Abstract
Aberrant immune response and changes in the gut microflora are the main causes of inflammatory bowel disease (IBD). Peptidoglycan recognition proteins (Pglyrp1, Pglyrp2, Pglyrp3, and Pglyrp4) are bactericidal innate immunity proteins that maintain normal gut microbiome, protect against experimental colitis, and are associated with inflammatory bowel disease in humans. Nod2 is an intracellular bacterial sensor and may be required for maintaining normal gut microbiome. Mutations in Nod2 are strongly associated with Crohn's disease, but the causative mechanism is not understood, and Nod2 role in ulcerative colitis is not known. Because IBD is likely caused by variable multiple mutations in different individuals, in this study we examined the combined role of Pglyrp3 and Nod2 in the development of experimental colitis in mice. We demonstrate that a combined deficiency of Pglyrp3 and Nod2 results in higher sensitivity to dextran sodium sulfate (DSS)-induced colitis compared with a single deficiency. Pglyrp3−/−Nod2−/− mice had decreased survival and higher loss of body weight, increased intestinal bleeding, higher apoptosis of colonic mucosa, elevated expression of cytokines and chemokines, altered gut microbiome, and increased levels of ATP in the colon. Increased sensitivity to DSS-induced colitis in Pglyrp3−/−Nod2−/− mice depended on increased apoptosis of intestinal epithelium, changed gut microflora, and elevated ATP. Pglyrp3 deficiency contributed colitispredisposing intestinal microflora and increased intestinal ATP, whereas Nod2 deficiency contributed higher apoptosis and responsiveness to increased level of ATP. In summary, Pglyrp3 and Nod2 are both required for maintaining gut homeostasis and protection against colitis, but their protective mechanisms differ.
INTRODUCTION
Mammalian gastrointestinal tract is inhabited by thousands of microbial species that exist in a mutually beneficial relationship with the host. The gut microflora influences normal development of the host immune system, prevents colonization and damage induced by opportunistic bacteria, and regulates development and repair of the intestinal mucosa (1-4). The host immune system along with environmental factors, in turn, shapes the composition of the microflora. Crohn's Disease (CD) and ulcerative colitis (UC) are common inflammatory bowel diseases (IBD) and are associated with deficiencies in the host immune system and with changes in gut microflora (5, 6). Host genetic factors involved in microbial recognition and immune responses are associated with IBD; however, the specific genes and their precise role in the development of both CD and UC remain mostly unknown (7-10). Similarly, the precise changes in the microflora and their role in the pathogenesis of IBD are not clearly understood (4-6).
Peptidoglycan Recognition Proteins (PGRP or Pglyrp) are innate immunity proteins that are conserved from insects to mammals, recognize the bacterial cell wall component peptidoglycan, and are antibacterial (11-13). Mammals have four Pglyrps, Pglyrp1, Pglyrp2, Pglyrp3, and Pglyrp4 (14, 15); Pglyrp1 is expressed in polymorphonuclear leukocytes, whereas Pglyrp2, Pglyrp3, and Pglyrp4 are expressed in epithelial cells of many organs, including salivary glands, throat, tongue, esophagus, stomach, and intestine (15-17). All mammalian Pglyrps are secreted proteins and are bactericidal for both Gram-negative and Gram-positive bacteria (12, 17-19). Pglyrp2 is also an amidase that hydrolyzes bacterial cell wall peptidoglycan (20, 21).
Pglyrps are important for maintaining a normal gut microbiome and for protection against experimental colitis (22). Pglyrp1−/−, Pglyrp2−/−, Pglyrp3−/−, and Pglyrp4−/− mice are all more sensitive to experimental (DSS-induced) colitis than WT mice (22). DSS-treated Pglyrp-deficient mice show a greater loss of body weight, more severe intestinal bleeding, more severe colon pathology, hyperplasia of the lamina propria, and loss of colonic epithelial cells (22). These changes are accompanied by higher production of IFN-γ and increased numbers of NK cells in the colon (22). In the absence of Pglyrps the gut flora changes to a more damaging, proinflammatory microbiome and these changes are responsible for the increased sensitivity to colitis in Pglyrp-deficient mice (22).
Pglyrps also modulate sensitivity to other inflammatory diseases. Pglyrp2 protects mice against psoriasis-like skin inflammation (23) and is required for the development of experimental arthritis (24), whereas Pglyrp3 and Pglyrp4 protect mice against atopic dermatitis (25). By contrast, Pglyrp1 has a pro-inflammatory effect in three mouse models of inflammatory skin diseases (psoriasis, atopic dermatitis, and contact dermatitis) (23, 25) and in experimentally induced asthma (26), but has anti-inflammatory effect in experimentally induced arthritis (24). Thus, each Pglyrp has a unique role in the development of different inflammatory diseases, and for this reason Pglyrps do not compensate for each other in mice deficient in a single Pglyrp. These unique effects of each Pglyrp are most likely based on their different effects on the microbiome (22). Moreover, genetic variants in human Pglyrp1, Pglyrp2, Pglyrp3, and Pglyrp4 genes are associated with CD and UC (27). Several of these Pglyrp polymorphisms result in missense amino acid changes, which may indicate a role of the Pglyrp proteins in modulating sensitivity to colitis in humans (27).
Nod2 is a member of the NLR family of cytosolic receptors, is expressed in antigen-presenting cells, such as monocytes (28) and dendritic cells (29), and also in structural cells such as intestinal epithelial cells (30), and is involved in host immune responses (31-33). Nod2 is an intracellular sensor for muramyl dipeptide, a bacterial peptidoglycan fragment (34, 35), and activation of Nod2 results in the activation of NF-κB and MAP kinase signaling cascades, followed by the synthesis and release of proinflammatory cytokines, chemokines, and antimicrobial peptides (33, 36). Nod2 was the first susceptibility gene identified for CD and common homozygous or compound heterozygous mutations in Nod2 increase susceptibility to CD by 20-fold (37-39). However, the functional role of Nod2 in CD remains unexplained, primarily because CD is characterized by increased inflammation in the intestine, whereas, Nod2 mutations associated with CD result in reduced activation of NF-κB and decreased production of inflammatory mediators after stimulation with muramyl dipeptide (35, 40). Studies in both mice and humans also present variable and contradictory roles of Nod2 in innate immune responses and in the development of intestinal inflammation. In few studies that examined the role of Nod2 in ulcerative colitis the data are also contradictory. Earlier results show no difference between WT and Nod2−/− mice in the sensitivity to DSS-induced colitis (41), whereas later studies show higher sensitivity of Nod2−/− mice to similar treatment (42, 43). Genome-wide association studies have demonstrated that CD and UC share many susceptibility loci, however a Nod2 loss-of-function variant, which is a risk allele for CD, may have a protective effect for UC (10). These results indicate similarities and difference in the mechanisms of disease development between CD and UC.
Here we tested the hypothesis that deficiencies in both Pglyrp3 and Nod2 synergize in increasing the sensitivity to experimental colitis, because both Pglyrp3 and Nod2 recognize the bacterial component peptidoglycan and play a role in maintaining normal intestinal microflora (22, 43-46). We demonstrate that Pglyrp3−/−Nod2−/− double knockout mice are more sensitive to DSS-colitis than Pglyrp3−/− and Nod2−/− single knockouts. The increased susceptibility to colitis in Pglyrp3−/−Nod2−/− mice is accompanied by increased apoptosis of epithelial cells in the colon, elevated expression of chemokines and cytokines, changes in gut bacteria, and higher levels of ATP in the colon. Our data indicate that Pglyrp3 deficiency is mainly responsible for the change to colitis-predisposing gut bacteria and higher ATP, whereas Nod2 deficiency is mainly responsible for the higher apoptosis in the colon and responsiveness to higher levels of ATP. Our data also show that the lack of Nod2 expression in bone marrow-derived cells combined with expression of Nod2 in structural (radio-resistant) cells predisposes Pglyrp3−/− mice to very severe colitis. We conclude that Pglyrp3 and Nod2 deficiencies enhance sensitivity to experimental colitis through different mechanisms, which are synergistic.
MATERIALS AND METHODS
Mice
Pglyrp3−/− (22) and Nod2−/− (24, 41) mice on BALB/c background were described previously. Pglyrp3−/− mice were crossed with Nod2−/− mice to generate the homozygous double knockout Pglyrp3−/−Nod2−/− mice. Deletion of the Pglyrp3 and Nod2 genes was confirmed by PCR analysis of genomic DNA (22, 24). Pglyrp3−/−Nod2−/− mice were viable, bred normally, and produced the expected male to female ratios and similar litter size as the WT mice. They had similar weight as WT mice and developed normally with no obvious defects. Their major internal organs had normal macroscopic appearance and normal histological appearance on hematoxylin/eosin stained sections. The original colony founder WT BALB/c mice were obtained from Harlan-Sprague-Dawly. WT germ-free mice (Swiss Webster female) were obtained from Taconic Farms (Hudson, NY).
All WT and knockout mice were on BALB/c background, female, 8-9 week-old, bred and kept under conventional pathogen-free conditions in the same room in our facility to minimize the influence of differences in the environment. For each experiment, mice from several different cages and breeder pairs were used. We did not use WT and homozygous knockout littermates from heterozygous breeding pairs for three reasons: first, this strategy cannot be used for double knockout mice; second, this strategy may skew the results to the particular microflora present only in this breeding pair; and third, the effect of Pglyrps on the composition of the microbiome is not instantaneous, but takes time, and stabilization of microbiome characteristic of a given mutant strain takes more than one generation. To avoid changes in microbiome that could accumulate over extended period of time, we backcross our mutant mice to WT females once every other year and re-derive our homozygous knockout breeding pairs. The latter strategy also minimized genetic drift in the population. The BALB/c background of knockout mice and their negative status for all common viral and bacterial pathogens and parasites (including negative PCR stool tests for mouse Norovirus) were confirmed as previously described (24). The Indiana University School of Medicine–Northwest Institutional Animal Care and Use Committee approved all experiments with mice.
Colitis model in conventional and germ-free mice
Experimental colitis was induced in WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice with 5% DSS (dextran sulfate sodium, MP Biomedical) in drinking water (47). DSS-induced intestinal inflammation is a well-established animal model for colitis and its manifestations include bloody diarrhea, weight loss, shortening of the colon, mucosal ulceration, and epithelial dysplasia. Manifestations such as predominant left-sided colitis, epithelial dysplasia and lack of granulomas are similar with ulcerative colitis; however, the complexity of the human disease is not completely reproduced in the DSS model (47). The development and severity of colitis was evaluated as previously described (22), using: (a) mortality; (b) weight loss; and (c) stool and rectal bleeding scores of 0-16. For evaluation of histopathology, untreated control mice and mice treated with 5% DSS were sacrificed on day 7, proximal and distal sections of the colon were fixed in 10% buffered formalin, embedded in paraffin and sectioned. Sections were stained with hematoxylin and eosin and scored for hyperplasia, loss of crypts, infiltration of immune cells, loss of epithelium, loss of goblet cells, and extent of ulceration, to evaluate severity of colitis. The scoring scale was from 0 to 5, with 0 being no change and 5 being the greatest change.
To determine the role of gut microflora in the development of colitis, female WT germ-free mice, 4 to 5 week-old, maintained under sterile conditions, were treated with sterile 4% DSS in drinking water and gavaged daily into the stomach with 12 mg stools from WT, Nod2−/−, Pglyrp3−/−, or Pglyrp3−/−Nod2−/− mice, prepared as previously described (22). Briefly, fresh stools were collected form 12 mice/strain, which were obtained from six different breeding parents (2 mice from 6 different parents for each strain), and kept in separate cages after weaning. This strategy minimizes the variability observed between different litters due to parent-to-parent and cage-to-cage differences. Stools were immediately resuspended in 0.2 ml of reduced anaerobic medium and frozen at −80oC until use. Development of colitis was monitored as described above.
