Abstract
Filamentous soil bacteria of the genus Streptomyces carry out complex developmental cycles that result in sporulation and production of numerous secondary metabolites with pharmaceutically important activities. To further characterize the molecular basis of these developmental events, we screened for mutants of Streptomyces coelicolor that exhibit aberrant morphological differentiation and/or secondary metabolite production. On the basis of this screening analysis and the subsequent complementation analysis of the mutants obtained we assigned developmental roles to a gene involved in methionine biosynthesis (metH) and two previously uncharacterized genes (SCO6938 and SCO2525) and we reidentified two previously described developmental genes (bldA and bldM). In contrast to most previously studied genes involved in development, the genes newly identified in the present study all appear to encode biosynthetic enzymes instead of regulatory proteins. The MetH methionine synthase appears to be required for conversion of aerial hyphae into chains of spores, SCO6938 is a probable acyl coenzyme A dehydrogenase that contributes to the proper timing of aerial mycelium formation and antibiotic production, and SCO2525 is a putative methyltransferase that influences various aspects of colony growth and development.
Streptomycetes are common soil bacteria that have an unusual multicellular lifestyle (7). Colonies grow vegetatively via formation of a branching substrate mycelium, which is composed of multigenomic hyphae that are only occasionally divided by cross walls. Morphological differentiation commences with the growth of a layer of hair-like hyphae, the aerial mycelium, in which individual filaments project from the surface of the colony into the air. The aerial hyphae are ultimately partitioned into uninucleoid compartments that mature into pigmented spores. Dispersal and subsequent germination of these spores complete the cycle. Metabolic changes that accompany this process of morphological differentiation result in the production of numerous useful secondary metabolites; the majority of antibiotics in current use, as well as drugs with anticancer, antifungal, and immunosuppressant activities, are obtained from Streptomyces species (5).
As a means of understanding the pathways responsible for differentiation in this important bacterial genus, over the past 30 years several screening analyses for developmental mutants have been carried out with the model species Streptomyces coelicolor (6, 15, 26, 32, 37). Most recently, an effective method for insertional mutagenesis of S. coelicolor has been developed (12), and this method continues to reveal novel loci involved in various aspects of this organism's growth and development (12, 13, 39). It is noteworthy that the vast majority of the developmental genes identified to date appear to encode proteins with gene-regulatory functions (sigma factors, response regulators, etc.) (reviewed in references 8 and 9). This is despite the fact that there is experimental evidence, at least on rich media such as R2YE medium, that there is an extensive cascade of extracellular signaling molecules that govern the early stages of S. coelicolor differentiation during which the aerial mycelium is formed (31, 44). The identities of the enzymes and/or structural proteins involved in the synthesis of and response to these proposed signals remain to be determined.
Using the insertional mutagenesis protocol (12), we continued the search for genes involved in the fundamental processes of aerial mycelium formation, sporulation, and antibiotic production in S. coelicolor. We discovered three loci not previously implicated in developmental events that encode a likely vitamin B12-dependent methionine synthase, an acyl coenzyme A (CoA) dehydrogenase, and a methyltransferase. The products of these genes are therefore promising targets for future investigations of the metabolic changes that occur in developing colonies and also possibly for delineation of the pathways responsible for biosynthesis of the previously proposed extracellular signaling molecules.
MATERIALS AND METHODS
Strains and growth conditions.
Prototrophic S. coelicolor strain M145 (SCP1− SCP2−) (4, 19) was used as the parent in mutagenesis experiments (see below). J2151 (ΔglkA119 bldM::hyg SCP1− SCP2−) was the source of the bldM::hyg allele (27). J1915 (ΔglkA119 SCP1− SCP2−) was the parent of the J2151 strain (18). Strain DB2321 (bldM::hyg SCP1− SCP2−) was one of eight identical transformants generated by transformation of M145 with genomic DNA isolated from J2151 (19, 33).
S. coelicolor strains were typically propagated on R2YE solid medium (19) at 30°C. Auxotrophy was assessed on solid minimal medium (19), and Escherichia coli-S. coelicolor conjugation experiments were conducted on MS-MgCl2 solid medium (11, 19).
Mutagenesis of S. coelicolor.
Insertional mutagenesis of M145 was conducted as described previously by in vitro transposition with Tn5apr of plasmid libraries of S. coelicolor DNA and subsequent introduction of the mutagenized libraries into M145 cells by either protoplast transformation or conjugation from E. coli (12). Transformants and exconjugants were selected by growth on R2YE and MS-MgCl2 media, respectively, flooded with apramycin (25 μg/ml). Colonies were visually screened for colonies with defects in the developmental process, and analysis was continued for the defective strains that also exhibited double-crossover marker replacement with a transposon-disrupted allele (apramycin-resistant, spectinomycin-sensitive colonies). In each case, linkage of the transposon insertion to the developmental mutation was tested by genomic DNA transformation (33). Genomic DNA was prepared from each mutant and used to transform M145 protoplasts (19). For the mutants described here, all of the resulting transformants exhibited phenotypes identical to that of the original mutant (DB2571, 3 transformants; DB5931, 4 transformants; AG1440, 285 transformants; NY247, 34 transformants; NY1868, 19 transformants). The chromosomal locations of the Tn5apr insertions were determined by sequencing the transposon-flanking DNA (12). The deduced chromosomal locations were verified by PCR amplification of the transposon-disrupted alleles.
