Abstract
Antigen-presenting cells (APCs) are critical in T-cell activation and in the induction of T-cell tolerance. Epigenetic modifications of specific genes in the APC play a key role in this process, and among them, histone deacetylases (HDACs) have emerged as key participants. HDAC6, one of the members of this family of enzymes, has been shown to be involved in regulation of inflammatory and immune responses. Here we show for the first time, that genetic or pharmacologic disruption of HDAC6 in macrophages and dendritic cells resulted in diminished production of the immunosuppressive cytokine IL-10, and induction of inflammatory APCs that effectively activate antigen-specific naïve T-cells and restore the responsiveness of anergic CD4+ T-cells. Mechanistically, we have found that HDAC6 forms a previously unknown molecular complex with STAT3, association that was detected in both the cytoplasmic and nuclear compartments of the APC. By using HDAC6 recombinant mutants we identified the domain comprising aminoacids 503-840 as being required for HDAC6 interaction with STAT3. Furthermore, by re-chromatin immunoprecipitation we confirmed that HDAC6 and STAT3 are both recruited to the same DNA sequence within the Il10 gene promoter. Of note, disruption of this complex by knocking down HDAC6 resulted in decreased STAT3 phosphorylation -but no changes in STAT3 acetylation- as well as diminished recruitment of STAT3 to the Il10 gene promoter region. The additional demonstration that a selective HDAC6 inhibitor disrupts this STAT3/IL-10 tolerogenic axis points to HDAC6 as a novel molecular target in APCs to overcome immune tolerance and tips the balance towards T-cell immunity.
INTRODUCTION
A better understanding of the molecular mechanism(s) regulating pro- and/or anti-inflammatory genes in the APC would provide important insights into how these cells influence T-cell responses and would unveil novel targets to overcome immune tolerance. Among those mechanisms, epigenetic control of gene expression has garnered increasing interest and significant effort is being devoted to elucidate the regulation of pro-inflammatory and anti-inflammatory genes in its natural setting, the chromatin substrate (1). Chromatin modification by acetylation/deacetylation of histone tails represents an important mechanism by which gene transcription is regulated, including genes involved in the inflammatory response (2). In general, histone acetylation is mediated by histone acetyl transferases (HATs), resulting in transcriptionally active chromatin. In contrast, histone deacetylation, mediated by histone deacetylases (HDACs) leads to inactive chromatin and gene repression (3).
HDACs are enzymes that are recruited by co-repressors or by multi-protein transcriptional complexes to gene promoters where they regulate gene expression through the removal of acetyl groups from lysine residues and/or acting as anchors for other transcriptional regulators (4, 5). Eighteen HDACs have been identified and they are subdivided in two families: the classical HDAC family of zinc-dependent metalloproteins, composed of classes I, II and IV; and the NAD+-dependent Class III sirtuin family of HDACs. Class I HDACs (HDAC1, 2, 3 and 8) are most closely related to the yeast deacetylase RPD3(4). Class II HDACs encompass HDAC 4, 5, 6, 7, 9, and 10, sharing homology with the yeast deacetylase HDA1(4). Finally, the newest HDAC identified, HDAC11, is the sole member of Class IV, and does not share homology with either RPD3 or HDA1(5). Although these proteins have been collectively named as “Histone Deacetylases”, it is now clear that some HDACs can also target a variety of non-histone proteins involved in several cellular processes, including proteins involved in structure (i.e. α-tubulin(6)), signaling (i.e. p53(7), STAT3(8), GATA1(9), E2F1(10)) as well as immune responses(11). Thus, the role of these proteins in cell biology, initially limited to their effects upon histones, now encompasses complex regulatory functions that are dependent on HDAC’s tissue expression profile, stage of cellular differentiation and subcellular compartment distribution(11, 12). As an example, initial characterization of HDAC6 assigned its localization and function to the cytosolic compartment(6, 13), however, recent reports have shown that HDAC6 is also present in the nucleus(14, 15). In the cytoplasm, HDAC6 is recognized as a key regulator of cytoskeleton, cell migration and cell-cell interactions(16) given its effect upon the acetylation status of α-tubulin, Hsp90, and cortactin(17). In the nucleus, HDAC6 has been implicated in the regulation of specific transcription factors and gene promoters(14, 15, 18-20). Importantly, accumulating evidence also points to HDAC6 as playing an important role in regulation of inflammatory and immune responses, in particular at the level of the APC/T-cell immune synapse(21), regulatory T-cell function(22), macrophage responses(23) and aberrant airway inflammation (24).
HDACs are the targets of a family of compounds known as HDAC inhibitors (HDACi)(12). Most of the HDACi currently available, including those already approved for the treatment of cancer patients (25-27) target multiple HDACs (pan-HDACi). Recently, we found that treatment of APCs with the pan-HDACi LAQ824 or LBH589 inhibits the production of the anti-inflammatory cytokine IL-10. This effect enabled the APCs to effectively prime naïve antigen-specific CD4+ T-cells and restore the responsiveness of tolerant T-cells isolated from tumor-bearing mice (28). These findings suggest that specific HDAC(s) likely participate in regulation of Il10 gene transcription and could be putative target(s) in the observed HDACi-mediated IL-10 inhibition. Supporting this concept, earlier studies in which different HDACs were genetically disrupted in murine and human APCs led us to demonstrate that HDAC11 is a negative regulator of Il10 gene transcriptional activity (29). Those earlier studies also suggested that another member of the HDAC family, HDAC6, could be involved in regulation of Il10 gene transcriptional activity. Here, we show for the first time that HDAC6 is indeed required for the production of IL-10 by macrophages and dendritic cells (DCs). Mechanistically, we have found that HDAC6 forms a previously unknown molecular complex with STAT3, association that was detected in both the cytoplasmic and nuclear compartments of the APC. Of note, genetic or pharmacologic inhibition of HDAC6 in APCs resulted in decreased STAT3 phosphorylation -but no changes in STAT3 acetylation- as well as diminished recruitment of STAT3 to the Il10 gene promoter region. HDAC6 inhibition represents therefore a novel molecular target to disrupt the anti-inflammatory STAT3/IL-10 axis in the APC and overcome tolerogenic mechanisms mediated by these cells.
METHODS
Mice
Male BALB/c or C57BL/6 mice (6- to 8-weeks old) were obtained from the National Institutes of Health (Frederick, MD). TCR transgenic mice expressing an αβ T-cell receptor specific for amino acids 110-120 from influenza hemagglutinin (HA) presented by I-Ed were a generous gift of H. von Boehmer (30). TCR transgenic mice (OT-II) expressing an αβ TCR specific for peptide 323-339 from Ovalbumin (OVA) presented by MHC class II, I-Ab (31) were provided by Dr. W. Heath (The Walter and Eliza Hall Institute of Medical Research, Victoria, Australia). HDAC6 KO mice (H-2b) were kindly provided by Dr. P. Matthias (32) (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland). Experiments involving the use of mice were performed in accordance with protocols approved by the Animal Care and Use Committee of the University of South Florida College of Medicine.
Cell lines
The macrophage cell line RAW264.7 has been described previously (29, 33). Cells were cultured in vitro in RPMI 1640 media, supplemented with 10% FCS, penicillin/streptomycin (50 U/ml), L-glutamine (2 mM), and β-mercaptoethanol (50 μM) (complete media), and grown at 37 °C and 5% CO2.
Isolation of macrophages and dendritic cells
BALB/c (H-2d), C57BL/6 (H-2b), or HDAC6 KO (H-2b) mice were injected intraperitoneally (ip) with 1 mL of thioglycollate (DIFCO Laboratories, Detroit, MI). Four days later, peritoneal elicited macrophages (PEM) were isolated by peritoneal lavage as previously described(34). DCs were generated from murine bone marrows using RPMI 1640 medium supplemented with 10%FCS, 20 ng/ml murine recombinant GM-CSF, and 10 ng/ml IL-4 (both from RDI, Flanders, NJ). The cultures were maintained at 37°C in 5% CO2. On day 3 of culture, floating cells were gently removed and fresh medium with cytokines was replaced. On day 5, cells were collected and DCs were enriched by centrifugation on metrizamide gradient (Accurate Chemicals, Westbury, NY).
Bone marrow derived macrophages (BMDM) were generated as described previously(33). Briefly, BM cells were differentiated in RPMI 1640 supplemented with 20% fetal bovine serum (BS), 100 U/ml mouse M-CSF, 100 U/ml of penicillin/streptomycin, and 2 mM L-glutamine. Cells were seeded on Petri dishes and incubated at 37°C in 5% CO2. Four days later an extra 10 ml of fresh medium was added to the plate and incubated for an additional 3 days. To harvest BMDM, the plate was washed once with sterile PBS. Then, 10 ml of ice-cold PBS was added to each plate and incubated at 4°C for 10 minutes. Cells were detached by gently pipetting. The re-isolated cells were then centrifuged at 200× g for 5 minutes and suspended in 10 ml of BMDM media (RPMI 1640, 10% FBS, 20 u/ml mouse M-CSF, and 2 mM L-glutamine). Cells were cultured in tissue culture plates for 12 hours before further experimental procedures.
Reagents
LPS (Escherichia coli 055:B5, catalog number L-2880) and shRNA lentiviral particles were purchased from Sigma-Aldrich (St. Louis, MO). Recombinant IL-10 was purchased from BD Biosciences (San Jose, CA). Lipofectamine 2000 and Lipofectamine LTX were from Invitrogen (Carlsbad, CA) The over-expression plasmid for IL-10 was a generous gift from Dr. Boisvert(35). HDAC6 mutants were kindly provided by Dr. Zhang’s lab at University of South Florida(36) .
Antibodies and Immunoblotting
Anti-HDAC6 (C0226) antibody was purchased from Assay Biotech. Anti-FLAG (F1804) antibody was from Sigma. Anti-GAPDH (sc-25778), anti-Tubulin (sc-32293) and anti-acetylated tubulin (sc-23950) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-acetylated H3 (06-599), anti-STAT3 (06-596), anti-JAK2 (06-255), anti-pJAK2 (04-1098) antibodies were purchased from Millipore (Billerica, MA). Anti-pSTAT3 Y705 (9138), anti-pSTAT3 S727 (9136) anti-acetyl-STAT3 (2523) and anti-pTYK2 (9321) antibodies were from Cell Signaling (Danvers, MA). Anti-TYK2 (PA5-34497) and anti-Lamin B (PA5-19468) antibodies were from Thermo Scientific.
Over-expression and knocking-down experiments
Adenoviruses and plasmids with FLAG-tag were used as vectors for HDAC6 and STAT3 over-expression as previously described (29). shRNA lentiviral transduction particles for murine HDAC6 (NM010413, TRCN0000008415), and non-target shRNA (SHC002V) were obtained from Sigma. Sequences for their respective targets are available from the company. For transduction of lentiviral particles for shRNA HDAC6 we followed the protocol provided by the manufacturer using a final MOI of 75. Lentiviral particles for non-target control were random sequences not present in the human or mouse genome. All assays were carried out in triplicate, and western blots were performed to evaluate protein expression.
