Summary
The circadian clock uses a widely expressed pair of clock activators to drive tissue-specific rhythms in target gene expression. A new study sheds light on this tissue-specificity by showing that binding of clock activators and tissue-specific transcription factors to closely associated target sites enables cooperative activation of target genes in different tissues.
In animals, different tissues have specialized physiological and metabolic functions such as the regulation of blood sugar by the pancreas, the absorption of nutrients by the intestine, and the elimination of toxins by the liver. These tissue-specific physiological and metabolic functions are coordinately controlled with respect to each other and the time of day. Such coordination is effected by circadian clocks, which use a conserved pair of basic-helix-loop-helix transcriptional activators, CLOCK-CYCLE (CLK-CYC) in Drosophila and CLOCK-BMAL1 (or NPAS2-BMAL1) in mammals, to drive rhythmic gene expression in a vast array of tissues [1, 2]. Clock regulation of different tissue-specific physiological and metabolic rhythms implies that CLK-CYC and CLOCK-BMAL1 activate a different set of target genes in each tissue. Indeed, accumulating evidence supports this view [2-4], but little is known about how different target genes are selected by CLK-CYC and orthologs in different tissues. New work by Alexander Stark and colleagues [5] takes an important step towards understanding tissue-specific rhythms in gene expression by showing that CLK-CYC collaborates with tissue-specific transcription factors bound at nearby cis-regulatory sequences to synergistically activate different sets of target genes in different tissues.
CLK-CYC and CLOCK-BMAL1 primarily bind consensus CACGTG E-box sequences to drive rhythmic transcription of target genes in most tissues. These target genes can be roughly divided into two groups; core clock genes that keep circadian time via feedback inhibition of CLK-CYC and CLOCK-BMAL1 in all clock-containing tissues, and clock output genes that control common processes in many clock-containing tissues or specialized processes in specific clock-containing tissues. Several lines of evidence suggest that CLK-CYC and CLOCK-BMAL1 collaborate with other factors to bind E-boxes and activate output gene transcription in different tissues. Since consensus CACGTG E-box sequences are (statistically) present about every 4kb, there are tens to hundreds of thousands of potential CLK-CYC and CLOCK-BMAL1 binding sites in Drosophila and mice, respectively. However, chromatin immunoprecipitation (ChIP) analysis demonstrates that there are only ~1500 CLK-CYC binding sites in Drosophila heads and ~6000 CLOCK-BMAL1 binding sites in the liver [3, 6-8], indicating that additional sequences and/or transcription factors contribute to CLK-CYC and CLOCK-BMAL1 binding. In Drosophila heads, rhythmic CLK-CYC binding only identified genes with cycling mRNAs ~7% of the time [3]. This poor correspondence between CLK-CYC binding rhythms and mRNA cycling can be explained by rhythmic binding to specific isoforms expressed in few cells or tissue-specific binding that is masked by high expression in other tissues. More direct evidence of tissue-specific mRNA cycling came from an early microarray study that interrogated cycling transcripts in Drosophila heads and bodies [4]. This study showed that there was little overlap in the cycling head and body mRNA populations besides core clock genes [4], and mirrored tissue-specific differences in cycling mRNAs that were being uncovered in mammals [9, 10]. These studies demonstrated that the clock drove different populations of rhythmic mRNAs in different tissues, which provided a wealth of information about how the clock regulates tissue-specific physiological and metabolic processes and set the stage for investigating how the clock activates a specific group of output genes in a given tissue.