To determine whether gut microflora from Pglyrp3−/− mice was also predisposing to colitis in Nod2−/− mice, an established non-germ-free model (48, 49) was used, in which Nod2−/−mice were depleted of their intestinal microflora with a 3-week treatment with antibiotics in drinking water (containing 0.33 mg/ml ciprofloxacin, 1.25 mg/ml metronidazole, and 20 mg/ml Kool-Aid mix), followed by 2 days of sterile drinking water, followed by treatment with sterile 4% DSS in drinking water and daily gavages into the stomach with 12 mg stools from Pglyrp3−/− or WT mice. Development of colitis was monitored as described above.
Intestinal permeability
WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice were given 5% DSS in drinking water and on days 0, 3, 6, and 7 mice were gavaged into the stomach with FITC-dextran solution (MW 4000, Sigma) at 0.6 g/kg body weight and sacrificed 4 h later. FITC-dextran in the serum was measured in duplicate samples with a fluorescence spectrophotometer. The concentration of FITC-dextran in μg/ml was determined using a standard curve generated with serial dilutions of FITC-dextran.
Cell proliferation
To determine the numbers of proliferating cells, WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/− Nod2−/− were given 5% DSS in drinking water and on days 0 and 4 mice were injected intraperitoneally with bromodexoyuridine (BrdU, Sigma) at 2 mg/mouse, twice at 12 h intervals. Mice were sacrificed 12 h after the second injection. Proximal sections of the colon were fixed in 10% buffered formalin and embedded in paraffin. Sections were stained with BrdU In-Situ Detection Kit (BD Pharmingen) and numbers of BrdU-positive cells per 500 epithelial cells were counted for each section.
TUNEL and cleaved caspase-3 staining and inhibition of cleaved caspase-3
The numbers of apoptotic cells in the colon were determined by TUNEL-labeling and by staining for cleaved caspase-3. WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice were given 5% DSS in drinking water and on days 0 and 4 mice were sacrificed. Proximal sections of the colon were fixed in 10% buffered formalin, embedded in paraffin, and sectioned. TUNEL-positive cells were detected by DeadEnd Colorimetric TUNEL System (Promega). Cleaved caspase-3 positive cells were detected using rabbit monoclonal antibody (clone 5A1E, Cell Signaling) and VECTASTAIN ABC Kit with DAB substrate Kit. TUNEL-positive or cleaved caspase-3-positive cells and total epithelial cells were counted on the entire section and percent of TUNEL-positive or cleaved caspase-3-positive cells were calculated.
To confirm the role of cleaved caspase 3 in the development of DSS-colitis, Pglyrp3−/− Nod2−/− mice were given 5% DSS in drinking water and gavaged daily into the stomach with the caspase inhibitor, Q-VD-Oph (R&D Systems, OPH001), at 80 µg/mouse/day or with 10% DMSO in PBS. The development of colitis was monitored as described in the Colitis Model section. To determine the effect of the caspase inhibitor, Q-VD-Oph, on apoptosis, Pglyrp3−/− Nod2−/− mice were treated with 5% DSS in drinking water and gavaged daily with caspase inhibitor (80 μg/mouse/day) or vehicle control (10% DMSO in PBS). On day 4, proximal and distal regions of the colon were fixed in 10% buffered formalin, embedded in paraffin, and sectioned. TUNEL-positive cells were detected by DeadEnd Colorimetric TUNEL System (Promega). TUNEL-positive cells and total epithelial cells were counted on the entire section and percent of TUNEL-positive were calculated.
DSS-treated gavaged mice show a delay in the onset of colitis and mortality compared with DSS-treated non-gavaged mice, because gavaged mice drink less DSS-containing water, and thus consume less DSS per day. Mice are reluctant to drink DSS-containing water and frequent gavaging provides mice with some fluid, and thus allows them to avoid drinking as much DSS-containing water as they would drink without gavaging. This shift in the onset of colitis and mortality did not affect our conclusions, because in each experiment we compared identically treated mice, i.e., all non-gavaged, or all on the same gavage regimen.
Inflammatory arrays
RNA was isolated from individual colons of WT, Nod2−/−, Pglyrp3−/− and Pglyrp3−/−Nod2−/−mice, untreated or treated with 5% DSS for 48, 72, and 96 h, using the TRIZOL method (InVitrogen), followed by digestion with RNAse-free DNAse (Qiagen) and purification on RNeasy spin columns using RNeasy Mini Kit (Qiagen) (22, 23, 26). Quantitative real-time reverse transcription (qRT-PCR) was used to quantify the amounts of mRNA in the colon using Mouse Inflammatory Arrays (Qiagen/SA Biosciences) or individual primer sets for some genes, with pooled cDNA from 3 mice/group, in 3 separate experiments (total 9 mice per group). For each gene, ΔCt was calculated followed by normalization to 5 housekeeping genes (Hsp90ab1, Gusb, Hprt1, Gapdh, and Actb) included in each array, followed by calculation of ΔΔCt for each gene: ΔΔCt = ΔCt1 – ΔCt2, where ΔCt1 is for DSS-treated mice and ΔCt2 is for untreated mice, using the program provided by Qiagen/SA Biosciences. This calculation gives the fold increase in expression of each gene in the treated mice versus untreated mice. The genomic DNA contamination controls, reverse transcription controls, and positive PCR controls were included in each array and were all passed. Additional controls to assure amplification from RNA, but not from possible contaminating DNA included parallel reaction sets from which reverse transcriptase was omitted, and which showed no amplification. The results were reported as mean fold increases after DSS treatment (treated/untreated) for all groups of mice and the entire data sets were deposited in NCBI GEO (accession number GSE47588). In some experiments, the expression of additional genes was measured by the same procedure using sets of qRT-PCR primers from SA Biosciences (IL-6, Nod2, Pglyrp1, Pglyrp3, Pglyrp4) or designed by us (24).
Stool flora analysis by qPCR
The abundance of specific bacterial groups in mouse stools was measured by qPCR using group-specific primers for 16S rRNA genes as previously described (22). Briefly, 200 mg of fresh stools (freshly defecated feces) were collected from female mice of each strain (WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/−) and immediately processed for DNA isolation or snap-frozen at -80ºC. Stools were collected from all strains and from total of 18 mice/strain at three time points throughout the entire period of this study (6 mice/strain each time in 2008, 2010, and 2013). Each time for each strain mice originated from 3 different litters from different parents, 2 mice per litter, weaned into separate cages, but all kept in the same room in our animal facility. DNA was isolated from stools from each mouse using Qiagen QIAamp DNA Stool Mini Kit. Abundance of all bacteria and specific bacterial groups was determined by qPCR using 20 ng DNA and common primers for all Eubacteria or primers specific for the following bacterial groups: Mouse Intestinal Bacteroides, Bacteroides sp., Eubacterium rectale/Clostridium coccoides, Clostridium leptum, Lactobacillus/Lactococcus, Segmented filamentous bacteria, Enterobacteriaceae, and Clostridium perfringens, with primer sequences described previously (22, 50, 51). The amounts of DNA for each bacterial group for each mouse were calculated using comparative cycle threshold method with common Eubacteria primers as a control. We combined the data from the three collection time points, because there were no statistically significant differences in the abundance of the bacterial groups tested for each strain between these three time points. The results for WT and Pglyrp3−/− mice with DNA collected in 2008 and 2010 were reported previously (22).
Bone marrow-derived macrophage isolation, colon and macrophage culture, activation, and cytokine assay
Untreated WT and Nod2−/− mice were sacrificed and bone marrow was flushed from femurs with cold RPMI. The cells were cultured overnight in RPMI-1640 supplemented with 10% FBS and antibiotic/antimycotic mix (Sigma) at 37°C, 5% CO2. The next day non-adherent cells were collected and cultured in complete medium with 10 ng/ml of murine colony stimulating factor (PeproTech Inc.), in 48-well plates at 0.5 × 106 cells per well and incubated for an additional 5 to 6 days or until they became confluent. For colon cultures, colons were excised from untreated WT and Nod2−/− mice, washed to remove fecal material and cut into 0.5 cm pieces. Cells and colon fragments were stimulated with diluted stools prepared from WT or Pglyrp3−/− mice and supernatants were collected and assayed for cytokines and chemokines, CCL-2, CXCL-9, CXCL-10, IL-6, and CXCL-1 by ELISA using paired capture and detection antibodies and standards from R&D Systems. For stimulation, 30 mg of fresh stools were collected from 6 WT or Pglyrp3−/− mice and immediately placed on ice and processed. For each strain, mice originated from 3 different litters from different parents (2 mice per litter), weaned into separate cages, but all kept in the same room in our animal facility. Stools were suspended in 5 ml of DPBS, sonicated on ice for 2 min, and centrifuged at 30g for 2 min to remove debris. The OD660 of the supernatants was adjusted to 1.4 and diluted superantants were used for cell activation experiments.
ATP measurement and colitis
Fresh stools were collected from WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice, 6 to 8 mice/group from different cages, 6 pellets per mouse, and resuspended in PBS by gently vortexing to avoid lysis of bacteria. The resuspended feces were centrifuged at 5,000 rpm, 4°C, for 15 min, and the amount of ATP in the supernatant was measured in triplicate samples from each mouse using the ATPlite System (Perkin Elmer). To assess the effect of ATP on the sensitivity to colitis, WT and Nod2−/− mice were treated with 5% DSS to induce colitis and gavaged daily into the stomach with 50 μg of α,β-ATP (Sigma-Aldrich) per mouse or with PBS. The development of colitis was monitored as described in the Colitis Model section.
Generation of chimeric mice with bone marrow transplantation
Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice, 6 to 8 mice/recipient group, were subjected to total body irradiation of 850 rads for marrow ablation, administered in 2 doses, 4 h apart. Bone marrow cells were obtained from the femurs and tibias of donor Pglyrp3−/− and Pglyrp3−/−Nod2−/−mice. 4 h after irradiation, 5 × 106 bone marrow cells were injected into the retro orbital vein of recipient mice. Four transplanted groups were generated: Pglyrp3−/− > Pglyrp3−/−, Pglyrp3−/− > Pglyrp3−/−Nod2−/−, Pglyrp3−/−Nod2−/− > Pglyrp3−/−Nod2−/−, Pglyrp3−/−Nod2−/− > Pglyrp3−/−. Transplanted mice were kept in a HEPA-filtered flow hood and given sterilized food and water (52). For the first four weeks mice were given filtered water with 15 g/L sucrose, 220 μg/ml neomycin and 25 μg/ml polymyxin B, and then switched to filtered water. To re-establish the recipient gut microflora, 24 h after the end of antibiotic treatment Pglyrp3−/− and Pglyrp3−/−Nod2−/−mice were colonized with microflora from Pglyrp3−/− or Pglyrp3−/−Nod2−/−, respectively, by gavaging with 0.2 ml of suspension containing 12 mg of preserved stools (22), three times over 6 days. Mice were then allowed to recover for one week before treatment with DSS to induce colitis. We used 4% DSS, rather than 5%, to avoid early mortality, which would obscure differences in sensitivity to colitis in these highly sensitive strains. The stool gavage procedure and measurement of severity of colitis were done as described in the Colitis Model section. Bone marrow reconstitution was verified at 6 weeks after irradiation and before the start of DSS. Blood was collected from individual mice, genomic DNA was isolated using the Qiagen Puregene Blood Core Kit and Nod2 WT and KO alleles were determined by PCR analysis (24, 41).