Over-expression of full length or mutants for HDAC6 was performed in RAW264.7 cells transfected with plasmids encoding for FLAG-tagged version of the respective proteins. Twenty-four hours later, cells expressing each protein variant were lysed and the respective recombinant proteins purified by anti-FLAG affinity columns. Cell extracts isolated from PEM or BMDM were incubated with the FLAG-tagged proteins previously purified and protein complexes were immunoprecipitated using anti-FLAG–agarose beads (Sigma A-2220). Immune complexes were eluted using 200 μg/ml FLAG peptide (Sigma F-3290) in whole-cell extract buffer and analyzed by immunoblot.
To over-express IL-10 in HDAC6KD RAW264.7 cells, PEM, or BMDCs from HDAC6KO mice, we used an expression vector coding for the murine IL-10(35) or control vector. Cells were transfected using Lipofectamine 2000 reagent for 24 or 48 hours according to manufacturer’s protocol (Invitrogen, Grand Island, NY).
Confocal studies
RAW264.7 cells or primary macrophages were stimulated with either LPS (1 μg/ml) or with IL-10 (10 ng/mL). Fixed cells were subjected to immunofluorescent staining using primary antibodies against HDAC6 (1:200, CO226, Assay Biotech) and STAT3 (1:25, 9139, Cell Signaling), secondary antibodies Alexa Fluor 488 donkey anti-mouse IgG (1:200, A-21202, Invitrogen) and Alexa Fluor 594 donkey anti-rabbit IgG (1:200, A-21207, Invitrogen), respectively. Samples were viewed with a Leica DMI6000 inverted microscope, TCS SP5 confocal scanner, and a 63X/1.40NA Plan Apochromat oil immersion objective (Leica Microsystems GmbH, Wetzlar, Germany). 405 Diode, Argon, and HeNe 594 laser lines were applied to excite the samples and a tunable AOBS was adjusted minimize crosstalk between fluorochrome emissions. Gain, offset, and pinhole settings were identical for all samples within the treatment group. Image were captured with photomultiplier detectors and prepared with the LAS AF software version 2.5.0 (Leica Microsystems GmbH, Wetzlar, Germany).
Phenotypic and functional analysis of APCs
The expression of B7.2 in macrophages was determined by staining with FITC-conjugated anti-CD86 (GL1 antibody, BD Bioscience San Jose, CA). Ten thousand gated events were collected on a FACScan (Becton Dickinson) and analyzed using Flow-Jo software. PEM were treated with HDAC6 inhibitor (Tubastatin A) alone, LPS alone or a combination thereof for 24 hours. Supernatants were then collected and production of IL-10 was determined by ELISA (R&D Systems, Minneapolis, MN). Similarly, the production of IL-10 by macrophages or DCs from HDAC6KO mice in response to stimulation with LPS was determined by ELISA. The expression of IL-10 mRNA by macrophages or DCs from HDAC6KO mice in response to stimulation with recombinant IL-10 was determined by quantitative RT-PCR.
Antigen-presentation studies
PEM (1×105/well) from the different experimental groups were treated with LPS alone or with LPS plus increasing concentrations of Tubastatin (Tub-A) for 24 hours. Cells were then washed and 5×104 purified naïve antigen-specific CD4+ T cells (isolated from the spleen of OVA TCR transgenic mice) or a similar number of tolerized antigen-specific CD4+ T-cells were added to the macrophage monolayer with and without OVA peptide323-339 (ISQAVHAAHAEINEAGR).
Similarly, WT, NT, or HDAC6KD RAW264.7 cells were treated with or without LPS for 24 hours. These cells were then cultured with 5×104 purified naïve CD4+ T cells from HA TCR transgenic mice or tolerized antigen-specific CD4+T cells with and without HA peptide110-120 (SFERFEIFPKE). After 48 hours, supernatants were collected and stored at −70°C until assayed for IFN-γ production by ELISA. Values for T-cells cultured in media alone are less than 10% of the values for antigen-stimulated T cells.
Quantitative Real time (qRT)-PCR analysis
RAW264.7, primary murine macrophages or DCs were plated at 2×106 cells per 35mm well and cultured under conditions detailed under each experiment. Total RNA was extracted using TriZol reagent (Invitrogen, NY) and cDNA obtained with the iScript cDNA synthesis kit (Bio-Rad,Hercules, CA). Target mRNA was quantified using MyIQ single color real time PCR detection system (Bio-Rad) and iQ SYBR green Supermix (Bio-Rad,Hercules,CA). Mouse I-10 (left, 5’-CAGGGATCTTAGCTAACGGAAA-3’; right, 5’-GCTCAGTGAATAAATAGAATGGGAAC-3’) were used for PCR amplification (cycling parameters 3 min 95°C, 15 secs 95 °C, 30 secs 60°C 40 reps, 1 min 95°C). Single product amplification was confirmed by melting curve analysis and primer efficiency was near 100% in all the experiments performed. Quantification is expressed in arbitrary units and target mRNA levels were normalized to GAPDH expression using the method of Pfaffl (37). Similar procedure was followed to assess the expression of Bcl-xL, c-Myc, Pim-1, cFos, p21, CDC25, SOCS3, VEGF, IL-1A and IL-6 in macrophages.
Chromatin Immunoprecipitation (ChIP) assays
ChIP studies were performed as previously described(29). All steps were carried out at 4°C. RAW264.7 or PEM grown on 100 mm dishes were incubated for 10 min with 1% formaldehyde under gentle agitation. Cross-linking was stopped by the addition of 0.125 M glycine for 5 min. Then, cells were washed with 10 ml of PBS, scraped off in the same volume of PBS, and collected by centrifugation at 1,000g for 5 min. The cell pellet was re-suspended in lysis buffer (50 mM HEPES [pH 7.8], 20 mM KCl, 3 mM MgCl2, 0.1% NP-40, and a cocktail of protease inhibitors) and incubated for 10 min on ice. The cell extract was collected by centrifugation at 1,000g for 5 min, resuspended in a sonication buffer (50 mM HEPES [pH 7.9], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, and a cocktail of protease inhibitors), and incubated for 10 min on ice. To reduce the length of the chromatin fragments to approximately 500 bp (confirmed by electrophoretic analysis and PCR), the extract was sonicated using a BioruptorTM (Diagenode). Eight pulses of 30 seconds with 30 seconds rest time were repeated six times at a frequency of 20 KHz. After centrifugation at 16,000g, the supernatant was collected, frozen in liquid nitrogen, and kept at −80°C. An aliquot was used for A260 measurements. Cross-linked extracts (6 U A260) were resuspended in a sonication buffer to a final volume of 500 μl. The samples were pre-cleared by incubation with normal mouse IgG plus protein A/G agarose (Santa Cruz Biotechnology) for 2 h at 4°C with agitation. After centrifugation at 1,000g for 5 min, the supernatant was collected and immunoprecipitated with specific antibodies as detailed for each experiment. The immunocomplexes (except from Flag/agarose conjugates) were recovered with the addition of 30 μl of protein-A agarose beads and subsequent incubation for 2 h at 4°C with agitation. The complexes were washed twice with sonication buffer plus 500 mM NaCl, twice with LiCl buffer (20 mM Tris-HCl [pH 8.0], 250 mM LiCl, 1 mM EDTA, and 0.5% Triton X-100), and twice with TE buffer (2 mM EDTA and 50 mM Tris-HCl [pH 8.0]). The solution was incubated at each washing for 5 min at 4°C. The protein-DNA complexes were eluted by incubation with 100 μl of elution buffer (50 mM NaHCO3 and 1% SDS) for 15 min at 65°C. After centrifugation at 1,000g for 5 min, the supernatant was collected and incubated with 10 μg of RNase A per ml for 1 h at 42°C. NaCl was added to the mixture to a final concentration of 200 mM and incubated at 65°C to reverse the cross-linking. The proteins were digested with 200 μg/ml of proteinase K for 2 h at 50°C. The DNA was recovered using Qiagen QuiaquickTM columns. Two different set of primers were used for IL-10: IL-10 (proximal region) sense 5’-GGAGGAGGAGCCTGAATAAC-3’ and antisense 5’-CTGTTCTTGGTCCCCCTTTT-3’, and IL-10 (distal region) sense 5’-AACTCAGCCTGGAACTGACC-3’ and antisense 5’-GCCTCTCCTCCTGACACTCTT -3’. All samples and Inputs were quantified using MyIQ single color real time PCR detection system (Bio-Rad) and iQ SYBR green Supermix (Bio-Rad). Single product amplification was confirmed by melting curve analysis and primer efficiency was near or close to 100% in all experiments performed. Quantification is expressed in arbitrary units indicating fold over non-stimulation condition, and target sequence levels were normalized to the input signal using the method of Pfaffl(37). All ChIP experiments were repeated twice starting from the crosslinking, and final quantitative real time PCR was done in triplicates.
The re-ChIP for HDAC6 and STAT3 was performed using the same experimental procedure described above, only adding an extra clearance step between the sequential immunoprecipitations. Briefly, the eluted fraction after the first immunoprecipitation was pre-cleared by incubation with protein A/G agarose (Santa Cruz Biotechnology) for 2h at 4°C.After centrifugation at 1,000g for 5 min, the supernatant was collected and immunoprecipitated using specific antibodies as detailed under each experiment.
Tolerance model
For induction of anergy of HA-specific CD4+ T-cells we used a well-established experimental model of A20HA tumor induced T-cell tolerance (38). Briefly, 2.5×106 naïve CD4+ transgenic T-cells specific for an MHC Class II epitope of influenza hemagglutin (HA) were injected intravenously into A20HA lymphoma-bearing mice. Twenty-one days after T-cell transfer, animals were sacrificed and T-cells were re-isolated from their spleens as previously described (38). Cytokine production by clonotypic CD4+ T-cells in response to HA-peptide110-120 presented by wild type, HDAC6KD RAW264.7 cells or non-target controls was then determined as described under antigen presentation studies. For induction of anergy of OVA-specific CD4+ T-cells, we used a model of high-dose intravenous injection of OVA-peptide into C57BL6 mice as previously described(39).
Phosphatase activity assay
The tyrosine phosphatase activity was determined using a tyrosine phosphatase assay system (Promega, WI) following manufacture’s protocol. Briefly, cell lysates from PEM isolated from WT or HDAC6KO mice were removed of endogenous phosphate by spin column. In parallel, 100ul of PPase 5× reaction buffer and 5 μl of 1mM phosphopeptide were added to each well in triplicate in a 96-well plate and incubated at 37°C for 3 minutes. Then, 37 μl of sample or standard were added to the wells and the plate was incubated at 37°C for 8 minutes. The reaction was stopped by adding 50 μl of Molybdate Dye Additive mixture to each wells, and the plate was incubated at room temperature for 20 minutes. Absorbance was then measured at 600nm and 630 nm using the Benchmark Plus microplate reader (BioRad).
Statistical Analysis
All experiments were repeated at least two times unless indicated otherwise. Unpaired t-tests were performed using Microsoft Excel Software (Microsoft, Redmond, WA) with significance at p < 0.05.