Since all rhythmic transcription in Drosophila stems directly or indirectly from CLK-CYC binding, Stark and colleagues first identified all CLK and CYC binding sites in DNA from fly heads and bodies. CLK and CYC bound sites containing conserved E-boxes in the promoters and introns of core clock genes in both heads and bodies as expected, but they also bound many sites that were different in heads and bodies [5]. Notably, the sites that were uniquely bound by CLK and CYC in the head identified genes that regulated neuronal function, whereas sites unique to bodies identified genes involved in metabolic functions. Having established that CLK and CYC bound many sites unique to head or body tissues, how do CLK and CYC select these tissue-specific targets? A computational approach was taken to identify sequence motifs associated with CLK and CYC binding sites unique to heads and bodies. This analysis revealed multiple sequence motifs situated nearby CLK and CYC sites that were enriched in heads or bodies, and several of these motifs corresponded to characterized transcription factor binding sites. Of these motifs, only five were required to correctly predict head- or body-specific CLK-CYC binding sites, but among these five motifs a GATA factor binding site was required to predict all CLK and CYC binding sites in the body. Given this remarkable predictive value, detailed analysis of a CLK-CYC site associated with a GATA site showed that one of the many Drosophila GATA factors, called SERPENT (SRP), bound the GATA sequence and synergistically activated transcription with CLK-CYC. Importantly, the vast majority of promoters containing GATA sites nearby CLK and CYC binding sites activated expression predominantly in body tissues. These results demonstrate that GATA factors play a key role in the tissue-specific activation of promoters bound by CLK-CYC in bodies and suggest that other tissue-specific factors activate expression in head tissues (Figure 1). Given the conservation of CLK-CYC and GATA factors, this model likely applies to tissue-specific activation of clock output genes in mammals.
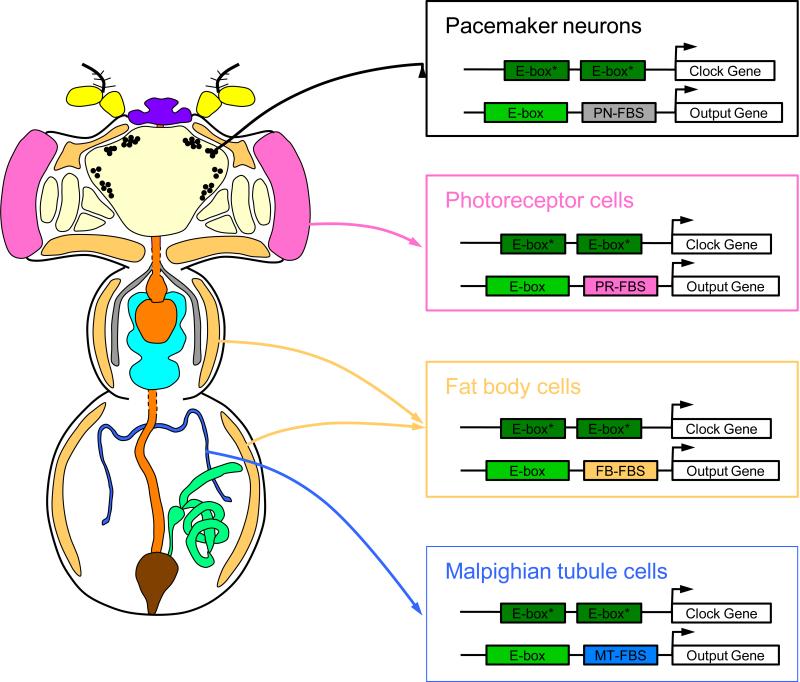
Figure 1.
Model for widespread and tissue-specific expression of CLK-CYC target genes. Clock gene-expressing tissues in an adult Drosophila (left). Yellow, antennae; cream, brain; tan, fat body; purple, proboscis; black, pacemaker neurons; pink, photoreceptors; orange, digestive tract; gray, salivary glands; aqua, ventral nerve chord; blue, Malpighian tubules; green, male reproductive tract; brown, rectum. Arrows denote regulation of clock gene and output gene expression in brain pacemaker neurons, photoreceptor cells, fat body cells and Malpighian tubules (right). A pair of closely-spaced E-boxes that bind CLK-CYC with high affinity are thought to promote transcription activation of clock genes in all tissues, whereas CLK-CYC bound to an E-box and a tissue-specific factor bound to a tissue-specific binding site cooperatively activate transcription of clock output genes in different tissues. Pacemaker neuron-specific factor binding site, PN-FBS; photoreceptor-specific factor binding site, PR-FBS; fat body-specific factor binding site, FB-FBS; Malpighian tubule-specific factor binding site, MT-FBS.
