The platinum hexahalides have an octahedral arrangement of six halogen atoms bound to a Pt centre, thus having an octahedral shape that could prove to be useful in interpreting poor electron-density maps. In a detailed characterization, PtI6 chemically transformed to a square-planar PtI3 complex bound to the Nδ atom of His15 of HEWL was also observed, which was not observed for PtBr6 or PtCl6.
Keywords: platinum hexahalides, hen egg-white lysozyme, PtI3 ligand bound to histidine, X-ray lasers
Abstract
This study examines the binding and chemical stability of the platinum hexahalides K2PtCl6, K2PtBr6 and K2PtI6 when soaked into pre-grown hen egg-white lysozyme (HEWL) crystals as the protein host. Direct comparison of the iodo complex with the chloro and bromo complexes shows that the iodo complex is partly chemically transformed to a square-planar PtI3 complex bound to the Nδ atom of His15, a chemical behaviour that is not exhibited by the chloro or bromo complexes. Each complex does, however, bind to HEWL in its octahedral form either at one site (PtI6) or at two sites (PtBr6 and PtCl6). As heavy-atom derivatives of a protein, the octahedral shape of the hexahalides could be helpful in cases of difficult-to-interpret electron-density maps as they would be recognisable ‘objects’.
1. Introduction
The platinum hexahalides (K2PtCl6, K2PtBr6 and K2PtI6) each consist of an octahedral arrangement of six halogen atoms bound to a platinum centre. As heavy-atom derivatives of a protein, the octahedral shape of the platinum hexahalides make them recognisable ‘objects’ in initial electron-density map interpretations compared with single-point metal atoms which may not be recognisable in noisy difference Patterson or Fourier maps (Helliwell, 2013b ▶). In particular, we envisage that they could become important, for example, in X-ray laser experiments that are striving to work with ever smaller samples, currently at the microcrystal/nanocrystal size but anticipated to transition to nanoclusters (see Helliwell, 2013a ▶). These compounds also offer a way to solve unknown protein structures by powder diffraction involving dispersive and/or isomorphous intensity differences (Helliwell et al., 2010 ▶).
K2PtCl6 and K2PtBr6 have both been studied before using soaking crystallization conditions into pre-grown hen egg-white lysozyme (HEWL) crystals; Sun and coworkers described a quick soak (∼10 min) approach and test using K2PtCl6 (Sun et al., 2002 ▶), while Helliwell and coworkers undertook a time-dependent analysis of K2PtBr6 binding to lysozyme studied by protein powder and single-crystal X-ray analysis (up to 3 h soak time; Helliwell et al., 2010 ▶). Both of these studies showed that these complexes bound to two sites on the protein. Site 1 is on a special position in a crevice between Arg14 in two symmetry-related molecules and site 2 is close to Ser86, Lys1 and Gln41 of one molecule in a crevice next to Pro79, Asn65 and Asn74 of a symmetry-related molecule. The platinum hexaiodide is probably of most interest as the most electron-dense complex and therefore this study scrutinizes in detail the chemical behaviour of K2PtI6 soaked into pre-grown HEWL crystals and also whether its chemical behaviour is the same as that of K2PtCl6 and K2PtBr6. The cases of K2PtCl6 and K2PtBr6 have also been studied under basically identical chemical and measurement conditions to the iodo form. An unexpected chemical behaviour of the iodo compound to form a PtI3 moiety bound to the His15 Nδ atom has been observed which was not observed with the chloro or bromo forms.
2. Methods
2.1. Crystallization conditions
HEWL crystals were prepared using a batch method as outlined by Blundell & Johnson (1976 ▶). 60 mg HEWL was dissolved in 1 ml 0.04 M acetate buffer pH 4.7 and 1 ml 10% NaCl was added to the solution. HEWL crystals were then soaked for 24 h in a 10 mM solution of either K2PtCl6, K2PtBr6 or K2PtI6. Each heavy-atom compound solution was obtained from a pre-made stock solution at 50 mM in acetate buffer.
