Abstract
Several dimorphic fungi are important human pathogens, but the origin and maintenance of virulence in these organisms is enigmatic, since an interaction with a mammalian host is not a requisite for fungal survival. Recently, Cryptococcus neoformans was shown to interact with macrophages, slime molds, and amoebae in a similar manner, suggesting that fungal pathogenic strategies may arise from environmental interactions with phagocytic microorganisms. In this study, we examined the interactions of three dimorphic fungi with the soil amoeba Acanthameobae castellanii. Yeast forms of Blastomyces dermatitidis, Sporothrix schenckii, and Histoplasma capsulatum were each ingested by amoebae and macrophages, and phagocytosis of yeast cells resulted in amoeba death and fungal growth. H. capsulatum conidia were also cytotoxic to amoebae. For each fungal species, exposure of yeast cells to amoebae resulted in an increase in hyphal cells. Exposure of an avirulent laboratory strain of H. capsulatum to A. castellanii selected for, or induced, a phenotype of H. capsulatum that caused a persistent murine lung infection. These results are consistent with the view that soil amoebae may contribute to the selection and maintenance of certain traits in pathogenic dimorphic fungi that confer on these microbes the capacity for virulence in mammals.
With the exception of Candida albicans, most common pathogenic fungi are considered saprophytic because they are free living and do not require an animal host for propagation (20). The mechanism(s) by which fungi acquire and maintain virulence for mammalian hosts in a life cycle that does not require animal parasitism is unexplained. Virulence for many environmental fungal pathogens is not constitutive, and passage of fungi in the laboratory can lead to virulence attenuation (4, 5, 14). Since virulence is almost always a complex trait, the pathogenicity of certain saprophytic fungi for mammals poses the question of how virulence is selected for and maintained in the environment (7, 35). The problem of fungal virulence is further complicated by the presence of specific virulence factors for mammalian infection including capsule, melanin, and adhesins (20) in many environmental fungi. Compounding the enigmatic question of the origin of virulence for environmental fungi is the fact that many fungi, such as Blastomyces dermatitidis, Sporothrix schenckii, and Histoplasma capsulatum, demonstrate no host specificity and can cause disease in multiple animal species.
Thermally dimorphic fungi such as B. dermatitidis, S. schenckii, and H. capsulatum can change from yeast to mycelial forms depending on temperature. Hence, they are believed to exist in the soil as distinct, multibranched, mycelial forms, but this has not been directly demonstrated. In fact, the available information found in the literature suggests otherwise, since B. dermatitidis in manure can exist in the yeast phase (33). However, even in the laboratory, changes in temperature do not induce a homogenous population switch. This observation, combined with soil temperatures reaching 37°C in tropical climates, strongly suggests that soil fungal populations are probably heterogeneous, containing mostly pseudohyphal fungal forms mixed with a nominal number of yeast forms. Disruption of soil or excrement contaminated with B. dermatitidis or H. capsulatum may result in the inhalation of mycelial cells and/or conidia resulting in an initial pulmonary infection that may disseminate (32). S. schenckii is found in soil and plant debris and does not typically cause systemic disease. However, inoculation of S. schenckii into the skin causes cutaneous lesions that frequently spread via the lymphatic system (6).
Recently, we reported that Cryptococcus neoformans, an environmental fungal pathogen that is macrophage-tropic during mammalian infection, can infect Acanthamoeba castellanii and amoeboid cells of the slime mold Dictyostelium discoideum in a manner comparable to infection of macrophages (36, 37). A. castellanii is an environmental soil amoeba that feeds on bacteria and fungi for sustenance and also serves as a host for both bacterial and fungal pathogens (30, 39). Since thermally dimorphic fungi are found primarily in the soil, we hypothesized that A. castellanii and other environmental phagocytic predators could place selective pressures on soil fungi. Here, we investigated the interaction of the dimorphic fungi B. dermatitidis, S. schenckii, and H. capsulatum with A. castellanii and compared them with the nonenvironmental fungus C. albicans and the nonpathogenic fungus Saccharomyces cerevisiae. The results indicate that B. dermatitidis, S. schenckii, and H. capsulatum are virulent for amoebae. Finally, we show that passage of an avirulent H. capsulatum strain through A. castellanii induces the fungal strain to become persistent in murine infection. The results suggest that soil amoebae such as A. castellanii could play a role in the environmental maintenance and selection of fungal virulence.
MATERIALS AND METHODS
Organisms and culture conditions.
A. castellanii strain 30324 was obtained from the American Type Culture Collection (ATCC) and cultured as adherent cells in peptone-yeast extract-glucose (PYG) broth (ATCC medium 354) at 28°C in the dark (2, 26). For experimental use and routine maintenance, A. castellanii was harvested from flasks, centrifuged at 220 × g for 10 min, and suspended in fresh PYG medium or 0.02 M phosphate-buffered saline (PBS) (0.137 M NaCl, 0.003 M sodium phosphate [pH 7.4]). A. castellanii was incubated at 37°C for 2 h prior to the addition of fungal cells to allow the amoeba cells to adjust to higher temperatures.
C. albicans SC5314 is a standard strain commonly used in the Candida field (34) that was a gift from M. Ghannoun (Cleveland, Ohio), and S. cerevisiae LM23-3az (12) was obtained from L. Marsh (Bronx, N.Y.). B. dermatitidis ATCC 26199 and H. capsulatum ATCC G217B were obtained from the ATCC. H. capsulatum CIB 1980 (an attenuated strain) and S. schenckii CIB 36989 were gifts from B. Gomez (London, United Kingdom) (16). Yeast cultures were maintained on brain heart infusion (BHI) agar slants incubated at 37°C. Yeast cultures were grown for 72 h at 37°C to stationary phase in BHI medium with rotary shaking at 150 rpm. Yeast cells were collected by centrifugation and washed three times with PBS.
H. capsulatum conidia were collected from two clinical isolates (J1 and J2) obtained from Jacobi Hospital, Bronx, N.Y., as previously described (25). In brief, cells were grown at room temperature on Sabouraud dextrose agar (Becton Dickinson Microbiology Systems, Sparks, Md.) for greater than 2 months. A solution of 0.85% NaCl in H2O was added to the agar, and manual manipulation with a loop was used to harvest the conidia. The conidia were centrifuged at 600 × g, washed with a 0.1% Tween 20 saline solution, and vortexed vigorously. The conidia were further washed and suspended with PBS and counted with a hemocytometer. Conidia viability was determined by trypan blue exclusion and confirmed by CFU counts on BHI agar.