Isolation of lamina propria cells and flow cytometry
Nod2−/− mice were gavaged once daily into the stomach with 50 μg of α,β-ATP per mouse or with PBS and given 5% DSS in drinking water to induce colitis. Mice were sacrificed on day 4 and colon lamina propria (LP) cells were isolated as described (53). Briefly, colons were excised, washed to remove fecal material and cut into 0.5 cm pieces. Colon pieces were shaken in HBSS with 5% FBS and 1 mM EDTA at 37°C, 2 times for 20 min. Epithelial cells were removed by straining through 100 μm filter and the colon tissue was digested with 2 mg/ml collagenase D (Worthington T3), 40 μg/ml DNAse I (Sigma DN25-1G) in RPMI-1640 supplemented with 10% FCS for 80 min. The colon LP cell suspensions were obtained by passing the digested tissue through 100 μm and then 40 μm filters, centrifuging, and resuspending in RPMI-1640 with 5% FBS. Colon LP cells were stained with the following fluorochrome-conjugated antibodies in different combinations: CD45-Vioblue, CD3-PE, CD8-APC from Miltenyi; CD4-APC, CD4-Pacific Blue, IFN-γ-PE, IL4-PE, IL10-PE, IL17-PE, and TGFβ-APC from Biolegend, and Foxp3-PE from eBioscience. All extracellular and intracellular staining was performed as described previously (25, 26). CD4-stained cells were fixed, permeabilized, and stained for Foxp3. Prior to staining for cytokines, CD4-stained cells were stimulated with TPA (25 ng/ml) and ionomycin (250 ng/ml) in the presence of the Golgi inhibitor, monensin, for 4 h at 37°C, 5% CO2, fixed, permeabilized, and stained for cytokines (25, 26). Cells were analyzed by flow cytometry using MACSQuant (Miltenyi) cytometer. CD3, CD4, and CD8 positive cells were measured within the CD45 gate. IFN-γ, IL-4, IL-10, IL-17, TGFβ, and Foxp3 positive cells were measured within the CD4 gate.
Statistical analysis
All quantitative results are presented as means ± SEM, with statistical significance of the differences between groups determined by the two-sample one-tailed Student's t-test, except for survival, which was analyzed using Chi-square test; p ≤ 0.05 was considered significant.
RESULTS
Pglyrp3−/−Nod2−/− mice are highly susceptible to DSS-induced colitis
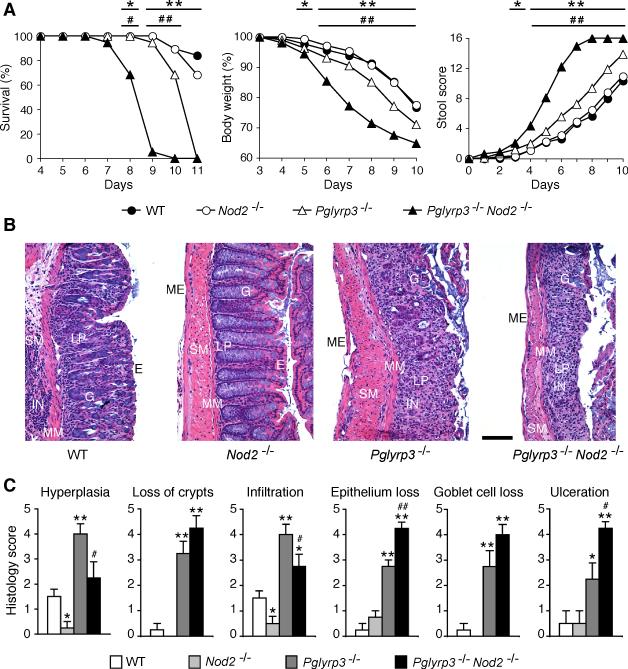
To test whether Pglyrp3 and Nod2 deficiencies synergize in increasing sensitivity to colitis, we constructed Pglyrp3−/−Nod2−/− double deficient mice and tested their sensitivity to colitis. Colitis was induced by oral administration of 5% DSS in drinking water in WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice. Pglyrp3−/−Nod2−/− mice developed more severe colitis than Pglyrp3−/− and Nod2−/− mice (Fig. 1). Pglyrp3−/−Nod2−/− mice had accelerated and significantly higher mortality, accelerated and significantly greater weight loss, and significantly higher diarrhea and intestinal bleeding than Pglyrp3−/−, Nod2−/−, and WT mice. Pglyrp3−/− mice, as shown previously, also developed severe DSS-induced colitis (22), however, they were less sensitive than Pglyrp3−/−Nod2−/− mice, as demonstrated by significantly higher survival, significantly lower weight loss, and significantly less severe intestinal bleeding than Pglyrp3−/−Nod2−/− mice (Fig. 1). By contrast, Nod2−/− mice were far less sensitive to colitis and manifested symptoms similar to WT mice (Fig. 1). We also evaluated histopathologic changes as a measure of the severity of colitis by scoring for hyperplasia, loss of crypts, infiltration with inflammatory cells, loss of epithelium, loss of goblet cells, and extent of ulceration on hematoxylin and eosin stained sections of the colon. These histopathologic changes are characteristic for colitis. Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice had significantly higher scores for hyperplasia, loss of crypts, infiltration with inflammatory cells, loss of epithelium, loss of goblet cells, and ulceration than WT and Nod2−/− mice (Fig. 1B and 1C). Furthermore, Pglyrp3−/−Nod2−/− mice had significantly higher scores for loss of crypts, loss of epithelial and goblet cells and ulceration than WT, Nod2−/−, and Pglyrp3−/− mice (Fig. 1B and 1C). Thus, our phenotypic and histopathologic data indicate that the double knockout Pglyrp3−/−Nod2−/− mice are more sensitive to DSS-induced colitis than single knockouts Pglyrp3−/− and Nod2−/− and WT mice.
Figure 1. Pglyrp3−/−Nod2−/− mice are more susceptible to DSS-induced colitis than Pglyrp3 and Nod2 single-deficient mice.
WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice were treated with 5% DSS in drinking water and development of colitis was evaluated. (A) DSS-treated mice were monitored over time for survival, change in body weight, and stool and rectal bleeding. (B and C) DSS-treated mice were sacrificed on day 7 of treatment and hematoxylin-eosin stained sections of the colon were (B) qualitatively and (C) quantitatively evaluated for severity of colitis. Epithelial cells (E), lamina propria (LP), goblet cells (G), muscularis mucosa (MM), submucosa (SM), muscularis externa (ME), and inflammatory cell infiltrations (IN) are indicated; size bar = 100 μm. The data are (A) mean ± SEM of 19-20 mice/group; and (B) representative images or (C) mean ± SEM from 4 mice/group. Significance of differences between Pglyrp3−/−Nod2−/− and WT mice is indicated by asterisks (*); and between Pglyrp3−/−Nod2−/− and Pglyrp3−/− is indicated by the number sign (#); *, #, P ≤ 0.05; **, ##, P ≤ 0.005.
Pglyrp3−/−Nod2−/− mice are deficient in cell proliferation and have higher apoptosis in the intestine
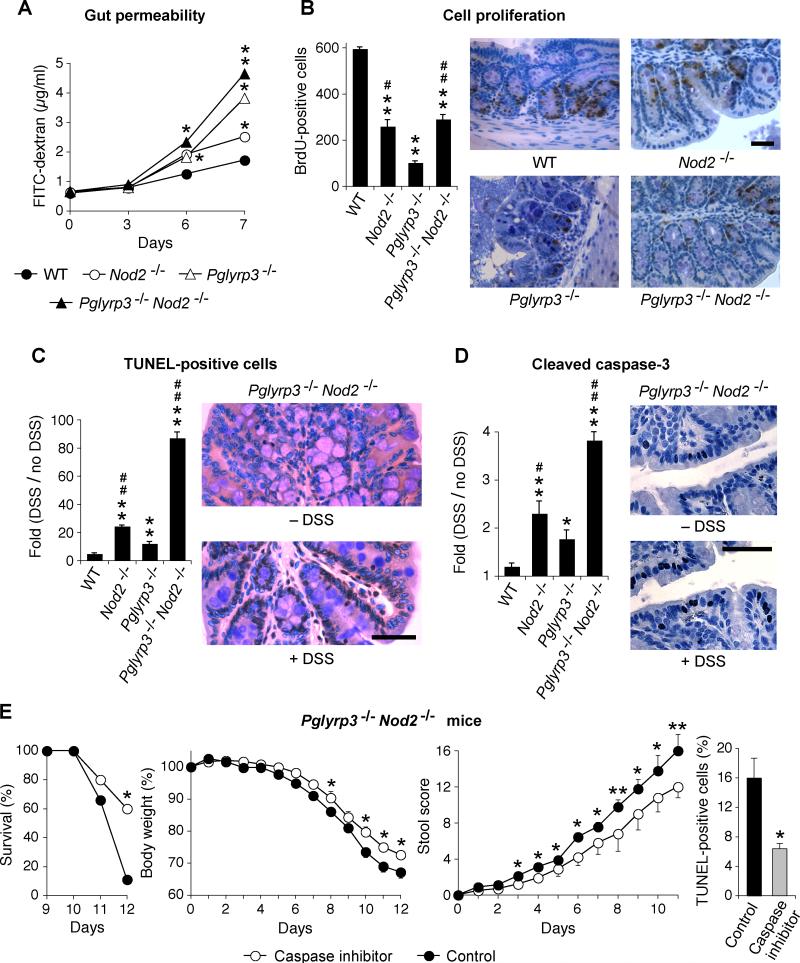
DSS-colitis is an epithelial injury model of IBD and we next determined whether the higher sensitivity of Pglyrp3−/−Nod2−/− mice to DSS-induced colitis is accompanied by changes in the permeability of the intestine and in the rates of proliferation and apoptosis of colon epithelial cells. In order to measure permeability changes, DSS-colitis was induced in WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice, and FITC-dextran was administered by gavage before sacrifice. Mice were sacrificed at different time points and the amount of FITC-dextran in the serum was measured. The concentration of FITC-dextran in the serum was significantly higher in Pglyrp3−/−Nod2−/− mice on days 6 and 7 of DSS treatment compared with WT, Nod2−/−, and Pglyrp3−/− mice (Fig. 2A). Pglyrp3−/− mice had significantly higher levels of FITC-dextran on day 7 than Nod2−/−mice and on days 6 and 7 than WT mice (Fig. 2A). These results indicate that both Pglyrp3 and Nod2 contribute to the integrity of the intestinal mucosa and a deficiency of both genes significantly increases damage to the intestine in DSS-treated mice. However, Pglyrp3−/− mice have more prolonged and severe damage than Nod2−/− mice.
Figure 2. Pglyrp3−/−Nod2−/− mice have higher permeability and increased apoptosis in the colon.
(A to D) WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice were treated with 5% DSS and (A) gavaged with FITC-dextran on days 0, 3, 6, and 7 and assayed for FITC-dextran in the serum; (B) 4 days later injected with BrdU and colon sections were stained for BrdU; (C and D) 4 days later colon sections were stained for (C) TUNEL; or (D) cleaved caspase-3. (E) Pglyrp3−/− Nod2−/− mice were treated with 5% DSS and gavaged daily with the caspase inhibitor Q-VD-Oph or with PBS for the control group and development of colitis was monitored by survival, change in body weight, and stool and rectal bleeding scores or 4 days later colon sections were stained for TUNEL. The data are mean ± SEM of (A) 12, (B, C, and D) 4 to 6, and (E) 9 to 10 mice/group for colitis and 6 mice/group for TUNEL staining; representative histological images are shown in (B-D). Significance of differences for (A to D) knockout versus WT mice or (E) inhibitor treated versus control group is indicated by asterisks (*); and between Pglyrp3−/−Nod2−/− or Nod2−/− versus Pglyrp3−/− is indicated by the number sign (#); *, #, P ≤ 0.05; **, ##, P ≤ 0.005.
