests were used: seven studies referred to the use of Panbio Dengue Early ELISA (Inverness, Brisbane, Australia),15–21 three to the Dengue NS1 Ag ELISA Kit (Standard Diagnostic Inc., Kyonggi-do, South Korea),15,19,22 11 to Platelia Dengue NS1 Ag-ELISA ([Au?7]),15–19,23–28 nine to Dengue NS1 Ag STRIP Kit ([Au?7]),15–18,27,29–32 two to SD BIOLINE Dengue Duo Strip Kit ([Au?7]),15,32 and three to NS1-based capture combined with IgM test15,20,32 for dengue. According to the QUADAS scale, the studies included were of very good methodological quality. The main characteristics of the studies included in this meta-analysis are summarized in Tables 1–3.
Table 1.
Diagnostic accuracy results of individual studies using the ELISA method, including the commercial kits Panbio Dengue Early ELISA, Dengue NS1 Ag ELISA, and Platelia Dengue NS1 Ag-ELISA
| Author (year) | Geography | Design | Size | Confirmed testsa | TP | FP | FN | TN | Sensitivity % (95% CI) | Specificity % (95% CI) | PPV % | NPV % | QUADAS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ramirez et al. (2009)b | South America (Venezuela) | Retrospective | 147 | 1 | 53 | 2 | 34 | 58 | 61 (50–71) | 97 (98–100) | 96 | 63 | 8 |
| Watthanaworawit et al. (2010)b | Southeast Asia (Thailand/Myanmar) | Prospective | 152 | 3 | 39 | 0 | 33 | 90 | 54 (42–66) | 100 (96–100) | 100 | 73 | 10 |
| Osorio et al. (2010)b | Southeast Asia (Colombia) [Au?16] | Retrospective | 400 | 1 | 155 | 10 | 63 | 82 | 71 (65–77) | 89 (81–95) | 94 | 57 | 11 |
| Lima et al. (2010)b | South America (Brazil) | Prospective | 550 | 1 | 159 | 0 | 61 | 230 | 72 (66–78) | 100 (88–100) | 100 | 79 | 9 |
| Pok et al. (2010)b | Europe (Singapore) [Au?16] | Prospective | 209 | 2 | 73 | 0 | 36 | 100 | 67 (57–76) | 100 (96–100) | 100 | 74 | 8 |
| Fry et al. (2011)b | Southeast Asia (Vietnam) | Prospective | 298 | 4 | 137 | 4 | 61 | 96 | 69 (62–76) | 96 (90–99) | 97 | 61 | 9 |
| Blacksell et al. (2012)b | Southeast Asia (Thailand), Asia (Sri Lanka) | Retrospective | 387 | 2 | 107 | 12 | 132 | 136 | 45 (38–51) | 92 (86–96) | 90 | 51 | 10 |
| Osorio et al. (2010)c | Southeast Asia (Colombia) [Au?16] | Retrospective | 400 | 1 | 150 | 5 | 68 | 87 | 69 (62–75) | 95 (88–96) | 97 | 56 | 11 |
| Wang et al. (2010)c | Southeast Asia (Malaya) | Prospective | 244 | 1 | 142 | 1 | 43 | 58 | 77 (70–83) | 98 (91–100) | 99 | 57 | 9 |
| Blacksell et al. (2012)c | Southeast Asia(Thailand), Asia (Sri Lanka) | Retrospective | 387 | 2 | 132 | 2 | 107 | 146 | 55 (49–62) | 99 (95–100) | 99 | 58 | 10 |
| Kumarasamy et al. (2007)d | Southeast Asia (Malaysia) | Retrospective | 567 | 2 | 199 | 0 | 4 | 354 | 96 (95–99) | 100 (96–100) | 100 | 99 | 8 |
| Lapphra et al. (2008)d | Southeast Asia (Thailand) | Prospective | 235 | 1 | 108 | 1 | 63 | 63 | 63 (55–70) | 96 (92–100) | 99 | 50 | 11 |
| Ramirez et al. (2009)d | South America (Venezuela) | Retrospective | 143 | 1 | 62 | 5 | 25 | 55 | 71 (61–80) | 92 (82–97) | 93 | 69 | 8 |
| Phuong et al. (2009)d | Southeast Asia (Vietnam) | Prospective | 459 | 2 | 20 | 2 | 34 | 403 | 37 (24–52) | 100 (96–100) | 91 | 92 | 8 |
| Osorio et al. (2010)d | Southeast Asia (Colombia) [Au?16] | Retrospective | 400 | 1 | 150 | 7 | 62 | 84 | 71 (64–77) | 92 (85–97) | 96 | 56 | 11 |
| Lima et al. (2010)d | South America (Brazil) | Prospective | 550 | 1 | 184 | 3 | 36 | 227 | 84 (78–88) | 99 (85–97) | 98 | 86 | 10 |
| Pok et al. (2010)d | Europe (Singapore) [Au?16] | Prospective | 209 | 2 | 89 | 0 | 20 | 100 | 82 (73–88) | 100 (96–100) | 100 | 45 | 8 |
| Duong et al.(2011)d | Southeast Asia (Cambodia) | Prospective | 359 | 5 | 150 | 0 | 110 | 79 | 58 (51–64) | 100 (95–100) | 100 | 42 | 8 |
| Najioullah et al. (2011)d | North America (Martinique) | Prospective | 537 | 3 | 156 | 0 | 99 | 271 | 61 (55–67) | 100 (96–100) | 100 | 73 | 10 |
| Kassim et al. (2011)d | Southeast Asia (Malaysia) | Retrospective | 208 | 3 | 60 | 7 | 77 | 64 | 44 (35–53) | 90 (81–96) | 90 | 45 | 7 |
| Blacksell et al. (2012)d | Southeast Asia(Thailand), Asia (Sri Lanka) | Retrospective | 387 | 2 | 107 | 12 | 132 | 136 | 45 (38–51) | 92 (86–96) | 90 | 51 | 9 |
TP, true positive; FP, false positive; FN, false negative; TN, true negative; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; QUADAS, Quality Assessment for Diagnostic Accuracy Studies.