Complementation of mutants.
Complementation plasmids were constructed by using the integrating vector pSET152S (13). To test the complementation of NY1868, the SCO6938 and SCO6939 genes were PCR amplified from M145 genomic DNA with the following primer pairs: 5′-GCTCTAGAGCTGGGCGACGCGTTGCAGGGT-3′ and 5′-GGAATTCCAGTGCACAGGTGATGGACCACATC-3′ (for SCO6938) and 5′-GCTCTAGAGCCATCAGGTCGGCGATCTTGCTTT-3′ and 5′-CGGAATTCCGTGAATTGCGCCGCCACCACTAA-3′ (for SCO6939). These genes were cloned between the XbaI and EcoRI sites of pSET152S to obtain pSET152S-SCO6938 and pSET152S-SCO6939. These plasmids were then introduced into NY1868 by conjugation from E. coli ET12567(pUB307) (11), and complementation in spectinomycin-resistant exconjugants was visually assessed after several days of growth on R2YE medium.
The complementation of NY247 was assessed in a similar manner except that primers 5′-GCTCTAGAACCAGGAAGTGCTCCTCGTACATGTC-3′ and 5′-GGAATTCTACAGACCTCGTTGCCCTGT-3′ were used for construction of pSET152S-SCO2525 and primers 5′-GCTCTAGAACCAGGAAGTGCTCCTCGTACATGTC-3′ and 5′-GGAATTCCGAAGACCAGCACGCTCACACCTCCT-3′ were used for construction of pSET152S-SCO2525+24.
Scanning electron microscopy.
Microscopy was performed with an environmental scanning electron microscope (FEI Quanta 400). Individual colonies were cut from R2YE agar plates and fixed in 4% glutaraldehyde in 0.1 M HEPES buffer (pH 7.2). Colonies were rinsed in HEPES buffer several times, incubated for 1 h in 1% osmium tetroxide, and then rinsed in progressively higher concentrations of ethanol (50 to 100%) (5 min each time). A final rinse for 5 min in hexamethyldisilazane was performed prior to air drying. Colonies were sputter coated with gold-palladium for 30 s (E5200 Polaron SEM autocoating unit). Microscopy was performed under a high vacuum at 10.0 kV.
RESULTS
Mutagenesis of S. coelicolor used to identify developmental genes.
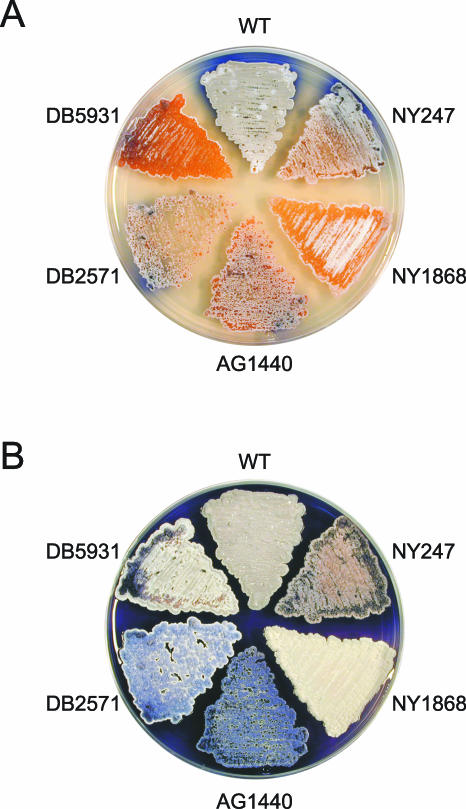
Insertional mutagenesis with an in vitro transposition system has proven to be an effective means of identifying novel developmental genes in S. coelicolor (12, 13, 39). We used this method to generate a new collection of mutants with abnormalities in aerial mycelium formation, sporulation, and secondary metabolite production. In each case, genomic DNA was isolated from the mutant and backcrossed into the wild-type strain to verify that a transposon insertion caused the phenotype observed. The phenotypes of the insertion mutants, both early and late in the development of an aerial mycelium, are shown in Fig. 1. Colonies of wild-type strain M145, which was used as the parent for mutagenesis, exhibited upon maturation a fluffy, grey aerial mycelium that had undergone abundant sporulation; colonies also produced large amounts of the blue-pigmented antibiotic actinorhodin. Insertion mutants NY1868 and DB5931 showed delayed progression through the developmental cycle. Mutant DB2571 produced an aerial mycelium but was unable to sporulate. Mutant AG1440, which was also a methionine auxotroph, formed a highly pigmented, blue aerial mycelium that failed to sporulate. Finally, insertion mutant NY247 exhibited a variety of aberrations in its aerial mycelium, pigmentation, and colony morphology (see below).
FIG. 1.
Comparison of wild-type S. coelicolor and five insertion mutants. The wild-type (WT) parent strain, M145, and the mutants were grown on solid R2YE medium plates at 30°C for 2 days (A) and 5 days (B). The mutant phenotypes were caused by transposon disruption of an unknown gene in the case of NY247 and NY1868, transposon disruption of metH in the case of AG1440, transposon disruption of bldM in the case of DB2571, and transposon disruption of bldA in the case of DB5931.