In order to identify the changes in the intestinal mucosa that result in increased permeability we determined the rate of colonic cell proliferation and apoptosis in DSS-treated and untreated mice. To measure the numbers of proliferating cells, DSS-treated or untreated WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice were gavaged with BrdU and proliferating cells were identified by immunohistochemistry. Pglyrp3−/−, Nod2−/−, and Pglyrp3−/−Nod2−/− mice all had significantly fewer BrdU-positive cells than WT mice (Fig. 2B). Also, Pglyrp3−/− mice had significantly fewer BrdU-positive cells than Nod2−/− and Pglyrp3−/−Nod2−/− mice (Fig. 2B). These results indicate that Pglyrp3 deficiency results in a severe inhibition of proliferation, which most likely contributes to the sensitivity to DSS-induced colitis in Pglyrp3−/− and Pglyrp3−/−Nod2−/−mice. By contrast, Nod2 deficiency caused less severe decrease in proliferation (Fig. 2B), which is reflected in less severe colitis in Nod2−/− than in Pglyrp3−/− mice (Fig. 1).
We next measured apoptosis in the colons of DSS-treated and untreated WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice using TUNEL immunohistochemistry. Pglyrp3−/−Nod2−/−mice had significantly more TUNEL-positive cells than Pglyrp3−/−, Nod2−/−, and WT mice and Nod2−/− mice had significantly more TUNEL-positive cells than Pglyrp3−/− and WT mice (Fig. 2C). To further evaluate apoptosis in the intestinal epithelium of DSS-treated Pglyrp3−/−Nod2−/− mice we assayed for cleaved caspase-3, which is the active form of caspase-3, an enzyme essential for apoptosis. We detected cleaved caspase-3 in the intestinal mucosa of untreated and DSS-treated WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice by immunohistochemistry. DSS-treated Pglyrp3−/−Nod2−/− mice had the highest increase of cleaved caspase-3 positive cells, significantly higher than Pglyrp3−/−, Nod2−/−, and WT mice (Fig. 2D). DSS-treated Nod2−/− mice had significantly higher numbers of cleaved caspase-3 positive cells than Pglyrp3−/− and WT mice (Fig. 2D). Our TUNEL and cleaved caspase-3 data indicate that a combined deficiency in both Pglyrp3 and Nod2 results in the highest apoptosis of colonic epithelial cells and that Nod2 deficiency has a greater contribution to this high apoptosis than Pglyrp3 deficiency.
Because Pglyrp3−/−Nod2−/− mice were highly sensitive to DSS-colitis and had significantly higher numbers of apoptotic epithelial cells in the colon, we next tested the effect of a caspase inhibitor on the development of colitis in these mice. Pglyrp3−/−Nod2−/− mice were treated with DSS and gavaged daily with the caspase inhibitor Q-VD-OPh or PBS as a control. The caspase inhibitor significantly decreased all clinical manifestations of colitis. Caspase inhibitor-treated Pglyrp3−/−Nod2−/− mice showed significantly higher survival and lower weight loss and stool scores than PBS-treated control mice in response to DSS (Fig. 2E). We next determined the effect of the caspase inhibitor Q-VD-OPh on apoptosis in the colon. The numbers of TUNEL-positive colon epithelial cells were significantly lower in caspase inhibitor-treated than control Pglyrp3−/−Nod2−/− mice (Fig. 2E). These results demonstrate that the caspase inhibitor Q-VD-OPh is effective in decreasing apoptosis of intestinal epithelial cells and that apoptosis is required for the full severity of DSS-induced colitis in Pglyrp3−/−Nod2−/− mice. However, the caspase inhibitor did not completely abrogate the symptoms of colitis, which indicates that other factors also contribute to the disease severity in DSS-treated Pglyrp3−/−Nod2−/− mice.
In summary, these results demonstrate that both Pglyrp3 and Nod2 are required for maintaining a functional and intact intestinal mucosa. A single deficiency of Pglyrp3, but not Nod2, is sufficient to make mice more sensitive to DSS-induced colitis. However, a combined deficiency of Pglyrp3 and Nod2 synergistically increases sensitivity to DSS-induced colitis, and increased apoptosis of intestinal epithelial cells is one of the factors responsible for this increased sensitivity.
Pglyrp3−/−Nod2−/− mice have increased expression of inflammatory molecules
We next measured the expression of inflammatory molecules in the colon using qRT-PCR microarrays. DSS-treated Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice had significantly higher, but not identical, expression of mRNA for several cytokines and chemokines (Fig. 3 and Table S1 with entire array data). Both Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice had higher expression of Th1 and NK cell markers, IFN-γ, CXCL-9, CXCL-10, and CXCL-11; Th2 markers, CCL-7 and CCL-8; Th17 markers, CCL-2, CXCL-1, and CXCL-5, than WT and Nod2−/− mice (Fig. 3). DSS-treated Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice also had elevated expression of the cytokines IL-1β, IL-6, and IL-11 (which are produced by many different cell types) compared with WT and Nod2−/− mice (Fig. 3). In addition, Pglyrp3−/−Nod2−/− mice had significantly higher expression of TNF-α, IL-1f6, and CCR-7 than Pglyrp3−/−, WT, and Nod2−/− mice (Fig. 3). These results indicate that colitis in Pglyrp3−/−Nod2−/− mice is accompanied by higher induction of several inflammatory mediators, many of which are shared with the Pglyrp3−/− inflammatory profile and some that are unique. Because expression of TNF-α, IL-1f6, and CCR-7 was higher in Pglyrp3−/−Nod2−/− than in Pglyrp3−/− mice, these molecules may contribute to the higher sensitivity of Pglyrp3−/−Nod2−/−mice to DSS-colitis. Several of these inflammatory gene responses in DSS-treated Pglyrp3−/− Nod2−/− mice were Nod2-independent, because they were similar or higher than in Pglyrp3−/−(Nod2+/+) mice (Fig. 3 and Table S1).
Figure 3. Pglyrp3−/−Nod2−/− mice express higher levels of chemokine and cytokine mRNA in the colon following DSS treatment.
WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice were treated with 5% DSS for indicated times and expression of inflammatory genes in the colon was measured by qRT-PCR. The results are means ± SEM of the ratio of the amount of mRNA in DSS-treated to untreated mice from 9 mice per group, 3 mice pooled per array. Significance of differences between knockouts and WT mice is indicated by asterisks (*) and between Pglyrp3−/− Nod2−/− and Pglyrp3−/− is indicated by the number sign (#); *, #, P ≤ 0.05; **, ##, P ≤ 0.005. The data are selected from the 96 inflammatory gene expression array shown in supplemental Table S1.
We determined that expression of Pglyrp1, Pglyrp2, Pglyrp3 and Pglyrp4 genes in tongue, esophagus, and colon was similar in Nod2−/− and WT mice (data not shown). We have previously shown that treatment with DSS increases the expression of Pglyrp3 in the colon (22). We further determined that the expression of Pglyrp3 in the colon in DSS-treated WT and Nod2−/− mice was similar, and also that the expression of Nod2 in the colon was similar in untreated or DSS-treated Pglyrp3−/− and WT mice (data not shown). These results indicate that the colitis phenotype in Pglyrp3−/−Nod2−/− double knockout mice is not due to changes in the expression of Pglyrp3 in a Nod2-deficient background or changes in the expression of Nod2 in a Pglyrp3-deficient background.
Pglyrp3-dependent, but not Nod2-dependent, intestinal microflora predisposes to DSS-colitis
Pglyrp3 is a secreted antibacterial protein and we have previously shown that Pglyrp3−/− mice have changes in the gut bacterial flora, which are responsible for the higher sensitivity of Pglyrp3−/− mice than WT mice to DSS-induced colitis (22). Nod2 is also involved in innate immune responses to bacteria and Nod2−/− mice also have an altered microbial population in the gut (43-46). Therefore, we next tested whether Pglyrp3−/−Nod2−/− mice have additional changes in the gut microflora and whether these cumulative changes are responsible for the higher sensitivity to colitis in the double-knockout mice. We compared the composition of bacterial flora in the stools of WT, Pglyrp3−/−, Nod2−/−, and Pglyrp3−/−Nod2−/− mice, by measuring the amounts of DNA for all Eubacteria and for eight major groups of mouse intestinal bacteria using qPCR. Single and double knockout mice had significant, but not identical changes in the composition of their bacterial flora in their stools. The amounts of bacteria in the following bacterial groups showed significant changes in knockout compared with WT mice: Lactobacillus and Clostridium perfringens groups in Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice; Eubacterium rectale, Clostridium leptum, and Enterobacteriaceae groups in Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice; and Segmented Filamentous bacteria in Pglyrp3−/−Nod2−/− mice (Fig. 4A).
Figure 4. Pglyrp3−/−Nod2−/− mice have changes in intestinal bacterial flora, however only Pglyrp3-dependent changes contribute to the increased sensitivity to colitis in Pglyrp3−/− Nod2−/− mice.
(A) Changes in the composition of bacterial flora in the stools of WT and knockout mice in all Eubacteria and the indicated bacterial groups, measured by qPCR; (B) WT germ-free mice were treated with 4% DSS and gavaged daily with stool homogenates from conventionally raised WT, Nod2−/−, Pglyrp3−/−, or Pglyrp3−/−Nod2−/− mice or (C) Nod2−/− mice were pre-treated with antibiotics for 3 weeks and then gavaged daily with stool homogenates from conventionally raised WT or Pglyrp3−/− and treated with 4% DSS. Development of colitis in B and C was evaluated by measuring survival, body weight, and stool score. Gross rectal bleeding in mice gavaged with stools from Pglyrp3−/− mice but not from WT mice is shown in (C). The results are means ± SEM of (A) 18 mice/group and (B and C) 6 mice/group from 2 experiments. Significance of differences for (A) *, knockout versus WT; (B) *, Pglyrp3−/− versus Nod2−/− and WT; #, Pglyrp3−/−Nod2−/− versus Nod2−/− and WT; ^, Pglyrp3−/−Nod2−/− versus Pglyrp3−/−; (C) *, Pglyrp3−/− versus WT; *, #, ^, P ≤ 0.05; **, ##, ^^, P ≤ 0.005.
We next tested whether these changes in the microflora in Pglyrp3−/−Nod2−/− mice are responsible for their increased sensitivity to DSS-colitis. We gavaged WT germ-free mice daily with stools from WT, Nod2−/−, Pglyrp3−/−, or Pglyrp3−/−Nod2−/− mice and we induced colitis with 4% DSS. Germ-free mice receiving stools from Pglyrp3−/− or Pglyrp3−/−Nod2−/− mice were more sensitive to DSS-colitis than mice receiving Nod2−/− or WT stools, but the sensitivity of mice receiving stools from Pglyrp3−/− or Pglyrp3−/−Nod2−/− mice was similar. Germ-free mice gavaged with stools from Pglyrp3−/−Nod2−/− or Pglyrp3−/− mice had significantly higher mortality, higher weight loss, and higher stool scores than mice gavaged with Nod2−/− or WT stools, and all these indicators were similar in germ-free mice gavaged with stools from Pglyrp3−/−Nod2−/− and Pglyrp3−/− mice (Fig. 4B). The sensitivity to colitis of germ-free mice gavaged with stools from WT or Nod2−/− mice was similar. These results confirm our previous results that the increased sensitivity to DSS colitis in Pglyrp3−/− mice is due to an altered gut microflora (22), and further demonstrate that this altered microflora contributes to a similar extent to the increased sensitivity to DSS colitis in Pglyrp3−/− mice and in Pglyrp3−/−Nod2−/− mice. These results also demonstrate that microflora from Nod2−/− mice does not predispose germ-free mice to colitis compared with microflora from WT mice. Thus, these results demonstrate that Pglyrp3−/− mice contribute colitis-predisposing flora to Pglyrp3−/−Nod2−/− mice, and that the additional changes in the microflora in Pglyrp3−/−Nod2−/− mice, compared with Pglyrp3−/− mice, do not further contribute to the increased sensitivity of Pglyrp3−/−Nod2−/− mice to colitis.