RESULTS
Genetic disruption of HDAC6 inhibits IL-10 gene expression and induces inflammatory APCs
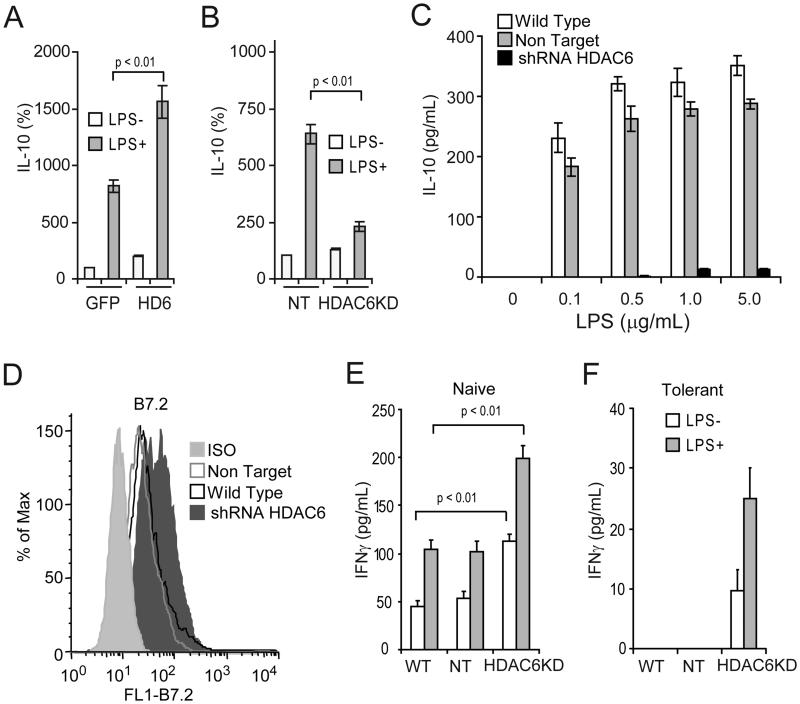
To better understand the role of HDAC6 in the regulation of Il10 gene expression, we first transduced the macrophage cell line RAW264.7 with adenovirus carrying a FLAG- and GFP-tagged version of HDAC6, and with control adenovirus carrying only GFP. Cells infected with control GFP adenovirus displayed minimal levels of IL-10 mRNA in the absence of in vitro stimulation with lipopolysaccharide (LPS) (Fig.1A, GFP, LPS−). LPS stimulation up regulated the expression of IL-10 mRNA in these control cells (Fig.1A, GFP, LPS+) and over-expression of HDAC6 further enhanced LPS-induced IL-10 mRNA (Fig. 1A, HD6, LPS+). Given these results, we next asked whether an opposite effect would be observed when the expression of HDAC6 is stably knocked down (HDAC6KD) in RAW264.7 cells by using lentiviral particles carrying specific shRNA for HDAC6. As compared with control cells transfected with non-target shRNA (Fig. 1B, NT, LPS+), HDAC6KD abrogated the response to LPS in RAW264.7 (Fig. 1B, shRNAHD6, LPS+). The efficiencies of the aforementioned genetic manipulations were confirmed by western blot analysis. As expected HDAC6KD resulted in increased tubulin acetylation (Supplemental Figure 1A).
Figure 1. Phenotypic and functional analysis of macrophages lacking HDAC6.
RAW264.7 cells were transduced in vitro with adenovirus carrying GFP-HDAC6 (HD6) or GFP alone (A), or with lentiviral particles containing HDAC6-specific shRNA (HDAC6KD) or non-targeting shRNA control (NT) (B). Forty-eight hours later cells were treated with or without LPS (1 μg/ml) for additional two hours. Then, total RNA was isolated and expression of IL-10 and GAPDH was measured by quantitative real-time RT-PCR. The results are expressed as percent over control cells transduced with GFP alone (panel A) or non-target shRNA (panel B) and data were normalized by GAPDH expression. Three experiments were performed with similar results. Error bars represent standard deviation from triplicates. (C) Wild type RAW264.7 (WT), non-target (NT) or HDAC6KD (shRNAHDAC6) cells were stimulated with increasing concentrations of LPS (0.1-5.0 μg/ml) for 24 hours. Then, IL-10 production was determined by ELISA. Two experiments were performed with similar results. Error bars represent standard deviation from triplicates. (D) The expression of B7.2 on LPS-stimulated macrophage clones was determined by flow cytometry. In a parallel experiment, 1×105/well RAW264.7 cells lacking HDAC6 or control cells were treated with or without LPS (1 μg/ml) for 24 hours (open bars: −LPS, gray bars: +LPS). Then, 5×104 purified naïve (E) or tolerized (F) anti-HA CD4+ T cells were added to the cultures in the presence of 12.5 μg/ml of cognate HA-peptide110-120 (SFERFEIFPKE). After 48 hours, IFN-γ production was determined by ELISA. Three experiments were performed with similar results. Error bars represent standard deviation from triplicates.
Next, we determined the phenotype and antigen-presenting capabilities of RAW264.7 cells lacking HDAC6. In response to increasing concentrations of LPS, (0.1-5.0 μg/ml), HDAC6KD cells produce minimal amounts of IL-10 protein as compared to wild type (WT) or non-target (NT) control cells (Figure 1C). In addition, HDAC6KD cells displayed an enhanced expression of the co-stimulatory molecule B7.2 (Fig. 1D). No changes in the expression of other co-stimulatory molecules were observed on these cells (data not shown). To assess the antigen-presenting capabilities of HDAC6KD cells, we co-cultured naïve CD4+ T-cells specific for a MHC Class II restricted epitope of influenza hemaglutinin (HA) with HDAC6KD RAW264.7, NT RAW264.7 or WT RAW264.7 cells in the presence of HA-peptide. As shown in Figure 1E, clonotypic T-cells encountering HA-peptide presented by cells lacking HDAC6 are better activated as evidenced by their increased production of IFN-γ relative to clonotypic T-cells recognizing antigen on WT or NT control cells. It is noteworthy that this enhancement in the APC function of HDAC6KD cells was more pronounced when they were stimulated with LPS (HDAC6KD, gray bars).
Previously we demonstrated that adoptive transfer of naïve anti-HA transgenic CD4+ T-cells into mice bearing tumors expressing HA as a model tumor antigen resulted in the induction of antigen-specific CD4+ T-cell tolerance (38). In this system, re-isolated T-cells were found to be anergic given their failure to be primed in vivo as well as by their diminished IFN-γ production in response to in vitro re-stimulation with cognate HA-peptide. As shown in Figure 1F, in vitro incubation of the same tolerant T-cells (re-isolated from tumor bearing mice) with HDAC6KD cells resulted in restoration of their ability to produce IFN-γ in response to cognate antigen (Fig. 1F, HDAC6KD). In sharp contrast, anergic T-cells encountering peptide on WT or NT cells remained unresponsive (Fig. 1F, WT and NT).
Macrophages and Dendritic cells (DCs) from HDAC6 knock-out (KO) mice are better activators of T-cells in vitro and in vivo
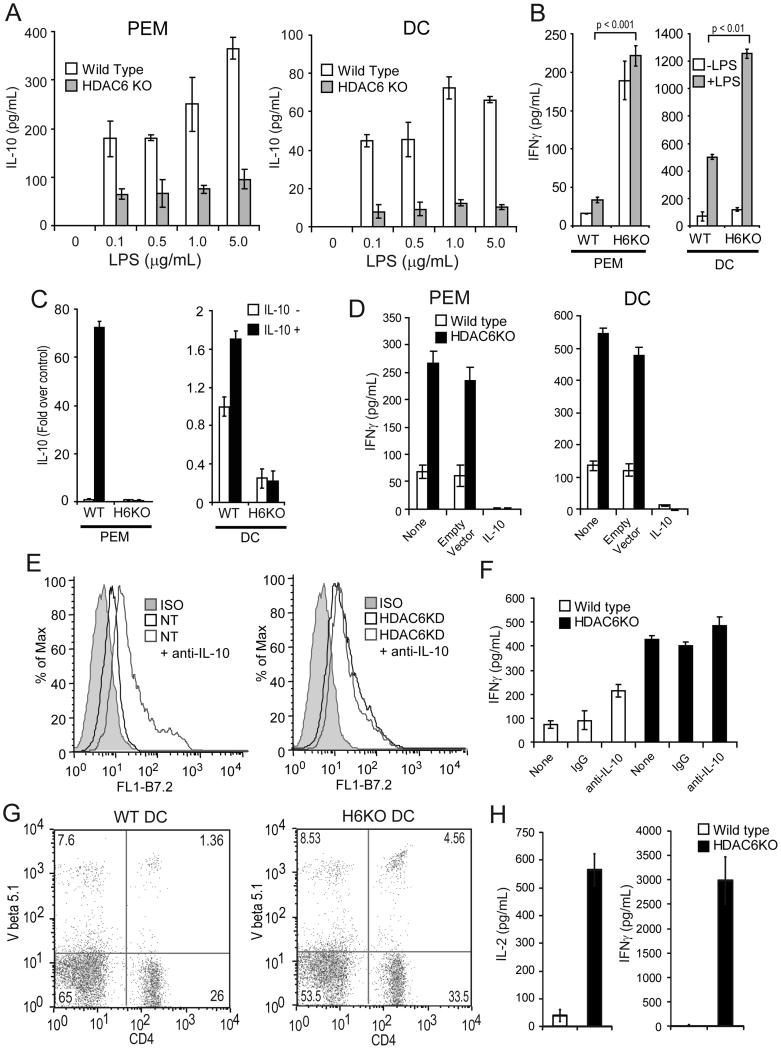
To confirm the above findings in primary APCs, we next determined the production of IL-10 by peritoneal elicited macrophages (PEM) or dendritic cells (DCs) isolated from mice with genetic disruption of HDAC6 (HDAC6 KO mice) (32). As shown in Figure 2A, PEM (Left panel) and DCs (Right panel) from HDAC6 KO mice produce less IL-10 in response to increasing concentrations of LPS (0.1-5.0 μg/ml) as compared to APCs from WT mice. Furthermore, they trigger a better activation of antigen-specific CD4+ T-cells (Fig. 2B) as naïve CD4+ T-cells specific for a MHC Class II restricted epitope of Ovalbumin (OVA) encountering OVA-peptide on PEM (left) or DCs (right) from HDAC6 KO mice (H-2b) produced significantly higher levels of IFN-γ relative to OVA-specific CD4+ T-cells recognizing antigen on APCs from WT mice.
Figure 2. APCs from HDAC6 KO mice are better activators of T-cells in vitro and in vivo.