Confirmed tests: (1) serological, nucleotide, or viral culture test; (2) serological or nucleotide test; (3) serological test; (4) nucleotide test; (5) serological, NS1 detection, nucleotide, or viral culture test.
Statistics retrieved from studies using the Panbio Dengue Early ELISA Kit.
Statistics retrieved from studies using the Dengue NS1 Ag ELISA Kit.
Statistics retrieved from studies usins the Platelia Densue NS1 As-ELISA
Table 3.
Diagnostic accuracy results of individual studies using NS1-based capture combined with IgM test (dengue if one of NS1 or IgM was positive and non-dengue if both were negative)
| Author (year) | Geography | Design | Size | Confirmed testsa | TP | FP | FN | TN | Sensitivity % (95% CI) | Specificity % (95% CI) | PPV % | NPV % | QUADAS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Watthanaworawit et al. (2010) | Southeast Asia (Thailand/Myanmar) | Prospective | 162 | 3 | 43 | 11 | 29 | 79 | 60 (47–71) | 88 (79–94) | 80 | 73 | 10 |
| Osorio et al. (2010) | Southeast Asia (Colombia) [Au?16] | Retrospective | 400 | 1 | 171 | 8 | 47 | 84 | 78 (72–84) | 91 (84–96) | 96 | 64 | 11 |
| Blacksell et al. (2011) | Southeast Asia (Thailand), Asia (Sri Lanka) | Retrospective | 250 | 2 | 92 | 18 | 7 | 142 | 93 (86–97) | 89 (83–93) | 84 | 95 | 10 |
| Blacksell et al. (2011) | Southeast Asia (Thailand), Asia (Sri Lanka) | Retrospective | 250 | 2 | 89 | 40 | 10 | 120 | 90 (82–95) | 75 (68–81) | 70 | 92 | 10 |
TP, true positive; FP, false positive; FN, false negative; TN, true negative; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; QUADAS, Quality Assessment for Diagnostic Accuracy Studies.
Confirmed tests: (1) serological, nucleotide, or viral culture test; (2) serological or nucleotide test; (3) serological test.
Diagnostic accuracy of the ELISA method, including Panbio Dengue Early ELISA Kit, Dengue NS1 Ag ELISA Kit, and Platelia Dengue NS1 Ag-ELISA Kit
Fourteen studies evaluated the diagnostic accuracy of the commercial kits Panbio Dengue Early ELISA, Dengue NS1 Ag ELISA Kit, and Platelia Dengue NS1 Ag-ELISA Kit for dengue (Table 1). The sensitivity and specificity of diagnosis for dengue ranged from 37% (95% CI 24–52%) to 96% (95% CI 95–99%) and 89% (95% CI 81–95%) to 100% (95% CI 95–100%), respectively. The summarized sensitivity and specificity were 67% (95% CI 59–74%) and 99% (98% CI 97–99%), respectively. The summarized diagnostic OR was 148 (95% CI 51–492) and HSROC was 0.92 (0.89–0.94) (Figure 2A). On the basis of these values, PPV and NPV were 0.98 (95% CI 0.96–0.99) and 0.73 (95% CI 0.72–0.74), respectively. There was statistically significant heterogeneity in diagnostic OR (Q = 55.47, p < 0.001, I2 = 96%). According to the meta-regression analysis, the accuracy of these two kits [Au?8] for detecting dengue was affected by study design, while the result was not changed (data not shown). Publication bias did not exist among these studies (p = 0.06).
Figure 2.
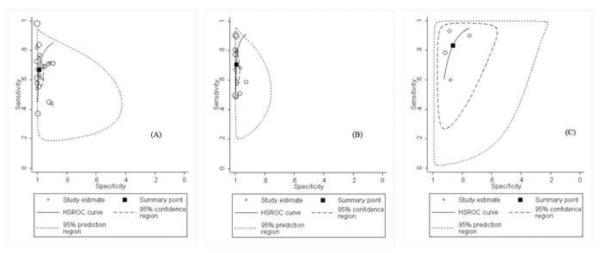
Hierarchical summarized receiver operating characteristic (HSROC) curves of NS1-based capture tests for dengue using: (A) the method of ELISA for dengue; (B) the method of immunochromatography for dengue; (C) NS1-based capture combined with IgM test for dengue. HSROCs for dengue were 0.92, 0.96, and 0.91, respectively.
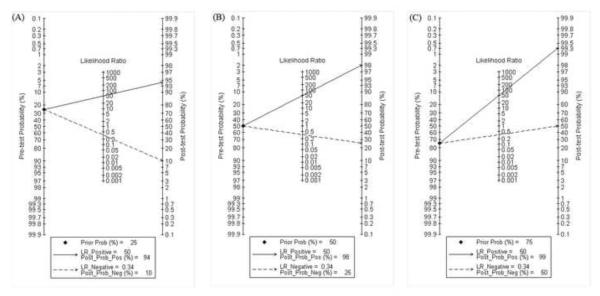
Fagan plot analysis demonstrated that the ELISA method was very informative, with 94% probability of correctly detecting dengue following a `positive' measurement when the pre-test probability was 25% and lowering the probability of disease to as low as 10% with a `negative' measurement. This diagnosis would be wrong in 25% and 50% of patients with a `negative' measurement when the pre-test probability was 50% and 75%, respectively, although the probability of a correct diagnosis following a `positive' measurement equaled 98% and 99% for dengue, respectively (Table 4, Figure 3).
Table 4.