We further tested the effect of microflora from Pglyrp3−/− mice on the development of DSS-colitis in a Nod2-deficient background. We treated Nod2−/− mice with antibiotics for 3 weeks to deplete their intestinal microflora. At the end of 3 weeks we gavaged the antibiotic-treated mice with stools from WT or Pglyrp3−/− mice and induced colitis with oral DSS. Nod2−/−mice receiving stools from Pglyrp3−/− mice were more sensitive to DSS-colitis than mice receiving WT stools (Fig. 4C). The more severe colitis in Nod2−/− mice gavaged with stools from Pglyrp3−/− mice manifested as significantly higher loss in body weight and stool scores and significantly lower survival (Fig. 4C). These results indicate that Pglyrp3−/− microflora is stable, Pglyrp3-dependent, and has colitis-predisposing characteristics on both Nod2−/− and Nod2+/+ backgrounds.
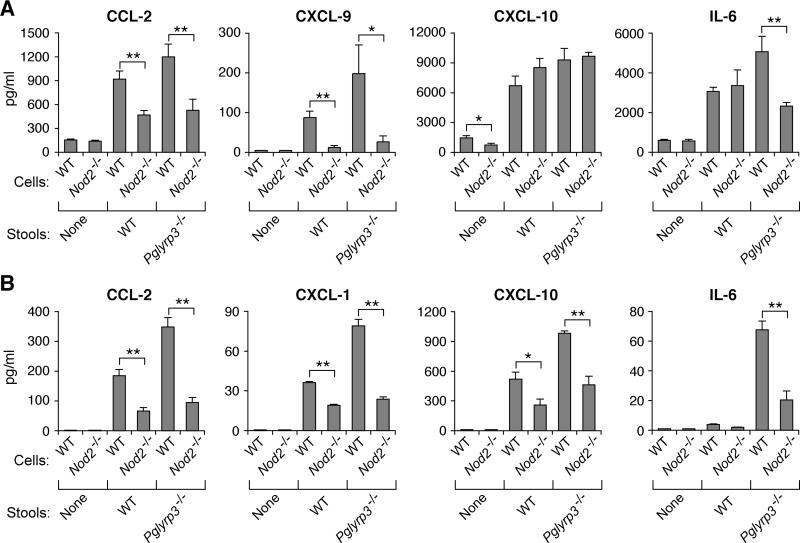
Our previous results demonstrated that the gut microflora from Pglyrp3−/− mice induces higher production of inflammatory molecules, IL-6 and CXCL-1, than WT microflora (22). Thus, we next tested whether the response to gut bacteria from Pglyrp3−/− mice is variable in Nod2−/− and Nod2+/+ backgrounds, because Nod2 is one of the bacterial recognition cell-activating pattern recognition receptors, and because Pglyrp3−/−Nod2−/− mice are more sensitive to colitis than Pglyrp3−/− mice. Colon cells and fragments or bone marrow macrophages from WT or Nod2−/− mice were stimulated with diluted stools from WT or Pglyrp3−/− mice, to determine whether cells from WT or Nod2−/− mice have differential responsiveness to colitis-promoting (Pglyrp3−/−) and non-promoting (WT) stools (Fig. 4 and ref. 22). Culture supernatants were then assayed for different cytokines and chemokines. Stools from Pglyrp3−/− mice induced higher production of CCL-2, CXCL-9, and IL-6 in cells from WT, but not from Nod2−/− colon cells and fragments (Fig. 5A) and higher production of CCL-2, CXCL-1, CXCL-10, and IL-6 in WT, but not Nod2−/− bone marrow macrophages (Fig. 5B) compared with stools from WT mice. These results confirm our previous data that stools from Pglyrp3−/− mice are more pro-inflammatory than stools from WT mice (22) and show that induction of these pro-inflammatory chemokines and cytokines is significantly higher on Nod2+/+ than on Nod2−/− background. The results in Fig. 5 also show that although the responses of cells from Nod2−/− mice are generally lower than from WT mice (as expected), the cells from Nod2−/− mice are still responsive to bacteria (presumably through other bacterial sensors), and also that all responses are not equally affected, showing that some responses are Nod2-dependent and some Nod2-independent and that cells from Nod2−/− mice are differentially responsive to stools from WT and Pglyrp3−/− mice. For example, CXCL-10 production was not reduced in colon from Nod2−/− mice, compared with WT mice (Fig. 5A), and bone marrow macrophages from both WT and Nod2−/− mice produced IL-6 in response to stools only from Pglyrp3−/− mice, but not WT mice (Fig. 5B). Moreover, several inflammatory gene responses to DSS in the entire colon in vivo involved additional Nod2-independent signals, because many of these responses were similar or higher in Pglyrp3−/−Nod2−/− compared with Pglyrp3−/− mice (Fig. 3 and Table S1).
Figure 5. WT (Nod+/+) but not Nod2−/− macrophages and colon cells produce more chemokines and cytokines in response to stools from Pglyrp3-deficient mice.
(A) Colon cells and colon fragments or (B) bone marrow-derived macrophages from WT or Nod2−/− mice were stimulated with stools from WT or Pglyrp3−/− mice or with PBS (no stimulant). Supernatants were assayed for the indicated chemokines or cytokines. The results are means ± SEM (N = 6 experiments); significance of differences for Nod2−/− versus WT cells; *, P ≤ 0.05; **, P ≤ 0.005.
Pglyrp3−/− and Pglyrp3−/− Nod2−/− mice have increased ATP in the colon, which increases inflammatory T cells and predisposes to colitis
Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice have altered gut microflora, which contributes to the increased sensitivity to DSS-colitis. In order to identify bacterial metabolites that may facilitate damage to the intestine, we assayed for ATP in mouse stools, because: (i) ATP is produced by gut microflora; (ii) ATP is known to increase sensitivity to T-cell-mediated colitis by increasing the numbers of inflammatory Th17 cells in the intestine (54); and (iii) we had increased expression of Th17 markers, CCL-2, CXCL-1, and CXCL-5 in the colon of DSS-treated Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice (Fig. 3).
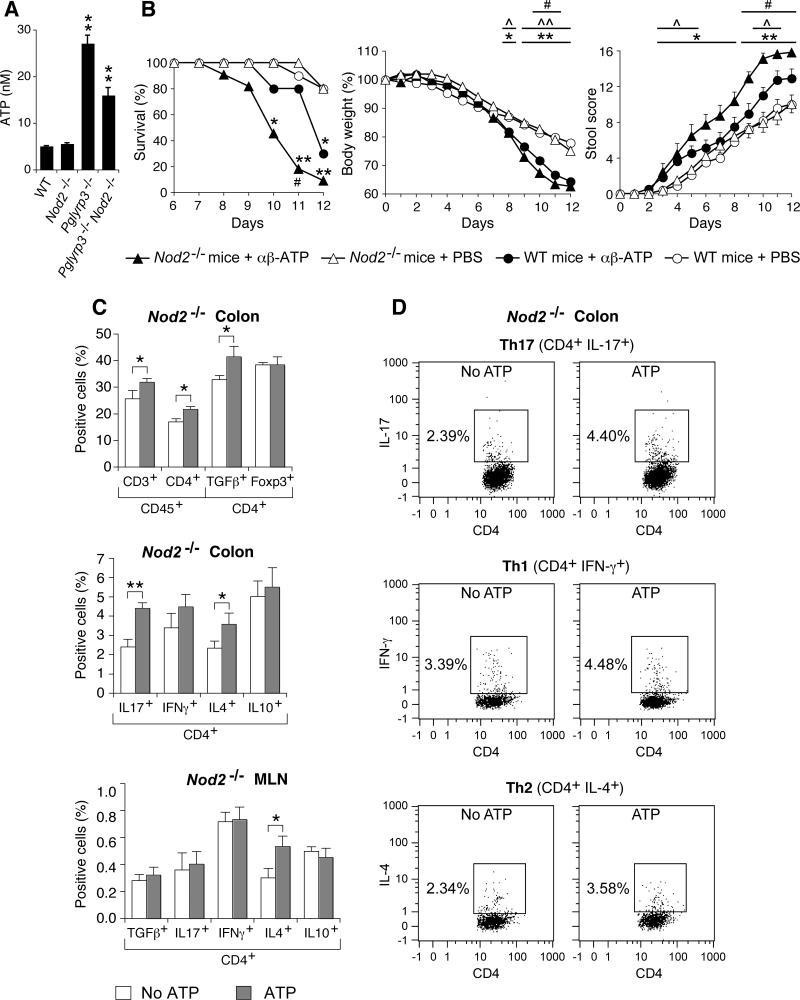
Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice had significantly higher levels of ATP in the stools than WT and Nod2−/− mice (Fig. 6A). Thus, we next tested whether elevated ATP increases the sensitivity to DSS-colitis in Nod2−/− and WT mice. We used Nod2−/− mice for these experiments to recreate the Nod2−/− genetic background of Pglyrp3−/−Nod2−/− mice and one aspect of Pglyrp3 deficiency (higher ATP) and we used WT mice as controls. Nod2−/− and WT mice were treated with DSS to induce colitis, gavaged daily into the stomach with the stable ATP analog, α,β-ATP or PBS (control), and assayed for the development of colitis. Nod2−/− and WT mice treated with α,β-ATP showed significantly higher mortality, significantly greater loss in body weight, and significantly higher stool scores, compared with PBS-treated Nod2−/− and WT mice, respectively (Fig. 6B). Furthermore, Nod2−/− mice treated with α,β-ATP had significantly higher mortality, significantly greater loss in body weight, and significantly higher stool scores, than WT mice treated with α,β-ATP. These results indicate that increased levels of ATP promote DSS-induced colitis in Nod2−/− and WT mice, and that Nod2 deficiency further enhances the promoting effect of ATP on the severity of colitis. Our results also suggest that the increased sensitivity to DSS-colitis in Pglyrp3−/−Nod2−/− mice can be partially explained by the elevated ATP in the intestine and that Pglyrp3 deficiency contributes the increased ATP, whereas Nod2 deficiency enhances sensitivity to ATP and severity of colitis.
Figure 6. Pglyrp3−/−Nod2−/− and Pglyrp3−/− mice have higher levels of ATP in the colon, and intestinal ATP preferentially increases sensitivity of Nod2−/− mice to DSS-colitis.
(A) Amounts of ATP in the stools of WT, Nod2−/−, Pglyrp3−/−, and Pglyrp3−/−Nod2−/− mice. (B) WT and Nod2−/− mice treated with 5% DSS and gavaged with the ATP analog, α,β-ATP, have lower survival and body weight, and higher stool scores than PBS-treated mice, and Nod2−/− mice are more sensitive to ATP than WT mice. (C) Increased percentages of the indicated cell types in colon lamina propria and mesenteric lymph nods (MLN) in Nod2−/− mice, gated for CD45 or CD4. (D) Representative dot plots for Th17, Th1, and Th2 cells for the data shown in (C). The results are means ± SEM of (A) 6-8, (B) 10-11, and (C) 6-7 mice/group from 2 experiments. Significance of differences for (A) knockouts versus WT; (B) *, **, ATP- versus PBS-treated Nod2−/− mice; ^, ^^, ATP- versus PBS-treated WT mice; #, Nod2−/− versus WT ATP-treated mice; or (C) ATP versus PBS; *, ^, #, P ≤ 0.05; **, ^^, P ≤ 0.001.
To further understand the role of elevated colon ATP levels in the increased sensitivity to DSS-colitis of Pglyrp3−/−Nod2−/− mice, we next compared the levels of inflammatory T cells in the colon lamina propria of DSS treated Nod2−/− mice in the presence and absence of the stable ATP analog, α,β-ATP. Again, we used Nod2−/− mice for these experiments to recreate Nod2−/− genetic background of Pglyrp3−/−Nod2−/− mice and one aspect of Pglyrp3 deficiency (higher ATP). The numbers of CD3+, CD4+, CD4+TGFβ+, CD4+IL-4+ (Th2), and CD4+IL-17+ (Th17) cells were significantly higher in DSS-treated Nod2−/− mice gavaged with α,β-ATP than in the control mice gavaged with PBS (Fig. 6C and D). There was no significant difference in CD4+ cells expressing Foxp3+ (Fig. 6C), which is the marker for the anti-inflammatory regulatory T (Treg) cells. These data also correlate with our mRNA expression microarray results, which demonstrate higher expression of Th2 and Th17 markers in Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice (Fig. 3 and Table S1).