By sequencing the DNA flanking the transposon insertion in each of the mutants and comparing this sequence information to the information for the complete S. coelicolor genome (4; http://www.sanger.ac.uk/Projects/S_coelicolor/), we identified the genes disrupted in each case (Table 1). Two of the insertions were in known developmental genes. DB5931 carried an insertion in the well-studied gene bldA, which encodes a leucyl-tRNA that is required in development (14, 23, 30, 40). The transposon in DB2571 was found to be inserted into the recently identified gene bldM, which has been described as a response regulator gene required for aerial mycelium formation (27). Consistent with the methionine auxotrophy of AG1440, this mutant carried an insertion in the coding region of the probable metH gene that encodes a methionine synthase (36). In the remaining mutants (NY247 and NY1868), the transposon disrupted the coding regions of genes that were novel and whose functions have not been characterized.
TABLE 1.
Genes identified
| Strain | Tn5apr insertion site and flanking sequencea | Gene |
|---|---|---|
| DB5931 | AAGGCGCGCGAAGCGGAGCCCATGGCCTG | bldA |
| DB2571 | GCGGAGGACGGCCATGACTTCCGTCCTCG | bldM |
| AG1440 | TTTGGAGGCGGCCGTGGTCTACGGCTACT | metH |
| NY1868 | CAACGCCAGTGCACAGGTGATGGACCACA | SCO6939 |
| NY247 | GTCGGCCAGAGCCTGGAGCCGTTCACCCA | SCO2525 |
The Tn5apr insertion site is indicated by boldface type.
Disruption of known developmental genes yields unexpected phenotypes.
Two of the mutant strains shown in Fig. 1 had insertions in known developmental genes. However, in both cases the phenotypes observed were different than those described for previously isolated mutants with mutations in the genes (23, 27). In the case of bldA, a deletion mutant (J1681) that exhibits phenotypes identical to those of bldA point mutants has been described (23). On R2YE medium, these strains do not form an aerial mycelium (thus the designation bald) and also do not produce the red- and blue-pigmented antibiotics. The bldA mutant isolated in this study, DB5931, showed a less severe disruption of the developmental cycle. Aerial mycelium formation and the subsequent sporulation were delayed approximately 2 days in a DB5931 culture compared to the aerial mycelium formation and sporulation of a culture of the M145 wild-type strain. However, after 4 to 5 days of growth grey spores were observed in the mutant culture; antibiotic production also occurred quite normally. The transposon insertion in DB5931 was located in the portion of the gene that encodes the 5′ region of the transcript that is absent in the mature tRNA (23). Perhaps a small amount of mature tRNA is produced in DB5931, allowing this mutant to escape with a delay rather than a complete block in aerial mycelium formation. The genetic background may also affect the severity of the observed bldA phenotype. J1681 was constructed in the J1501 background (hisA1 uraA1 strA1), while DB5931 was generated in the prototrophic M145 background (19). Our attempts to construct a bldA deletion mutant in M145 were unsuccessful.
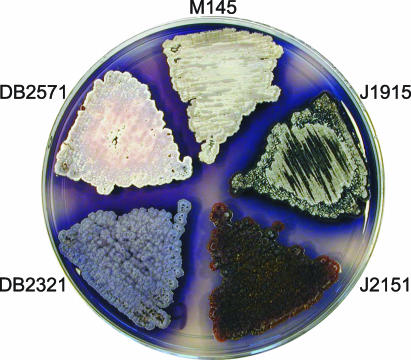
In the case of DB2571, which has a disruption in bldM, we also observed a developmental phenotype different from that reported for a previously constructed knockout strain (J2151) (27). J2151 had a bld phenotype with a complete block in differentiation and a lack of aerial mycelium formation (Fig. 2) (27), like J3146, a bldM derivative of M600 (M. Elliot and M. Buttner, personal communication). In contrast, DB2571 formed an aerial mycelium, but the aerial hyphae did not initiate sporulation (Fig. 2 and data not shown). To confirm this difference, the bldM::hyg allele from J2151 was moved to the M145 background by genomic transformation. The resulting strain, DB2321, could likewise form an aerial mycelium but could not sporulate (Fig. 2 and data not shown). The originally isolated bldM point mutants, also generated in M145, had phenotypes similar to those of DB2571 and DB2321, and the locus was originally designated whiK (37). Thus, although bldM is clearly essential for complete differentiation, the genetic background appears to have a strong influence on the bldM null mutant phenotype.
FIG. 2.
Comparison of bldM alleles in different genetic backgrounds. The strains were grown on R2YE solid medium at 30°C for 4 days. A bldM::hyg allele was previously introduced into the J1915 genetic background to obtain bald strain J2151, which is blocked for aerial mycelium formation (27). Transfer of the bldM::hyg allele to M145 by genomic transformation yielded strain DB2321, which produces an aerial mycelium but fails to sporulate. Strain DB2571, which bears a bldM::Tn5apr allele and was generated in the M145 genetic background, also produces a nonsporulating aerial mycelium.
Primary metabolism gene is required for sporulation.