J774.16 is a murine macrophage-like cell line, which has properties similar to peritoneal macrophages (29). Briefly, cells were cultured as an adherent monolayer in Dulbecco's modified Eagle medium (Life Technologies) with 10% NCTC-109 medium (Life Technologies), 10% heat inactivated fetal calf serum (Gemini Bio-products, Woodland, Calif.), and 1% nonessential amino acids (Cellgro; Mediatech, Washington, D.C.).
Macrophage and amoeba phagocytosis assays.
B. dermatiditis, S. schenckii, and H. capsulatum cells were washed, harvested, and suspended in PBS, and cell numbers were determined with a hemocytometer. Fungal cells were labeled with Oregon green-fluorescein isothiocyanate (FITC) (Molecular Probes, Leiden, The Netherlands) as previously described (40). Fungal cells were suspended at 2 × 108 cells/ml in a microcentrifuge tube, Oregon green-FITC was added to a final concentration of 5 × 10−4 g/ml, and the suspension was incubated at room temperature for 30 min. The fungal cells were washed three times with PBS. Labeling did not affect viability as determined by CFU counts on BHI agar. Next, the labeled fungal cells were suspended in Dulbecco's modified Eagle medium to 1.6 × 108 cells/ml, and 400 μl was added to each monolayer of J774.16 or amoeba cells consisting of 3.2 × 107 cells in eight-chamber glass culture slides (Falcon). The final effector (e.g., phagocytic cell)-to-target (e.g., fungal cell) ratio was 1:2. The tissue culture slides were incubated for 2 h at 37°C. The medium was aspirated, wells were washed with PBS, and 400 μl of 1-mg/ml trypan blue was added for 15 min to each well to quench extracellular fungal fluorescence. The wells were washed with PBS and fixed with 1% paraformaldehyde at 4°C for 30 min. Coverslips were mounted with a mounting solution of 0.1% n-propyl gallate and 50% glycerol in PBS, and the slides were viewed with an Olympus IX 70 microscope (Olympus America, Melville, N.Y.) equipped with a standard FITC filter. The phagocytosis index was determined by counting the total number of fluorescent fungal cells per 100 amoebae or macrophages. Five wells were counted per experimental condition, and each experiment was repeated.
Transmission electron microscopy (TEM).
A. castellanii and the B. dermatiditis, S. schenckii, and H. capsulatum conidia or yeast cells were washed, harvested, suspended in PBS, and counted on a hemocytometer. Amoebae and fungal cells were mixed together in a 1.5-ml microcentrifuge tube at a 1:1 effector-to-target ratio with 107 cells of each cell type/ml. The tubes were incubated for 2 or 24 h at 37°C and then centrifuged at 600 × g for 10 min. The medium was aspirated, and the cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate at room temperature overnight. The samples were processed for electron microscopy as previously described (37). The samples were viewed with a model 102 electron microscope (Siemens, Berlin, Germany), and representative micrographs were taken of each fungal species at both 2 and 24 h.
Fungal killing assays.
A. castellanii cells were removed from tissue culture flasks (Corning, Corning, N.Y.), washed with PBS, and counted with a hemocytometer. The cells were suspended to 105 cells/ml in PBS, and 100 μl was added to 96-well tissue culture plates. The plates were incubated at 37°C for 2 h prior to adding fungal cells to allow for A. castellanii acclimation. A. castellanii viability was determined by trypan blue staining, and the initial viability was always greater than 98% (data not shown). Fungal cells were suspended at 105 cells/ml, which was confirmed by CFU determination on BHI agar plates incubated at 37°C. Fungal cells were added to the acclimated cultures of A. castellanii at a 1:1 effector-to-target ratio and incubated at 37°C. At 0, 24, and 48 h, the number of viable yeast cells was determined by CFU. At each time interval, the 96-well plates were placed on ice for 10 min to loosen the cells from the bottoms of the plates. The A. castellanii cells were lysed by shear stress induced by pulling the suspension through a 27-gauge needle five to seven times (26). Fungal viability was unaffected by this procedure, as determined by comparison of initial hemocytometer determinations and CFU counts. For each well, serial dilutions were plated onto BHI agar plates, which were then incubated at 37°C for 48 h. At each time, a minimum of 4 tissue culture wells per strain were used to ascertain CFU, and each experiment was repeated at least one time. Conidial killing assays were performed as described above with two differences. For conidial assays, incubation was done at 30°C, and because of low conidial yields, the conidium-to-amoeba ratio was 1:4 with 2.5 × 103 conidia to 1 × 104 amoebae per well.
Amoeba killing.
Trypan blue exclusion assays were used to determine the number of viable A. castellanii cells at 0, 24, and 48 h. Amoebae and fungal cells were incubated in PBS in 96-well tissue culture plates at a 1:1 ratio. For the conidial assays, the ratio of 1 conidium to 4 amoebae was used. At each time interval, the medium was aspirated and the cultures were incubated with a 1:10 dilution of trypan blue in PBS. The 96-well plates were viewed at a magnification of ×200, and the percentage of dead amoebae was ascertained by counting the number of amoeba cells unable to exclude the dye per total amoebae counted. At each time interval, five wells per culture condition were counted and experiments were repeated at least one additional time.
Cytotoxicity of fungal supernatants.
The effect of fungal supernatants on amoeba toxicity was examined by using cell culture inserts that allowed separation of fungal cells from amoeba cells. These experiments were modeled on similar experiments done to evaluate the cytotoxicity of C. neoformans var. gattii, which is toxic to A. castellanii despite minimal phagocytosis (24). For these experiments, 0.4 μm of translucent cell culture inserts (Falcon; Becton Dickinson Labware, Franklin Lakes, N.J.) were used so that cells of B. dermatitidis, S. schenckii, or H. capsulatum could be incubated in fluid continuity, but not in direct contact, with A. castellanii. For these experiments, A. castellanii cells were collected, washed, and suspended at a density of 105 cells/ml in PBS. A volume of 500 μl of A. castellanii cell suspension was added to each well in a 24-well tissue culture plate (Falcon; Becton Dickinson Labware), with cell culture inserts placed in the appropriate wells, and the plates were incubated at 28°C for 1 h. Experiments were done whereby fungal cells were cultured either alone or in combination with amoeba cells. With fungal cells alone, the organisms were washed, suspended, and added to the insert at a 10:1 fungal cell-to-A. castellanii ratio. For the coculture experiments, the fungal cells were similarly prepared, but A. castellanii cells were added both above and below the inserts. Trypan blue exclusion was used to determine the viability of the A. castellanii under the different conditions at 0, 24, and 48 h.
Passage of an avirulent H. capsulatum strain with amoebae.