In conclusion, a deficiency in Pglyrp3 results in high levels of ATP in the colon, and high ATP results in increased numbers of inflammatory Th17 and Th2 cells in the colon and causes higher sensitivity to DSS-colitis. These results suggest that in Pglyrp3−/−Nod2−/− mice, Pglyrp3-deficiency contributes the elevated ATP and that Nod2−/− mice respond to higher concentration of ATP with increased numbers of inflammatory Th17 and Th2 cells and higher sensitivity to DSS-colitis. These results offer one possible mechanism of microbiota-induced sensitivity to colitis in Nod2−/− mice, which are generally less responsive to bacteria than WT mice (Fig. 5), yet are more sensitive to colitis induced by microflora from Pglyrp3−/− mice (Fig. 4C).
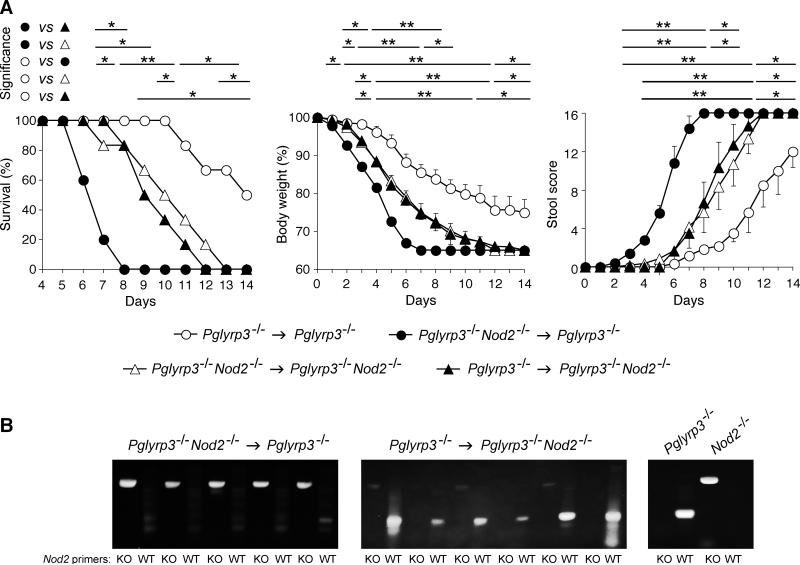
Lack of Nod2 in bone marrow-derived cells combined with presence of Nod2 in structural cells predisposes Pglyrp3−/− mice to severe colitis
Pglyrp3 is expressed in epithelial cells, whereas Nod2 is expressed in both bone marrow-derived and structural (epithelial and stromal) cells. We next determined whether the lack of Nod2 expression in bone marrow-derived cells or structural cells was important for enhanced sensitivity to DSS-induced colitis in Pglyrp3−/−Nod2−/− mice. We generated Pglyrp3−/− > Pglyrp3−/−Nod2−/− and Pglyrp3−/−Nod2−/− > Pglyrp3−/− bone marrow chimeric mice, as well as control mice, which received bone marrow cells from the same strain of mice: Pglyrp3−/− > Pglyrp3−/− and Pglyrp3−/−Nod2−/− > Pglyrp3−/−Nod2−/−. Chimerism was confirmed by PCR of genomic DNA and showed that blood cells from Pglyrp3−/−Nod2−/− > Pglyrp3−/− mice contained only Nod2 KO allele, whereas blood cells from Pglyrp3−/− > Pglyrp3−/−Nod2−/− mice contained only Nod2 WT allele (Fig. 7B). All groups of mice were treated with DSS to induce colitis and development of colitis was monitored. Pglyrp3−/−Nod2−/− > Pglyrp3−/− chimeras developed the most severe colitis compared with Pglyrp3−/− > Pglyrp3−/−Nod2−/− chimeras and the control Pglyrp3−/− > Pglyrp3−/− and Pglyrp3−/−Nod2−/− > Pglyrp3−/−Nod2−/− mice, with significantly higher mortality, significantly higher stool scores, and significantly greater loss of body weight (Fig. 7A). Pglyrp3−/− > Pglyrp3−/−Nod2−/− and Pglyrp3−/−Nod2−/− > Pglyrp3−/−Nod2−/− mice had intermediate sensitivity to colitis, and Pglyrp3−/− > Pglyrp3−/− mice developed the least severe colitis.
Figure 7. Lack of Nod2 in bone marrow-derived cells combined with presence of Nod2 in structural cells predisposes Pglyrp3−/− mice to severe colitis.
(A) Chimeric (Pglyrp3−/− > Pglyrp3−/−Nod2−/− and Pglyrp3−/−Nod2−/− > Pglyrp3−/−) and transplanted control (Pglyrp3−/− > Pglyrp3−/− and Pglyrp3−/−Nod2−/− > Pglyrp3−/− Nod2−/−) mice were treated with 4% DSS in drinking water and development of colitis was evaluated. DSS-treated mice were monitored over time for survival, change in body weight, and stool and rectal bleeding. The results are means ± SEM of 5-6 mice/group. Statistically significant differences between the indicated groups are shown as *, P ≤ 0.05; **, P ≤ 0.005. (B) Presence of Nod2 KO (1 kb amplified fragment) or Nod2 WT (300 bp amplified fragment) alleles was assessed by PCR of genomic DNA from blood cells of chimeric (Pglyrp3−/−Nod2−/− > Pglyrp3−/− and Pglyrp3−/− > Pglyrp3−/−Nod2−/−) or non-transplanted Pglyrp3−/− and Nod2−/− mice.
These results show that the lack of Nod2 expression in bone marrow-derived cells combined with expression of Nod2 in structural (radio-resistant) cells (Pglyrp3−/−Nod2−/− > Pglyrp3−/−) predisposes Pglyrp3−/− mice to very severe colitis, more severe than the lack of Nod2 in both bone marrow-derived and structural cells (Pglyrp3−/−Nod2−/− > Pglyrp3−/−Nod2−/−) or the lack of Nod2 only in structural cells (Pglyrp3−/− > Pglyrp3−/−Nod2−/−). Also, expression of Nod2 in all cells protects from colitis, because Pglyrp3−/− > Pglyrp3−/− mice were less sensitive to colitis than Nod2-chimeric or fully Nod2-deficient mice, which confirms our other results in this study. Altogether, these results further indicate that expression of Nod2 is protective against colitis, and suggest that expression of Nod2 in bone marrow-derived cells is more important for protection against colitis than Nod2 expression in structural cells. These results also suggest that expression of Nod2 in structural cells has a colitis-promoting effect, but only when Nod2 is not expressed in marrow-derived cells.
DISCUSSION
Inflammatory bowel disease is caused by aberrant host immune responses, altered gut microbiome, and environmental factors. Thus, there is no single cause for IBD and the exact genetic and environmental factors that result in IBD most likely differ in different individuals. In this study we report that a combined deficiency of two innate immunity genes, Pglyrp3 and Nod2, results in increased sensitivity to an experimental model of colitis. We show that Pglyrp3−/−Nod2−/− double deficient mice are more sensitive to DSS-induced colitis than Pglyrp3−/−, Nod2−/−, and WT mice. We also confirmed our previous results showing increased sensitivity to DSS-colitis in Pglyrp3−/− mice (22). Nod2−/− mice were significantly less sensitive to DSS-colitis than Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice and had similar sensitivity to WT mice, which confirms the findings of Kobayashi et al. (41), but differs from two other studies (42, 43). These conflicting results with Nod2−/− mice are difficult to explain and may result from differences in genetic backgrounds, gut bacteria, environment, and/or other unknown factors.
Pglyrp3−/−Nod2−/− mice treated with DSS had high mortality, severe loss in body weight and high stool scores. These phenotypic manifestations were accompanied by several histopathologic changes including severe hyperplasia, loss of crypts, infiltration with inflammatory cells, loss of epithelium, loss of goblet cells, and ulcers, which are characteristic for colitis. Additional changes associated with colitis in Pglyrp3−/−Nod2−/− mice were increased gut permeability, decreased colonic epithelial cell proliferation, increased colonic epithelial cell apoptosis, increased expression of chemokines and cytokines, changes in gut bacteria, and increased ATP in the colon. Our results demonstrate that Nod2 deficiency contributes significantly to higher apoptosis and that Pglyrp3 deficiency contributes to higher ATP in the colon, which causes increased numbers of inflammatory T cells and increased sensitivity to DSS-colitis. These results suggest that the enhanced sensitivity of Pglyrp3−/−Nod2−/− mice to DSS-colitis is due to multiple factors and that Pglyrp3 and Nod2 have both overlapping and unique contributions toward sensitivity to colitis in the double-knockout mice.
Pglyrp3−/−Nod2−/− mice have significantly higher apoptosis in the colon, which is required for the increased sensitivity to colitis of Pglyrp3−/−Nod2−/− mice. Deficiency in both Pglyrp3 and Nod2 contributed to the enhanced apoptosis in the double-knockouts, however, Nod2 deficiency had a stronger effect than Pglyrp3 deficiency. The increased sensitivity of Pglyrp3−/−Nod2−/− mice to DSS-colitis required the protease caspase-3, an important regulatory enzyme that activates downstream execution caspases during apoptosis. Apoptosis is crucial for the rapid turnover of intestinal mucosal cells and for homeostasis of the gut, and the role of caspase-3 in this process is well documented (55). The mechanism by which Nod2 regulates apoptosis is not known. However, our results also suggest that Nod2 deficiency in bone marrow-derived cells highly increases sensitivity to colitis, which suggests that higher sensitivity to apoptosis of epithelial cells in Nod2−/− mice may result from the lack of protective effects of cytokines or other factors from bone marrow-derived cells. A recent report demonstrated that Nod2 is involved in cross talk with the apoptosis protein, Bid, a member of the Bcl2 family, and that this interaction is required for the pro-inflammatory function of Nod2, which involves activation of NF-κB and increased expression of chemokines and cytokines (56). However, data showing Nod2-Bid interaction have not been replicated and it is not known whether this interaction regulates apoptosis. The role of mammalian Pglyrps in apoptosis is novel for these proteins and increased apoptotic cells may be an indirect consequence of mucosal damage by changed microflora in Pglyrp-deficient background.
Pglyrp3−/−Nod2−/− and Pglyrp3−/− mice treated with DSS had higher expression of Th1, Th2, and Th17 markers and higher expression of IL-1β, IL6, and IL-11, which are produced by many cell types. These changes are most likely due to a deficiency of Pglyrp3 and altered microflora, as they are observed in both Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice. These results also confirm our previous discovery of a predominant role of IFN-γ and IFN-γ-regulated chemokines, CXCL9, CXCL-10, CXCL-11, and CCL-2 in DSS-colitis in Pglyrp-deficient mice (22). CCR-7, TNF-α, IL-1β, and IL-1f6 were significantly higher in the colon of Pglyrp3−/−Nod2−/− than Pglyrp3−/− mice and may indicate a role for Nod2 in their regulation. CCR7 is the receptor for CCL19 and CCL21 and influences both immune responses and tolerance in the gut by regulating migration of T cells to lymphoid organs (57). The role of CCR7 in Pglyrp3−/−Nod2−/− mice during DSS-colitis is not understood, but may indicate greater trafficking of T cells in the damaged intestine. TNF-α is a common pathogenic factor in many models of colitis and is also known to trigger apoptosis of epithelial cells (58). Higher expression of TNF-α in Pglyrp3−/−Nod2−/− mice correlates with higher apoptosis and may be one of the contributing factors to the increased mucosal cell death in the double knockout mice. Synthesis of IL-1β is induced by TLRs in response to bacteria and bacterial fragments (59) and the increased expression of IL-1β in DSS-treated Pglyrp3−/−Nod2−/− mice may be in response to the altered gut microflora due to Pglyrp3 deficiency. IL-1f6 is a new member of the IL-1 family of cytokines and its elevated expression is associated with the inflammatory disease psoriasis and with psoriasis-like symptoms (60). Our results indicate that IL-1f6 expression is increased in a Nod−/− but not a Nod+/+ background and may have a role in sensitivity to DSS-colitis in Pglyrp3−/−Nod2−/− mice.