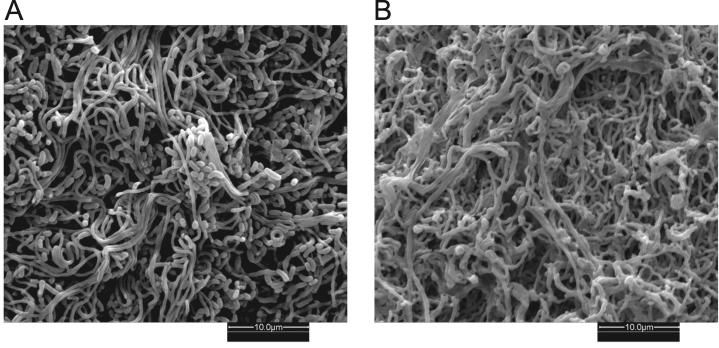
Auxotrophic strain AG1440 displayed several developmental abnormalities when it was cultured on rich medium that permitted vegetative growth (Fig. 1). Colonies developed an immature aerial mycelium composed of kinky filaments (Fig. 3). Very few, if any, spores were produced from these aerial filaments. The aerial mycelium was also bright blue, presumably due to overproduction of actinorhodin. AG1440 could not grow on minimal medium unless it was supplemented with methionine; this auxotrophy was rationalized by the location of the insertion in the coding region of metH (Table 1) (36). The metH gene (SCO1657 or SCI46.02c) is inferred to encode the vitamin B12-dependent methionine synthase that catalyzes the final step in methionine biosynthesis, the conversion of homocysteine to methionine. The S. coelicolor genome also contains a metE homolog (SCO0985 or SCBAC19F3.12) encoding an apparent vitamin B12-independent methionine synthase (4). Presumably, in the absence of MetH activity, the methionine levels in the cell are not sufficient for growth on minimal medium and sporulation on rich medium.
FIG. 3.
Scanning electron micrographs showing the colony surfaces of wild-type S. coelicolor and a metH mutant. The aerial mycelium of wild-type strain M145 produces abundant spores (A), while the aerial hyphae of metH mutant AG1440 appear to be wrinkled and lack any visible signs of sporulation (B). Colonies were grown on solid R2YE medium for 4 days at 30°C prior to scanning electron microscopy.
Probable acyl-CoA dehydrogenase participates in proper timing of differentiation and antibiotic production.
Mutant NY1868 was characterized by a pronounced delay in aerial mycelium formation and blue pigment production. Aerial mycelium formation and the subsequent sporulation were delayed by at least 1 day compared to aerial mycelium formation and sporulation in the parent strain (Fig. 1). Once under way, the delayed development appeared to be normal; aerial hyphae underwent the coiling characteristic of the wild-type hyphae and eventually produced abundant spores. Production of the blue antibiotic actinorhodin was also delayed by a couple of days; morphological mutants of S. coelicolor often have concurrent defects in secondary metabolism (5).
Sequencing revealed disruption of a previously uncharacterized gene in NY1868, SCO6939 (SC1G8.11c), which encodes a protein whose function is unknown (Fig. 4A). The four genes immediately upstream of, and overlapping, SCO6939 (cvnA8 to cvnD8) have been designated a conservon to reflect the conserved clustering and probable operon structure of these genes and the 12 other paralogous clusters in the genome (4), and the cvn9 cluster has recently been implicated in aerial mycelium development in both S. coelicolor and Streptomyces griseus (22). Analysis of the sequence of the SCO6939 product by a BLAST search (2) indicated that its closest homologs were in Streptomyces, including SCO6798 (SC1A2.07) in S. coelicolor (whose corresponding gene is also located immediately downstream of a conservon, cvnA7 to cvnD7).
FIG. 4.
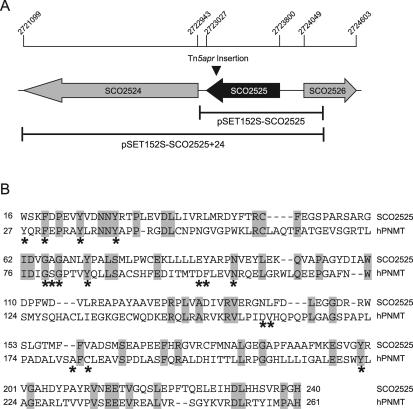
Genomic location of the insertion in mutant NY1868 and complementation of the insertion. (A) The Tn5apr transposon was found to be inserted into the 3′ end of the SCO6939 gene in the chromosome of NY1868. This gene and the flanking genes are shown, and the scale at the top indicates the base pair positions of these genes in the S. coelicolor chromosome sequence determined by the Sanger Centre genome sequencing project (4). The Sanger Centre annotation is indicated below each gene. The DNA regions that were PCR amplified, cloned into pSET152S, and used in complementation experiments are indicated at the bottom. (B) The delay in development exhibited by NY1868 was not complemented by a pSET152S derivative containing SCO6939 but was complemented by a construct containing SCO6938. The strains were grown on R2YE solid medium at 30°C for 2 days. WT, wild type.
The insertion in SCO6939 occurred in the 3′ end of the gene, just 41 bp from the end of the coding region and 124 bp from the start of the SCO6938 (SC1G8.10c) coding region. It was therefore possible that this insertion disrupted not only the final gene in the putative cvn8 operon but also transcription of the downstream gene(s). Complementation experiments revealed that indeed, the delayed development defects observed in NY1868 were not complemented by DNA corresponding to SCO6939 alone. However, complementation plasmids containing the downstream SCO6938 gene allowed the mutant to produce an aerial mycelium and actinorhodin in a timely fashion (Fig. 4B). The SCO6938 gene product showed strong similarity to acyl-CoA dehydrogenases, a family of enzymes whose members participate in oxidation of fatty acids and branched-chain amino acids (25).