Summarized accuracy of NS1-based capture tests for dengue
| NS1 detection method | Studies, n | Sensitivity % (95% CI) | Specificity % (95% CI) | Diagnostic OR (95% CI) | PPV (95% CI) | NPV (95% CI) | HSROC [Au?17] | I2 a | p-Valuea | Pre-test probability (%) | Post-test probability (+) (%) | Post-test probability (−) (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ELISA | 14 | 67 (59–74) | 99 (97–99) | 148 (51– 429) |
0.98 (0.96– 0.99) |
0.73 (0.72– 0.74) |
0.92 (0.89– 0.94) |
96% | 0.001 | 25 | 94 | 10 |
| 50 | 98 | 25 | ||||||||||
| 75 | 99 | 50 | ||||||||||
| Panbio Dengue Early ELISA Kit |
7 | 63 (56–70) | 99 (93–100) | 127 (20– 804) |
0.98 (96– 1.00) |
0.71 (0.70– 0.73) |
0.80 (0.76– 0.83) |
89% | 0.001 | 25 | 94 | 11 |
| 50 | 98 | 27 | ||||||||||
| 75 | 99 | 53 | ||||||||||
| Platelia Dengue NS1 Ag-ELISA |
11 | 69 (54–81) | 99 (96–100) | 228 (36– 1456) |
0.98 (0.97– 1.00) |
0.75 (0.74– 0.76) |
0.95(0.92– 0.96) |
94% | 0.001 | 25 | 96 | 9 |
| 50 | 99 | 24 | ||||||||||
| 75 | 100 | 48 | ||||||||||
| Immunochromatography | 9 | 71 (61–79) | 99 (98–100) | 328 (103– 1046) |
0.99 (0.98– 1.00) |
0.75 (0.75– 0.76) |
0.96(0.95– 0.98) |
88% | 0.001 | 25 | 97 | 9 |
| 50 | 99 | 23 | ||||||||||
| 75 | 100 | 52 | ||||||||||
| Dengue NS1 Ag STRIP Kit |
8 | 71 (64–82) | 99 (98–100) | 486 (124– 1902) |
0.99 (0.98– 1.00) |
0.78 (0.77– 0.78) |
0.96 (0.94– 0.98) |
88% | 0.001 | 25 | 98 | 8 |
| 50 | 99 | 21 | ||||||||||
| 75 | 100 | 44 | ||||||||||
| NS1-based capture combined with IgM test |
3 | 83 (68–92) | 86 (79–91) | 31 (14–70) | 0.84 (0.80– 0.89) |
0.82 (0.77– 0.87) |
0.91 (0.89– 0.93) |
91% | 0.001 | 25 | 67 | 6 |
| 50 | 86 | 16 | ||||||||||
| 75 | 95 | 37 |
CI, confidence interval; OR, odds ratio; PPV, positive predictive value; NPV, negative predictive value; HSROC, hierarchical summary receiver operating characteristic.
An I2 value of more than 50% or a p-value of 0.10 was considered substantial heterogeneity.
Figure 3.
Fagan plot analysis to evaluate the clinical utility of the ELISA method. (A) Pre-test probability = 25%; with a pre-test probability of dengue of 25%, the post-test probabilities of dengue, given positive and negative results (post-positive and post-negative probability) were 94% and 10%, respectively. (B) Pre-test probability = 50%; with a pre-test probability of dengue of 50%, the post-test probabilities of dengue, given positive and negative results (post-positive and post-negative probability) were 98% and 25%, respectively. (C) Pre-test probability = 75%; with a pre-test probability of dengue of 75%, the post-test probabilities of dengue, given positive and negative results (post-positive and post-negative probability) were 99% and 50%, respectively. The Fagan plot consists of a vertical axis on the left with the pre-test probability, an axis in the middle representing the likelihood ratio, and a vertical axis on the right representing the post-test probability (LR Negative, negative likelihood ratio; LR Positive, positive likelihood ratio).
Seven studies evaluated the diagnostic accuracy of the commercial kit Panbio Dengue Early ELISA (Table 1 and Table 4) for dengue. The summarized sensitivity and specificity were 67% (95% CI 59–74%) and 99% (98% CI 97–99%), respectively. The summarized diagnostic OR was 127 (20–804) and HSROC was 0.80 (0.76–0.83). On the basis of these values, PPV and NPV were 0.98 (95% CI 0.98–1.00) and 0.71 (95% CI 0.76–0.83), respectively. Fagan plot analysis demonstrated that this method was very informative, with 94% probability of correctly detecting dengue following a `positive' measurement when the pre-test probability was 25% and lowering the probability of disease to as low as 11% with a `negative' measurement. This diagnosis would be wrong in 27% and 53% of patients with a `negative' measurement when the pre-test probability was 50% and 75%, respectively, although the probability of a correct diagnosis following a `positive' measurement equaled 98% and 99% for dengue, respectively (Table 4).
Eleven studies evaluated the diagnostic accuracy of the commercial kit Platelia Dengue NS1 Ag-ELISA (Table 1 and Table 4) for dengue. The summarized sensitivity and specificity were 66% (95% CI 54–81%) and 99% (95% CI 96–100%), respectively. The summarized diagnostic OR was 228 (36–1456) and HSROC was 0.95 (0.92–0.96). On the basis of these values, PPV and NPV were 0.98 (95% CI 0.97–1.00) and 0.75 (95% CI 0.74–0.76), respectively. Fagan plot analysis demonstrated that this method was very informative, with 94% probability of correctly detecting dengue following a `positive' measurement when the pre-test probability was 25% and lowering the probability of disease to as low as 9% with a `negative' measurement. This diagnosis would be wrong in 24% and 48% of patients with a `negative' measurement when the pre-test probability was 50% and 75%, respectively, although the probability of a correct diagnosis following a `positive' measurement equaled 98% and 99% for dengue, respectively (Table 4).
