Abstract
This paper presents a novel theoretical basis for accurately calculating the isotope cluster of polypeptides. In contrast to previous approaches to this problem, which consider exhaustive or near exhaustive combinations of isotopic species, the program, Neutron Cluster, groups probabilities to yield highly accurate information without elucidating any fine structure within a nominal mass unit. This is a fundamental difference from any previously described algorithm for calculating the isotope cluster. As a result of this difference, the accurate isotope clusters for high molecular weight polypeptides can be calculated rapidly without any pruning. When applied to isotope enriched polypeptides, the algorithm introduces “grouping error”, which is described, quantified, and avoided by using probability partitioning.
Proper interpretation of mass spectrometric data requires some understanding of the isotope cluster for the analyte. When considering analysis of peptides and proteins, prediction of the expected isotope cluster facilitates the recognition of contamination and potential instrumental malfunction [1–3]. Prediction of the change in the isotope cluster due to enrichment leads to identification of enriched molecules, a method on which mass spectrometric quantification and quantitative proteomic discovery relies [4–13]. Finally, accurate calculation of the isotope cluster may enable better Fourier transform deconvolution schemes for complex electrospray envelopes of macromolecules [14–18].
At the most basic level, the isotope cluster is a distribution of neutrons among the various elements present in a molecule. To date, the intensities of peaks in the isotope cluster have been estimated by the enumeration of the isotopic contributions of each element, with pruning introduced for practicality. The most classical examples of such an approach are the polynomial algorithms [19–23], first described by Yergey [23], which combine polynomial distributions for all the atoms in a molecule. Separately, Hsu described a calculation of the isotope cluster by means of convoluting multiple large Diophantine equations [24], and Kubinyi [25] introduced the concept of “splitting” isotopic distributions to yield a final cluster. These two methods are notable because they provide theoretical advantages in preserving the abundance accuracy of the cluster. However, since the number of combinations for large molecules precludes infinitely resolved exhaustive enumeration by any means, the convolution algorithms, either by polynomial expansion, peak splitting, or large elemental Diophantine equations, must implement probability thresholds to prune the number of species to satisfy computational time and memory demands.
Recently, dynamic programming [26] and Fourier transform [13, 27] methods have emerged as powerful tools for calculating fine isotopic structure. Since the fine isotopic structure obtained from these comprehensive algorithms is beyond the resolution of most contemporary instruments, the details are generally combined into nominal mass bins that resemble real measurements. The implementation of dynamic programming, as Snider [26] has done, offers the best means introduced so far for obtaining highly accurate predictions of the isotope cluster. This algorithm represents a genuine advancement in the field but, in its presently available rendering, it faces a fundamental problem similar to the polynomial algorithms since highly resolved isotopic combinations or “states” must be elaborated before generating a theoretical spectrum. While this is a desirable feature for users who may be interested in fine isotopic structure, it calculates more detail than is necessary for a theoretical spectrum that can be subsequently compared with data from a standard instrument.
To date, accurate determination of peak intensities has entailed more robust strategies for handling species enumeration. In contrast, this paper introduces a novel approach, which calculates the accurate probability for each number of neutrons added to the monoisotopic composition. At the outset, the algorithm—Neutron Cluster—calculates global neutronic probabilities rather than individual atomic distributions or isotopic combinations. Such probability grouping greatly simplifies the calculations and completely eliminates the need for pruning. It is the first algorithm described to date that uses grouped probabilities to obviate time- and memory-consuming species elucidation. As will be shown below, this approach calculates accurate intensities in nominal mass bins and yields helpful information towards elucidating fine isotopic structure after, rather than before, generating a theoretical spectrum.
Theory
Neutronic Distribution
Isotopes and equatransneutronic isotopes
For each element, E, each isotope, A, occurs at a natural abundance, PAE, which has been measured by other methods. These natural abundances are normalized for all the isotopes of each element:
| (1) |
To calculate the isotope cluster, which is the distribution of neutrons in a molecule, it is necessary to define the concept of equatransneutronic (ETN) isotopes, groups of isotopes differing from their element's most abundant isotope by the same number of neutrons. For the following derivation, isotopes in ETN group Gx differ from the most abundant isotope by dGx. For example, peptides, which contain C, H, N, O, and S, have 3 ETN groups: dG = 1, isotopes with 1 additional neutron (13C, 2H, 15N, 17O, 33S); dG = 2, those with 2 additional neutrons (18O, 34S), and one isotope with dG = 4 additional neutrons (36S).