Six-to-eight-week-old, female, BALB/c mice (National Cancer Institute, Bethesda Md.) were used to study H. capsulatum virulence. H. capsulatum CIB 1980 (attenuated laboratory strain) were consecutively passaged four times with or without A. castellanii for 48 h per passage in PYG media at 37°C. The fungal cells were collected by centrifugation, and remaining amoeba cells were lysed via shear stresses by pulling the solution through a 27-gauge needle five times. The fungal cells were then washed four times with PBS and suspended in PBS. Mice were anesthetized with 125 mg of ketamine/kg of body weight and 10 mg of xylazine/kg of body weight, and 10 mice per group were infected intranasally with 1.25 × 107 cells in 20 μl (lethal dose) of either passaged or unpassaged H. capsulatum by using the intranasal model (10). The infectious dose was confirmed by CFU. The mice were killed by CO2 overdose followed by cervical dislocation at day 14, and the lungs were removed, homogenized, and plated onto BHI with penicillin and streptomycin (Gibco BRL) for CFU determinations (9). For histological analysis, the right upper lobe of the lung was removed, fixed in 10% buffered formalin, and embedded in paraffin. Five-micrometer sections were stained with Gomori's methenamine silver and viewed by light microscopy. The experiment was done twice.
Statistical analysis.
Student's t test was used for statistical analyses. Both the statistical analysis and the graphs were compiled in Microsoft Excel 2000 (Redmond, Wash.).
RESULTS
Phagocytosis of dimorphic fungi by macrophages and amoebae.
The phagocytosis indexes of both A. castellanii and J774.16 macrophage-like cells for B. dermatitidis ATCC 26199, S. schenckii CIB 36989, and H. capsulatum strain ATCC G217B or CIB 1980 were determined (Fig. 1). Both amoeba and J774.16 cells phagocytosed each of the fungi. The number of phagocytic events by either amoebae or macrophages followed a parallel trend. Fungal cells that were poorly phagocytosed by amoebae were also phagocytosed inefficiently by macrophages. For instance, B. dermatitidis induced the lowest number of phagocytic events for either macrophages or amoebae, with the phagocytic index averaging only 2.2%. Alternatively, S. schenckii and both H. capsulatum strains were phagocytosed at higher rates by both phagocytic cells. The greatest difference in amoeba and macrophage internalization was evident in H. capsulatum CIB 1980, where 81% of the amoebae phagocytosed fungal cells and only 70% of the macrophages phagocytosed the fungal cells.
FIG. 1.
Phagocytosis of H. capsulatum, B. dermatitidis, and S. schenckii cells by A. castellanii cells and J774.16 macrophages. Bars represent the phagocytic index by either amoebae (solid bars) or macrophages (hatched bars), and each error bar denotes one standard deviation. The phagocytosis index was determined by counting the total number of fluorescent fungal cells per 100 amoebae or macrophages. For each experimental condition, the number of repetitions was five. This experiment was repeated on different days and yielded similar results.
Electron microscopy of amoeba-fungus interactions.
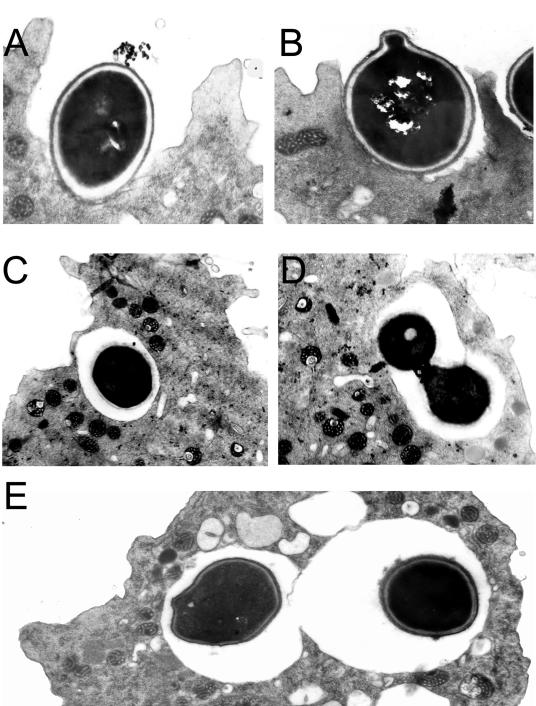
The interactions between amoebae and B. dermatitidis, S. schenckii, and H. capsulatum were studied by TEM. The panels in Fig. 2 are representative for both S. schenckii and the H. capsulatum strains. Fungal cells are phagocytosed (Fig. 2A and B) and enclosed in membrane-bound vacuoles (Fig. 2C and D). Several amoebae had more than one internalized fungal cell indicative of either separate phagocytic events or intracellular replication (Fig. 2D and E).
FIG. 2.
TEM of H. capsulatum cells interacting with A. castellanii. (A and B) Two separate phagocytic events at 2 h postincubation with amoebae. (C and D) Yeast cells in a membrane-bound vacuole surrounding the fungal cell 2 h after infection of the amoeba suspension with fungal cells. (E) Two individual H. capsulatum fungal cells in separate phagocytic compartments indicating two independent phagocytic events. Magnification, ×15,000 (A, B, and E) and ×12,000 (C and D).
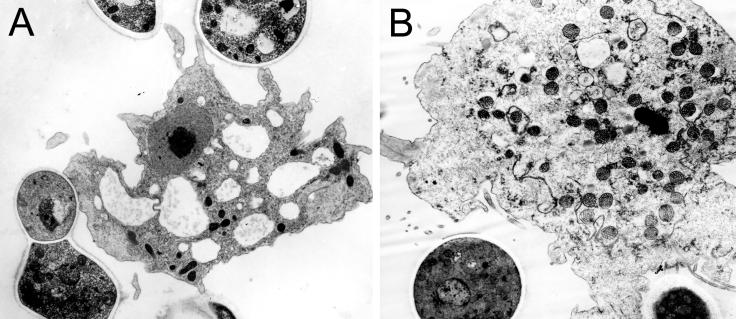
Consistent with the low phagocytic index of B. dermatitidis (Fig. 1), few B. dermatitidis fungal cells are found internalized by A. castellanii. However, B. dermatitidis cells are associated with the A. castellanii cells (Fig. 3). The interaction of B. dermatititdis with the amoebae results in swelling of mitochondria (data not shown) and reduced cytoplasmic electron density (Fig. 3B), consistent with amoeboid cell injury (9, 23).
FIG. 3.
TEM of B. dermatitidis cells interacting with A. castellanii cells. (A) An apparently intact A. castellanii cell with pseudopodia interacting with a B. dermatitidis cell. (B) B. dermatitidis cells engaged with an amoeba manifest reduced cytoplasmic electron density and electron-dense mitochondria in amoeba cells at 24 h postinfection. Magnification, ×9,000 (A) and ×12,000 (B).