Pglyrp3−/−Nod2−/− mice had significant differences in the major groups of gut bacteria compared with WT mice, and some of these changes were also common for Pglyrp3−/− and Nod2−/− mice. Our results using germ-free mice gavaged with stools form WT, Nod2−/−, Pglyrp3−/−, or Pglyrp3−/−Nod2−/− mice demonstrate that Pglyrp3 deficiency, but not Nod2 deficiency, contributes colitis-promoting microflora to the double-knockout mice. These data suggest that a deficiency in Pglyrp3, but not in Nod2, is responsible for the colitis-predisposing dysbiosis in Pglyrp3−/− Nod2−/− mice, and that Nod2 deficiency contributes other factors further predisposing these mice to colitis. Pglyrp3-dependent microflora also increases sensitivity to colitis in a Nod2-deficient background, which further indicates that Pglyrp3-dependent microflora is required for the increased sensitivity to DSS-colitis in Pglyrp3−/−Nod2−/− mice.
Our results demonstrating differences in gut microbial populations between Nod2−/− and WT mice are in agreement with other studies (43-46). However, our results showing that microflora from Nod2−/− mice does not increase the sensitivity to colitis in WT germ-free mice differ from the results of Courtier-Maillard et al., who used co-housed WT mice (43). Moreover, differences in gut microbial populations between F2 WT and Nod2−/− littermates were recently shown to be dependent on maternal lineage and not on the Nod2−/− deficiency (61). It was not determined, however, whether Nod2−/− genotype influences microflora over a longer period of time. We did not use F2 littermates in our experiments because a change to colitis-predisposing microflora due a Pglyrp-deficiency does not happen immediately or over a few weeks, but is established over more than one generation (Dziarski, unpublished data). Therefore, all mice used in our experiments were derived from knockout strains that were established after backcrossing to the same WT stock at least six months earlier and bred in the same facility. Furthermore, in our experiments with gut microflora we used 18 mice/strain, obtained from 9 different breeding pairs for each strain (2 mice/breeding pair) and kept in separate cages after weaning. This strategy allows stabilization of microflora and minimizes the variability observed between different litters due to different parents and different cages. Stools for microflora testing were collected at three time points over a 5-year period, and there were no statistically significant differences in the abundance of the bacterial groups tested for each strain between these three time points, which indicates stable and genotype-dependent stool microflora over the entire 5-year period of the study. Moreover, our results with the transfer of the predisposition to colitis to germ-free mice with stool microflora from either Pglyrp3−/− or Pglyrp3−/−Nod2−/− mice show that Pglyrp3−/− microflora is stable, Pglyrp3-dependent, and has colitis-predisposing characteristics on both Nod2−/− and Nod2+/+ backgrounds.
Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice had higher levels of ATP in the colon than Nod2−/−and WT mice. Administration of a stable ATP analog to Nod2−/− and WT mice made them more sensitive to DSS-colitis. However, Nod2 deficiency further increased sensitivity to DSS-induced colitis in the presence of elevated ATP. Increased colitis in ATP-treated Nod2−/− mice was accompanied by higher numbers of CD3, CD4, Th17, and Th2 cells in the colon lamina propria. Extracellular levels of ATP are tightly controlled and elevated amounts serve as a danger signal that modulates immune functions (54, 62, 63). High extracellular ATP induces differentiation of lamina propria T cells into Th17 cells, which causes colitis (54) and drives inflammation in asthma (62, 63). High concentrations of extracellular ATP also induce activation of caspase-3 and apoptosis in macrophages (64), gingival epithelial cells (65), and anti-inflammatory Treg cells (66). ATP in the colon may be released from injured epithelial cells or from gut bacteria, which are known to secrete large amount of ATP (67). The most likely source of elevated ATP in Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice is the altered microbiota and not injured epithelial cells, because elevated ATP is present in untreated mice, which have a healthy gut mucosa (22). Accordingly, changes in the gut bacterial populations are known to modify the amount of ATP produced in the colon (54). Collectively, these studies suggest a possible mechanism for the role of ATP in increased sensitivity to DSS-colitis in Pglyrp3−/− and Pglyrp3−/−Nod2−/− mice, which includes ATP-induced apoptosis of intestinal epithelial cells and lamina propria Treg cells, and increased differentiation of lamina propria Th17 cells. Our data demonstrate that Nod2−/− mice treated with DSS and a stable analog of ATP are more sensitive to colitis and have increased numbers of Th17, but unchanged numbers of Treg cells in the colon lamina propria compared with PBS-treated mice. Thus, in our model of ulcerative colitis, ATP may regulate sensitivity to DSS-colitis through increased apoptosis of epithelial cells and increased differentiation of Th17 cells, but not through increased apoptosis of Treg cells. Thus, our data suggest that a Pglyrp3 deficiency results in elevated ATP in the colon, which drives differentiation of inflammatory Th17 cells and increased sensitivity to DSS-induced colitis in Nod2−/− mice.
Because both immune and colon structural (epithelial and stromal) cells express Nod2, we performed bone marrow transplant experiments and generated Pglyrp3−/− > Pglyrp3−/−Nod2−/−and Pglyrp3−/−Nod2−/− > Pglyrp3−/− chimeric mice to identify the source of Nod2-deficient cells that mediate increased sensitivity to DSS-induced colitis. Our data indicate that an unbalanced expression of Nod2 due to the lack of Nod2 expression in bone marrow-derived cells combined with the expression of Nod2 in structural (radio-resistant) cells in a Pglyrp3-deficient background results in the highest sensitivity to colitis. On the contrary, the lack of Nod2 expression in structural cells combined with the expression of Nod2 in bone marrow-derived cells results in intermediate sensitivity to colitis, and similar to the sensitivity of mice lacking Nod2 in all cells. Finally, expression of Nod2 in all cells results in the lowest sensitivity to colitis. These results suggest that expression of Nod2 in bone marrow-derived cells is more important for protection against colitis than Nod2 expression in structural cells, and also that expression of Nod2 in structural cells has a colitis-promoting effect, but only when Nod2 is not expressed in bone marrow-derived cells. The reasons for the protection from colitis from bone marrow-derived Nod2 and the susceptibility to colitis from structural cells-derived Nod2 (but only when bone marrow-derived Nod2 is missing) are not clear and may include protective effects of cytokines from bone marrow-derived cells or other cross-talk between these two cell types.
The role of Nod2 in intestinal disease is complex, often contradictory, and not clearly understood. Different models have been proposed to explain the role of Nod2 in CD and in animal models of colitis. One possible explanation is an impaired response of intestinal mucosa and immune cells in Nod2-deficient individuals to intestinal bacteria resulting in an inability to control microflora-induced intestinal damage. A second possible explanation is dysbiosis in Nod2-deficient mice (43-46) and humans with Nod2 mutations (45), resulting in more damaging bacteria in the terminal ileum. A third possible explanation is that Nod2 is a negative regulator of TLR-signaling and that Nod2 deficiency results in increased TLR-mediated responses to bacteria, which leads to damaging inflammation (42, 68, 69). However, an inhibitory role of Nod2 in TLR-signaling is controversial and has been shown by only one research group, whereas several investigators have determined that during acute responses, Nod2 activation synergizes with TLRs to increase production of proinflammatory cytokines (41, 70-72).
Our data support the first explanation and show that Pglyrp3-controlled changes in intestinal microflora and Pglyrp3-controlled increases in colonic ATP promote colitis on Nod2−/− background. Thus, Nod2 deficiency results in less efficient control of microflora-induced damage, and one contributing factor is the higher sensitivity to apoptosis and ATP in the colon on Nod2−/− than on Nod2+/+ background. Our data do not support the second and third explanations, because Nod2 deficiency did not contribute to colitis-predisposing intestinal microflora (Fig. 4), and because cytokine and chemokine responses of macrophages or colonic cells to gut microflora were lower on Nod2−/− than on Nod2+/+ background (Fig. 5).
In summary, the two innate immunity genes, Pglyrp3 and Nod2, synergistically protect mice in an experimental model of colitis. A combined deficiency of Pglyrp3 and Nod2 results in higher sensitivity to DSS-colitis due to a combination of factors, including higher apoptosis, altered gut microbiota, and increased ATP in the colon. Pglyrp3 deficiency contributes colitispredisposing intestinal microflora and increased intestinal ATP, whereas Nod2 deficiency contributes higher apoptosis and higher sensitivity to increased ATP and to microflora.
Supplementary Material
Acknowledgements
We thank Julie Cook for maintaining and breeding our mice.
This work was supported by USPHS Grants R01AI028797 and R01AI073290 from NIH Abbreviations: CD, Crohn's disease; DSS, dextran sodium sulfate; IBD, inflammatory bowel disease; LP, lamina propria; Nod, nucleotide-binding oligomerization domain; Pglyrp, peptidoglycan recognition protein; qRT-PCR, quantitative real time RT-PCR; Treg, regulatory T; UC, ulcerative colitis; WT, wild type.