Elemental, isotopic, and equatransneutronic probabilities
The abundances of elements in a molecule are known from the molecular formula, so the probability, PEmolecule, that a randomly selected atom is element E in a molecule having N atoms of which NE are element E can be calculated as follows.
| (2) |
Thus, the probability PAEmolecule that a randomly selected atom in the molecule is the isotope AE is calculated in a straightforward manner:
| (3) |
Further, the probability, PETNGx, of selecting an atom from the molecule, which is a member of a particular ETN group Gx is obtained from summing the probabilities of each PAEmolecule for each isotope in the ETN group. If Ei is the designation of an element of which isotope AEi is a member of ETN group Gx, and j is the number of elements in the ETN group, then this probability is represented as follows.
| (4) |
Because each ETN group contains atoms that have the same difference, dGx, in the number of neutrons from the most abundant isotope, PETNGx, is the probability that such a difference in neutrons will be present in a randomly selected atom of the molecule. This probability for each ETN group will be combined into absolute probabilities that a molecule contains a specific number of extra neutrons.
The Diophantine equation1,2 for obtaining combinations of equatransneutronic groups
To calculate the desired probability—the probability that a molecule differs by n neutrons from the monoisotopic composition—it is necessary to consider all the combinations of the ETN groups that can yield n, and to sum all of those contributions together. Since dGx, the neutronic difference between ETN group and the monoisotopic molecule must be a positive integer, and kx, the number of ETN group Gx members in the molecule, must also be a positive integer, the equation for n is the following Diophantine equation, where f is the number of ETN groups considered.
| (5) |
The solutions for k1 . . . kf to this Diophantine equation represent all the combinations of ETN groups that can yield the addition of n neutrons to the monoisotopic composition. Since the Diophantine equation is dependent only on the ETN groups that are being considered and not on the actual elements or isotopes in those groups, the equation can be solved once a priori and stored in a lookup table. The Diophantine can be set up and solved easily for any desired combination of ETN groups and any number of neutrons. For this paper, which focuses on polypeptides, the Diophantine was solved for dG = {1,2,4}, and for n = {1 . . . 500}. As will be shown below, this simple lookup table is sufficient to calculate the correct isotope cluster for proteins > 500 kDa.
Combining the contributions of equatransneutronic groups to the number of neutrons
The solutions to the Diophantine are sets of integers which, when substituted into the equation, yield a desired n. For example, if dG = {1,2,4}, and n = 1, then there is only one solution, {k1 = 1; k2 = 0; k3 = 0}. However, for the same dG if n = 2, then there are two solutions: {k1 = 2; k2 = 0; k3 = 0} and {k1 = 0; k2 = 1; k3 = 0}. The probability Psolution that one of these solutions will occur is the product of the probabilities Pkx that atoms from each ETN group Gx will occur kx times.
| (6) |
The probability Pkx can be calculated by the binomial formula where N is the total number of atoms, and PETNGx has is calculated in eq 4.
| (7) |
While the binomial distribution is used here to calculate Pkx, this algorithm differs substantially from other algorithms that use the binomial distribution to calculate the isotope cluster because the values for each variable in the equation are fundamentally different. As a consequence, this algorithm uses the binomial formula fewer times, and with smaller integers, than other algorithms. These differences are readily apparent from the above derivation and will be discussed below.
Calculating the abundance of a peak in the cluster
Equations 6 and 7 yield the probability that a particular combination of ETN groups, with a total addition of n neutrons to the monoisotopic composition, is present in the molecule. The abundance of the nth peak in the isotope cluster, Pn, is the sum of probabilities for each of the t combinations of ETN groups that add n neutrons to the monoisotopic composition. The abundance of the nth peak in the cluster is shown in eq 8.
| (8) |
Since there is no permutation threshold, Pn can be calculated correctly and rapidly for any number of neutrons for any molecule. For practical reasons, the algorithm allows the user to determine a fraction of the total isotope cluster to be characterized (i.e., 0.999999), and the algorithm stops after achieving enough probability peaks to explain that fraction of the cluster. This fraction is merely an indication of how much of the cluster is desired; it is not a permutation or abundance threshold and thus does not change the run time significantly.
Multiplicity
If each of the f ETN groups contain jx equatransneutronic isotopes, and kx is the number of atoms from ETN group Gx in solution s of the Diophantine, then the total multiplicity, M, for the nth peak in the isotope cluster is given in eq 9.
| (9) |
Since the algorithm here does not calculate individual atomic combinations, it does not implement or need permutation or abundance thresholds. Thus, prima facie, multiplicity is irrelevant. As will be discussed below, multiplicity is calculated for two reasons: (1) to determine the contribution of pruning error in the difference between this algorithm and others, and (2) to determine the number of combinations that would exist if fine isotopic combinations were desired.
Neutronic Mass
Each ETN group contains isotopes with different of nuclear binding energies. Table 1 lists the average mass per neutron between ETN isotopes and the most abundant isotope for each of the three ETN groups that are encountered in a peptide. Equation 7 yields Pkx, the contribution of each ETN combination to the abundance of the nth peak in the isotope cluster. Equation 10 incorporates this probability into a weighted average that yields the average mass difference between the nth peak in the isotope cluster and the mass of the monoiso-topic composition.
| (10) |
The total mass of the nth peak in the cluster is simply the monoisotopic mass plus the mass change as calculated in eq. 10.