(A) PEM (left) or bone marrow derived DCs (right) were isolated from HDAC6 KO or WT mice and stimulated with increasing concentrations of LPS (0.1-5.0 μg/mL) for 24 hours. The production of IL-10 was then determined by ELISA. (B) 1×105/well PEM or DCs from HDAC6 KO or WT mice were treated with or without LPS (1.0 μg/ml) for 24 hours. Then, 5×104 purified naïve anti-OVA CD4+ T cells were added to the cultures in the presence of 3 μg/ml of cognate OVA-peptide323-339. After 48 hours, IFN-γ was measured by ELISA. Three experiments were performed with similar results. Error bars represent standard deviation from triplicates. (C) PEM or DCs as described in (B) were treated with or without IL-10 (10ng/ml) for 2 hours. Then total RNA was isolated and expression of IL-10 mRNA and GAPDH was measured by quantitative RT-PCR. The results are expressed as fold over non treated control cells normalized by GAPDH expression. Three independent experiments were performed with similar results. (D) The antigen-presenting function of HDAC6 KO macrophages or DCs transfected with a plasmid expressing IL-10 was evaluated as described in (B). Data is from a representative experiment of two independent experiments with similar results. (E) Expression of B7.2 on WT (left) or HDAC6KD (right) macrophages in the presence (or not) of anti-IL-10 neutralizing antibodies. (F) Production of IFN-γ by T cells co-cultured with macrophages from WT or HDAC6KO mice in the presence of anti-IL-10 neutralizing antibodies or isotype control (IgG). Data (E-F) is from a representative experiment of two independent experiments with similar results. (G) DCs were isolated from WT and HDAC6KO mice and pulsed with OVA peptide for 24 hours, then stimulated with LPS (1.0 μg/ml) for an additional 24 hours. These DCs were then injected intravenously into C67BL/6 recipients that had previously received antigen-specific OT-II CD4+ T-cells (2.5 × 106 given iv one day before the injection of DCs). On day 5 after DC transfer, mice were sacrificed and T-cells were purified from their spleens. The percent of double positive CD4 and V-beta 5.1 TCR population was determined by flow cytometry. (H) In parallel, 5×104 purified T-cells from these mice were cultured with splenocytes from C57BL/6 mice in the presence of 3 μg/ml of OVA323-339 peptide. After 48 hours, supernatants were collected and IL-2 (left panel) and IFN-γ (right panel) production were determined by ELISA
To determine whether this lack of IL-10 production by HDAC6-deficient APCs could be observed in response to stimuli other than LPS, we treated PEM and DCs with recombinant IL-10 which has been shown to induce Il10 gene expression in an autocrine fashion(40). As shown in Figure 2C, minimal IL-10 mRNA was observed in HDAC6KO PEM or DCs treated with recombinant murine IL-10 as compared to WT APCs. Similarly, no IL-10 mRNA expression was observed in HDAC6KO APCs stimulated with IL-6, a cytokine that also induces Il10 gene expression (data not shown). Taken together, HDAC6 is required for Il10 gene transcriptional activity in APCs in response to different stimuli.
To assess whether the observed inhibition of IL-10 production represents the major explanation for the improved APC function of PEM and DCs devoid of HDAC6, we next transduced HDAC6KO cells with a plasmid expressing IL-10 (IL-10) or with an empty vector control (35). In the absence of LPS stimulation, cells transduced with plasmid-IL-10 produced higher levels of IL-10 (350-550 pg/ml) relative to cells transduced with empty vector control (data not shown). HDAC6KO PEM or DCs with or without empty vector trigger effective activation of CD4+ T-cells (Fig. 2D, black bars, None and Empty vector). When these same APCs were transduced with a plasmid-IL-10, they lost their ability to trigger IFN-γ production by CD4+ T-cells (Fig. 2D, HDAC6KO, IL-10). This abrogation of IFN-γ production may have been the result of the significantly higher levels of IL-10 produced by APCs transduced with the IL-10 plasmid. However, the production of IFN-γ by T-cells was also diminished when DCs from HDAC6KO mice were cultured with OT-II CD4+T-cells in the presence of lower concentrations of recombinant IL-10 (range 10-1000 pg/ml) (Supplemental Figure 1B). These concentrations of IL-10 were within the range of IL-10 detected in the supernatants of LPS-stimulated WT APCs (range 50-400 pg/ml). Given the known autocrine effects of IL-10 produced by the APC, we determined next the phenotypic and functional changes in APCs lacking HDAC6 or control cells when they were cultured in the presence of neutralizing anti-IL-10 antibodies. As shown in Figure 2E (left panel), addition of IL-10 neutralizing antibodies to control APCs resulted in increased B7.2 expression. However, this effect was not seen in HDAC6KD macrophages (Fig. 2E, right panel). We also evaluated the production of IFN-γ by T cells co-cultured with macrophages from WT or HDAC6KO mice in the presence of anti-IL-10 antibodies. As expected, IFN-γ production by T-cells cultured with WT macrophages was enhanced in the presence of IL-10 neutralizing antibodies (Figure 2F, white bars). However, this effect was not observed when T-cells were co-cultured with HDAC6KO APCs (Figure 2F, black bars). Taken together, these findings suggest that the improved T-cell activating function of APCs lacking HDAC6 is mainly mediated by the regulatory effect of this HDAC upon IL-10 production.
Next, we determined the in vivo T-cell activating function of APCs lacking HDAC6. DCs from HDAC6 KO or WT mice were pulsed in vitro with OVA peptide and stimulated with LPS for 24 hours. Then, these DCs were injected intravenously (iv) into WT C57BL/6 mice that had previously received antigen-specific OTII CD4+ T-cells. Animals were sacrificed after 5 days, and T-cells were isolated from their spleens. As shown in Figure 2G, a higher expansion of antigen-specific CD4+ T-cells was observed when these cells were exposed in vivo to HDAC6KO DCs as compared to WT DCs (4.56% versus 1.36% respectively). Furthermore, antigen-specific CD4+ T-cells exposed to HDAC6 KO DCs were better primed in vivo since they produced higher amounts of IL-2 (Fig. 2H, left) and IFN-γ (Fig. 2H, right) upon in vitro re-stimulation with cognate OVA-peptide.
Treatment of APCs with an isotype-specific HDAC6 inhibitor in vitro and in vivo also improves T cell activation via decreased IL-10 production
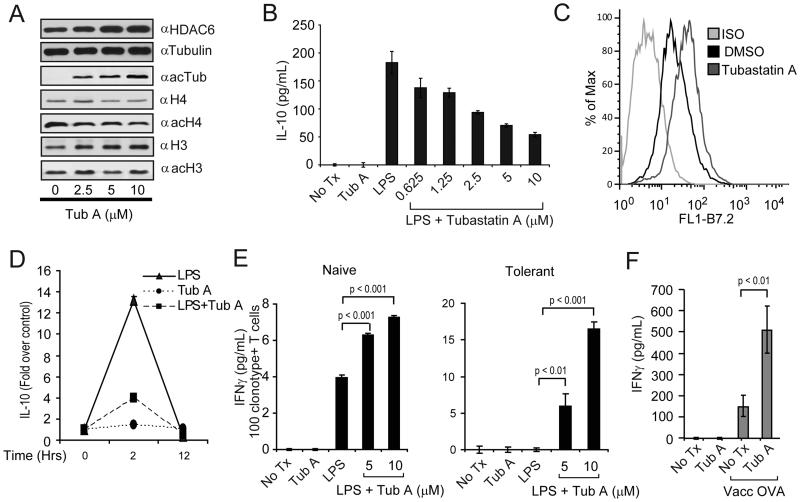
Given the above results, we next asked whether similar effects could be observed when APCs are treated with the isotype-selective HDAC6 inhibitor, Tubastatin A (Tub-A)(41). First, Tub-A did not affect the viability of APCs (data not shown) and induced acetylation of α-tubulin but not of histones H3 or H4 (Fig. 3A), indicating that at the concentrations used, Tub-A selectively inhibits HDAC6. Second, reminiscent of the changes observed in APCs lacking HDAC6, treatment of PEM from WT mice with Tub-A resulted in a dose-dependent inhibition of IL-10 production in response to LPS (Fig. 3B). Tub-A treated macrophages also displayed an enhanced expression of the co-stimulatory molecules B7.2 (Fig. 3C).
Figure 3. Phenotype and function of APCs treated with the isotype-selective HDAC6 inhibitor, Tubastatin A.
(A) PEM were treated with increasing concentrations of Tubastatin A (Tub-A) for 24 hours. Protein extracts were prepared and subjected to SDS-PAGE and immunoblotting with α–HDAC6, α-tubulin, α-acetylated tubulin, α-histone 4 (H4), α-acetylated H4, α-histone 3 (H3), α-acetylated H3 specific antibodies. (B) PEM (1×105/well) were treated with LPS (1 μg/ml) alone or LPS plus increasing concentrations of Tub-A for 24 hours. Then, supernatants were collected and the production of IL-10 was determined by ELISA. (C) Expression of B7.2 on macrophages treated with Tub-A (5 μM) was determined by flow cytometry. (D) PEM (2×106/well) were treated with 1ug/ml of LPS (triangles), 5μM of Tub-A (circles), or the combination thereof (squares). Cells were harvested at 0, 2 and 12 hours and total RNA was isolated. The IL-10 expression by quantitative RT-PCR is expressed as fold over non treated control cells and normalized by GAPDH expression. Data (A-D) is from a representative experiment of three experiments with similar results. (E) PEM (1×105/well) were treated as in (B). Then, cells were washed and 5×104 purified naïve (left) or tolerized (right) anti-HA CD4+T-cells were added to the cultures in the presence of 12.5 μg/ml of cognate HA-peptide110-120. After 48 hours, supernatants were collected and assayed for IFN-γ production by ELISA. Data is from a representative experiment of three independent experiments with similar results. (F) C57BL/6 mice were adoptively transferred with 2.5×106 anti-OVA TCR transgenic CD4+ T-cells given intravenously (day zero). Animals were then treated with either Tub-A (25mg/kg) or vehicle control given intraperitoneally (ip) for 10 days (day zero to +9). On day +9 half the mice in each group were immunized s.c. with 1×107 pfu of vaccOVA. Animals were sacrificed six days later and T-cells were purified from their spleens as indicated in Methods. T-cells were then cultured with splenocytes from C57BL/6 mice in the presence of 3 μg/ml of OVA323-339 peptide. After 48 hours, supernatants were collected and IFN-γ production was determined by ELISA. Data represent mean ± SE of triplicate cultures from 8 mice in each group and is representative of two independent experiments with similar results. Values for T-cells cultured without OVA-peptide were below the limit of detection.
Next, we evaluated the kinetics of IL-10 mRNA expression in PEM treated with LPS, Tub-A or LPS plus Tub-A. In response to LPS stimulation, IL-10 mRNA expression peaked at two hours, followed by a rapid decline, and was back to baseline by 12 hours (Fig. 3D-triangles). However, in PEM treated with LPS plus Tub-A there was a decrease in IL-10 mRNA induction (Fig. 3D-squares). No changes in IL-10 mRNA levels were observed in PEM treated with Tub-A alone (Fig. 3D-circles). Therefore, pharmacologic inhibition of HDAC6 resulted in decreased Il10 gene transcriptional activity in response to LPS.
To assess the antigen-presenting capabilities of Tub-A treated PEM, these cells as well as untreated PEM were cultured in vitro with naïve or tolerant anti-HA CD4+T-cells in the presence (or not) of cognate HA-peptide. Compared to untreated PEM, PEM treated with Tub-A triggered a better activation of naïve HA-specific CD4+ T-cells given their increased production of IFN-γ (Fig 3E, left). More importantly, Tub-A treated PEM were able to restore the responsiveness of tolerant CD4+ T-cells isolated from tumor bearing mice (Fig. 3E, right).