2.2. X-ray diffraction data collection, protein structure solution and model refinement
A crystal from each soaking condition was scooped into a loop using Paratone as a cryoprotectant. All X-ray diffraction (XRD) data were measured on a Bruker APEX II home-source diffractometer at an X-ray wavelength of 1.5418 Å and a fixed temperature of 100 K (Table 1 ▶). The XRD data-collection strategy used led to a high completeness of unique data, high anomalous difference completeness and a reasonable level of data redundancy. All XRD data were processed using the PROTEUM2 software package (Bruker AXS, Madison, WI, USA).
Table 1. X-ray crystallographic data and final protein model-refinement statistics for HEWL crystals soaked in K2PtI6 .
Values in parentheses are for the last shell.
| PDB code | 4owc |
| Data-collection temperature (K) | 100 |
| Data reduction | |
| Space group | P43212 |
| Unit-cell parameters () | a = b = 78.69, c = 36.94 |
| Crystal-to-detector distance (mm) | 50 |
| Observed reflections | 135236 |
| Unique reflections | 14174 |
| Resolution () | 27.821.62 (1.651.62) |
| Completeness (%) | 97.8 (79.0) |
| R merge (%) | 0.0893 (0.1779) |
| I/(I) | 16.2 (3.7) |
| Multiplicity | 8.7 (2.4) |
| Refinement | |
| Cruickshank DPI () | 0.09 |
| No. of atoms | |
| Protein atoms | 1001 |
| Water molecules | 105 |
| Pt and halogen atoms | 14 |
| Other bound molecules or ions† | 1 |
| Average B factors (2) | |
| Protein atoms | 16.6 |
| Water molecules | 23.3 |
| Pt and halogen atoms | 16.8 |
| Other bound molecules or ions† | 11.5 |
| R factor/R free (%) | 17.4/19.3 |
| R.m.s.d., bonds ()/angles () | 0.02/1.81 |
| Ramachandran values (%) | |
| Most favoured | 96.1 |
| Additional allowed | 3.9 |
| Disallowed | 0 |
The other bound atom to the protein is an Na ion.
The crystal structures were solved using Phaser (McCoy et al., 2007 ▶) followed by rigid-body and restrained refinement with REFMAC5 in CCP4 (Murshudov et al., 2011 ▶), using the previously reported lysozyme structure with PDB code 2w1y as the molecular search model (Cianci et al., 2008 ▶). The use of Phaser was probably not required as 2w1y is relatively isomorphous to these crystals. Model building, adjustment and refinement were carried out using Coot (Emsley & Cowtan, 2004 ▶) and REFMAC5 (Murshudov et al., 2011 ▶) in CCP4. Ligand-binding occupancies were calculated using SHELXL (Sheldrick, 2008 ▶). The crystallographic and molecular model-refinement parameters for K2PtI6 are summarized in Table 1 ▶ and those for K2PtCl6 and K2PtBr6 are given in Supplementary Table S11. All figures were produced using CCP4mg (McNicholas et al., 2011 ▶).