Table 1.
The elements (Ei), isotopes (AE), exact mass, and the average mass of an added neutron for each of the ETN groups encountered in a polypeptide
| dG | Ei | AE | Mass (Da) | ΔMass/neutron (Da) | (Da) |
|---|---|---|---|---|---|
| 0 | H | 1 | 1.007825 | – | – |
| C | 12 | 12.00000 | – | ||
| N | 14 | 14.00307 | – | ||
| O | 16 | 15.99491 | – | ||
| S | 32 | 31.97297 | – | ||
| 1 | H | 2 | 2.014102 | 1.006277 | 1.00188 |
| C | 13 | 13.00335 | 1.003354 | ||
| N | 15 | 15.00011 | 0.997038 | ||
| O | 17 | 16.99913 | 1.004220 | ||
| S | 33 | 32.97146 | 0.99849 | ||
| 2 | O | 18 | 17.99916 | 1.002125 | 0.99979 |
| S | 34 | 33.96787 | 0.99745 | ||
| 4 | S | 36 | 35.96708 | 0.99853 | 0.99853 |
Enrichment
The equations and algorithm up to this point have been developed under the assumption that ETN groups share a similar distribution. While it will be shown below that such an assumption succeeds well with natural abundances for C, H, N, O, and S, enriched isotopes of these elements or elements with a different natural distribution than those commonly present in organic molecules (for example Sn or Hg) cannot be included in the same ETN group without the introduction of “grouping error,” which will be illustrated later. To avoid grouping error with these distributions, the algorithm groups similar distributions and partitions divergent ones. The treatment for a single distribution is shown above, and the different distributions are combined in a straightforward manner. If distribution D1 has b peaks from the monoisotopic to the bth peak, and distribution D2 has c peaks from the monoisotopic to the cth peak, then the final distribution D’ is calculated by multiplying each intensity in D1 by each intensity in D2. The total number of neutrons added to the monoisotopic composition must be less than bc since D1 and D2 contain overlapping numbers of neutrons.
Graphical Output
While the algorithm only calculates mass and intensity for peaks in the cluster that correspond to additional neutrons, a graphical representation or “theoretical spectrum” is often desired to compare predicted results to measurements. For this paper, the graphical output is rendered by a Gaussian approximation for the combination of instrumental and combinatorial variance around the neutronic mass. Each peak is used to generate a Gaussian bell, where the resolution, R, and the peak mass, m, are converted into the Gaussian standard deviation σ by eq 11.
| (11) |
Values obtained from eq 11 are placed into appropriately spaced bins and graphed on any commercially available graphing software.
Computational Features and Requirements
The algorithm above is implemented in Perl and is available in the supplemental material. The CPAN Perl modules BigInt and BigFloat were implemented for computational accuracy on factorial values above and exponential values below the validated computational range for Perl. The calculations in this paper run the BigFloat module with a precision of six significant figures, although greater precision can be introduced if desired. All run times discussed below are on a Mac-Book with a 2.16 GHz Intel 2 Core Duo Processor and 4 GB RAM.
Results and Discussion
Comparison with Existing Algorithms
To compare existing algorithms to the one described here, results for a published peptide, bovine insulin, with the elemental composition C254H377N65O75S6, were analyzed using IsoPro, a widely available embodiment of the standard polynomial algorithm, and IsoDalton [26], a well-implemented open source dynamic programming algorithm. These programs were operated with reasonable parameters: a 10−6 permutation threshold for IsoPro and a 10−8 state cut-off for IsoDalton. The abundances were all set to the same values as defined by NIST. Comparisons of intensity and multiplicity are shown in Table 2. The minor (1.1%) negative difference between Neutron Cluster and IsoDalton at the [M + 1] peak is a result of using absolute abundances, which add to unity; because IsoDalton's intensities are lower than those obtained in Neutron Cluster for the addition of more neutrons, the intensity of the [M + 1] peak is slightly higher. IsoPro does not normalize intensities to unity, and the effect of pruning error is seen dramatically in the higher mass peaks.
Table 2.