Putative methyltransferase is required for proper aerial mycelium formation and colony morphology.
Insertion mutant NY247 had a number of abnormalities in morphological differentiation compared to the morphological differentiation of the corresponding wild-type strain (Fig. 1). An aerial mycelium formed at the proper time. However, under low magnification the hyphae appeared to be shorter and/or more matted than those of the wild-type strain (data not shown); these aerial hyphae also developed a lavender pigmentation, in contrast to the progression from white to grey observed in normal sporulating hyphae. These unusual hyphae were capable of forming spores. Another striking phenotype of this mutant was that by 4 days of growth, cracks began to develop throughout the colony surface. Overall, this mutant appeared to be quite similar to a strain that has been described previously, AG24 (12), which also produced short, purple aerial hyphae and cracked colonies.
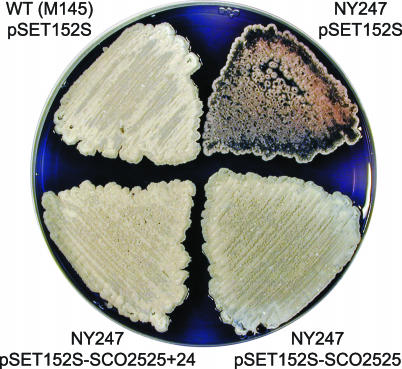
Sequencing revealed that the insertion in NY247 was different from that in AG24 that was characterized previously (12). In the case of NY247, the transposon disrupted the SCO2525 (SCC121.28c) gene (Fig. 5A). To confirm that this gene is necessary for normal growth and development in S. coelicolor, NY247 was transformed with integrating plasmids containing either SCO2525 DNA alone or a combination of this gene and its downstream open reading frame (SCO2525+24). The multiple defects observed in NY247 were complemented by SCO2525 either alone or in combination with the downstream gene (Fig. 6).
FIG. 5.
Genomic location of the insertion in mutant NY247. The Tn5apr transposon was found to be inserted into gene SCO2525 in the chromosome of NY247. (A) SCO2525 gene and flanking genes. The scale bar at the top indicates the base pair positions in the S. coelicolor genome sequence (4). SCO2525 and SCO2524 have been annotated as unknown hypothetical proteins, while SCO2526 has been annotated as a possible acetyltransferase by the Sanger Centre. The DNA regions that were PCR amplified, cloned into pSET152S, and shown to complement the defects in NY247 are indicated at the bottom. (B) Partial alignment of SCO2525 with human PNMT (hPNMT). Identical residues are shaded, and residues that participate in binding to the SAM cofactor in human PNMT are indicated by asterisks (24). These active site residues are well conserved in the two proteins.
FIG. 6.
Requirement of SCO2525 for proper aerial mycelium formation and colony morphology. The defects apparent in NY247 (upper right) were complemented by introduction of pSET152S derivatives bearing SCO2525 either alone or in combination with the downstream gene (lower half). The strains were grown on R2YE solid medium for 5 days at 30°C. WT, wild type.
The SCO2525 gene product has not been assigned a function (4), and no apparent bacterial homologs were revealed by a BLAST search (2). However, SCO2525 did show some similarity to the NNMT/PNMT/TEMT family of vertebrate methyltransferases. A crystal structure has recently been determined for human phenylethanolamine N-methyltransferase (PNMT), which catalyzes transfer of a methyl group from an S-adenosyl-l-methionine (SAM) cofactor to the primary amine of noradrenaline, yielding the final product adrenaline (24). A partial alignment of the 257-amino-acid S. coelicolor SCO2525 protein and the 282-amino-acid human PNMT is shown in Fig. 5B. Strikingly, a considerable number of residues are identical or show conservative substitution in the two proteins (22% overall identity), including 75% of the active site residues that have been implicated in SAM cofactor binding (24). On the basis of this alignment, we propose that SCO2525 encodes a methyltransferase required for morphological differentiation in S. coelicolor.
DISCUSSION
We used insertional mutagenesis to identify a collection of both known and novel genes involved in the development of the important bacterial model organism S. coelicolor. Mutants defective in aerial mycelium formation, sporulation, and antibiotic production have been known for over 30 years. However, the signals and molecular pathways that elicit the developmental events remain poorly defined. Most of the previously identified bld and whi genes appear to encode regulatory proteins (reviewed in references 8 and 9), which raises the question of what genes and subsequent biochemical activities must be properly regulated in order to achieve differentiation. Here we identified three likely biosynthetic enzymes with roles in development: the methionine synthase MetH, the putative methyltransferase SCO2525, and the probable acyl-CoA dehydrogenase SCO6938.
The methionine auxotroph isolated here, AG1440, not only failed to grow vegetatively on minimal medium but on rich medium developed an aerial mycelium that could not sporulate. A methionine requirement for sporulation has also been described previously for a Streptomyces fradiae mutant (42). This strain was not an auxotroph but did require addition of methionine for sporulation to occur. The authors speculated that this mutant was defective in conversion of homocysteine to methionine, the reaction catalyzed by the metH gene product that is disrupted in AG1440, since biosynthetic intermediates upstream of methionine did not allow sporulation.