3. Results
3.1. HEWL + K2PtI6
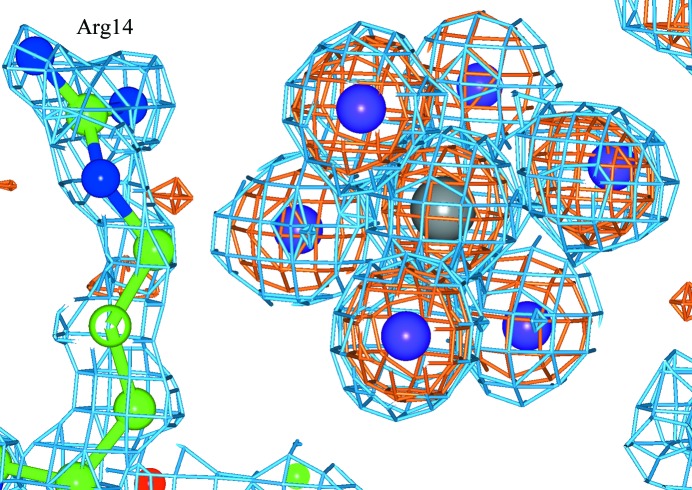
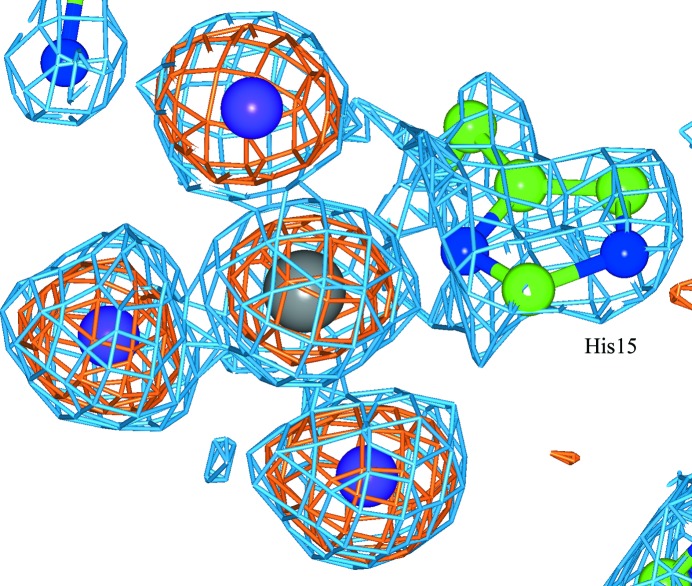
The binding of PtI6 to HEWL shows both similarities and differences to the previous studies involving PtCl6 (Sun et al., 2002 ▶) and PtBr6 (Helliwell et al., 2010 ▶). PtI6 binds to site 1, a crevice between two Arg14 residues in symmetry-related molecules (Fig. 1 ▶). This site is located at a special position, with a twofold axis passing through the Pt atom and two iodines. Individually refined heavy-atom occupancies, as well as whole group refined occupancies, are given in Supplementary Tables S2–S4 for each hexahalide complex. An octahedral PtI6 molecule is not bound in site 2, a crevice between Ser86, Lys1 and Gln41 of chain A next to Pro79, Asn65 and Asn74 of a symmetry-related molecule. Most interestingly, there is a new binding site that is chemically distinct comprising a PtI3 moiety bound to the Nδ atom of His15, forming a square-planar complex (Fig. 2 ▶).
Figure 1.
PtI6 binding in a special position between two Arg14 residues in symmetry-related molecules. The 2F o − F c electron-density map (blue) and the anomalous difference electron-density map (orange) are shown. The Pt atom is in grey and I atoms are in purple.
Figure 2.
A PtI3 moiety bound to the Nδ atom of His15. The 2F o − F c electron-density map (blue) and the anomalous difference electron-density map (orange) are shown. The Pt atom is in grey and I atoms are in purple.
3.2. HEWL + K2PtBr6 and HEWL + K2PCl6
The PtBr6 and PtCl6 complexes again showed binding at two sites on the HEWL protein as seen in the previously published short soaking studies of 10 min and 3 h with HEWL (Sun et al., 2002 ▶; Helliwell et al., 2010 ▶). However, no binding to His15 was observed for these complexes in this or the previous studies.
4. Discussion
Of most novel interest is the PtI3 moiety bound to His15. This is reminiscent of Zeise’s salt (PtCl3C2H4; Black et al., 1969 ▶). This also seems to be somewhat similar, but not identical, to the results of Messori et al. (2013 ▶), who soaked PtI2(NH3)2 into pre-grown HEWL crystals for two months in DMSO medium; they described a PtI2(NH3) molecule alternating between two different binding modes.
In our companion study (Tanley & Helliwell, 2014 ▶) of cisplatin and carboplatin binding to HEWL in NaI crystallization conditions, both cisplatin and carboplatin were partially converted to transiodoplatin bound at the Nδ binding site of His15. This is also similar to the results that we obtained under NaBr crystallization conditions for carboplatin (Tanley et al., 2014 ▶) and cisplatin (Tanley & Helliwell, 2014 ▶) and in NaCl conditions, where carboplatin was partially converted to cisplatin (Tanley et al., 2013 ▶).