Utilizing an adoptive transfer system of antigen-specific CD4+ T-cells into C57BL/6 mice, we previously demonstrated that vaccination of these mice with recombinant vaccinia encoding a model antigen (vacc-OVA) resulted in antigen-specific T-cells that fulfill several functional criteria indicative of effective priming by APCs in vivo: clonal expansion and differentiation into effector cells capable of producing IFN-γ (38, 42). We therefore used this system to determine whether treatment of C57BL/6 mice with Tub-A might increase APC’s function and their response to vacc-OVA immunization in vivo. Briefly, on day zero C57BL/6 mice were adoptively transferred with antigen-specific OTII CD4+ T-cells. Mice were then treated with Tub-A (25mg/kg) or with vehicle control from day zero to +9. On day +9 mice were given 1×107 pfu of vacc-OVA given s.c. Six days later, animals were sacrificed and T-cells were isolated from their spleen and their function was evaluated. First, antigen-specific CD4+ T-cells isolated from vaccinated C57BL/6 mice produced higher levels of IFN-γ, (Fig 3F, No Tx, vaccOVA) relative to unvaccinated mice (No Tx) or mice treated with Tubastatin-A alone (Tub A), following in vitro re-stimulation with OVA-peptide. Second, the magnitude of this antigen-specific T-cell response was further augmented in mice treated with Tub-A in combination with vaccination. Indeed, indicative of enhanced priming by APCs in vivo, clonotypic T-cells from Tub-A treated mice produce significantly higher level of IFN-γ in response to in vitro re-stimulation with OVA-peptide (Fig. 3F, Tub A, Vacc OVA).
Therefore, reminiscent of our results with genetic disruption of HDAC6, the HDAC6 selective inhibitor, Tub-A also inhibits IL-10 production resulting in inflammatory APCs that are better activators of CD4+ T-cells in vitro as well as in vivo.
STAT3 phosphorylation but not acetylation is decreased in APCs with genetic or pharmacologic inhibition of HDAC6
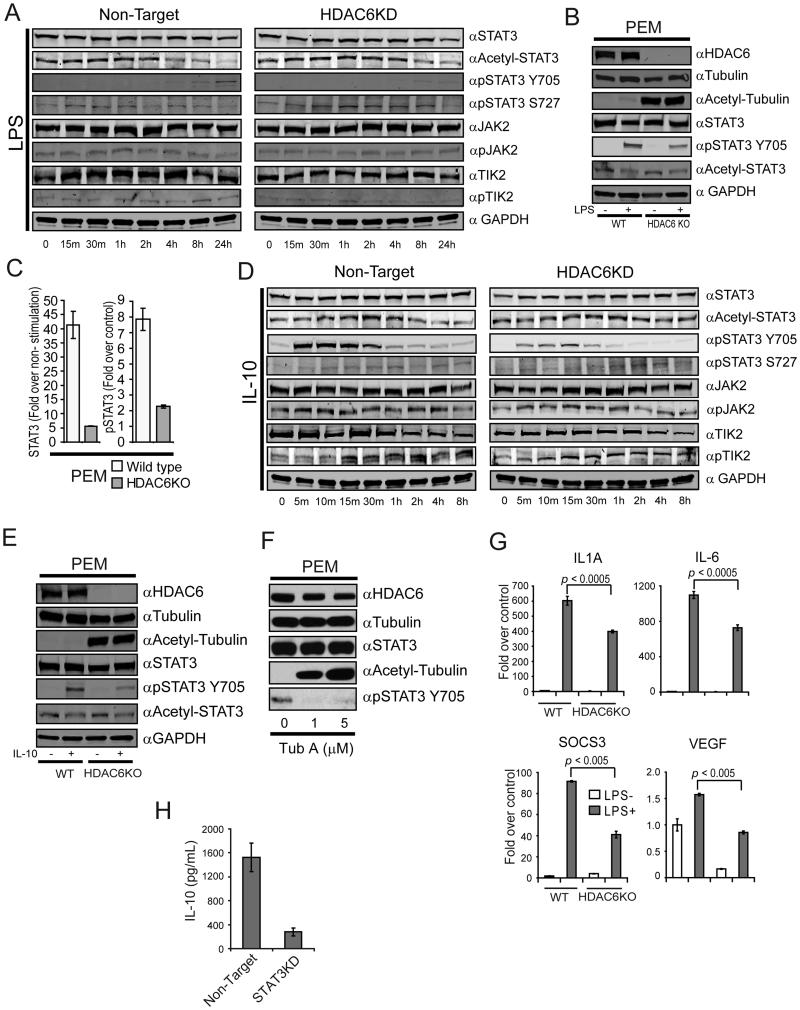
Previous studies have shown the important role of STAT3 in the regulation of Il10 gene expression and in tolerance induction mediated by APCs (39) (40). The demonstration by Yuan et al. that acetylation of STAT3 regulates its function(8) and that STAT3 can partner with HDACs to influence gene expression(43, 44) led us therefore to explore whether HDAC6 might influence Il10 gene transcriptional activity through regulation of STAT3 function in the APC. As shown in Figure 4A (row 1) STAT3 protein expression was similar in HDAC6KD clones and in non-target controls. However, no differences in STAT3 acetylation was found among HDAC6KD macrophages and non-target control cells (Fig. 4A, row 2). These results led us to explore next the phosphorylation status of STAT3 in these cells. While phosphorylation of STAT3 Y705 was observed in control cells after 4 hours of LPS stimulation, notably less phosphorylation was observed in macrophages lacking HDAC6 (Fig. 4A, row 3). Unlike tyrosine phosphorylation, analysis of serine 727 phosphorylation revealed no differences among cells lacking HDAC6 and non-target controls (Fig. 4A, row 4). In order to assess the potential participation of HDAC6 upon activation of upstream components of the STAT3 pathway, we evaluated the phosphorylation status of JAK2 and TYK2, two well-known activators of STAT3 signaling(45). As shown in Figure 4A, no difference in the expression of total JAK2, phospho-JAK2, total TYK2 and phospho-TYK2 were observed between non-target and HDAC6KD cells, suggesting that disruption of HDAC6 is not affecting these two upstream components of the STAT3 pathway in the APC.
Figure 4. Genetic or pharmacologic disruption of HDAC6 decreased phosphorylation but not acetylation of STAT3 in APCs.
(A) Non-target or HDAC6KD RAW264.7 macrophages were stimulated with LPS (1.0 μg/mL). After the indicated time points, cells were lysed and immunoblotted using specific antibodies against STAT3, acetylated-STAT3, phospho-STAT3 Y705, phospho-STAT3 S727, JAK2, phospho-JAK2, TYK2, phospho-TYK2 and GAPDH. (B) PEM were isolated from WT or HDAC6 KO mice and stimulated or not with LPS (1.0 μg/mL). After 24 hours, cells were lysed and immunoblotted using antibodies against HDAC6, tubulin, acetylated-tubulin, STAT3, pSTAT3 Y705, ac-STAT3 and GAPDH. (C) PEM from HDAC6 KO or WT mice were treated with LPS as in (B) and then subjected to ChIP analysis using α-STAT3 and α-p-STAT3 antibodies. (D) Non-target or HDAC6KD RAW264.7 cells were stimulated with IL-10 (10 ng/mL). After the indicated time points, cells were lysed and immunoblotted using specific antibodies as indicated on panel A. (E) PEM isolated from WT or HDAC6 KO mice were stimulated with IL-10 or left untreated. After 30 minutes, cells were lysed and immunoblotted using the indicated antibodies (F) PEM isolated from C57BL/6 mice were treated with Tub-A for 24 hours. Cells were then lysed and immunoblotted using the indicated antibodies. (G) PEM from HDAC6 KO as well as WT mice were stimulated with LPS (1.0 μg/ml) for 2 hr or remained untreated. Then, total RNA was isolated and analyzed by qRT-PCR for expression of Socs3, Vegf, Il1A and Il6 genes. GAPDH was used as control. The results are expressed as fold over non treated control cells normalized by GAPDH expression according to the Pfaffl equation(37). Three experiments were performed with similar results. (H) Production of IL-10 by non-target or STAT3KD macrophages in response to LPS. Data displayed (A-H) is from a representative experiment of three independent experiments with similar results.
The results above were then confirmed in PEM isolated from HDAC6KO mice in which we observed a diminished STAT3 Y705 phosphorylation (Fig. 4B, row 5), but no changes in STAT3 acetylation relative to PEM from WT mice (Fig. 4B, row 6). In addition, in these HDAC6KO macrophages we found a decreased recruitment of STAT3 and p-STAT3 to the Il10 gene promoter (Fig. 4C). This impairment in STAT3 phosphorylation and its recruitment to the Il10 gene promoter could explain the diminished IL-10 production observed in HDAC6KO macrophages in response to LPS (Fig. 2A).
To determine whether this decrease in STAT3 phosphorylation also occurred in response to stimuli other than LPS, macrophages lacking HDAC6 and non-target (NT) controls were treated with IL-10, cytokine that induces STAT3 phosphorylation within minutes of binding to its receptor(46). As shown in Figure 4D, phosphorylation of STAT3 Y705 in NT controls occurs at five minutes post-stimulation and peaks by 30-60 minutes (Non-Target, row 2). In contrast, Y705 phosphorylation was both delayed and diminished in HDAC6KD cells (HDAC6KD, row 2). Analysis of S727 phosphorylation and STAT3 acetylation revealed no differences (rows 3 and 4 respectively). Reminiscent of our findings with LPS stimulation, the phosphorylation status of JAK2 and TYK2 was similar in HDAC6KD and control macrophages treated with IL-10. (Fig. 4D, row 5-8). Studies in PEM from HDAC6KO mice or control mice treated with IL-10 in vitro confirmed that STAT3 Y705 phosphorylation, but not STAT3 acetylation, is diminished in the absence of HDAC6 (Fig. 4E, rows 5 and 6).
To assess whether similar changes in STAT3 phosphorylation could be seen with pharmacologic inhibition of HDAC6, we treated PEM with Tub-A. No changes in HDAC6, tubulin and STAT3 proteins were observed in untreated or Tub-A treated PEM (Fig. 4F, rows 1, 2 and 3 respectively). However, treatment with Tub-A was associated with an increased in tubulin acetylation (row 4) and almost complete absence in STAT3 Y705 phosphorylation (Fig. 4F, row 5).
Next, we determined the impact of the absence of HDAC6 upon downstream STAT3-regulated genes other than Il10. Macrophages lacking HDAC6 display decreased mRNA expression of Bclxl, Cmyc, Pim1, Cfos, P21 and Cdc25 in response to LPS (Supplemental Figure 1C). Interestingly, immunologically relevant genes known to influence the T-cell activating properties of APCs, such as Socs3, Vegf, Il1A, and Il6 were also down regulated in response to LPS in macrophages lacking HDAC6 (Figure 4G). However, no significant differences were observed in the expression of Socs3, Il1A, and Il6 at the basal state (absence of LPS stimulation) among control and HDAC6KO macrophages.