Though the Meireles-Filho et al. study has provided valuable insights into tissue-specific regulation of clock output genes, it also raises several questions. First, how do CLK-CYC or CLOCK-BMAL1 cooperate with GATA factors to activate target genes in a specific tissue? When Meireles-Filho and colleagues carried out luciferase reporter gene assays in cultured Drosophila cells, low levels of either CLK-CYC or GATA factor alone were unable to activate transcription, but the same levels of CLK-CYC and GATA factor together were able to bind their DNA consensus sequences and synergistically activate transcription. This synergistic activation does not preclude the possibility that one factor may be silently bound to DNA and primes the binding of a second factor that then recruits the transcriptional machinery. The recent characterization of circadian rhythms in permissive chromatin modifications at CLOCK-BMAL1 DNA binding sites [6, 11], along with the role GATA factors play as ‘pioneer transcription factors’ (i.e. factors that can bind target sites in closed chromatin) [12] would support such a hypothesis. In any case, while cooperation between transcription factors is not unprecedented and has been described in other systems [13, 14], the precise mechanisms that mediate cooperative interactions remains enigmatic and warrants further investigation.
In contrast to the many tissue-specific clock output genes, genes encoding core clock components are expressed in all clock-containing tissues (Figure 1). How do mechanisms that drive core clock gene expression differ from those that control tissue-specific clock outputs? A 69bp per enhancer fragment is capable of driving rhythmic expression in all Drosophila head and body tissues that contain clocks [15], which argues against the presence of multiple tissue specific binding sites that enable CLK-CYC binding and transcription in all clock tissues. A recent study on widespread and tissue-specific expression of estrogen receptor (ER) targets suggests a mechanism through which CLK-CYC and CLOCK-BMAL1 could drive some target genes in all clock tissues and others in specific clock tissues. ER acts as a pioneer transcription factor to drive expression in multiple tissues if high-affinity estrogen response elements (EREs) are present, but can drive expression in specific target tissues in collaboration with other factors when low-affinity EREs are present [16]. Three pieces of evidence suggest that E-box binding site affinity could account for differences in the expression of core clock genes and clock output genes. First, the top CLK-CYC and CLOCK-BMAL1 binding sites target core clock genes in both Drosophila and mice, respectively [3, 6-8]. Second, the core clock gene targets of CLK-CYC and CLOCK-BMAL1 contain dual E-box elements that drive higher levels of transactivation than single E-boxes [8, 17], suggesting that they are high-affinity target sites. Third, Drosophila Clk is able to activate both core clock gene expression and circadian oscillator function when expressed in novel locations [18], implying that clock genes have a ‘special status’ that allows them to be activated by CLK-CYC even in ectopic cells. Further studies are required to test whether CLK-CYC and CLOCK-BMAL1 drive target gene expression globally or tissue-specifically depending on target site affinity. To extend the insights from Meireles-Filho et al. further it is also necessary to identify sites bound by CLK-CYC and CLOCK-BMAL1 in specific tissues to accelerate the computational identification of associated binding sites that can be tested for their impact on tissue-specific expression. Identifying factors that bind these nearby sites will enable detailed biochemical studies of factor binding affinity, binding order, and cooperative interactions that promote tissue-specific transcription and provide a more complete picture of how CLK-CYC and CLOCK-BMAL1 select and activate target gene transcription.
References
- 1.Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol. 2013;23:724–731. doi: 10.1016/j.conb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mireles-Filho ACA, Bardet AF, Yanez-Cuna JO, Stampfel G, Stark A. Cis-regulatory Requirements for Tissue-specific Programs of the Circadian Clock. Curr Biol. 2013 doi: 10.1016/j.cub.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 10.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 11.Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez GJ, Rao A. Immunology. Cooperative transcription factor complexes in control. Science. 2012;338:891–892. doi: 10.1126/science.1231310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, Karagianni P, Brazma A, Adams DJ, Talianidis I, Marioni JC, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154:530–540. doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao H, Glossop NR, Lyons L, Qiu J, Morrish B, Cheng Y, Helfrich-Forster C, Hardin P. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J Neurosci. 1999;19:987–994. doi: 10.1523/JNEUROSCI.19-03-00987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, Cooper GM, Reddy TE, Crawford GE, Myers RM. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52:25–36. doi: 10.1016/j.molcel.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paquet ER, Rey G, Naef F. Modeling an evolutionary conserved circadian cis-element. PLoS Comput Biol. 2008;4:e38. doi: 10.1371/journal.pcbi.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, Allada R. Drosophila Clock Can Generate Ectopic Circadian Clocks. Cell. 2003;113:755–766. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]