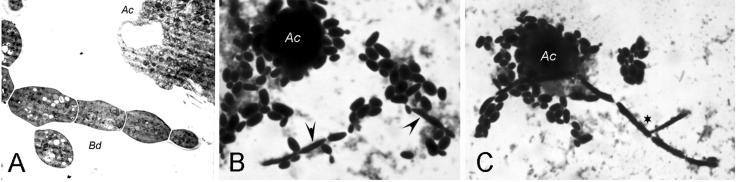
Interestingly, incubation of A. castellanii with yeast cells of B. dermatitidis, S. schenckii, or H. capsulatum resulted in increased numbers of hyphal and pseudohyphal cells, despite the fact that the experiment was done at 37°C. Within 24 h of incubation in the presence of amoebae, suspensions of B. dermatitidis (Fig. 4A), H. capsulatum (Figs. 4b and c), and S. schenckii (data not shown) contained large numbers of pseudohyphal or hyphal cells, despite the fact that the cultures were maintained at 37°C. For H. capsulatum, approximately 11.5% of cells were pseudohyphal and 1% were hyphal at 24 h. At 48 h, 18% were pseudohyphal and 4.5% were hyphal. There was no noticeable increase in pseudohyphal or hyphal cells when B. dermatitidis, S. schenckii, or H. capsulatum were incubated in PBS at 37°C.
FIG. 4.
Transformation of dimorphic fungi from yeast forms to pseudohyphal or hyphal forms. (A) TEM (magnification, ×15,000) of a representative hyphal form from a 24-h coculture of B. dermatitidis (Bd) yeast with A. castellanii (Ac). Pseudohyphal (arrowhead) and hyphal forms (asterisk) were present in cocultures of H. capsulatum yeast with A. castellanii (Ac) after 24 h (B) and 48 h (C). The cells were stained with Gram's crystal violet (magnification, ×1,000).
Growth of fungi in presence of A. castellanii.
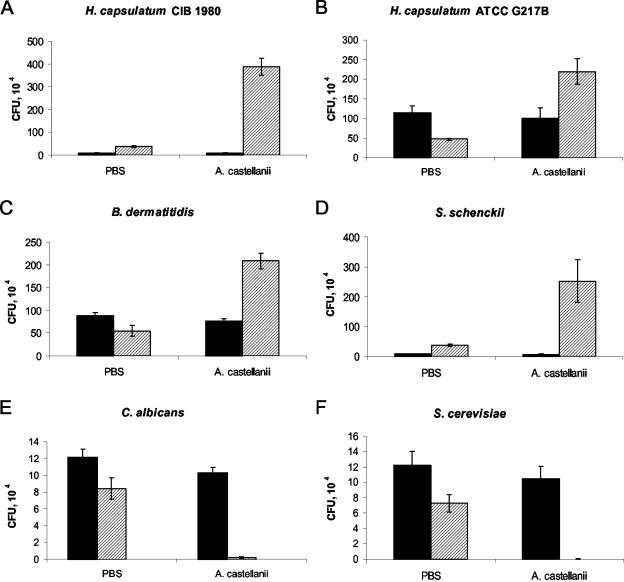
Neither fungi nor amoebae replicated significantly when incubated in PBS alone, presumably as a consequence of nutritional starvation (Fig. 5 and 6), although both S. schenckii and H. capsulatum CIB 1980 had some growth in PBS, possibly resulting from the completion of their growth cycle. Incubation of the dimorphic fungi with amoeba cells resulted in significant CFU increases ranging between 3- and 45-fold (Fig. 5). Increases in CFU for each of the dimorphic fungi in the presence of amoebae at 48 h were significant compared to the fungi alone (P ≤ 0.001). For H. capsulatum CIB 1980, incubation with A. castellanii resulted in a 45-fold increase in CFU while incubation in PBS resulted in only a doubling of CFU (Fig. 5A). As depicted in Fig. 5B and C, respectively, H. capsulatum ATCC G217B and B. dermatitidis ATCC 26199 incubated with A. castellanii each had a CFU increase of fourfold at 48 h compared to the PBS-alone condition. Finally, S. schenckii grown with amoebae had a significant CFU increase of almost 35-fold at 48 h. This CFU increase translated to a sevenfold difference in CFU compared to S. schenckii in PBS at 48 h (Fig. 5D).
FIG. 5.
Fungal cell counts after incubation with or without amoebae in PBS. Bars represent CFU at different times: solid bars denote CFU at 0 h and hatched bars denote CFU at 48 h. The error bars each represent one standard deviation. There are significant differences (P ≤ 0.001) between each of the fungi incubated with amoebae and the corresponding cells in PBS at 48 h. (A) H. capsulatum CIB 1980 (an attenuated strain); (B) H. capsulatum ATCC G217B; (C) B. dermatitidis; (D) S. schenckii; (E) C. albicans; (F) S. cerevisiae. Initial numbers of CFU vary, since the experiment with all fungi was done simultaneously. Each experiment was done at least twice with similar results.
FIG. 6.
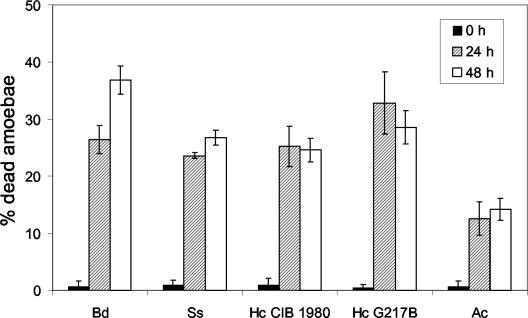
Viability of amoebae (Ac) after incubation with three different dimorphic fungi, B. dermatitidis (Bd), S. schenckii (Ss), and H. capsulatum (Hc) strains CIB 1980 and ATCC G217B. Amoeba cell viability was inferred by the ability of the cell to exclude trypan blue dye. Error bars each denote one standard deviation. At 24 and 48 h, the P value was ≤0.001 when comparing amoebae incubated with any of the dimorphic fungal species to amoebae alone. The experiment was done at least twice with similar results.
C. albicans and S. cerevisiae were used to study the interaction of a human-adapted fungus and a nonpathogenic fungus, respectively, with A. castellanii. As shown in Fig. 5E and F, the amoebae killed both C. albicans and S. cerevisiae.
Amoebae are killed by B. dermatitidis, S. schenckii, and H. capsulatum.