Diagnostic accuracy of the immunochromatography method, including Dengue NS1 Ag STRIP Kit and SD BIOLINE Dengue Duo Strip Kit for dengue
Nine studies evaluated the diagnostic accuracy of the commercial kits Dengue NS1 Ag STRIP Kit and SD BIOLINE Dengue Duo Strip Kit for dengue (Table 2). The sensitivity and specificity of diagnosis for dengue ranged from 48% (95% CI 38–59%) to 90% (95% CI 87–93%) and 93% (95% CI 87–96%) to 100% (95% CI 93–100%), respectively. The summarized sensitivity and specificity were 71% (95% CI 61–79%) and 99% (95% CI 98–100%), respectively. The summarized diagnostic OR was 328 (95% CI 103–1046) and HSROC was 0.96 (0.95–0.98) (Figure 2B). On the basis of these values, PPV and NPV were 0.99 (95% CI 0.98–1.00) and 0.75 (95% CI 0.75–0.76), respectively. There was statistically significant heterogeneity in diagnostic OR (Q = 17.05, p < 0.001, I2 = 88%). However, according to the meta-regression analysis, the accuracy of these two kits for detecting dengue was affected by study design and publication year, while the result was not changed (data not shown). Publication bias existed among these studies (p = 0.02).
Table 2.
Diagnostic accuracy results of individual studies using the immunochromatography method, including the commercial kit Dengue NS1 Ag STRIP Kit and SD BIOLINE Dengue Duo Strip Kit
| Author (year) | Geography | Design | Size (time) | Confirmed testsa | TP | FP | FN | TN | Sensitivity % (95% CI) | Specificity % (95% CI) | PPV % | NPV % | QUADAS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shu et al. (2009)b | Asia | Retrospective | 162 (15/30 min) | 4 | 82 | 0 | 40 | 50 | 67 (58–75) | 100 (93–100) | 100 | 56 | 7 |
| Ramirez et al. (2009)b | South America (Venezuela) | Retrospective | 147 (30 min) | 1 | 59 | 2 | 28 | 58 | 68 (57–77) | 97 (88–100) | 97 | 67 | 8 |
| Zainah et al. (2009)b | Southeast Asia (Malaya) | Retrospective | 533 (15/30 min) | 1 | 284 | 1 | 30 | 218 | 49 (43–56) | 100 (97–100) | 100 | 88 | 9 |
| Osorio et al. (2010)c | Southeast Asia (Colombia) [Au?16] | Retrospective | 400 (NR) | 1 | 111 | 3 | 107 | 89 | 51 (44–58) | 97 (91–99) | 97 | 45 | 11 |
| Lima et al. (2010)b | South America (Brazil) | Prospective | 550 (15/30 min) | 1 | 197 | 2 | 23 | 228 | 90 (85–93) | 99 (97–100) | 99 | 91 | 10 |
| Pok et al. (2010)b | Europe (Singapore) [Au?16] | Prospective | 209 (NR) | 2 | 86 | 1 | 23 | 99 | 79 (70–86) | 99 (95–100) | 99 | 81 | 8 |
| Chaterji et al. (2011)b | Europe (Singapore) [Au?16] | Prospective | 354 (15 min) | 3 | 119 | 0 | 35 | 200 | 77 (70–86) | 100 (98–100) | 100 | 85 | 10 |
| Chaterji et al. (2011)b | Europe (Singapore) [Au?16] | Prospective | 354 (30 min) | 3 | 124 | 0 | 30 | 200 | 77 (70–84) | 100 (98–100 | 100 | 87 | 10 |
| Najioullah et al. (2011)b | North America (Martinique) | Prospective | 537 (30 min) | 4 | 125 | 0 | 128 | 272 | 81 (73–86) | 100 (99–100) | 100 | 68 | 10 |
| Blacksell et al. (2011)b | Asia (Sri Lanka) | Prospective | 259 (NR) | 2 | 58 | 2 | 41 | 158 | 90 (87–93) | 99 (96–100) | 97 | 79 | 10 |
| Blacksell et al. (2011)b | Asia (Sri Lanka) | Prospective | 259 (NR) | 2 | 58 | 12 | 41 | 148 | 59 (48–68) | 93 (87–96) | 83 | 78 | 10 |
| Blacksell et al. (2011)c | Asia (Sri Lanka) | Prospective | 259 (NR) | 2 | 48 | 1 | 51 | 159 | 48 (38–59) | 99 (97–100) | 98 | 76 | 10 |
TP, true positive; FP, false positive; FN, false negative; TN, true negative; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; QUADAS, Quality Assessment for Diagnostic Accuracy Studies; NR, not reported.
Confirmed tests: (1) serological, nucleotide, or viral culture test; (2) serological or nucleotide test; (3) viral culture or nucleotide test; (4) nucleotide test.
Statistics retrieved from studies using the Dengue NS1 Ag STRIP Kit.
Statistics retrieved from studies using the SD BIOLINE Dengue Duo Strip Kit.
Fagan plot analysis demonstrated that the method of immunochromatography was very informative, with 97% probability of correctly detecting dengue following a `positive' measurement when the pre-test probability was 25% and lowering the probability of disease to as low as 9% with a `negative' measurement. This diagnosis would be wrong in 23% and 52% of patients with a `negative' measurement when the pre-test probability was 50% and 75%, respectively, although the probability of a correct diagnosis following a `positive' measurement equaled 99% and 100% for dengue, respectively (Table 4, Figure 4).
Figure 4.
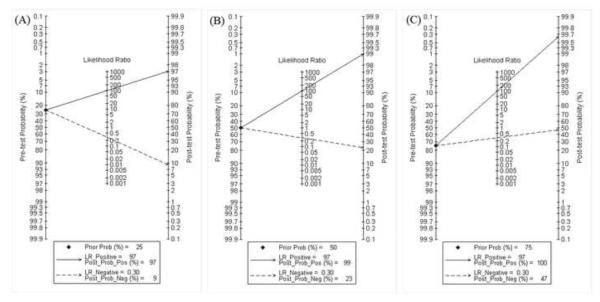
Fagan plot analysis to evaluate the clinical utility of the immunochromatography method. (A) Pre-test probability = 25%. (B) Pre-test probability = 50%. (C) Pre-test probability = 75%. [Au?18]
Eight studies evaluated the diagnostic accuracy of the commercial kit Dengue NS1 Ag STRIP Kit for dengue (Table 2 and Table 4). The summarized sensitivity and specificity were 71% (95% CI 64–82%) and 99% (95% CI 98–100%), respectively. The summarized diagnostic OR was 486 (124–1902) and HSROC was 0.96 (0.94–0.98). On the basis of these values, PPV and NPV were 0.99 (95% CI 0.98–1.00) and 0.78 (95% CI 0.77–0.78), respectively. Fagan plot analysis demonstrated that this method was very informative, with 94% probability of correctly detecting dengue following a `positive' measurement when the pre-test probability was 25% and lowering the probability of disease to as low as 8% with a `negative' measurement. This diagnosis would be wrong in 21% and 44% of patients with a `negative' measurement when the pre-test probability was 50% and 75%, respectively, although the probability of a correct diagnosis following a `positive' measurement equaled 98% and 99% for dengue, respectively (Table 4).