The discovery that disruption of metH blocks sporulation in aerial hyphae of S. coelicolor provides further evidence that the levels of methionine and/or SAM play an important role in both morphological differentiation and secondary metabolite production. It was recently demonstrated that in both S. coelicolor and Streptomyces lividans increased levels of SAM due to either overexpression of the SAM synthetase gene metK or exogenous addition can inhibit sporulation of aerial mycelia and cause overproduction of actinorhodin (20, 34). The metH mutant AG1440 likewise did not sporulate and appeared to produce increased amounts of blue actinorhodin. SAM is synthesized from methionine and ATP via the action of the SAM synthetase, and in E. coli SAM serves as a corepressor of the methionine biosynthetic genes (43). Repression of SAM synthetase activity by methionine has been demonstrated in Streptomyces hygroscopicus (21). If the regulation of methionine biosynthesis in S. coelicolor is similar to the regulation of methionine biosynthesis in these other organisms, then high intracellular levels of SAM may cause methionine concentrations to be reduced in the cell and vice versa. This would explain how both metK overexpression (high SAM levels) and metH disruption (low methionine levels) could cause the same developmental phenotype. The overproduction of actinorhodin elicited by high SAM concentrations has been explained by increased transcription of the actII-ORF4 gene, whose product activates transcription of the actinorhodin biosynthetic genes (20, 34). It will be interesting to determine if the transcription of actII-ORF4 and other genes that regulate morphological differentiation is likewise altered in this metH mutant.
We also identified a putative methyltransferase, an enzyme that uses SAM as a cofactor to transfer a methyl group to a substrate molecule, which is required for proper colony morphology, aerial mycelium formation, and pigmentation. There is a high degree of conservation between the active-site residues of human PNMT and SCO2525. PNMT catalyzes the final step in the biosynthetic pathway of the hormone adrenaline, in which tyrosine is hydroxylated to form 3,4-dihydroxyphenylalanine (DOPA), DOPA is decarboxylated to give dopamine, dopamine is hydroxylated to give noradrenaline, and finally noradrenaline is N methylated to form adrenaline (29). Does the SCO2525 methyltransferase participate in a similar pathway for production of a metabolite or signal molecule required for proper streptomycete colony growth and development? And if so, what is the enzyme's physiological substrate? The SCO2525 sequence contains, in addition to many of the SAM binding residues, several of the amino acid residues that have been modeled to contact the human PNMT noradrenaline substrate (24). These include residues that interact with the aromatic ring and the amine of noradrenaline (human PNMT F182, N39, and Y222) but not the residues that interact with the β-hydroxyl group that arises from hydroxylation of the dopamine precursor (human PNMT E219 and D267).
S. coelicolor may produce molecules whose structures are related to the structures of adrenaline pathway intermediates that could serve as substrates for the SCO2525 methyltransferase. Streptomyces species typically are able to synthesize DOPA from tyrosine, and the DOPA is then utilized in melanin pigment biosynthesis (17). While S. coelicolor does not make melanin, its genome does contain the melC2 gene, whose product is a monophenol monooxygenase (tyrosinase) that should be capable of catalyzing DOPA production (35). It is also possible that S. coelicolor possesses a DOPA decarboxylase that could convert DOPA to dopamine. The first bacterial example of an l-DOPA decarboxylase was found in Sorangium cellulosum (28), and the product of the S. coelicolor SCO2782 (SCC105.13) gene is very similar to this myxobacterial enzyme (although BLAST searches have indicated that it is more similar to 2,4-diaminobutyrate decarboxylases and glutamate decarboxylases than to aromatic amino acid decarboxylases [38]). S. coelicolor also possesses a homolog of the S. griseus epoA (SCO6712 or SC4C6.22) gene, which encodes a phenol oxidase that uses DOPA as a substrate to produce an unidentified compound that can stimulate aerial mycelium formation in both S. griseus and S. coelicolor (10). Finally, certain Streptomyces species have been found to produce large amounts of the aromatic compound phenylethylamine, which lacks the β-hydroxyl and catechol groups of noradrenaline and may be generated from the decarboxylation of phenylalanine (1, 41). Biochemical studies are necessary to determine if SCO2525 can catalyze methyl transfer to phenylethylamine, DOPA, derivatives of DOPA, or some other molecule.
If the SCO2525 gene product is necessary for methylation of a cellular metabolite that participates in colony development, it might be expected that this gene is not expressed during vegetative growth but rather is expressed during aerial mycelium formation. Huang and colleagues performed a global analysis of gene expression of S. coelicolor grown in liquid culture (16). While aerial mycelium formation and sporulation do not occur under these culture conditions, antibiotic production does take place, and some of the genes involved in morphological differentiation were shown to be upregulated upon the switch from primary to secondary metabolism as the culture progressed toward the stationary phase. Notably, expression of the SCO2525 methyltransferase gene increased more than threefold during the stationary phase compared to expression during the logarithmic growth. The timing and magnitude of SCO2525 induction observed were comparable to the timing and magnitude of induction of the actinorhodin biosynthetic genes. The gene encoding the SCO2782 pyridoxal-dependent decarboxylase likewise showed induction during the stationary phase.