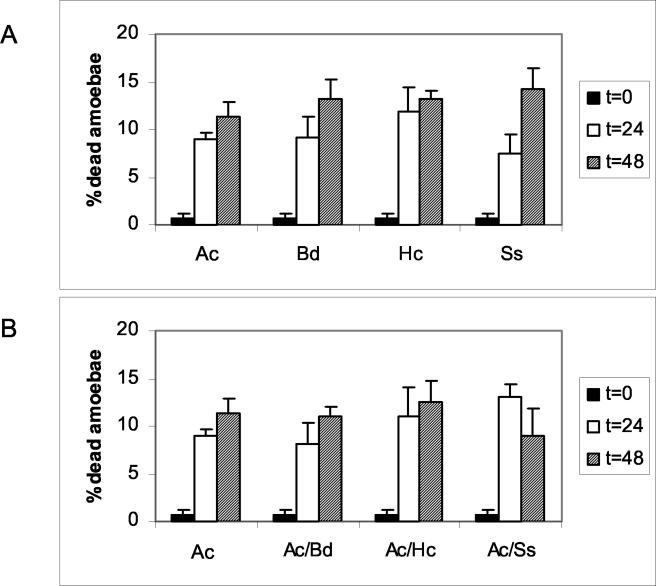
Trypan blue exclusion assays were used to determine the percentage of amoebae alive after incubation with the dimorphic fungi. The results, depicted in Fig. 6, show that a significant proportion of amoebae exposed to B. dermatitidis, S. schenckii, or H. capsulatum were killed. At the beginning of the assay, 99% of the amoebae were alive and excluded the dye. At 48 h, a large proportion of amoeba cells were no longer viable, as indicated by an inability to exclude dye. B. dermatitidis, S. schenckii, and H. capsulatum killed 25 to 37% of the A. castellanii cells. B. dermatitidis had the highest killing rate of 37%. The greatest increase in amoeba death occurred within the first 24 h for all the strains; however, B. dermatitidis was the only fungus to continue to cause amoeba death through 48 h. The cytotoxic effect on A. castellanii required amoeba-fungal cell contact, since the incubation of B. dermatitidis, S. schenckii, or H. capsulatum with A. castellanii under conditions where the cells were separated by porous inserts did not result in increased amoeba cell death (Fig. 7).
FIG. 7.
Viability of amoebae (Ac) after incubation in wells where B. dermatitidis (Bd), H. capsulatum (Hc), or S. schenckii (Ss) and amoeba cells were separated by permeable inserts (A) or where amoebae were separated from coincubated fungal and amoeba cells (B). There were no significant differences in viability for A. castellanii cells incubated in PBS alone and under the other conditions.
Conidial killing assays.
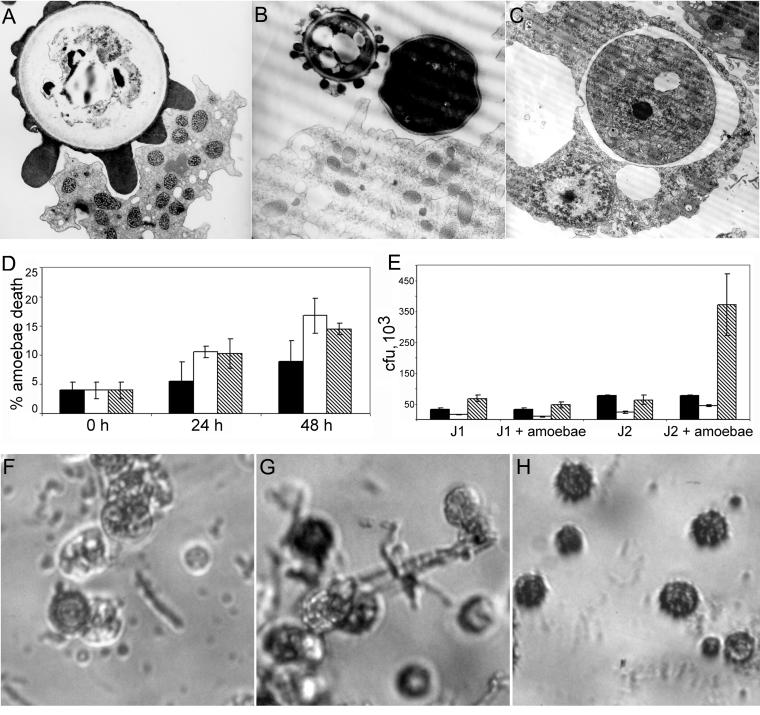
Conidial forms were collected from two clinical H. capsulatum isolates and used in fungal killing assays, trypan blue assays, and microscopy (Fig. 8). The rationale for studying conidia was that the dimorphic fungi are primarily mycelial in the environment and, presumably, phagocytic predators in soil encounter these forms. The interactions between the conidia and amoebae were studied by TEM (Fig. 8A, B, and C). The amoebae made multiple contacts with the conidia during phagocytosis (Fig. 8A). Within 24 h, the H. capsulatum conidia exposed to amoebae were internalized into phagocytic vacuoles and some had transformed to yeast forms (Fig. 8C). J1 and J2 strains of H. capsulatum interacted differently with the amoebae, evidenced by the results of the killing assays (Fig. 8E). J1 conidia had minimal growth in the presence of amoebae, whereas J2 conidia germinated to increase fungal cell numbers by almost fivefold in the presence of amoebae (Fig. 8E). Growth of J2 cells correlated with a change in morphology of the H. capsulatum cells from mostly conidial forms to a mixture of hyphal, pseudohyphal, and yeast forms of the fungi by 24 and 48 h (Fig. 8F and G). However, conidia incubated in PBS remained predominantly conidial forms, with rare cells demonstrating a conversion to hyphal, pseudohyphal, or yeast forms (Fig. 8H).
FIG. 8.
Interactions of H. capsulatum conidia with amoebae. (A) TEM illustrating A. castellanii phagocytosing a conidial cell after 2 h of incubation. (B) Micrograph depicting a conidial cell and a yeast cell in proximity to an amoeba cell after 24 h of incubation. (C) Micrograph illustrating an internalized H. capsulatum yeast cell surrounded by a membrane-bound vacuole after 24 h of incubation. (D) Trypan blue exclusion assay results illustrating the percentage of dead amoebae after incubation with and without H. capsulatum conidia at 0, 24, and 28 h. Solid bars indicate amoebae in PBS only, open bars represent amoebae with J1 conidia, and hatched bars represent J2 conidia with amoebae. Error bars each represent one standard deviation. At 24 h, the P value was 0.028 when comparing amoebae alone to both J1 and J2 conidia with amoebae, and at 48 h, death of amoebae incubated with J1 and J2 was also significant compared to amoebae alone (P = 0.015 and 0.022, respectively). (E) Changes in CFU of J1 and J2 conidia after incubation with amoebae. Solid bars represent CFU at 0 h, open bars represent CFU at 24 h, and hatched bars represent CFU at 48 h. Error bars each denote one standard deviation. Both the 24- and 48-h results for J2 conidia with amoebae are significant compared to J2 in PBS alone (P ≤ 0.001 for both). (F and G) Micrograph depicting morphology changes of J2 conidia after 24 h of incubation with amoebae illustrating conidial, yeast, and hyphal forms of H. capsulatum J2. (H) Micrograph of J2 conidia after 24 h of incubation in PBS. Magnification, ×15,000 (A), ×12,000 (B and C), and ×200 (F, G, and H).