Diagnostic accuracy of a commercial NS1-based capture kit combined with IgM test for dengue
Three studies evaluated the diagnostic accuracy of NS1-based capture combined with IgM test for dengue (Table 3). The sensitivity and specificity of diagnosis for dengue ranged from 60% (95% CI 47–71%) to 93% (95% CI 86–97%) and 75% (95% CI 68–81%) to 91% (95% CI 84–96%), respectively. The summarized sensitivity and specificity were 83% (95% CI 68–92%) and 86% (95% CI 79–91%), respectively. The summarized diagnostic OR was 31 (95% CI 14–70) and HSROC was 0.91 (0.89–0.93) (Figure 2C). On the basis of these values, PPV and NPV were 0.84 (95% CI 0.80–0.89) and 0.82 (95% CI 0.77–0.87), respectively. There was statistically significant heterogeneity in diagnostic OR (Q = 23.12, p < 0.001, I2 = 91%). However, according to the meta-regression analysis, the accuracy of this method for detecting dengue was affected by publication year. As a result of only three studies being included, the subgroup could not be analyzed. No publication bias existed among these studies (p = 0.29).
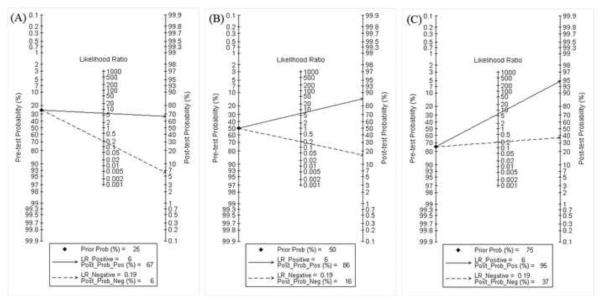
Fagan plot analysis demonstrated that this method was very informative, with 67% probability of correctly detecting dengue following a `positive' measurement when the pre-test probability was 25% and lowering the probability of disease to as low as 6% with a `negative' measurement. This diagnosis would be wrong in 16% and 37% of patients with a `negative' measurement when the pre-test probability was 50% and 75%, respectively, although the probability of a correct diagnosis following a `positive' measurement equaled 86% and 95% for dengue, respectively (Table 4, Figure 5).
Figure 5.
Total accuracy of typing of dengue using NS1-based captured tests. (A) Pre-test probability = 25%. (B) Pre-test probability = 50%. (C) Pre-test probability = 75%. [Au?19]
Accuracy of serotyping for dengue by NS1-based capture tests
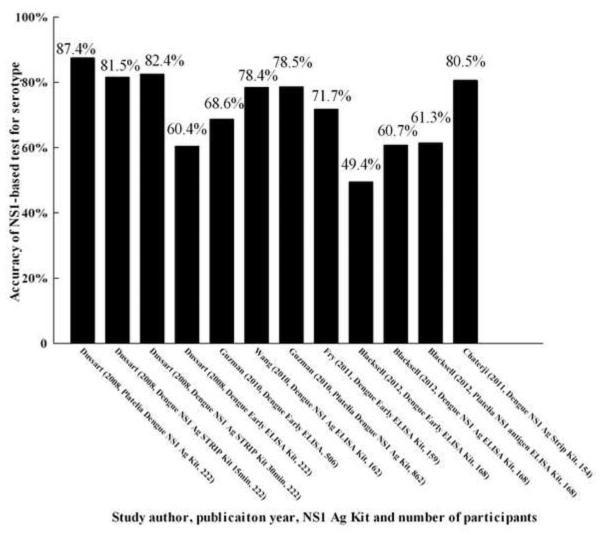
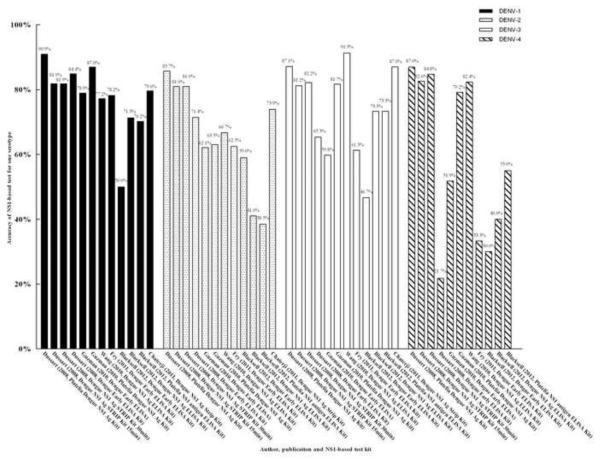
Six studies evaluated the accuracy of serotyping for dengue by NS1-based capture test.13,14,19,21,22,31 The accuracy of total serotyping for dengue ranged from 49.4% to 87.4% (Figure 5). The highest accuracy for serotyping was found in two studies: 81.5%, 82.4%, and 80.5% by Dengue NS1 Ag STRIP Kit [Au?9].13,31 The percentage correct for DENV-1 was 50.0–90.9%, for DENV-2 was 38.5–85.7%, for DENV-3 was 46.7–91.3%, and for DENV-4 was 21.7–87.0% (Figure 6). The highest correct percentages for DENV-1 (81.9% and 79.6%), DENV-2 (81.0% and 73.9%), DENV-3 (81.2%, 82.2%, and 87.0%), and DENV-4 (82.6% and 84.8%) found in the above studies were also acquired using the Dengue NS1 Ag STRIP Kit [Au?10].