We have also seen a PtI3 X moiety bound to a symmetry-related molecule in the cisplatin and carboplatin NaI conditions, as shown by three anomalous difference electron-density peaks bound to the Pt centre (Tanley & Helliwell, 2014 ▶).
5. Conclusions
For the octahedral PtI6 hexahalide molecule bound to HEWL, we see a chemical transformation to a square-planar PtI3 moiety bound to the Nδ atom of His15. This showed a different chemical behaviour to that of either the PtBr6 or the PtCl6 hexahalide complexes.
For the anticipated use with the X-ray laser, and other possible challenging-to-interpret electron-density map situations, as described in §1, PtI6 preserved its octahedral shape at one binding site but also appeared as a chemically transformed square-planar PtI3 moiety bound to a histidine N atom. This is acceptable of course when interpreting an electron-density map, but one has to know in advance that a complex of this shape is to be looked out for as well as an octahedron.
6. Related literature
The following references are cited in the Supporting Information for this article: Moreno-Gordaliza et al. (2009 ▶, 2010 ▶).
Supplementary Material
Supporting Information.. DOI: 10.1107/S2053230X14014009/no5054sup1.pdf
PDB reference: 4owe
PDB reference: 4owh
PDB reference: 4owc
Acknowledgments
JRH is grateful to the University of Manchester for general support, to the EPSRC for a PhD studentship to SWMT, to the Thailand Government for studentship support to SK, to the School of Chemistry for crystallization and computing facilities and to the Faculty of Life Sciences for X-ray diffraction facilities.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: NO5054).
References
- Black, M., Mais, R. H. B. & Owston, P. G. (1969). Acta Cryst. B25, 1753–1759.
- Blundell, T. L. & Johnson, L. N. (1976). Protein Crystallography London: Academic Press.
- Cianci, M., Helliwell, J. R. & Suzuki, A. (2008). Acta Cryst. D64, 1196–1209. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Helliwell, J. R. (2013a). Science, 339, 146–147. [DOI] [PubMed]
- Helliwell, J. R. (2013b). Acta Cryst. A69, s144.
- Helliwell, J. R., Bell, A. M. T., Bryant, P., Fisher, S., Habash, J., Helliwell, M., Margiolaki, I., Kaenket, S., Watier, Y., Wright, J. P. & Yalamanchilli, S. (2010). Z. Kristallogr. 225, 570–575.
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McNicholas, S., Potterton, E., Wilson, K. S. & Noble, M. E. M. (2011). Acta Cryst. D67, 386–394. [DOI] [PMC free article] [PubMed]
- Messori, L., Marzo, T., Gabbiani, C., Valdes, A. A., Quiroga, A. G. & Merlino, A. (2013). Inorg. Chem. 52, 13827–13829. [DOI] [PubMed]
- Moreno-Gordaliza, E., Cañas, B., Palacios, M. A. & Gómez-Gómez, M. M. (2009). Anal. Chem. 81, 3507–3516. [DOI] [PubMed]
- Moreno-Gordaliza, E., Cañas, B., Palacios, M. A. & Gómez-Gómez, M. M. (2010). Analyst, 135, 1288–1298. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sun, P. D., Radaev, S. & Kattah, M. (2002). Acta Cryst. D58, 1092–1098. [DOI] [PubMed]
- Tanley, S. W. M., Diederichs, K., Kroon-Batenburg, L. M. J., Levy, C., Schreurs, A. M. M. & Helliwell, J. R. (2014). Acta Cryst. F70, 1135–1142. [DOI] [PMC free article] [PubMed]
- Tanley, S. W. M., Diederichs, K., Kroon-Batenburg, L. M. J., Schreurs, A. M. M. & Helliwell, J. R. (2013). J. Synchrotron Rad. 20, 880–883. [DOI] [PMC free article] [PubMed]
- Tanley, S. W. M. & Helliwell, J. R. (2014). Acta Cryst. F70, 1127–1131. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.. DOI: 10.1107/S2053230X14014009/no5054sup1.pdf
PDB reference: 4owe
PDB reference: 4owh
PDB reference: 4owc