H. capsulatum virulence is enhanced by passage in A. castellanii.
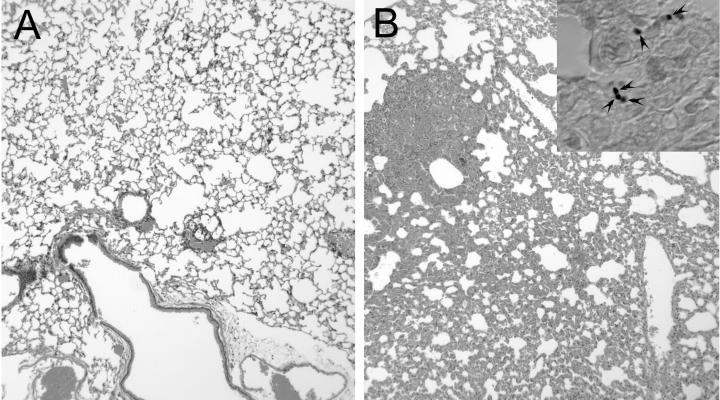
We postulated that if amoebae are involved in the maintenance of virulence in environmental fungi, virulence could also be affected by passage of fungal cells through amoeba cultures. To examine this possibility, we passaged the avirulent strain of H. capsulatum, CIB 1980, with and without amoebae and then used these fungal cells to infect mice. No CFU were recovered at day 14 from mice infected with CIB 1980 grown in medium alone. However, H. capsulatum was recovered at day 14 from each mouse infected with H. capsulatum CIB 1980 cells grown in the presence of amoebae. The total number of CFU isolated per lung from mice infected with passaged H. capsulatum was relatively low, 61.1 ± 19.2 cells per lung. The lungs of infected mice were examined histopathologically. H. capsulatum cells were present only in lung tissue from mice infected with passaged fungal cells (Fig. 9, inset). Mouse lungs infected with passaged H. capsulatum showed granulomatous inflammation and a loss of alveolar air space, whereas control lungs appear normal, indicating the ability of the passaged cells to cause host damage (Fig. 9).
FIG. 9.
Hematoxylin and eosin staining of lung tissue from mice 14 days after infection with H. capsulatum CIB 1980 grown in PYG media without (A) or with (B) amoebae. Magnification, ×250. The inset in panel B shows the tissue stained with Gomori's methenamine silver. The arrowheads indicate H. capsulatum yeast cells.
DISCUSSION
Three recent studies with C. neoformans suggest that environmental predators such as amoebae, slime mold, and nematodes may exert powerful selection pressures on environmental populations to generate variants with the potential for mammalian pathogenicity (27, 36, 37). Consequently, we investigated the interaction of A. castellanii with B. dermatitidis, S. schenckii, and H. capsulatum and compared these interactions with those of A. castellanii with C. albicans and S. cerevisiae. Our results suggest that amoebae are potential hosts for dimorphic fungi. The interactions of the dimorphic fungi with A. castellanii result in killing of A. castellanii and growth of the fungi, which utilize A. castellanii as a nutritional source. In addition, an avirulent H. capsulatum strain increased in virulence after exposure to A. castellanii. The demonstration that H. capsulatum conidia were cytotoxic to amoeba cells provides evidence for the potential environmental significance of these results.
Coincubation of the amoebae and fungi resulted in amoeba death and fungal growth. To explore the interactions between the fungi and amoebae, we needed to identify conditions where neither the fungi nor the amoebae could replicate. Since both the fungi and the amoebae are autotrophs for many essential nutrients, we could not identify a medium that would sustain the amoebae without also allowing the fungi to grow. Hence, we used PBS, a solution in which neither organism could replicate and where nutrients could only come from amoeba predation of fungi or fungal destruction of amoebae. Although we recognize the artificial nature of this system, we point out that soils inhabited by these organisms are also likely to be nutrient poor. Under these starvation conditions, each of the dimorphic fungi was able to kill amoebae, and the amoebae killed C. albicans and S. cerevisiae. The different outcomes resulting from the interaction different fungal species and amoebae provided confidence that the system discriminated between interactions pathogenic to amoebae and those that were not.
Interestingly, incubation of B. dermatitidis, S. schenckii, or H. capsulatum yeast cells with A. castellanii resulted in an increase in hyphal and pseudohyphal cells. This observation echoes reports that dimorphic fungi can manifest morphological changes in response to macrophages. In this regard, Coccidioides immitis arthroconidia grow as spherules and mycelia in the presence and absence of leukocytes, respectively (15); murine macrophages block the dimorphic transition of B. dermatitidis (38); S. schenckii filament in response to macrophages (31); and phagocytosis of H. capsulatum results in the emergence of unusual morphologies, including elongated variants with short mycelial extensions (11). For H. capsulatum, both yeast cells maintained at 37°C and conidia grown at room temperature replicated by using amoebae as the only nutritional source. The interactions between the H. capsulatum conidia and the amoebae caused the conidia to change morphology to pseudohyphal, hyphal, and yeast forms. Similar conversions presumably occur in patients, since morphological transformations are induced by temperature and environmental changes (41). However, on occasion, temperature alone is insufficient to cause complete transition of dimorphic fungi. For example, in cavitary coccidioidal lesions, the saprophytic form of the fungus can be found in up to 73% of patients (28), hyphal forms of H. capsulatum have been described in endovascular lesions (21), and there are numerous reports of B. dermatitidis filamentous forms in infected tissues (1, 17, 18, 22). Although the biochemical basis of this effect is not known, and is beyond the scope of this study, this observation suggests a fertile area for future investigation.