Finally, we asked whether disruption of STAT3 in macrophages (STAT3KD) would recapitulate the effects of HDAC6 knock-down upon IL-10 production. Reminiscent of our findings in STAT3 KO PEM(39), STAT3 KD PEM produce significantly lower levels of IL-10 in response to LPS as compared to non-target PEMs (Figure 4H). Taken together, we have found that HDAC6 is required for STAT3 phosphorylation and activation of STAT3 downstream target genes in macrophages.
HDAC6 physically interacts with STAT3 in the cytoplasm and nuclei of the APC
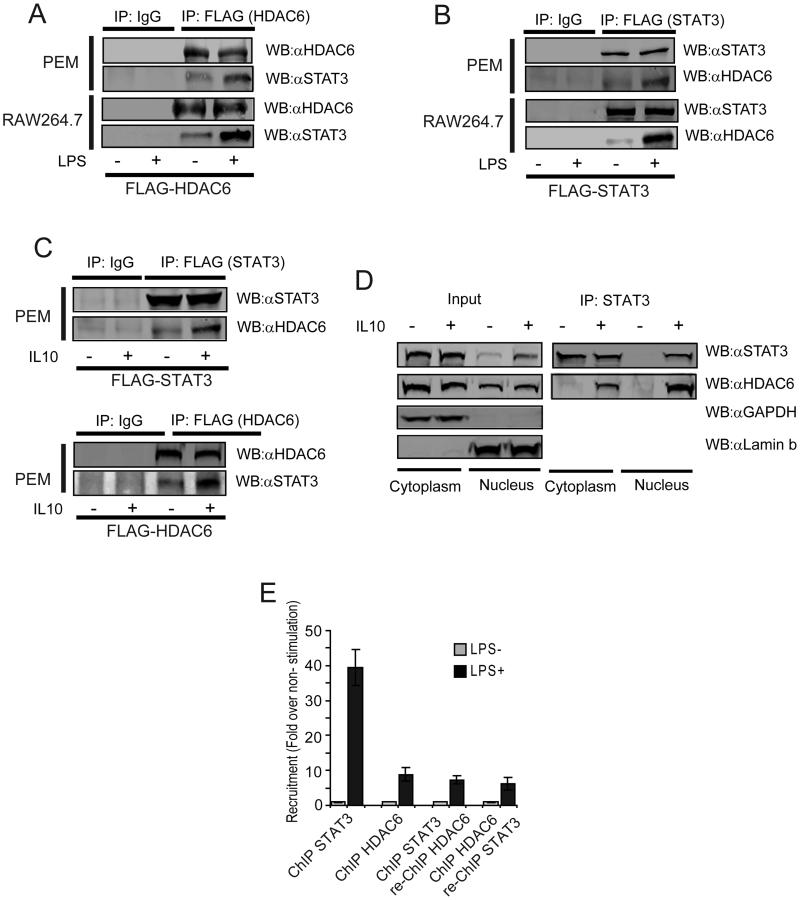
Next, we asked whether a physical interaction between HDAC6 and STAT3 is required for their effects upon Il10 gene expression in APCs. First, PEM (Fig. 5A top) or RAW264.7 cells (Fig. 5A bottom) transfected with a plasmid encoding FLAG-tagged HDAC6 were subjected to immunoprecipitation and immunoblot analysis. Endogenous STAT3 co-immunoprecipitated with FLAG-HDAC6 in the absence of LPS stimulation (Fig 5A, −LPS) and this interaction was further increased in response to LPS stimulation (Fig 5A, +LPS). Similarly, HDAC6 was identified in the immunoprecipitated fraction after the reverse immunoprecipitation was performed with a FLAG-tagged STAT3 construct (Figure 5B). This STAT3-HDAC6 interaction was also demonstrated in primary macrophages stimulated with IL-10 when either a FLAG-tagged STAT3 (Fig. 5C top) or a FLAG-tagged HDAC6 (Fig. 5C bottom) were used.
Figure 5. HDAC6 physically interacts with STAT3 in the cytoplasm and nuclei of the APC.
(A) PEM (top) or RAW264.7 macrophages (bottom) were transfected with a plasmid encoding FLAG-HDAC6. 48 hours after infection, cells were treated with LPS (1.0 μg/mL). 24 hours later cellular extracts were obtained and immunoprecipitated (IP) with an anti-FLAG antibody. The IP fraction was then subjected to WB to evaluate for the presence of STAT3. (B) PEM (top) or RAW264.7 macrophages (bottom) were infected with adenovirus encoding FLAG-STAT3. 48 hours after infection, cells were treated with LPS (1.0 μg/mL) and then cellular extracts were obtained and immunoprecipitated (IP) with an anti-FLAG antibody. The IP fraction was then subjected to WB to evaluate for the presence of HDAC6. (C) PEM were transfected with a plasmid encoding FLAG-STAT3 (top) or FLAG-HDAC6 and then stimulated (or not) with IL-10 (60ng/mL) for 30 min. Cellular extracts were obtained and immunoprecipitated with an anti-FLAG antibody. The presence of HDAC6 and STAT3 was then evaluated in the IP fraction using the indicated antibodies. (D) Nuclear and cytosolic fractions from macrophages (PEM or RAW cells) treated or not with IL-10 (60ng/mL) for 30 min were subjected to immunoblotting to evaluate the intracellular distribution of HDAC6 and STAT3 Immunoblotting of GAPDH and laminin B were used as controls for cytosolic and nuclear fractions respectively (left: input). In parallel, endogenous STAT3 was immunoprecipitated from the nuclear and cytoplasmic fractions and then the presence of HDAC6 was evaluated by western blot. Data is from a representative experiment of two independent experiments with similar results. (E) Sequential chromatin immunoprecipitation (re-ChIP) of HDAC6 and STAT3 on the Il10 gene promoter. Data (A-E) is from a representative experiment of two independent experiments with similar results.
To determine the subcellular fraction of the APC in which the STAT3-HDAC6 interaction might be occurring, we first evaluated the presence of STAT3 and HDAC6 in immunoblots of cytosolic and nuclear extracts from macrophages treated with IL-10. In the absence of stimulation, both STAT3 and HDAC6 were detected in the cytoplasm (Fig. 5D, Input, −IL10). In the nuclear fraction of untreated cells,STAT3 was almost undetectable, while HDAC6 was detected. In response to IL-10 treatment, a significant increase in STAT3 was observed in the nucleus, while cytosolic STAT3, cytosolic HDAC6 and nuclear HDAC6 remain unchanged (Fig. 5D, Input, +IL10). Anti-GAPDH and anti-Lamin b antibodies were used as controls for cytosolic and nuclear localization respectively (Fig. 5D). Next, co-immunoprecipitation of endogenous STAT3 from cyosolic and nuclear fractions of macrophages treated with IL-10 demonstrated that STAT3 and HDAC6 interact in both subcellular fractions, however, this association seems to be increased in the nuclear compartment of the APC (Fig. 5D, IP: STAT3)
To further assess the role of this nuclear STAT3/HDAC6 interaction upon Il10 gene expression, we asked whether HDAC6 and STAT3 might be recruited to the same DNA sequence within the Il10 promoter. We therefore performed a re-ChIP (re-chromatin immunoprecipitation or co-ChIP) for HDAC6 and STAT3. Re-ChIP is a two-step sequential chromatin immunoprecipitation, where the first antibody is directed against HDAC6 and the second antibody against STAT3, and vice-versa(47). As shown in Figure 5E, both proteins were recruited to the il-10 gene promoter in response to LPS stimulation (black bars, ChIP STAT3 and ChIP HDAC6 respectively). Following this first ChIP, and after proper pre-clearance, samples were subjected to a second immunoprecipitation with antibody against the second protein. Regardless of the order of the antibodies, both re-ChIP approaches demonstrated that STAT3 and HDAC6 are recruited to the same DNA sequence within the il-10 gene promoter (Fig. 5E, black bars, ChIP STAT3, re-ChIP HDAC6 and ChIP HDAC6, re-ChIP STAT3, respectively). These results suggest that in addition to its role in regulating STAT3 phosphorylation, HDAC6 might also function as a transcriptional cofactor of STAT3 at the level of the Il10 gene promoter.
Finally, confocal microscopy studies performed in RAW264.7 and PEM stimulated with either LPS (Supplemental Fig. 2A) or IL-10 (Supplemental Fig. 2B) also showed that HDAC6 and STAT3 co-localize in both the cytoplasmic and nuclear compartment of these cells.
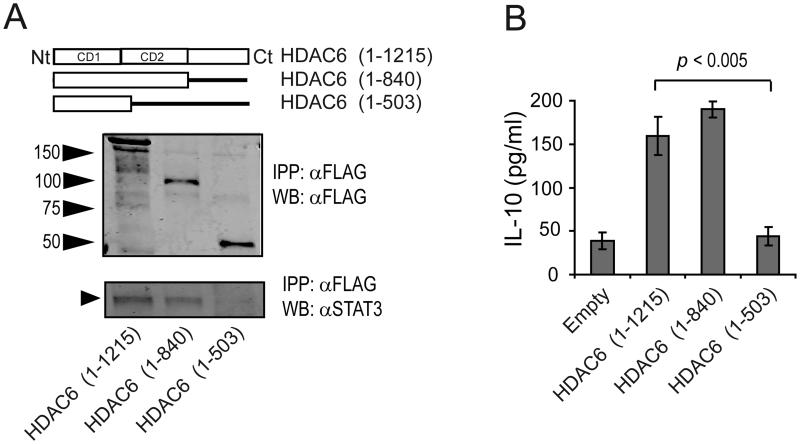
HDAC6 domain comprising aminoacids 503-840 is required for its interaction with STAT3 and IL-10 production
Next, we investigated which segment of the HDAC6 protein is necessary for their physical interaction with STAT3. We over-expressed two FLAG-tagged HDAC6 constructs (HD6 1-840 or HD6 1-503) and full length HD6 1-1215 (Figure 6A, top) in macrophages. Figure 6A (middle) shows the electrophoretic migration of these HDAC6 proteins on WB probed with anti-FLAG antibody. Following purification, these recombinant mutants were incubated in vitro with protein extracts from primary murine macrophages to determine the presence of STAT3 in the IP fraction. As shown in Figure 6A (bottom), STAT3 from murine macrophages co-immunoprecipitated with full-length HDAC6 (1-1215) and HDAC6 mutant 1-840 (lanes 1 and 2), but not with HDAC6 mutant 1-503 (lane 3). These results point to the segment comprising aminoacids 503-840 of HDAC6 as being required for its physical interaction with STAT3.
Figure 6. HDAC6 domain comprising aminoacids 503-840 is required for IL-10 production by macrophages.
(A) Cell lysates from PEM were incubated with recombinant proteins for FLAG-HDAC6 full length or truncated forms (top), and subjected to immunoprecipitation using a FLAG antibody (middle). The IP fraction was then assayed with antibodies against FLAG and STAT3 (bottom). (B) The aforementioned HDAC6 deletion mutants were expressed in PEM obtained from HDAC6 KO mice. Cells were then treated with LPS (1.0 μg/mL) for 24 hours and the production of IL-10 was measured by ELISA. Data is from a representative experiment of two independent experiments with similar results.