Comparison of absolute abundance (A) and multiplicity (M) for the isotope cluster obtained involving n neutrons from this algorithm—called NeutronCluster (NC)—Isodalton (ID), and IsoPro (IP) for bovine insulin (C254H377N65O75S6)
| Neutron Cluster |
IsoDalton |
IsoPro |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mass (Da) | NC (A) | NC (M) | ID (A) | ID (M)* | NC-ID (%A) | NC-ID (M) | Mass NC-IP (ppm) | IP (A) | IP (M)* | NC-IP (%A) | NC-IP (M) |
| 0 | 5729.606 | 0.030 | 1 | 0.030 | 1 | 0.0 | 0 | 1.0 | 0.029 | 1 | 3.3 | 0 |
| 1 | 5730.610 | 0.092 | 5 | 0.093 | 5 | –1.1 | 0 | 1.2 | 0.092 | 5 | 0 | 0 |
| 2 | 5731.613 | 0.160 | 17 | 0.160 | 17 | 0.0 | 0 | 1.3 | 0.16 | 16 | 0 | 1 |
| 3 | 5732.617 | 0.190 | 45 | 0.190 | 45 | 0.0 | 0 | 1.7 | 0.19 | 33 | 0 | 12 |
| 4 | 5733.617 | 0.180 | 104 | 0.180 | 104 | 0.0 | 0 | 1.4 | 0.18 | 57 | 0 | 47 |
| 5 | 5734.622 | 0.140 | 216 | 0.140 | 216 | 0.0 | 0 | 2.0 | 0.14 | 81 | 0 | 135 |
| 6 | 5735.624 | 0.097 | 416 | 0.096 | 416 | 1.0 | 0 | 2.0 | 0.097 | 107 | 0 | 309 |
| 7 | 5736.629 | 0.059 | 752 | 0.059 | 752 | 0.0 | 0 | 2.7 | 0.059 | 121 | 0 | 631 |
| 8 | 5737.629 | 0.033 | 1294 | 0.032 | 1294 | 3.0 | 0 | 2.4 | 0.032 | 132 | 3 | 1162 |
| 9 | 5738.634 | 0.017 | 2134 | 0.016 | 2134 | 5.9 | 0 | 3.1 | 0.016 | 131 | 5.9 | 2003 |
| 10 | 5739.635 | 0.0080 | 3398 | 0.0074 | 3396 | 7.5 | 2 | 3.0 | 0.0073 | 112 | 8.8 | 3286 |
| 11 | 5740.640 | 0.0035 | 5246 | 0.0032 | 5246 | 8.6 | 0 | 3.7 | 0.0030 | 92 | 14.3 | 5154 |
| 12 | 5741.640 | 0.0015 | 7888 | 0.0013 | 7884 | 13.3 | 4 | 3.4 | 0.0011 | 66 | 26.7 | 7822 |
| 13 | 5742.645 | 0.00057 | 11584 | 0.0005 | 11564 | 12.3 | 20 | 4.2 | 0.0003 | 28 | 47.3 | 11556 |
| 14 | 5743.646 | 0.00022 | 16664 | 0.0002 | 16592 | 9.1 | 72 | 4.0 | 0.0001 | 11 | 54.5 | 16653 |
| 15 | 5744.651 | 0.00010 | 23528 | 0.0001 | 23358 | 0.0** | 170 | – | – | 0 | – | 23528 |
Because Isodalton does not produce multiplicity in its standard output, the numbers here are maximal estimates for multiplicity, which were generated from NC by removing the contribution to multiplicity for all ETN combinations that were below ID's threshold of 1 × 10–8; an ETN combination represents at least 1 isotopic species, so the true multiplicity for ID is at most the number here. Thus, differences in multiplicity between ID and NC are minimal estimates.
No difference can be calculated within the single significant figure that ID yields for this n.
As described elsewhere [20, 25], pruning has an enormous effect on multiplicity. The results in Table 2 reiterate previous findings. The approach described differs fundamentally from other isotope cluster algorithms because neutron clustering allows for the calculation of the exact probability of adding neutrons to a molecule without elucidating every possible atomic location of these neutrons. Thus, multiplicity is a calculated rather than counted value, and all the low abundance species are intrinsic to the probability of adding a certain number of neutrons. While the algorithm avoids calculating the fine isotopic structure altogether, it yields the probability of each equatransneutronic combination, and this data could be used to calculate fine isotopic structure if desired. Such computations will be examined in a later paper.