As described here, an acyl-CoA dehydrogenase (SCO6938) has been implicated in initiation of aerial mycelium formation and actinorhodin production in S. coelicolor. Acyl-CoA dehydrogenases involved in the catabolism of branched-chain fatty acids have been described as enzymes that are important for generating acyl-CoAs used to synthesize antibiotics in Streptomyces (45). It therefore seems possible that the dehydrogenase identified in this study participates in the formation of acyl-CoA species necessary for synthesis of a secondary metabolite that stimulates aerial mycelium formation and actinorhodin production in S. coelicolor. It has also previously been shown that in S. coelicolor an acyl-CoA synthetase, another enzyme involved in fatty acid catabolism, is necessary for timely actinorhodin production (3).
In the screening analysis described in this paper we identified new genes whose products contribute to the developmental process in S. coelicolor. Not surprisingly, we also identified previously described developmental loci. However, disruption of these previously described loci gave phenotypes that were perceptibly different from those observed previously in other genetic backgrounds, highlighting the importance of considering the background when the role of putative developmental genes is investigated. The new genes described here, unlike many previously described developmental genes, encode proteins with proposed enzymatic activities that may contribute to synthesis of cellular metabolites that promote aerial mycelium formation and subsequent sporulation. The phenotypes of the corresponding mutants were too leaky to assess the position of the new genes in the previously described extracellular signaling cascade that initiates aerial mycelium formation (i.e., none of the mutants exhibited a true aerial mycelium-negative, bld phenotype [44]). However, future biochemical characterization of the encoded enzymes should provide insight into the molecular events that govern the progression of a streptomycetes colony from vegetative growth to aerial mycelium formation and finally culminate in sporulation.
Acknowledgments
We thank Joel Schmid and Nancy Piatczyc at Williams College for assistance with the scanning electron microscopy and Mark Buttner for supplying strains J2151 and J1915. We also thank Mark Buttner, Keith Chater, and Marie Elliot for helpful comments on the manuscript.
This work was supported by grant MCB-0110090 to R.L. from the National Science Foundation. A.M.G. was supported in part by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship (grant DRG-1524).
REFERENCES
- 1.Abel, C. B. L., J. C. Lindon, D. Noble, B. A. M. Rudd, P. J. Sidebottom, and J. K. Nicholson. 1999. Characterization of metabolites in intact Streptomyces citricolor culture supernatants using high-resolution nuclear magnetic resonance and directly coupled high-pressure liquid chromatography-nuclear magnetic resonance spectroscopy. Anal. Biochem. 270:220-230. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banchio, C., and H. Gramajo. 2002. A stationary-phase acyl-coenzyme A synthetase of Streptomyces coelicolor A3(2) is necessary for the normal onset of antibiotic production. Appl. Environ. Microbiol. 68:4240-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Keiser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Keiser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Champness, W. 2000. Actinomycete development, antibiotic production, and phylogeny: questions and challenges, p. 11-31. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, D.C.
- 6.Champness, W. C. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chater, K. F. 2000. Developmental decisions during sporulation in the aerial mycelium in Streptomyces, p. 33-48. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, D.C.
- 8.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 9.Chater, K. F., and S. Horinouchi. 2003. Signaling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 10.Endo, K., K. Hosono, T. Beppu, and K. Ueda. 2002. A novel extracytoplasmic phenol oxidase of Streptomyces: its possible involvement in the onset of morphogenesis. Microbiology 148:1767-1776. [DOI] [PubMed] [Google Scholar]
- 11.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergenic conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 12.Gehring, A. M., J. R. Nodwell, S. M. Beverley, and R. Losick. 2000. Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. Proc. Natl. Acad. Sci. USA 97:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gehring, A. M., N. J. Yoo, and R. Losick. 2001. RNA polymerase sigma factor that blocks morphological differentiation by Streptomyces coelicolor. J. Bacteriol. 183:5991-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gramajo, H. C., E. Takano, and M. J. Bibb. 1993. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7:837-845. [DOI] [PubMed] [Google Scholar]
- 15.Hopwood, D. A., H. Wildermuth, and H. M. Palmer. 1970. Mutants of Streptomyces coelicolor defective in sporulation. J. Gen. Microbiol. 61:397-408. [DOI] [PubMed] [Google Scholar]
- 16.Huang, J., C.-J. Lih, K.-H. Pan, and S. N. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda, K., T. Masujima, K. Suzuki, and M. Sugiyama. 1996. Cloning and sequence analysis of the highly expressed melanin-synthesizing gene operon from Streptomyces castaneoglobisporus. Appl. Microbiol. Biotechnol. 45:80-85. [DOI] [PubMed] [Google Scholar]