To further explore the functional role of the HDAC6 segment 503-840 in regulation of IL-10 production, we asked whether this cytokine would be produced when macrophages expressing endogenous STAT3 -but lacking HDAC6- were transfected with the HDAC6 mutants described above. Briefly, HDAC6KO macrophages were transfected with empty vector, full length HDAC6 or mutant HDAC6. HDAC6KO macrophages transfected with empty vector produced minimal amounts of IL-10 (Figure 6B, Empty), whereas those cells transfected with full length HDAC6 or HDAC6 mutant 1-840 produced significant amounts of this cytokine. However, in cells transfected with the mutant 1-503, the production of IL-10 was minimal and similar to the levels observed in empty vector-transfected cells. These findings indicate that the HDAC6 domain comprising aa 503-840 is required for its direct interaction with STAT3 and regulation of IL-10 production by macrophages.
DISCUSSION
Accumulating evidence indicates that epigenetic modifications of specific genes play a key role in influencing the inflammatory status of the APC and T-cell activation versus T-cell anergy (48, 49). The immunosuppressive action of the cytokine IL-10 is critical in the generation of APCs with tolerogenic properties (50, 51) and in the prevention of self-tissue damage (52-54). In this study we have shown for the first time that genetic or pharmacologic disruption of HDAC6 in macrophages and dendritic cells resulted in diminished production of the immunosuppressive cytokine IL-10, and generation of pro-inflammatory APCs able of better activate naïve antigen-specific T-cells in vitro as well as in vivo. More importantly, APCs lacking HDAC6 are capable of restoring the responsiveness of anergic CD4+ T-cells.
Initially recognized as a cytoplasmic protein, HDAC6 is a 131KDa protein with key regulatory roles upon cytoskeleton, cell migration and cell-cell interactions(16) given its effect upon the acetylation status of α-tubulin, Hsp90, and cortactin(17). Accumulating evidence also indicates that HDAC6 is present in the nucleus(14, 15) where it can function as cofactor of specific transcription factors to influence gene promoter activity (14, 15, 18-20). Of note, HDAC6 has been also found to regulate inflammatory and immune responses (21-24). Expanding upon these properties, we have found that HDAC6 regulates IL-10 production in APCs by influencing the function of STAT3, a transcriptional activator of Il10 gene activity. First, HDAC6 forms a previously unknown molecular complex with STAT3. Such a complex was found in both the cytoplasm and nucleus, with a stronger protein interaction occurring in the nuclear compartment of stimulated APCs. Second, by re-ChIP, we demonstrated that HDAC6 and STAT3 are both recruited to the same DNA sequence within the Il10 gene promoter. Third, disruption of this complex, by either knocking down HDAC6 or by studies in HDAC6KO APCs showed a significant impairment in STAT3 phosphorylation -but no changes in STAT3 acetylation- as well as diminished recruitment of STAT3 to the Il10 gene promoter region in response to stimulation with either IL-10 or LPS.
Our observation that the impairment of STAT3 phosphorylation in APCs lacking HDAC6 occurs within minutes after IL-10 stimulation suggests a proximal (cytosolic) role for HDAC6 in STAT3 regulation. However, the findings that HDAC6 and STAT3 become more nuclear in WT APCs after similar stimulation, together with the demonstration that both proteins are recruited to the same DNA sequence within the Il10 promoter, also suggest a nuclear role for HDAC6 in influencing Il10 gene promoter activity. Therefore, the diminished IL-10 production observed in APCs lacking HDAC6 could be explained by the impairment in STAT3 phosphorylation and its recruitment to the Il10 gene promoter and by the absence of this HDAC in the nuclei where it could be functioning as a transcriptional cofactor of STAT3 at the level of the Il10 gene promoter (as suggested by the re-CHIP data). Needless to say, we are currently determining which one of these two mechanisms could be the dominant one in influencing IL-10 production by the APC.
Regardless of the mechanism(s), the above findings provided the rationale to target HDAC6 with specific inhibitors (55)’ (41, 56) and determine whether such an approach could also disrupt the tolerogenic STAT3/IL-10 axis in the APC. Proof of concept was provided in APCs treated with the HDAC6 selective inhibitor, Tubastatin A, in which we observed a diminished production of IL-10 in response to LPS stimulation. These APCs were also rendered more inflammatory in vitro and in vivo and were able to effectively activate naïve antigen-specific CD4+ T-cells and restore the responsiveness of anergic T-cells in vitro. Thus, HDAC6 represents a novel target to therapeutically tip the balance towards the generation of inflammatory APCs able of inducing T-cell activation rather than T-cell unresponsiveness.
Unlike IL-10 stimulation, which results in STAT3 phosphorylation within minutes, LPS triggers STAT3 phosphorylation at later time points (after 4 hours). This kinetics suggests an indirect mechanism for LPS-induced STAT3 phosphorylation, perhaps mediated by cytokines produced by macrophages in response to LPS. Nevertheless, in the absence of HDAC6 this mechanism of LPS-induced STAT3 phosphorylation is also disrupted. Of note, such a disruption was accompanied by a decreased expression of several STAT3-regulated genes in addition to Il10. Among these genes it is important to highlight VEGF and SOCS3 since they have been shown to negatively influence T-cell activation (46, 57, 58). However, our data points to the regulatory effects of HDAC6 upon the STAT3/IL-10 axis as the major explanation for the increased inflammatory phenotype and improved T cell priming elicited by APCs lacking HDAC6. First, transfection of HDAC6KO APCs with a plasmid expressing IL-10 resulted in lost of their improved T-cell priming function. Second, adding back recombinant IL-10 to cultures of APCs lacking HDAC6 with antigen-specific T-cells also yielded similar results, i.e. diminished IFN-γ production by T-cells. Third, the addition of anti-IL-10 neutralizing antibodies attenuated IL-10 production, B7.2 upregulation and the T-cell priming function of wild type APCs, but had minimal effects upon APCs lacking HDAC6.
Although previous reports have indicated that the deacetylase activity of type I HDACs, in particular HDAC1 and HDAC3, play a role in regulating the function and nuclear trafficking of STAT3(8, 44, 59), we did not find any changes in STAT3 acetylation in the absence of HDAC6. Instead, our studies are the first linking HDAC6 with regulation of STAT3 phosphorylation. Given its lack of kinase activity, it is plausible that HDAC6 could influence the function of upstream tyrosine kinases known to regulate STAT3 phosphorylation. However, we did not find significant differences in the expression of total JAK2, phospho-JAK2, total TYK2 and phospho-TYK2 between wild type or APCs lacking HDAC6. Alternatively, HDAC6 might influence the function of phosphatase(s) that regulate phosphorylation of STAT3. Supporting this possibility, Gupta et al. reported that treatment of malignant cells with the pan-HDACi LBH589, which inhibits the enzymatic activity of HDAC6 among other HDACs, resulted in increased protein phosphatase activity and Akt dephosphorylation (60). More recently, the McKinsey group has shown that class I HDAC inhibitors suppress ERK1/2 signaling through the induction of an ERK-specific phosphatase, DUSP5 (61). Furthermore, Togi et al. found that HDAC3 interacts with the phosphatase PP2A to regulate phosphorylation of target proteins (59). The particular association of HDAC6, HDAC1 and HDAC10 with phosphatases has been previously reported in transformed cells (62). Of note, treatment of these cells with the pan-HDACi TSA disrupted the association of HDACs with phosphatases, suggesting that their enzymatic activity is necessary for this association. In line with these reports, we have recently found an important increase in the global Tyr phosphatase activity in HDAC6KO PEM when compared to wild type PEM (Supplementary Figure 2B), suggesting that the diminished STAT3 phosphorylation observed in these APCs could be a consequence of an enhanced phosphatase activity in the absence of HDAC6. Experiments to identify which STAT3-specific phosphatase(s) might be regulated/associated with HDAC6 are currently ongoing in our laboratory.
In summary, we have unveiled a previously unknown mechanism for the transcriptional regulation of Il10 that involves HDAC6 and STAT3. To our knowledge, this is the first demonstration of such an interaction that is necessary for STAT3 phosphorylation and Il10 gene expression. A dynamic interaction between HDAC6 and STAT3 that occurs both in the cytoplasmic and nuclear compartments of the APC, and as a result, dynamic changes in the expression of the tolerogenic IL-10 cytokine, might explain the intrinsic plasticity of these cells to determine T-cell activation versus T-cell tolerance, a critical decision with important implications to the autoimmunity, cancer immunotherapy and transplantation fields.
Supplementary Material
Acknowledgements
This work was supported by PHS Grants CA100850, CA87583, CA134807 (EMS) and 1R21CA153246 (AV).