Footnotes
qRT-PCR array data were deposited in NCBI GEO (accession number GSE47588), confidential reviewer link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=fjghxccmwqucupy&acc=GS
REFERENCES
- 1.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 2.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Ann. Rev. Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 6.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterol. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 7.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D'Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D'Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagace C, Li C, Green T, Stevens CR, Beauchamp C, Fleshner PR, Carlson M, D'Amato M, Halfvarson J, Hibberd ML, Lordal M, Padyukov L, Andriulli A, Colombo E, Latiano A, Palmieri O, Bernard EJ, Deslandres C, Hommes DW, de Jong DJ, Stokkers PC, Weersma RK, Consortium NIG, Sharma Y, Silverberg MS, Cho JH, Wu J, Roeder K, Brant SR, Schumm LP, Duerr RH, Dubinsky MC, Glazer NL, Haritunians T, Ippoliti A, Melmed GY, Siscovick DS, Vasiliauskas EA, Targan SR, Annese V, Wijmenga C, Pettersson S, Rotter JI, Xavier RJ, Daly MJ, Rioux JD, Seielstad M. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagace C, Scott R, Amininejad L, Bumpstead S, Baidoo L, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Colombel JF, Denson LA, De Vos M, Dubinsky M, Edwards C, Ellinghaus D, Fehrmann RS, Floyd JA, Florin T, Franchimont D, Franke L, Georges M, Glas J, Glazer NL, Guthery SL, Haritunians T, Hayward NK, Hugot JP, Jobin G, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, McGovern DP, Milla M, Montgomery GW, Morley KI, Mowat C, Ng A, Newman W, Ophoff RA, Papi L, Palmieri O, Peyrin-Biroulet L, Panes J, Phillips A, Prescott NJ, Proctor DD, Roberts R, Russell R, Rutgeerts P, Sanderson J, Sans M, Schumm P, Seibold F, Sharma Y, Simms LA, Seielstad M, Steinhart AH, Targan SR, van den Berg LH, Vatn M, Verspaget H, Walters T, Wijmenga C, Wilson DC, Westra HJ, Xavier RJ, Zhao ZZ, Ponsioen CY, Andersen V, Torkvist L, Gazouli M, Anagnou NP, Karlsen TH, Kupcinskas L, Sventoraityte J, Mansfield JC, Kugathasan S, Silverberg MS, Halfvarson J, Rotter JI, Mathew CG, Griffiths AM, Gearry R, Ahmad T, Brant SR, Chamaillard M, Satsangi J, Cho JH, Schreiber S, Daly MJ, Barrett JC, Parkes M, Annese V, Hakonarson H, Radford-Smith G, Duerr RH, Vermeire S, Weersma RK, Rioux JD. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, International IBDGC, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat. Rev. Immunol. 2011;11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 12.Kashyap DR, Wang M, Liu LH, Boons GJ, Gupta D, Dziarski R. Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nat. Med. 2011;17:676–683. doi: 10.1038/nm.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat. Rev. Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 14.Kang D, Liu G, Lundstrom A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. USA. 1998;95:10078–10082. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem. 2001;276:34686–34694. doi: 10.1074/jbc.M105566200. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Wang S, Wang H, Gupta D. Differential expression of peptidoglycan recognition protein 2 in the skin and liver requires different transcription factors. J. Biol. Chem. 2006;281:20738–20748. doi: 10.1074/jbc.M601017200. [DOI] [PubMed] [Google Scholar]
- 17.Lu X, Wang M, Qi J, Wang H, Li X, Gupta D, Dziarski R. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 2006;281:5895–5907. doi: 10.1074/jbc.M511631200. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Liu LH, Wang S, Li X, Lu X, Gupta D, Dziarski R. Human peptidoglycan recognition proteins require zinc to kill both gram-positive and gram-negative bacteria and are synergistic with antibacterial peptides. J. Immunol. 2007;178:3116–3125. doi: 10.4049/jimmunol.178.5.3116. [DOI] [PubMed] [Google Scholar]
- 19.Kashyap DR, Rompca A, Gaballa A, Helmann JD, Chan J, Chang CJ, Hozo I, Gupta D, Dziarski R. Peptidoglycan recognition proteins kill bacteria by inducing oxidative, thiol, and metal stress. PLoS Pathogens. 2014 doi: 10.1371/journal.ppat.1004280. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelius E, Persson C, Karlsson J, Steiner H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem. Biophys. Res. Comm. 2003;306:988–994. doi: 10.1016/s0006-291x(03)01096-9. [DOI] [PubMed] [Google Scholar]
- 21.Wang ZM, Li X, Cocklin RR, Wang M, Wang M, Fukase K, Inamura S, Kusumoto S, Gupta D, Dziarski R. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 2003;278:49044–49052. doi: 10.1074/jbc.M307758200. [DOI] [PubMed] [Google Scholar]
- 22.Saha S, Jing X, Park SY, Wang S, Li X, Gupta D, Dziarski R. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-γ. Cell Host Microbe. 2010;8:147–162. doi: 10.1016/j.chom.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SY, Gupta D, Hurwich R, Kim CH, Dziarski R. Peptidoglycan recognition protein Pglyrp2 protects mice from psoriasis-like skin inflammation by promoting regulatory T cells and limiting Th17 responses. J. Immunol. 2011;187:5813–5823. doi: 10.4049/jimmunol.1101068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha S, Qi J, Wang S, Wang M, Li X, Kim YG, Nunez G, Gupta D, Dziarski R. PGLYRP-2 and Nod2 are both required for peptidoglycan-induced arthritis and local inflammation. Cell Host Microbe. 2009;5:137–150. doi: 10.1016/j.chom.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SY, Gupta D, Kim CH, Dziarski R. Differential effects of peptidoglycan recognition proteins on experimental atopic and contact dermatitis mediated by Treg and Th17 cells. PloS One. 2011;6:e24961. doi: 10.1371/journal.pone.0024961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Jing X, Gupta D, Dziarski R. Peptidoglycan recognition protein 1 enhances experimental asthma by promoting Th2 and Th17 and limiting regulatory T cell and plasmacytoid dendritic cell responses. J. Immunol. 2013;190:3480–3492. doi: 10.4049/jimmunol.1202675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zulfiqar F, Hozo I, Rangarajan S, Mariuzza R, Dziarski R, Gupta D. Genetic association of peptidoglycan recognition protein variants with inflammatory bowel disease. PloS One. 2013;8(6):e67393. doi: 10.1371/journal.pone.0067393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 29.Tada H, Aiba S, Shibata K, Ohteki T, Takada H. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect. Immun. 2005;73:7967–7976. doi: 10.1128/IAI.73.12.7967-7976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hisamatsu T1, Suzuki M, Reinecker HC, Nadeau WJ, McCormick BA, Podolsky DK. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124:993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 31.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Shaw MH, Kamada N, Warner N, Kim YG, Nunez G. The ever-expanding function of NOD2: autophagy, viral recognition, and T cell activation. Trends Immunol. 2011;32:73–79. doi: 10.1016/j.it.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 35.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 36.Strober W, Watanabe T. NOD2, an intracellular innate immune sensor involved in host defense and Crohn's disease. Mucosal Immunol. 2011;4:484–495. doi: 10.1038/mi.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 38.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 39.Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am. J. Gastroenterol. 2004;99:2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 40.van Heel DA, Ghosh S, Butler M, Hunt KA, Lundberg AM, Ahmad T, McGovern DP, Onnie C, Negoro K, Goldthorpe S, Foxwell BM, Mathew CG, Forbes A, Jewell DP, Playford RJ. Muramyl dipeptide and toll-like receptor sensitivity in NOD2-associated Crohn's disease. Lancet. 2005;365:1794–1796. doi: 10.1016/S0140-6736(05)66582-8. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, Kitani A, Strober W. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J. Clin. Invest. 2008;118:545–559. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, Gaillot O, Schreiber S, Lemoine Y, Ryffel B, Hot D, Nunez G, Chen G, Rosenstiel P, Chamaillard M. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. USA. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, Schreiber S, Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 46.Mondot S, Barreau F, Al Nabhani Z, Dussaillant M, Le Roux K, Dore J, Leclerc M, Hugot JP, Lepage P. Altered gut microbiota composition in immune-impaired Nod2−/− mice. Gut. 2012;61:634–635. doi: 10.1136/gutjnl-2011-300478. [DOI] [PubMed] [Google Scholar]
- 47.Elson CO, Sartor RB, Tennyson GS, Riddell RH. Experimental models of inflammatory bowel disease. Gastroenterol. 1995;109:1344–1367. doi: 10.1016/0016-5085(95)90599-5. [DOI] [PubMed] [Google Scholar]
- 48.Kang SS, Bloom SM, Norian LA, Geske MJ, Flavell RA, Stappenbeck TS, Allen PM. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr., Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duran-Struuck R, Dysko RC. Principles of bone marrow transplantation (BMT): providing optimal veterinary and husbandry care to irradiated mice in BMT studies. J. Am. Assoc. Lab. Anim. Sci. 2009;48:11–22. [PMC free article] [PubMed] [Google Scholar]
- 53.Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, Karp CL, Pulendran B. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J. Immunol. 2011;187:733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. ATP drives lamina propria TH17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 55.Grossmann J, Mohr S, Lapentina EG, Fiocchi C, Levine AD. Sequential and rapid activation of select caspases during apoptosis of normal intestinal epithelial cells. Am. J. Physiol. 1998;274:G1117–1124. doi: 10.1152/ajpgi.1998.274.6.G1117. [DOI] [PubMed] [Google Scholar]
- 56.Yeretssian G, Correa RG, Doiron K, Fitzgerald P, Dillon CP, Green DR, Reed JC, Saleh M. Non-apoptotic role of BID in inflammation and innate immunity. Nature. 2011;474:96–99. doi: 10.1038/nature09982. [DOI] [PubMed] [Google Scholar]
- 57.Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 58.Ruemmele FM, Dionne S, Levy E, Seidman EG. TNFα-induced IEC-6 cell apoptosis requires activation of ICE caspases whereas complete inhibition of the caspase cascade leads to necrotic cell death. Biochem. Biophys. Res. Comm. 1999;260:159–166. doi: 10.1006/bbrc.1999.0734. [DOI] [PubMed] [Google Scholar]
- 59.Burns K, Martinon F, Tschopp J. New insights into the mechanism of IL-1β maturation. Curr. Op. Immunol. 2003;15:26–30. doi: 10.1016/s0952-7915(02)00017-1. [DOI] [PubMed] [Google Scholar]
- 60.Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N, Kanaly ST, Towne JE, Willis CR, Kuechle MK, Sims JE, Peschon JJ. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J. Exp. Med. 2007;204:2603–2614. doi: 10.1084/jem.20070157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robertson SJ, Zhou JY, Geddes K, Rubino SJ, Cho JH, Girardin SE, Philpott DJ. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 2013;4:222–231. doi: 10.4161/gmic.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Idzko M, Hammad H, van Nimwegen M, Kool M, Willart MA, Muskens F, Hoogsteden HC, Luttmann W, Ferrari D, Di Virgilio F, Virchow JC, Jr., Lambrecht BN. Extracellular ATP triggers and maintains asthmatic airway inflammation by activating dendritic cells. Nat. Med. 2007;13:913–919. doi: 10.1038/nm1617. [DOI] [PubMed] [Google Scholar]
- 63.Riteau N, Gasse P, Fauconnier L, Gombault A, Couegnat M, Fick L, Kanellopoulos J, Quesniaux VF, Marchand-Adam S, Crestani B, Ryffel B, Couillin I. Extracellular ATP is a danger signal activating P2X7 receptor in lung inflammation and fibrosis. Am. J. Resp. Crit. Care Med. 2010;182:774–783. doi: 10.1164/rccm.201003-0359OC. [DOI] [PubMed] [Google Scholar]
- 64.Noguchi T, Ishii K, Fukutomi H, Naguro I, Matsuzawa A, Takeda K, Ichijo H. Requirement of reactive oxygen species-dependent activation of ASK1-p38 MAPK pathway for extracellular ATP-induced apoptosis in macrophage. J. Biol. Chem. 2008;283:7657–7665. doi: 10.1074/jbc.M708402200. [DOI] [PubMed] [Google Scholar]
- 65.Yilmaz O, Yao L, Maeda K, Rose TM, Lewis EL, Duman M, Lamont RJ, Ojcius DM. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cell. Microbiol. 2008;10:863–875. doi: 10.1111/j.1462-5822.2007.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aswad F, Kawamura H, Dennert G. High sensitivity of CD4+CD25+ regulatory T cells to extracellular metabolites nicotinamide adenine dinucleotide and ATP: a role for P2X7 receptors. J. Immunol. 2005;175:3075–3083. doi: 10.4049/jimmunol.175.5.3075. [DOI] [PubMed] [Google Scholar]
- 67.Ivanova EP, Alexeeva YV, Pham DK, Wright JP, Nicolau DV. ATP level variations in heterotrophic bacteria during attachment on hydrophilic and hydrophobic surfaces. Int. Microbiol. 2006;9:37–46. [PubMed] [Google Scholar]
- 68.Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat. Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 69.Watanabe T, Kitani A, Murray PJ, Wakatsuki Y, Fuss IJ, Strober W. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity. 2006;25:473–485. doi: 10.1016/j.immuni.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 70.van Heel DA, Ghosh S, Hunt KA, Mathew CG, Forbes A, Jewell DP, Playford RJ. Synergy between TLR9 and NOD2 innate immune responses is lost in genetic Crohn's disease. Gut. 2005;54:1553–1557. doi: 10.1136/gut.2005.065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Netea MG, Ferwerda G, de Jong DJ, Jansen T, Jacobs L, Kramer M, Naber TH, Drenth JP, Girardin SE, Kullberg BJ, Adema GJ, Van der Meer JW. Nucleotide-binding oligomerization domain-2 modulates specific TLR pathways for the induction of cytokine release. J Immunol. 2005;174:6518–6523. doi: 10.4049/jimmunol.174.10.6518. [DOI] [PubMed] [Google Scholar]
- 72.Hedl M, Li J, Cho JH, Abraham C C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci U S A. 2007;104:19440–19445. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.