Calculation of High Molecular Weight Isotope Clusters
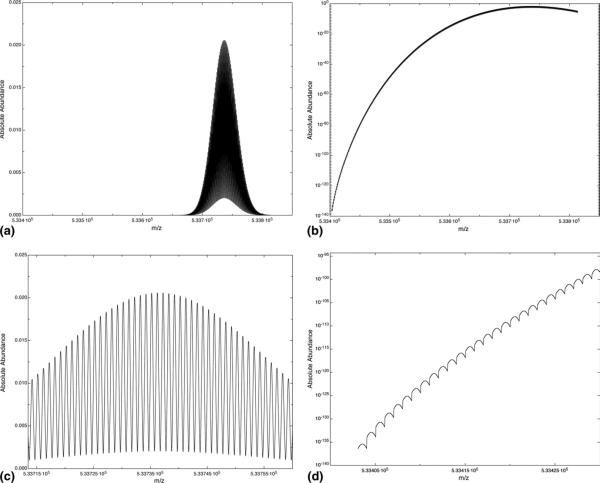
As an example of the algorithm's function at high molecular weight, 0.9999 of the isotope cluster for human dynein heavy chain 1, with the molecular formula C23,832H37,816N6528O7031S170, the monoisotopic mass of 533403.57, and the average of 533741.15 was calculated without any pruning in 39.95s. This entailed the addition of 400 neutrons; when plotted on a linear intensity scale, as in Figure 1a, the low end of the visible portion of the cluster is at 286 Da above the monoiso-topic mass and spans 98 Da. Figure 1b shows the same mass range on a logarithmic intensity scale. Comparison of these two figures highlights the low abundance of all peaks in the isotope cluster; the highest peak in the cluster, shown in Figure 1c, has an absolute intensity of 0.0206, and the monoisotopic peak, shown in Figure 1d, has an abundance of 4.0 × 10–136. These figures demonstrate that the range of absolute intensities required to calculate the full isotope cluster of a high molecular weight protein requires an algorithm that does not have any cutoffs or thresholds. For example, if the most abundant peak, the 337 neutron peak, were pruned to only include equatransneutronic combinations with probabilities greater than 10−8, only 105 out of the 7225 possible equatransneutronic combinations would remain. An error of ~3% would result on this peak alone.
Figure 1.
The theoretical spectrum for the isotope cluster for the microtubule associated protein dynein heavy chain 1 (accession KIAA0325) with a monoisotopic mass of 533403.57 Da and a resolution of 105. The cluster seen here explains .9999 of the absolute cluster. With absolute abundance plotted on a linear scale (a), only values close to the average mass of 533741.15 appear while the same data plotted on a logarithmic scale (b) demonstrate the very small peaks that lead up to the most abundant peaks. A close view of most abundant peaks (c) can be achieved on a linear scale. The monoisotopic and immediately subsequent peaks are rendered accurately by the algorithm and can be seen plotted on a low intensity logarithmic scale (d).
Computational time for isotope cluster calculations has remained a significant hurdle in the routine determination of accurate intensities for high molecular weight polypeptides, principally because the probabilities for individual atomic or isotopic combinations had to be elucidated before the intensity of the peak could be calculated. This algorithm presents a means of calculating the peaks while considering individual isotopes only once: in the calculation that obtains the global equatransneutronic probabilities (eq 4). For peptides, the number of equatransneutronic combinations is much less than the number of isotopic, and even atomic, combinations, so the algorithm presents a significant time advantage, as seen in Table 3. As stated before, these run times represent the time required to calculate the cluster without any pruning.
Table 3.
Run times needed for the algorithm to calculate the unpruned isotope cluster of some biologically relevant polypeptides. In the case of Angiotensin II, which has no sulfur, only 2 ETN groups were necessary, so Diophantine solutions in 2 variables (dG = {1,2}) were used
| Accession | Common name | Molecular formula | Monoisotopic mass (Da) | Time (s) |
|---|---|---|---|---|
| – | Angiotensin II | C50H71N13O12 | 1045.534 | 0.004 |
| 550085A | Bovine insulin | C254H377N65O75S6 | 5729.606 | 0.009 |
| AAA59179 | Human insulin | C520H817N139O147S8 | 11616.855 | 0.013 |
| P02144 | Human myoglobin | C744H1224N210O222S5 | 17172.957 | 0.017 |
| P27352 | Human intrinsic factor | C2023H3208N524O619S20 | 45387.020 | 0.109 |
| P02769 | Bovine serum albumin | C2934H4615N781O897S39 | 66389.890 | 0.151 |
| P05023 | Human Na/K ATPase, Renal isoform, subunit | C5047H8014N1338O1495S48 | 112823.910 | 0.787 |
| Q8WWZ7 | Human ATP binding cassette protein | C8574H13378N2092O2392S77 | 186386.849 | 3.056 |
| O60494 | Human intrinsic factor-hydroxocobalamin receptor | C17600H26474N4752O5486S197 | 398470.499 | 20.174 |
| KIAA0325 | Human dynein heavy chain | C23832H37816N6528O7031S170 | 533403.568 | 39.950 |
Since each neutronic peak in the isotope cluster represents an enormous multiplicity of isotopic species, and the mass assigned to each peak is the weighted average of the equatransneutronic groups, some mass error is expected in the mass assignments of these peaks. One way to estimate this error is to compare the mass assigned to the most abundant peak in the calculated isotope cluster against the mass calculated from average atomic masses. The results of such calculations are shown in Table 4. At higher masses, the theoretical average mass converges more with the most abundant peak because the number of added neutrons is greater, so the probability distribution of lesser abundant isotopes, which go into the average atomic mass, becomes more significant.