Figure 6.
Overall accuracy of NS1-based tests for the diagnosis dengue infection. [Au?20]
Discussion
The dramatic increase in the global dengue burden has promoted social interest in improving dengue diagnosis. The most common methods for confirming dengue in the laboratory are currently serology, viral isolation, and nucleotide detection; these tests have their own limitations. Soluble NS1 detected in the serum or plasma of a DENV-infected patient may represent recent infection. Although, DENV NS1 antigen capture ELISAs have been reported to be promising tools for the diagnosis of acute dengue infections,22,33–35 a recent study showed that NS1 tests were of low quality for the diagnosis of dengue infection.36 Thus, in this meta-analysis, we evaluated the performance of NS1-based captured tests in dengue diagnosis.
Our results indicate that NS1-based capture tests have a high accuracy for dengue detection. The HSROC for dengue diagnosis using these tests ranged from 0.80 (95% CI 0.76–0.83) to 0.96 (95% CI 0.95–0.98). The summarized specificity (0.99, 95% CI 0.93–1.00, and 0.99, 95% CI 0.96–1.00 [Au?11]) and PPV (0.99, 95% CI 0.97–0.99, and 0.99, 95% CI 0.98–1.00 [Au?11]) were extremely high. Fagan plot analysis showed that these could be used to diagnose dengue (when the pre-test probability is 25%), with 94%, 94%, 96%, 97%, and 98% probability of correctly diagnosing dengue following a `positive' measurement. When the pre-test probability is 50% or 75%, the post-test probability was found to remain satisfactory for diagnosing dengue. Furthermore, a `negative' measurement was also informative, as dengue was present in only 10%, 11%, 9%, 9%, and 8%. In this respect, the NS1-based capture method is reliable, promising, and worth using in clinical practice to confirm dengue infection. This test could be integrated into the method for dengue detection.
Although the summarized specificity for dengue was 99%, the summarized sensitivity was relatively low: 67% (95% CI 59–74%), 63% (56–70%), 69% (54–81%), 71% (95% CI 61–79%), and 71% (64–82%). At this point, NS1-based capture tests may not be sufficient to detect dengue alone. Nevertheless, taking into account the good performance of the NS1-based capture test, it could be a useful tool for diagnosing and treating patients with dengue.
According to the analysis of the different kits (Panbio Dengue Early ELISA Kit, Platelia Dengue NS1 Ag-ELISA, and Dengue NS1 Ag STRIP Kit), it appears that the Dengue NS1 Ag STRIP Kit is the best method for confirming dengue due to its higher summarized sensitivity, specificity, PPV, NPV, HSROC, etc. (Table 4). This is followed by Platelia Dengue NS1 Ag-ELISA and then the Panbio Dengue Early ELISA Kit. This might mean that the Dengue NS1 Ag STRIP Kit is the most efficient in confirming dengue infections by capturing NS1 antigen in the serum or plasma of the infected patients. Moreover, this kit is more convenient to use as the results can be achieved in 30 min at the most. It is also easy to perform without the need for special laboratory equipment. Of note, these kits gave different accuracies for confirming dengue infection. This phenomenon is caused by the samples being taken late after illness onset, patients with secondary infections, and those with different serotype infections, all of which will influence the accuracy of NS1-based diagnostic tools.15,37
To improve the sensitivity of single NS1-based tests, NS1-based capture combined with an IgM test was also evaluated. The summarized sensitivity and NPV were actually increased to 0.83 (95% CI 0.68–0.92) and 0.82 (95% CI 0.77–0.87), much higher than results using single NS1-based capture methods; however the summarized specificity was decreased to 86% (95% CI 79–91%), approximately 10% lower than that of single NS1 tests. The reason for this decrease in specificity is that a positive IgM result in a single sample does not confirm dengue,15 which means false positives with the IgM test. The HSROC for dengue diagnosis with the combined detection was almost the same as with the single NS1 detection method (0.92 and 0.96 vs. 0.91). However, the post-test probability in the combined detection was not better than that in the single test method according to the same pre-test probability (data in Table 4). So, the single NS1-based capture method could be used to confirm dengue infection, while the combined test could be used in the screening of dengue infection.
The results of this meta-analysis are in accordance with those of most of the previous studies which have shown the single NS1-based test to be suitable for diagnosing dengue infection, while the sensitivity is increased in combination with the IgM test.15,17–24,28,31,32 Although the result of the combined test was better than the single NS1-based test, the diagnosis of dengue still needs to be improved in terms of sensitivity and other indicators. A study has reported the use of a recombinant non-structural protein 3 (rNS3) from all serotypes of dengue virus in the diagnosis of dengue, and the results showed excellent agreement with commercial kits and IgM/IgG respectively.38 So, an NS1 antigen combined with rNS3 protein test might provide better results in the future.
Because of the wide range of total accuracy of serotyping for dengue, the NS1-based capture test is not a stable method for serotyping. However, this method is suitable to distinguish the two serotypes DENV-1 and DENV-3 from the other serotypes based on the accuracies for DENV-1, DENV-2, DENV-3, and DENV-4. Due to the wide range of percentages for DENV-2 and DENV-4, this method is not applicable to distinguish these two serotypes (DENV-2 and DENV-4) from other serotypes. Hence the total accuracy of serotyping for dengue by this method is probably affected by the poor accuracy with regard to DENV-2 and DENV-4. However, for the Dengue NS1 Ag STRIP Kit, the overall accuracy for serotyping dengue infection was approximately 80%, and this did not vary as much as with the other kits (Figures 6 and 7) [Au?12]. Thus, the Dengue NS1 Ag STRIP Kit may be the better test for serotyping dengue.
Figure 7.
Accuracy of serotyping dengue infection. [Au?20]
As significant heterogeneity and publication bias were present in the analysis, caution must be taken when interpreting the results. In addition, our study explored factors that may be the source of heterogeneity using meta-regression analysis. Although three specific covariates were examined, the study design and publication year were definitely the source of heterogeneity. However the results did not change.