- 18.Kelemen, G. H., K. A. Plaskitt, C. G. Lewis, K. C. Findlay, and M. J. Buttner. 1995. Deletion of DNA lying close to the glkA locus induces ectopic sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 17:221-230. [DOI] [PubMed] [Google Scholar]
- 19.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 20.Kim, D.-J., J.-H. Huh, Y.-Y. Yang, C.-M. Kang, I.-H. Lee, C.-G. Hyun, S.-K. Hong, and J.-W. Suh. 2003. Accumulation of S-adenosyl-l-methionine enhances production of actinorhodin but inhibits sporulation in Streptomyces lividans TK23. J. Bacteriol. 185:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, W.-S., Y. Wang, A. Fang, and A. L. Demain. 2000. Methionine interference in rapamycin production involves repression of demethylrapamycin methyltransferase and S-adenosylmethioine synthetase. Antimicrob. Agents Chemother. 44:2908-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu, M., Y. Kuwahara, A. Hiroishi, K. Hosono, T. Beppu, and K. Ueda. 2003. Cloning of the conserved regulatory operon by its aerial mycelium-inducing activity in an amfR mutant of Streptomyces griseus. Gene 306:79-89. [DOI] [PubMed] [Google Scholar]
- 23.Leskiw, B. K., R. Mah, E. J. Lawlor, and K. F. Chater. 1993. Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2). J. Bacteriol. 175:1995-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, J. L., J. Begun, M. J. McLeish, J. M. Caine, and G. L. Grunewald. 2001. Getting the adrenaline going: crystal structure of the adrenaline-synthesizing enzyme PNMT. Structure 9:977-985. [DOI] [PubMed] [Google Scholar]
- 25.Matsubara, Y., Y. Indo, E. Naito, H. Ozasa, R. Glassberg, J. Vockley, Y. Ikeda, J. Kraus, and K. Tanaka. 1989. Molecular cloning and nucleotide sequence of cDNAs encoding the precursors of rat long chain acyl-coenzyme A, short chain acyl-coenzyme A, and isovaleryl-coenzyme A dehydrogenases: sequence homology of four enzymes of the acyl-CoA dehydrogenase family. J. Biol. Chem. 264:16321-16331. [PubMed] [Google Scholar]
- 26.Merrick, M. J. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96:299-315. [DOI] [PubMed] [Google Scholar]
- 27.Molle, V., and M. J. Buttner. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36:1265-1278. [DOI] [PubMed] [Google Scholar]
- 28.Müller, R., K. Gerth, P. Brandt, H. Blöcker, and S. Beyer. 2000. Identification of an L-dopa decarboxylase gene from Sorangium cellulosum So ce90. Arch. Microbiol. 173:303-306. [DOI] [PubMed] [Google Scholar]
- 29.Nagatsu, T. 1991. Genes for human catecholamine-synthesizing enzymes. Neurosci. Res. 12:315-345. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen, K. T., J. Tenor, H. Stettler, L. T. Nguyen, L. D. Nguyen, and C. J. Thompson. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 185:7291-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nodwell, J. R., and R. Losick. 1998. Purification of an extracellular signaling molecule involved in production of aerial mycelium by Streptomyces coelicolor. J. Bacteriol. 180:1334-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nodwell, J. R., M. Yang, D. Kuo, and R. Losick. 1999. Extracellular complementation and the identification of additional genes involved in aerial mycelium formation in Streptomyces coelicolor. Genetics 151:569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh, S.-H., and K. F. Chater. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto, S., A. Lezhava, T. Hosaka, Y. Okamoto-Hosoya, and K. Ochi. 2003. Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in Streptomyces coelicolor A3(2). J. Bacteriol. 185:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism, Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redenbach, M., H. M. Keiser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 37.Ryding, N. J., M. J. Bibb, V. Molle, K. C. Findlay, K. F. Chater, and M. J. Buttner. 1999. New sporulation loci in Streptomyces coelicolor A3(2). J. Bacteriol. 181:5419-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandmeier, E., T. I. Hale, and P. Christen. 1994. Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 221:997-1002. [DOI] [PubMed] [Google Scholar]
- 39.Sprusansky, O., L. Zhou, S. Jordan, J. White, and J. Westpheling. 2003. Identification of three new genes involved in morphogenesis and antibiotic production in Streptomyces coelicolor. J. Bacteriol. 185:6147-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takano, E., M. Tao, F. Long, M. J. Bibb, L. Wang, W. Li, M. J. Buttner, M. J. Bibb, Z. X. Deng, and K. F. Chater. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 50:475-486. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi, T., K. Ogawa, H. Iinuma, H. Suda, K. Ukita, T. Nagatsu, M. Kato, and H. Umezawa. 1973. Monomaine oxidase inhibitors isolated from fermented broths. J. Antibiot. 26:162-167. [DOI] [PubMed] [Google Scholar]
- 42.Vargha, G., V. Zs-Nagy, G. Lustyik, and G. Szabó. 1986. Methionine requirement of sporulation in a Streptomyces fradiae mutant. J. Gen. Microbiol. 132:2931-2936. [Google Scholar]
- 43.Weissbach, H., and N. Brot. 1991. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 5:1593-1597. [DOI] [PubMed] [Google Scholar]
- 44.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y.-X., C. D. Denoya, D. D. Skinner, R. W. Fedechko, H. A. I. McArthur, M. R. Morgenstern, R. A. Davies, S. Lobo, K. A. Reynolds, and C. R. Hutchinson. 1999. Genes encoding acyl-CoA dehydrogenase (AcdH) homologues from Streptomyces coelicolor and Streptomyces avermitilis provide insights into the metabolism of small branched-chain fatty acids and macrolide antibiotic production. Microbiology 145:2323-2334. [DOI] [PubMed] [Google Scholar]