Table 4.
Relationship between the mass of the most abundant peak in the isotope cluster and the calculated average mass of the polypeptides in Table 3
| Common name | Molecular formula | Predicted average mass (Da) | Most abundant peak (Da) | Error (ppm) | Absolute intensity of most abundant peak |
|---|---|---|---|---|---|
| Angiotensin II | C50H71N13O12 | 1045.53* | 1045.53 | 0.0 | 0.528 |
| Bovine insulin | C254H377N65O75S6 | 5733.54 | 5732.62 | 16.0 | 0.186 |
| Human insulin | C520H817N139O147S8 | 11624.50 | 11623.88 | 5.3 | 0.135 |
| Human myoglobin | C774H1224N210O222S5 | 17183.71 | 17182.98 | 4.3 | 0.116 |
| Human intrinsic factor | C2023H3208N524O619S20 | 45415.86 | 45415.10 | 1.7 | 0.696 |
| Bovine Serum Albumin | C2934H4615N781O897S39 | 66432.73 | 66432.01 | 1.1 | 0.0567 |
| Human Na/K ATPase, renal isoform, subunit | C5047H8014N1338O1495S48 | 112895.58 | 112895.12 | 0.4 | 0.0442 |
| Human ATP binding Cassette Protein | C8574H13378N2092O2392S77 | 186506.81 | 186506.19 | 0.3 | 0.0342 |
| Human intrinsic factor-hydroxocobalamin receptor | C17600H26474N4752O5486S197 | 398724.57 | 398724.21 | 0.1 | 0.0233 |
| Human dynein heavy chain | C23832H37816N6528O7031S170 | 533737.29 | 533736.52 | 0.1 | 0.0206 |
Because of its small mass, the predicted peak for Angiotensin II is the monoisotopic, rather than the average mass, so there is no mass error.
Perturbation of the Isotopic Composition
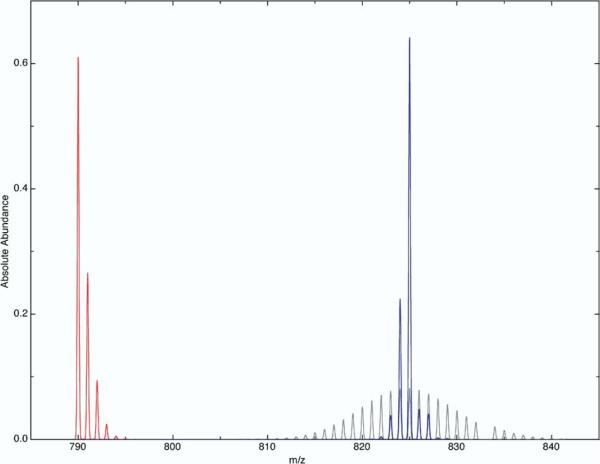
Stable isotope enrichment comprises the most reliable strategy for mass spectrometric quantification. Molecules labeled with stable isotopes, usually 13C or 2H, are used to quantify an analyte. Given the extreme importance of quantitative mass spectrometric measurements, any isotope clustering algorithm for organic molecules should provide accurate intensity values for enriched atoms. The algorithm in this paper simplifies calculations immensely by grouping intensities. While this allows for high accuracy (Table 2) and speed (Table 3), the trade-off is grouping error, which enters into the cluster intensities if members in the ETN group widely divergent distributions. Figure 2 shows an extreme example of grouping error for the peptide DARWIM (C35H54N10O16S1) when the abundance of 13C is 99%. The enriched abundance of carbon in this example differs so significantly from the natural abundances of 2H,15N,16O, and 33S that placing carbon in the same ETN group as these low abundance isotopes effectively averages 13C into the cluster. This results in an incorrectly broad distribution (the gray trace in Figure 2). Partitioning 13C and multiplying its distribution by the cluster generated from clustering all the other elements yields the correct distribution (the blue trace in Figure 2). Because all the other elements retain their clustering, the effect of this partitioning is negligible. Since the theoretical basis for both the clustered and partitioned distributions is the same, no pruning is introduced in the process of partitioning an enriched distribution.
Figure 2.
An example of grouping error for the peptide DARWIM (C35H54N10O9S1). The natural abundance distribution is shown in red, and the correct 99% enriched distribution, calculated by partitioning carbon from the grouped cluster, is shown in blue. At this enrichment, the difference between the correct cluster and the grouped cluster, shown in gray, is obvious. All the intensities are absolute.
Table 5 calculates the effect of grouping on isotope cluster calculations. It can serve as a general guide for applying this algorithm to enrichment experiments. If the instrumental cluster variance is less than the error on the cluster for a certain enrichment, then calculation of the theoretical cluster should involve partitioning of the enriched isotope. While partitioning reverts to previously described polynomial combination methods, the computational time is much less and does not require pruning because partitioning results in a combination of much fewer clusters, in this case the grouped cluster and the partitioned cluster, than would be encountered in the calculation of individual clusters for C, H, N, O, and S.