Some limitations of this study should be taken into consideration. First, the standard methods were serological test, viral culture, and nucleotide detection, but not all the studies used the same means to detect samples. Second, only four studies evaluated the performance of NS1-based capture combined with the IgM test, limiting the robustness of the conclusions that can be drawn. Third, as a result of heterogeneity and publication bias, the results were based on a random effect model so that the strength of the evidence became weak.
In conclusion, our meta-analysis suggests that the single NS1-based capture method could be used as a good dengue diagnosis method with a high summarized specificity, and if combined with an IgM test with a high sensitivity, it could be used as a screening method. The NS1-based capture tests can be applied to distinguish DENV-1 and DENV-3 from other serotypes. Moreover, the Dengue NS1 Ag STRIP Kit may be the best kit for confirming and serotyping dengue infection. Large-scale, international, multicenter prospective studies are needed to further evaluate the potential role of NS1-based capture methods in the diagnosis of dengue.
Acknowledgement
This work was supported by grants from the National Science Foundation of China (30771899) to Y.P. Zhou, NIH (AI083202-02) to X.G. Chen, and SMU Crossing Sciences to X.G. Chen and Y.P. Zhou.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare that they have no competing interests.
References
- 1.Rigau-Pérez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, Vorndam AV. Dengue and dengue haemorrhagic fever. Lancet. 1998;352:971–7. doi: 10.1016/s0140-6736(97)12483-7. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . Dengue: guidelines for diagnosis, treatment, prevention and control. WHO; Geneva: 2009. [PubMed] [Google Scholar]
- 3.Peeling RW, Artsob H, Pelegrino JL, Buchy P, Cardosa MJ, Devi S, et al. Evaluation of diagnostic tests: dengue. Nat Rev Microbiol. 2010;8:S30–8. doi: 10.1038/nrmicro2459. [DOI] [PubMed] [Google Scholar]
- 4.Young PR, Hilditch PA, Bletchly C, Halloran W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J Clin Microbiol. 2000;38:1053–7. doi: 10.1128/jcm.38.3.1053-1057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alcon S, Talarmin A, Debruyne M, Falconar A, Deubel V, Flamand M. Enzyme-linked immunosorbent assay specific to dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J Clin Microbiol. 2002;40:376–81. doi: 10.1128/JCM.40.2.376-381.2002. [Au?13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dussart P, Labeau B, Lagathu G, Louis P, Nunes MR, Rodrigues SG, et al. Evaluation of an enzyme immunoassay for detection of dengue virus NS1 antigen in human serum. Clin Vaccine Immunol. 2006;13:1185–9. doi: 10.1128/CVI.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H, Di B, Pan YX, Qiu LW, Wang YD, Hao W, et al. Serotype 1-specific monoclonal antibody-based antigen capture immunoassay for detection of circulating nonstructural protein NS1: implications for early diagnosis and serotyping of dengue virus infections. J Clin Microbiol. 2006;44:2872–8. doi: 10.1128/JCM.00777-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smidt N, Deeks JM. Guide to the contents of a Cochrane review and protocol for diagnostic test accuracy. The Cochrane Collaboration. 2008 [Google Scholar]
- 9.Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med. 2001;20:2865–84. doi: 10.1002/sim.942. [DOI] [PubMed] [Google Scholar]
- 10.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–93. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Harbord R. Metandi: Stata module for meta-analysis of diagnostic accuracy. 2008. Au?14. [Google Scholar]
- 12.Dwamena B. Midas: a program for meta-analytical integration of diagnostic accuracy studies in Stata. 2007. Au?14. [Google Scholar]
- 13.Dussart P, Petit L, Labeau B, Bremand L, Leduc A, Moua D, et al. Evaluation of two new commercial tests for the diagnosis of acute dengue virus infection using NS1 antigen detection in human serum. PLoS Negl Trop Dis. 2008;2:e280. doi: 10.1371/journal.pntd.0000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzman MG, Jaenisch T, Gaczkowski R, Ty HV, Sekaran SD, Kroeger A, et al. Multi-country evaluation of the sensitivity and specificity of two commercially-available NS1 ELISA assays for dengue diagnosis. PLoS Negl Trop Dis. 2010;4 doi: 10.1371/journal.pntd.0000811. Au?15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osorio L, Ramirez M, Bonelo A, Villar LA, Parra B. Comparison of the diagnostic accuracy of commercial NS1-based diagnostic tests for early dengue infection. Virol J. 2010;7:361. doi: 10.1186/1743-422X-7-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lima MR, Nogueira RM, Schatzmayr HG, Dos SF. Comparison of three commercially available dengue NS1 antigen capture assays for acute diagnosis of dengue in Brazil. PLoS Negl Trop Dis. 2010;4:e738. doi: 10.1371/journal.pntd.0000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez AH, Moros Z, Comach G, Zambrano J, Bravo L, Pinto B, et al. Evaluation of dengue NS1 antigen detection tests with acute sera from patients infected with dengue virus in Venezuela. Diagn Microbiol Infect Dis. 2009;65:247–53. doi: 10.1016/j.diagmicrobio.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 18.Pok KY, Lai YL, Sng J, Ng LC. Evaluation of nonstructural 1 antigen assays for the diagnosis and surveillance of dengue in Singapore. Vector Borne Zoonotic Dis. 2010;10:1009–16. doi: 10.1089/vbz.2008.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blacksell SD, Jarman RG, Gibbons RV, Tanganuchitcharnchai A, Mammen MJ, Nisalak A, et al. Comparison of seven commercial antigen and antibody enzyme-linked immunosorbent assays for detection of acute dengue infection. Clin Vaccine Immunol. 