Footnotes
Disclosures
The authors have no financial conflict of interest
References
- 1.Georgopoulos K. From immunity to tolerance through HDAC. Nat Immunol. 2009;10:13–14. doi: 10.1038/ni0109-13. [DOI] [PubMed] [Google Scholar]
- 2.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 3.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 4.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Ruijter AJM, van Gennip AH, Caron HN, Kemp S, van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Roeder RG. Activation of p53 Sequence-Specific DNA Binding by Acetylation of the p53 C-Terminal Domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 8.Yuan Z.-l., Guan Y.-j., Chatterjee D, Chin YE. Stat3 Dimerization Regulated by Reversible Acetylation of a Single Lysine Residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 9.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 10.Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F Family Members Are Differentially Regulated by Reversible Acetylation. J Biol Chem. 2000;275:10887–10892. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 11.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 13.Verdel A, Curtet S, Brocard MP, Rousseaux S, Lemercier C, Yoshida M, Khochbin S. Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 14.Palijan A, Fernandes I, Bastien Y, Tang L, Verway M, Kourelis M, Tavera-Mendoza LE, Li Z, Bourdeau V, Mader S, Yang XJ, White JH. Function of Histone Deacetylase 6 as a Cofactor of Nuclear Receptor Coregulator LCoR. J Biol Chem. 2009;284:30264–30274. doi: 10.1074/jbc.M109.045526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toropainen S, Väisänen S, Heikkinen S, Carlberg C. The Down-regulation of the Human MYC Gene by the Nuclear Hormone 1[alpha],25-dihydroxyvitamin D3 is Associated with Cycling of Corepressors and Histone Deacetylases. J Mol Biol. 2010;400:284–294. doi: 10.1016/j.jmb.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 16.Aldana-Masangkay GI, Sakamoto KM. The role of HDAC6 in cancer. J Biomed Biotechnol. 2010;2011:875824. doi: 10.1155/2011/875824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valenzuela-Fernández A, Cabrero JR, Serrador JM, Sánchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends in Cell Biology. 2008;18:291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Jung KH, Noh JH, Kim JK, Eun JW, Bae HJ, Chang YG, Kim MG, Park WS, Lee JY, Lee SY, Chu IS, Nam SW. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology. 2012;56:644–657. doi: 10.1002/hep.25699. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Peng L, Seto E, Huang S, Qiu Y. Modulation of histone deacetylase 6 (HDAC6) nuclear import and tubulin deacetylase activity through acetylation. J Biol Chem. 2012;287:29168–29174. doi: 10.1074/jbc.M112.371120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng F, Lienlaf M, Perez-Villarroel P, Wang HW, Lee C, Woan K, Woods D, Knox T, Bergman J, Pinilla-Ibarz J, Kozikowski A, Seto E, Sotomayor EM, Villagra A. Divergent roles of histone deacetylase 6 (HDAC6) and histone deacetylase 11 (HDAC11) on the transcriptional regulation of IL10 in antigen presenting cells. Mol Immunol. 2014;60:44–53. doi: 10.1016/j.molimm.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrador JM, Cabrero JR, Sancho D, Mittelbrunn M, Urzainqui A, Sanchez-Madrid F. HDAC6 deacetylase activity links the tubulin cytoskeleton with immune synapse organization. Immunity. 2004;20:417–428. doi: 10.1016/s1074-7613(04)00078-0. [DOI] [PubMed] [Google Scholar]
- 22.de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, Bradner JE, Mazitschek R, Kozikowski AP, Matthias P, Hancock WW. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol. 2011;31:2066–2078. doi: 10.1128/MCB.05155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halili MA, Andrews MR, Labzin LI, Schroder K, Matthias G, Cao C, Lovelace E, Reid RC, Le GT, Hume DA, Irvine KM, Matthias P, Fairlie DP, Sweet MJ. Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J Leukoc Biol. 2010;87:1103–1114. doi: 10.1189/jlb.0509363. [DOI] [PubMed] [Google Scholar]
- 24.Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, Cervo M, Yao H, Chung AL, Mizumura K, An CH, Shan B, Franks JM, Haley KJ, Owen CA, Tesfaigzi Y, Washko GR, Quackenbush J, Silverman EK, Rahman I, Kim HP, Mahmood A, Biswal SS, Ryter SW, Choi AM. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest. 2013;123:5212–5230. doi: 10.1172/JCI69636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann BS, Johnson JR, He K, Sridhara R, Abraham S, Booth BP, Verbois L, Morse DE, Jee JM, Pope S, Harapanhalli RS, Dagher R, Farrell A, Justice R, Pazdur R. Vorinostat for Treatment of Cutaneous Manifestations of Advanced Primary Cutaneous T-Cell Lymphoma. Clin Cancer Res. 2007;13:2318–2322. doi: 10.1158/1078-0432.CCR-06-2672. [DOI] [PubMed] [Google Scholar]
- 26.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotech. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 27.Campas-Moya C. Romidepsin for the treatment of cutaneous T-cell lymphoma. Drugs Today (Barc) 2009;45:787–795. doi: 10.1358/dot.2009.45.11.1437052. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Cheng F, Woan K, Sahakian E, Merino O, Rock-Klotz J, Vicente-Suarez I, Pinilla-Ibarz J, Wright KL, Seto E, Bhalla K, Villagra A, Sotomayor EM. Histone Deacetylase Inhibitor LAQ824 Augments Inflammatory Responses in Macrophages through Transcriptional Regulation of IL-10. J Immunol. 2011;186:11. doi: 10.4049/jimmunol.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, Nguyen D, Wright KL, Atadja PW, Bhalla K, Pinilla-Ibarz J, Seto E, Sotomayor EM. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10:92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex-restricted receptor. J Exp Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, Lombard D, Mizeracki A, Matthias G, Alt FW, Khochbin S, Matthias P. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vicente-Suarez I, Takahashi Y, Cheng F, Horna P, Wang HW, Wang HG, Sotomayor EM. Identification of a novel negative role of flagellin in regulating IL-10 production. Eur J Immunol. 2007;37:3164–3175. doi: 10.1002/eji.200737306. [DOI] [PubMed] [Google Scholar]
- 34.Sotomayor EM, Fu YX, Lopez-Cepero M, Herbert L, Jimenez JJ, Albarracin C, Lopez DM. Role of tumor-derived cytokines on the immune system of mice bearing a mammary adenocarcinoma. II. Down-regulation of macrophage-mediated cytotoxicity by tumor-derived granulocyte-macrophage colony-stimulating factor. J Immunol. 1991;147:2816–2823. [PubMed] [Google Scholar]
- 35.Han X, Kitamoto S, Wang H, Boisvert WA. Interleukin-10 overexpression in macrophages suppresses atherosclerosis in hyperlipidemic mice. FASEB J. 2010;24:2869–2880. doi: 10.1096/fj.09-148155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang X-J, Dent SR, Yao T-P, Lane WS, Seto E. HDAC6 Modulates Cell Motility by Altering the Acetylation Level of Cortactin. Molecular Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staveley-O' Carroll K, Sotomayor E, Montgomery J, Borrello I, Hwang L, Fein S, Pardoll D, Levitsky H. Induction of antigen-specific T cell anergy: An early event in the course of tumor progression. Proc Natl Acad Sci U S A. 1998;95:1178–1183. doi: 10.1073/pnas.95.3.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng F, Wang HW, Cuenca A, Huang M, Ghansah T, Brayer J, Kerr WG, Takeda K, Akira S, Schoenberger SP, Yu H, Jove R, Sotomayor EM. A critical role for Stat3 signaling in immune tolerance. Immunity. 2003;19:425–436. doi: 10.1016/s1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- 40.Saraiva M, O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:11. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 41.Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational Design and Simple Chemistry Yield a Superior, Neuroprotective HDAC6 Inhibitor, Tubastatin A. Journal of the American Chemical Society. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sotomayor EM, Borrello I, Tubb E, Rattis FM, Bien H, Lu Z, Fein S, Schoenberger S, Levitsky HI. Conversion of tumor-specific CD4+ T-cell tolerance to T-cell priming through in vivo ligation of CD40. Nat Med. 1999;5:780–787. doi: 10.1038/10503. [DOI] [PubMed] [Google Scholar]
- 43.Wang R, Cherukuri P, Luo J. Activation of Stat3 Sequence-specific DNA Binding and Transcription by p300/CREB-binding Protein-mediated Acetylation. J Biol Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 44.Ray S, Lee C, Hou T, Boldogh I, Brasier AR. Requirement of histone deacetylase1 (HDAC1) in signal transducer and activator of transcription 3 (STAT3) nucleocytoplasmic distribution. Nucleic Acids Res. 2008;36:4510–4520. doi: 10.1093/nar/gkn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murray PJ. The JAK-STAT Signaling Pathway: Input and Output Integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 46.Niemand C, Nimmesgern A, Haan S, Fischer P, Schaper F, Rossaint R, Heinrich PC, Muller-Newen G. Activation of STAT3 by IL-6 and IL-10 in primary human macrophages is differentially modulated by suppressor of cytokine signaling 3. J Immunol. 2003;170:3263–3272. doi: 10.4049/jimmunol.170.6.3263. [DOI] [PubMed] [Google Scholar]
- 47.Villagra A, Cruzat F, Carvallo L, Paredes R, Olate J, van Wijnen AJ, Stein GS, Lian JB, Stein JL, Imbalzano AN, Montecino M. Chromatin Remodeling and Transcriptional Activity of the Bone-specific Osteocalcin Gene Require CCAAT/Enhancer-binding Protein beta-dependent Recruitment of SWI/SNF Activity. J. Biol. Chem. 2006;281:22695–22706. doi: 10.1074/jbc.M511640200. [DOI] [PubMed] [Google Scholar]
- 48.Woan KV, Sahakian E, Sotomayor EM, Seto E, Villagra A. Modulation of antigen-presenting cells by HDAC inhibitors: implications in autoimmunity and cancer. Immunol Cell Biol. 2012;90:55–65. doi: 10.1038/icb.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 50.Grütz G. New insights into the molecular mechanism of interleukin-10-mediated immunosuppression. Journal of Leukocyte Biology. 2005;77:3–15. doi: 10.1189/jlb.0904484. [DOI] [PubMed] [Google Scholar]
- 51.Wakkach A, Fournier N, Brun V, Breittmayer JP, Cottrez F, Groux H. Characterization of dendritic cells that induce tolerance and T regulatory 1 cell differentiation in vivo. Immunity. 2003;18:605–617. doi: 10.1016/s1074-7613(03)00113-4. [DOI] [PubMed] [Google Scholar]
- 52.Li MO, Flavell RA. Contextual regulation of inflammation: a duet by transforming growth factor-beta and interleukin-10. Immunity. 2008;28:468–476. doi: 10.1016/j.immuni.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, Jr., Muller W, Rudensky AY. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 54.Murai M, Turovskaya O, Kim G, Madan R, Karp CL, Cheroutre H, Kronenberg M. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozikowski AP, Tapadar S, Luchini DN, Kim KH, Billadeau DD. Use of the Nitrile Oxide Cycloaddition (NOC) Reaction for Molecular Probe Generation: A New Class of Enzyme Selective Histone Deacetylase Inhibitors (HDACIs) Showing Picomolar Activity at HDAC6. Journal of Medicinal Chemistry. 2008;51:4370–4373. doi: 10.1021/jm8002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bergman JA, Woan K, Perez-Villarroel P, Villagra A, Sotomayor EM, Kozikowski AP. Selective histone deacetylase 6 inhibitors bearing substituted urea linkers inhibit melanoma cell growth. J Med Chem. 2012;55:9891–9899. doi: 10.1021/jm301098e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabrilovich DI, Chen HL, Girgis KR, Cunningham HT, Meny GM, Nadaf S, Kavanaugh D, Carbone DP. Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells. Nat Med. 1996;2:1096–1103. doi: 10.1038/nm1096-1096. [published erratum appears in Nat Med 1996 Nov;2(11):1267] [DOI] [PubMed] [Google Scholar]
- 58.Brender C, Tannahill GM, Jenkins BJ, Fletcher J, Columbus R, Saris CJ, Ernst M, Nicola NA, Hilton DJ, Alexander WS, Starr R. Suppressor of cytokine signaling 3 regulates CD8 T-cell proliferation by inhibition of interleukins 6 and 27. Blood. 2007;110:2528–2536. doi: 10.1182/blood-2006-08-041541. [DOI] [PubMed] [Google Scholar]
- 59.Togi S, Kamitani S, Kawakami S, Ikeda O, Muromoto R, Nanbo A, Matsuda T. HDAC3 influences phosphorylation of STAT3 at serine 727 by interacting with PP2A. Biochem Biophys Res Commun. 2009;379:616–620. doi: 10.1016/j.bbrc.2008.12.132. [DOI] [PubMed] [Google Scholar]
- 60.Gupta M, Ansell SM, Novak AJ, Kumar S, Kaufmann SH, Witzig TE. Inhibition of histone deacetylase overcomes rapamycin-mediated resistance in diffuse large B-cell lymphoma by inhibiting Akt signaling through mTORC2. Blood. 2009;114:2926–2935. doi: 10.1182/blood-2009-05-220889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferguson BS, Harrison BC, Jeong MY, Reid BG, Wempe MF, Wagner FF, Holson EB, McKinsey TA. Signal-dependent repression of DUSP5 by class I HDACs controls nuclear ERK activity and cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A. 2013;110:9806–9811. doi: 10.1073/pnas.1301509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brush MH, Guardiola A, Connor JH, Yao T-P, Shenolikar S. Deactylase Inhibitors Disrupt Cellular Complexes Containing Protein Phosphatases and Deacetylases. J Biol Chem. 2004;279:7685–7691. doi: 10.1074/jbc.M310997200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.