Table 5.
Calculated grouping error for 13C abundances from natural to 99% enrichment. The error is calculated as a sum of the absolute error for the intensities of the peaks in the grouped cluster against the intensities for the peaks in the cluster calculated by partitioning carbon
| Abundance of 13C (% carbon) | Total error (%) |
|---|---|
| 1.1 | 0.070 |
| 5.0 | 1.2 |
| 10 | 3.6 |
| 20 | 7.2 |
| 50 | 25 |
| 99 | 140 |
Conclusion and Future Directions
The theory and calculations in this paper present a novel basis for simplifying accurate calculations of the isotope cluster by grouping. Since each neutronic probability is calculated separately, the algorithm is the most conducive to parallel processing that has been described to date, and this benefit will be explored. Additionally, the detailed probabilities for each ETN combination can serve as a starting point for calculation of the fine isotopic structure when desired. Finally, by partitioning enriched isotopes, the theoretical spectra from quantitative experiments for a known enrichment can be obtained accurately and rapidly.
Acknowledgments
The authors acknowledge support for this research by the Intramural Research Program of the National Institute of Child Health and Human Development. The authors gratefully acknowledge the helpful comments and discussion of Jonathan A. Epstein (OSD/NICHD/NIH).
Footnotes
A Diophantine equation is a linear equation with positive integer coefficients and a finite multitude of positive integer solutions. In the case described in the text we are interested in the number of ways 1, 2, and 4 neutrons can combine to yield n neutrons. For a similar but less scientific example, the reader is encouraged to consider all the ways a cashier may dispense 16¢ using various numbers of 1¢, 5¢, and 10¢ coins. The solutions to this problem are the solutions to the Diophantine equation: 1n1+5n5+10n10=16.
It should be clear that Diophantine equations are a kind of equation rather than a specific equation. Thus, while Diophantine equations are not new to isotope cluster calculations24, the element-independent Diophantine equations here are much simpler, and their context is much different-than the Diophantine equations used previously. Additionally, they have been precomputed to enhance efficiency.
References
- 1.Zubarev RA, Demirev PA, Hakansson P, Sundqvist BU. R. Approaches and Limits for Accurate Mass Characterization of Large Biomolecules. Anal. Chem. 1995;67(20):3793–3798. [Google Scholar]
- 2.Blank PS, Sjomeling CM, Backlund PS, Yergey AL. Use of Cumulative Distribution Functions to Characterize Mass Spectra of Intact Proteins. J. Am. Soc. Mass Spectrom. 2002;13(1):40–46. doi: 10.1016/S1044-0305(01)00338-5. [DOI] [PubMed] [Google Scholar]
- 3.Havilio M, Haddad Y, Smilansky Z. Intensity-Based Statistical Scorer for Tandem Mass Spectrometry. Anal. Chem. 2003;75(3):435–444. doi: 10.1021/ac0258913. [DOI] [PubMed] [Google Scholar]
- 4.Spellman DS, Deinhardt K, Darie CC, Chao MV, Neubert TA. Stable Isotopic Labeling by Amino Acids in Cultured Primary Neurons: Application to Brain-Derived Neurotrophic Factor-Dependent Phosphotyrosine-Associated Signaling. Mol. Cell. Proteom. 2008;7(6):1067–1076. doi: 10.1074/mcp.M700387-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor EC, Rott AS, Zeder M, Juttner F, Dorn S. C-13-Labeling Patterns of Green Leaf Volatiles Indicating Different Dynamics of Precursors in Brassica Leaves. Phytochemistry. 2008;69(6):1304–1312. doi: 10.1016/j.phytochem.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Xiao GG, Garg M, Lim S, Wong D, Go VL, Lee WNP. Determination of Protein Synthesis in Vivo Using Labeling from Deuterated Water and Analysis of MALDI-TOF Spectrum. J. App. Physiol. 2008;104(3):828–836. doi: 10.1152/japplphysiol.00976.2007. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, Sasaki R, Ogata Y, Nakamura Y, Sakurai N, Kitajima M, Takayama H, Kanaya S, Aoki K, Shibata D, Saito K. Metabolic Profiling of Flavonoids in Lotus japonicus Using Liquid Chromatography Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Phytochemistry. 2008;69(1):99–111. doi: 10.1016/j.phytochem.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Pascal BD, Chalmers MJ, Busby SA, Mader CC, Southern MR, Tsinoremas NF, Griffin PR. The Deuterator: Software for the Determination of Backbone Amide Deuterium Levels from H/D Exchange MS Data. BMC Bioinformatics. 