2012;19:804–10. doi: 10.1128/CVI.05717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watthanaworawit W, Turner P, Turner CL, Tanganuchitcharnchai A, Jarman RG, Blacksell SD, Nosten FH. A prospective evaluation of diagnostic methodologies for the acute diagnosis of dengue virus infection on the Thailand–Myanmar border. Trans R Soc Trop Med Hyg. 2011;105:32–7. doi: 10.1016/j.trstmh.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fry SR, Meyer M, Semple MG, Simmons CP, Sekaran SD, Huang JX, et al. The diagnostic sensitivity of dengue rapid test assays is significantly enhanced by using a combined antigen and antibody testing approach. PLoS Negl Trop Dis. 2011;5:e1199. doi: 10.1371/journal.pntd.0001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang SM, Sekaran SD. Evaluation of a commercial SD dengue virus NS1 antigen capture enzyme-linked immunosorbent assay kit for early diagnosis of dengue virus infection. J Clin Microbiol. 2010;48:2793–7. doi: 10.1128/JCM.02142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duong V, Ly S, Lorn TP, Tuiskunen A, Ong S, Chroeung N, et al. Clinical and virological factors influencing the performance of a NS1 antigen-capture assay and potential use as a marker of dengue disease severity. PLoS Negl Trop Dis. 2011;5:e1244. doi: 10.1371/journal.pntd.0001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phuong HL, Thai KT, Nga TT, Giao PT, Hung LQ, Binh TQ, et al. Detection of dengue nonstructural 1 (NS1) protein in Vietnamese patients with fever. Diagn Microbiol Infect Dis. 2009;63:372–8. doi: 10.1016/j.diagmicrobio.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Kumarasamy V, Wahab AH, Chua SK, Hassan Z, Chem YK, Mohamad M, Chua KB. Evaluation of a commercial dengue NS1 antigen-capture ELISA for laboratory diagnosis of acute dengue virus infection. J Virol Methods. 2007;140:75–9. doi: 10.1016/j.jviromet.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Lapphra K, Sangcharaswichai A, Chokephaibulkit K, Tiengrim S, Piriyakarnsakul W, Chakorn T, et al. Evaluation of an NS1 antigen detection for diagnosis of acute dengue infection in patients with acute febrile illness. Diagn Microbiol Infect Dis. 2008;60:387–91. doi: 10.1016/j.diagmicrobio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Najioullah F, Combet E, Paturel L, Martial J, Koulmann L, Thomas L, et al. Prospective evaluation of nonstructural 1 enzyme-linked immunosorbent assay and rapid immunochromatographic tests to detect dengue virus in patients with acute febrile illness. Diagn Microbiol Infect Dis. 2011;69:172–8. doi: 10.1016/j.diagmicrobio.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 28.Kassim FM, Izati MN, TgRogayah TA, Apandi YM, Saat Z. Use of dengue NS1 antigen for early diagnosis of dengue virus infection. Southeast Asian J Trop Med Public Health. 2011;42:562–9. [PubMed] [Google Scholar]
- 29.Shu PY, Yang CF, Kao JF, Su CL, Chang SF, Lin CC, et al. Application of the dengue virus NS1 antigen rapid test for on-site detection of imported dengue cases at airports. Clin Vaccine Immunol. 2009;16:589–91. doi: 10.1128/CVI.00475-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zainah S, Wahab AH, Mariam M, Fauziah MK, Khairul AH, Roslina I, et al. Performance of a commercial rapid dengue NS1 antigen immunochromatography test with reference to dengue NS1 antigen-capture ELISA. J Virol Methods. 2009;155:157–60. doi: 10.1016/j.jviromet.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 31.Chaterji S, Allen JJ, Chow A, Leo YS, Ooi EE. Evaluation of the NS1 rapid test and the WHO dengue classification schemes for use as bedside diagnosis of acute dengue fever in adults. Am J Trop Med Hyg. 2011;84:224–8. doi: 10.4269/ajtmh.2011.10-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blacksell SD, Jarman RG, Bailey MS, Tanganuchitcharnchai A, Jenjaroen K, Gibbons RV, et al. Evaluation of six commercial point-of-care tests for diagnosis of acute dengue infections: the need for combining NS1 antigen and IgM/IgG antibody detection to achieve acceptable levels of accuracy. Clin Vaccine Immunol. 2011;18:2095–101. doi: 10.1128/CVI.05285-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hang VT, Nguyet NM, Trung DT, Tricou V, Yoksan S, Dung NM, et al. Diagnostic accuracy of NS1 ELISA and lateral flow rapid tests for dengue sensitivity, specificity and relationship to viraemia and antibody responses. PLoS Negl Trop Dis. 2009;3:e360. doi: 10.1371/journal.pntd.0000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh CJ, Chen MJ. The commercial dengue NS1 antigen-capture ELISA may be superior to IgM detection, virus isolation and RT-PCR for rapid laboratory diagnosis of acute dengue infection based on a single serum sample. J Clin Virol. 2009;44:102. doi: 10.1016/j.jcv.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Alcon S, Talarmin A, Debruyne M, Falconar A, Deubel V, Flamand M. Enzyme-linked immunosorbent assay specific to dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J Clin Microbiol. 2002;40:376–81. doi: 10.1128/JCM.40.2.376-381.2002. Au?13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felix AC, Romano CM, Centrone CD, Rodrigues CL, Villas-Boas L, Araujo ES, et al. Low sensitivity of NS1 tests evidenced during a dengue 2 outbreak in Santos, Brazil, 2010. Clin Vaccine Immunol. 2012;19:1972–6. doi: 10.1128/CVI.00535-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang CH, Kuo LL, Yang KD, Lin PS, Lu PL, Lin CC, et al. Laboratory diagnostics of dengue fever: an emphasis on the role of commercial dengue virus nonstructural protein 1 antigen rapid test. J Microbiol Immunol Infect. 2013;46:358–65. doi: 10.1016/j.jmii.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez-Rodriguez LM, Ramos-Ligonio A, Rosales-Encina JL, Martinez-Cazares MT, Parissi-Crivelli A, Lopez-Monteon A. Expression, purification, and evaluation of diagnostic potential and immunogenicity of a recombinant NS3 protein from all serotypes of dengue virus. J Trop Med. 2012;2012:956875. doi: 10.1155/2012/956875. [DOI] [PMC free article] [PubMed] [Google Scholar]