The outcome of the interaction between A. castellanii and the two H. capsulatum strains, CIB 1980 and ATCC G217B, was similar despite the fact that these strains differed in virulence for mice. H. capsulatum CIB 1980 is an attenuated laboratory strain whose virulence was unable to be reconstituted by animal passage (B. Gomez, London, United Kingdom, personal communication). Nevertheless, the H. capsulatum CIB 1980 strain is cytotoxic to the amoebae and is able to grow with amoebae as a nutritional source. The discrepancy between virulence for mice and amoebae killing by these strains of H. capsulatum may reflect differences in the fungal traits used to interact with these two hosts. For example, certain sets of traits important for virulence may be used in the interaction with mice and amoebae, whereas other determinants of pathogenicity could preferentially be used in conjunction with each host. In this regard, it is noteworthy that mice differ from amoebae in that the former have immune systems that can respond to fungal cells and that over exuberant responses are capable of mediating host damage. In this regard, the pathogenesis of H. capsulatum in mammals is characterized by both microbial and immune-mediated damage (8). Hence, one should not expect one-to-one correspondence in the behavior of each strain with the different hosts. Nevertheless, our results showed that passage of H. capsulatum strain CIB 1980 in amoebae was able to increase virulence, as measured by the ability of this strain to persist in vivo. When inoculated into mice, nonpassaged strain CIB 1980 is avirulent, as evidenced by the fact that no disease is apparent in mice. Furthermore, the infection is cleared, as indicated by the absence of organisms in host tissues. Passaging H. capsulatum CIB 1980 with amoebae produced a clear change in the virulence phenotype, since amoeba-passaged yeast acquired the ability to persist in lung tissue after inoculation in mice. Furthermore, histological analysis of infected tissues revealed yeast cells and profuse inflammation. Given that we have recently reported that C. neoformans virulence can be increased by passage in D. discoideum (36), this experiment provides a second example whereby fungal virulence can be altered by the interaction with another soil organism. Several mechanisms could be responsible for this phenomenon, including the induction of H. capsulatum virulence genes by A. castellanii or the selection of H. capsulatum cells with increased overall fitness to infect the mice.
B. dermatitidis caused the greatest amount of amoeba death but replicated the least compared to the other dimorphic fungi. The slower replication rate can be explained by the normal growth rate of B. dermatitidis, which is approximately 17 h in optimal media (3). Electron microscopy revealed that although few amoebae had phagocytosed B. dermatitidis cells, the overwhelming majority of fungal cells were adherent to the amoeba cell membrane. Although electron microscopy analysis cannot be used to unequivocally determine cell viability, the decrease in cellular density, the alterations in mitochondria, and the disruption of the cellular membranes are each indicative of cell death. Therefore, based on these criteria and the trypan blue exclusion studies, it appears that B. dermatitidis is toxic to amoeba cells as a consequence of attaching to the amoebic cell membrane. These cellular killing characteristics were not observed when A. castellanii was incubated with either H. capsulatum or S. schenckii. The B. dermatitidis killing results parallel previous findings in macrophage studies (3). B. dermatitidis expresses a cell surface protein, Bad1, which is an adhesin required for virulence and is responsible for adhering the fungal cell to the outside of macrophages (13). The adherence of B. dermatitidis to macrophages is cytotoxic (13). Therefore, the two systems indicate similar results with adherence of the fungal cells leading to cytotoxicity. Furthermore, for B. dermatitidis, contact between fungal cells and amoebae was required for toxicity to the amoebae, strongly arguing against a soluble toxin.
The observation that fungi reproduced only in the presence of amoebae and the trypan blue results that showed a reduction in viable amoebae are interpreted to indicate that each of the dimorphic fungi killed amoebae and used them for food. The mechanism of killing of amoeba cells required contact between the fungal and amoeba cells and probably included both intracellular and extracellular cytotoxic effects. Electron microscopy revealed that both H. capsulatum and S. schenckii were enclosed in a membrane-bound vacuole after ingestion by amoebae. For these fungi, internalized fungal cells exploit the amoebae after ingestion and possibly gain nutrients by feeding from the remains of the killed host cells. For the B. dermatitidis-amoeba interaction, phagocytosis was rare and the cytotoxic effect was probably extracellular.
Our results demonstrate that A. castellanii can serve as a host system for the dimorphic fungi B. dermatitidis, S. schenckii, and H. capsulatum. We propose that phagocytic predators in the environment exert selective pressures, which favor fungal attributes that confer survival advantages in animal hosts. We do not imply that amoebae are the sole selective pressure for the emergence and maintenance of virulence in the environment. Clearly, the ability to grow at 37°C would seem to be an important requirement for mammalian virulence. In fact, recent studies suggest that nematodes (27) and slime molds (36) could provide additional selection pressures for the acquisition of virulence factors. Bacteria may also contribute to the selection of traits associated with mammalian pathogenesis (19). Furthermore, we note that there are many types of amoebae, and it is possible that other amoeboid species are able to efficiently kill these fungi. Nevertheless, the similarity of interactions between amoebae and macrophages and the observation that fungal virulence can be enhanced by exposure to amoebae strongly link these phagocytic predators to the phenomenon of fungal virulence for animals.
Editor: T. R. Kozel
REFERENCES
- 1.Atkinson, J. B., and T. L. McCurley. 1983. Pulmonary blastomycosis: filamentous forms in an immunocompromised patient with fulminating respiratory failure. Hum. Pathol. 14:186-188. [DOI] [PubMed] [Google Scholar]
- 2.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64:668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandhorst, T. T., M. Wuthrich, T. Warner, and B. Klein. 1999. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J. Exp. Med. 189:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass, C., C. M. Volkmann, D. E. Philpott, H. P. Klein, C. J. Halde, and D. A. Stevens. 1982. Spontaneous mutant of Blastomyces dermatitidis attenuated in virulence for mice. Sabouraudia 20:145-158. [PubMed] [Google Scholar]
- 5.Brummer, E., A. Restrepo, L. H. Hanson, and D. A. Stevens. 1990. Virulence of Paracoccidiodes brasiliensis: the influence of in vitro passage and storage. Mycopathologia 109:13-17. [DOI] [PubMed] [Google Scholar]