2007;8:156–167. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hotchko M, Anand GS, Komives EA, Ten Eyck LF. Automated Extraction of Backbone Deuteration Levels from Amide H/(2) H Mass Spectrometry Experiments. Protein Sci. 2006;15(3):583–601. doi: 10.1110/ps.051774906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snijders APL, de Koning B, Wright PC. Perturbation and Interpretation of Nitrogen Isotope Distribution Patterns in Proteomics. J. Proteome Res. 2005;4(6):2185–2191. doi: 10.1021/pr050260l. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XM, Hines W, Adamec J, Asara JM, Naylor S, Regnier FE. An Automated Method for the Analysis of Stable Isotope Labeling Data in Proteomics. J. Am. Soc. Mass Spectrom. 2005;16(7):1181–1191. doi: 10.1016/j.jasms.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Johnson KL, Muddiman DC. A Method for Calculating O-16/O-18 Peptide Ion Ratios. J. Am. Soc. Mass Spectrom. 2004;15(4):437–445. doi: 10.1016/j.jasms.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Rockwood AL, Van Orman JR, Dearden DV. Isotopic Compositions and Accurate Masses of Single Isotopic Peaks. J. Am. Soc. Mass Spectrom. 2004;15(1):12–21. doi: 10.1016/j.jasms.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Prebyl BS, Cook KD. Use of Fourier Transform for Deconvolution of the Unresolved Envelope Observed in Electrospray Ionization Mass Spectrometry of Strongly Ionic Synthetic Polymers. Anal. Chem. 2004;76:127–136. [Google Scholar]
- 15.Valkenborg D, Jansen I, Burzykowski T. A Model-Based Method for the Prediction of the Isotopic Distribution of Peptides. J. Am. Soc. Mass Spectrom. 2008;19(5):703–712. doi: 10.1016/j.jasms.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Aizikov K, O'Connor PB. Use of the Filter Diagonalization Method in the Study of Space Charge Related Frequency Modulation in Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2006;17(6):836–843. doi: 10.1016/j.jasms.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 17.Maleknia SD, Downard KM. Charge Ratio Analysis Method to Interpret High Resolution Electrospray Fourier Transform-Ion Cyclotron Resonance Mass Spectra. Int. J. Mass Spectrom. 2005;246(1/3):1–9. [Google Scholar]
- 18.Meija J, Caruso JA. Deconvolution of Isobaric Interferences in Mass Spectra. J. Am. Soc. Mass Spectrom. 2004;15(5):654–658. doi: 10.1016/j.jasms.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-de-Cossio J, Gonzalez LJ, Satomi Y, Betancourt L, Ramos Y, Huerta V, Amaro A, Besada V, Padron G, Minamino N, Takao T. Isotopica: A Tool for the Calculation and Viewing of Complex Isotopic Envelopes. Nucleic Acids Res. 2004;32:W674–W678. doi: 10.1093/nar/gkh423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roussis SG, Proulx R. Reduction of Chemical Formulas from the Isotopic Peak Distributions of High-Resolution Mass Spectra. Anal. Chem. 2003;75(6):1470–1482. doi: 10.1021/ac020516w. [DOI] [PubMed] [Google Scholar]
- 21.Horn DM, Zubarev RA, McLafferty FW. Automated Reduction and Interpretation of High Resolution Electrospray Mass Spectra of Large Molecules. J. Am. Soc. Mass Spectrom. 2000;11(4):320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 22.Datta BP. Polynomial Method of Molecular Isotopic Abundance Calculations: A Computational Note. Rapid Commun. Mass Spectrom. 1997;11(16):1767–1774. [Google Scholar]
- 23.Yergey JA. A General Approach to Calculating Isotopic Distributions for Mass-Spectrometry. Int. J. Mass Spectrom. 1983;52:337–349. doi: 10.1002/jms.4498. [DOI] [PubMed] [Google Scholar]
- 24.Hsu CS. Diophantine Approach to Isotopic Abundance Calculations. Anal. Chem. 1984;56(8):1356–1361. [Google Scholar]
- 25.Kubinyi H. Calculation of Isotope Distributions in Mass Spectrometry. A Trivial Solution for a Non-Trivial Problem. Anal. Chim. Acta. 1991;247:107–119. [Google Scholar]
- 26.Snider RK. Efficient Calculation of Exact Mass Isotopic Distributions. J. Am. Soc. Mass Spectrom. 2007;18(8):1511–1515. doi: 10.1016/j.jasms.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rockwood AL, Kushnir MM, Nelson GJ. Dissociation of Individual Isotopic Peaks: Predicting Isotopic Distributions of Product Ions in MSn. J. Am. Soc. Mass Spectrom. 2003;14(4):311–322. doi: 10.1016/S1044-0305(03)00062-X. [DOI] [PubMed] [Google Scholar]