- 6.Bustamante, B., and P. E. Campos. 2001. Endemic sporotrichosis. Curr. Opin. Infect. Dis. 14:145-149. [DOI] [PubMed] [Google Scholar]
- 7.Casadevall, A., J. D. Nosanchuk, and J. N. Steenbergen. 2003. ‘Ready-made’ virulence and ‘dual-use’ virulence factors in pathogenic enviromental fungi-the Cryptococcus neoformans paradigm. Curr. Opin. Microbiol. 112:1164-1175. [DOI] [PubMed] [Google Scholar]
- 8.Casadevall, A., and L. Pirofski. 1999. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect. Immun. 67:3703-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotter, D. A., and R. P. George. 1975. Germination and mitochondrial damage in spores of Dictyostelium discoideum following supraoptimal heating. Arch. Microbiol. 103:163-168. [DOI] [PubMed] [Google Scholar]
- 10.Deepe, G. S., Jr., and R. Gibbons. 2001. Protective efficacy of H antigen from Histoplasma capsulatum in a murine model of pulmonary histoplasmosis. Infect. Immun. 69:3128-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eissenberg, L. G., S. Poirier, and W. E. Goldman. 1996. Phenotypic variation and persistence of Histoplasma capsulatum yeasts in host cells. Infect. Immun. 64:5310-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elia, L., and L. Marsh. 1998. A role for a protease in morphogenic responses during yeast cell fusion. J. Cell Biol. 142:1473-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkel-Jimenez, B., M. Wuthrich, and B. S. Klein. 2002. BAD1, an essential virulence factor of Blastomyces dermatitidis, suppresses host TNF-alpha production through TGF-beta-dependent and -independent mechanisms. J. Immunol. 168:5746-5755. [DOI] [PubMed] [Google Scholar]
- 14.Franzot, S. P., J. Mukherjee, R. Cherniak, L. Chen, J. S. Hamdan, and A. Casadevall. 1998. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect. Immun. 66:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galgiani, J. N., R. Hayden, and C. M. Payne. 1982. Leukocyte effects on the dimorphism of Coccidioides immitis. J. Infect. Dis. 146:56-63. [DOI] [PubMed] [Google Scholar]
- 16.Gomez, B. L., J. I. Figueroa, A. J. Hamilton, S. Diez, M. Rojas, A. Tobon, A. Restrepo, and R. J. Hay. 1999. Detection of the 70-kilodalton Histoplasma capsulatum antigen in serum of histoplasmosis patients: correlation between antigenemia and therapy during follow-up. J. Clin. Microbiol. 37:675-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardin, H. F., and D. I. Scott. 1974. Blastomycosis. Occurence of filamentous forms in vivo. Am. J. Clin. Pathol. 62:104-106. [DOI] [PubMed] [Google Scholar]
- 18.Herd, A. M., S. B. Greenfield, G. W. Thompson, and R. C. Brunham. 1990. Miliary blastomycosis and HIV infection. CMAJ 143:1329-1330. [PMC free article] [PubMed] [Google Scholar]
- 19.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 20.Hogan, L. H., S. M. Levitz, and B. S. Klein. 1996. Virulence factors of medically important fungi. Clin. Microbiol. Rev. 9:469-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutton, J. P., J. B. Durham, D. P. Miller, and E. D. Everett. 1985. Hyphal forms of Histoplasma capsulatum. A common manifestation of intravascular infections. Arch. Pathol. Lab. Med. 109:330-332. [PubMed] [Google Scholar]
- 22.Kaufmann, A. F., W. Kaplan, and D. E. Kraft. 1979. Filamentous forms of Ajellomyces (blastomyces) dermatitidis in a dog. Vet. Pathol. 16:271-273. [DOI] [PubMed] [Google Scholar]
- 23.Kong, H. H., and D. I. Chung. 1998. Ultrastructural changes of Acanthamoeba cyst of clinical isolates after treatment with minimal cysticidal concentration of polyhexamethylene biguanide. Korean J. Parasitol. 36:7-13. [DOI] [PubMed] [Google Scholar]
- 24.Malliaris, S. D., J. N. Steenbergen, and A. Casadevall. 2004. Cryptococcus neoformans var. gattii can exploit Acanthamoeba castellanii for growth. Med. Mycol. 42:149-158. [DOI] [PubMed] [Google Scholar]
- 25.McDonough, E. S., T. R. Wisniewski, L. A. Penn, D. M. Chan, and W. J. McNamara. 1976. Preliminary studies on conidial liberation of Blastomyces dermatitidis and Histoplasma capsulatum. Sabouraudia 14:199-204. [PubMed] [Google Scholar]
- 26.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mylonakis, E., F. M. Ausubel, J. R. Perfect, J. Heitman, and S. B. Calderwood. 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 99:15675-15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puckett, T. F. 1954. Hyphae of Coccidioides immitis in tissues of the human host. Am. Rev. Tuberc. 70:320-327. [DOI] [PubMed] [Google Scholar]
- 29.Ralph, P., J. Prichard, and M. Cohn. 1975. Reticulum cell sarcoma: an effector cell in antibody-dependent cell-mediated immunity. J. Immunol. 114:898-905. [PubMed] [Google Scholar]
- 30.Rodriguez-Zaragoza, S. 1994. Ecology of free-living amoebae. Crit Rev. Microbiol. 20:225-241. [DOI] [PubMed] [Google Scholar]
- 31.Ryter, A., and H. Fromentin. 1985. Ultrastructural study of the interaction of the fungi Sporothrix schenckii and Ceratocystis stenoceras with bone-marrow-derived murine macrophages. Ann. Inst. Pasteur Microbiol. 136B:9-27. [DOI] [PubMed] [Google Scholar]
- 32.San Blas, G., and F. San Blas. 1984. Molecular aspects of fungal dimorphism. Crit Rev. Microbiol. 11:101-127. [DOI] [PubMed] [Google Scholar]
- 33.Sarosi, G. A., and D. S. Serstock. 1976. Isolation of Blastomyces dermatitidis from pigeon manure. Am. Rev. Respir. Dis. 114:1179-1183. [DOI] [PubMed] [Google Scholar]
- 34.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhauser. 2002. Host versus in vitro signals and intrastrain allelic differences in the expression of a Candida albicans virulence gene. Mol. Microbiol. 44:1351-1366. [DOI] [PubMed] [Google Scholar]
- 35.Steenbergen, J. N., and A. Casadevall. 2003. The origin and maintenance of virulence for the human pathogenic fungus Cryptococcus neoformans. Microbes Infect. 5:667-675. [DOI] [PubMed] [Google Scholar]
- 36.Steenbergen, J. N., J. D. Nosanchuk, S. D. Malliaris, and A. Casadevall. 2003. Cryptococcus neoformans virulence is enhanced after intracellular growth in the genetically malleable host Dictyostelium discoideum. Infect. Immun. 71:4862-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steenbergen, J. N., H. A. Shuman, and A. Casadevall. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 18:15245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugar, A. M., and M. Picard. 1991. Macrophage- and oxidant-mediated inhibition of the ability of live Blastomyces dermatitidis conidia to transform to the pathogenic yeast phase: implications for the pathogenesis of dimorphic fungal infections. J. Infect. Dis. 163:371-375. [DOI] [PubMed] [Google Scholar]
- 39.Visvesvara, G. S., and W. Balamuth. 1975. Comparative studies on related free-living and pathogenic amebae with special reference to Acanthamoeba. J. Protozool. 22:245-256. [DOI] [PubMed] [Google Scholar]
- 40.Walenkamp, A. M., J. Scharringa, F. M. Schramel, F. E. Coenjaerts, and I. M. Hoepelman. 2000. Quantitative analysis of phagocytosis of Cryptococcus neoformans by adherent phagocytic cells by fluorescence multi-well plate reader. J. Microbiol. Methods 40:39-45. [DOI] [PubMed] [Google Scholar]
- 41.Woods, J. P. 2002. Histoplasma capsulatum molecular genetics, pathogenesis, and responsiveness to its environment. Fungal Genet. Biol. 35:81-97. [DOI] [PubMed] [Google Scholar]