Two alternate crystal forms of the human CD44 hyaluronan-binding domain, one hexagonal and one monoclinic, have been determined at 1.60 and 1.08 Å resolution, respectively. A short peptide found in the monoclinic form may identify a site of CD44 for protein–protein interactions.
Keywords: CD44, hyaluronan-binding domain
Abstract
Two new crystal structures of the extracellular hyaluronan-binding domain of human CD44 are described at high resolution. A hexagonal crystal form at 1.60 Å resolution and a monoclinic form at 1.08 Å resolution both have two molecules in the asymmetric unit arranged about a similar noncrystallographic twofold axis of symmetry. These structures are compared with those previously reported at 2.20 Å resolution to show that the fold is quite resistant to structural deformation in different crystal environments. Unexpectedly, a short peptide is found in the monoclinic crystals at a site remote from the known hyaluronan-binding groove. The peptide with a valine at the carboxy-terminus must have co-purified from the bacterial expression host and binds on the opposite side of the domain from the known hyaluronan-binding groove. This opportunistic binding may identify a site of interaction used as CD44 assembles with other proteins to accomplish effective signaling regarding changes to the extracellular environment.
1. Introduction
CD44 is a type I transmembrane receptor that is presented on the outer surface of many kinds of immune and cancer cells and helps these cells to sense and respond to changes in the tissue microenvironment (Zöller, 2011 ▶). These receptors use the ectodomain to communicate with extracellular matrix components such as hyaluronan, collagen, growth factors, cytokines and proteases. It also serves as a platform for signal transduction by assembling protein complexes with receptor kinases and membrane proteases (Toole, 2009 ▶). More recently, a number of exogenous peptides have been identified that bind CD44 to regulate cell motility and malignancies (Hibino et al., 2004 ▶; Piotrowicz et al., 2011 ▶; Park et al., 2012 ▶; Ugarte-Berzal et al., 2014 ▶), although little is known regarding the details of these interactions (Zöller, 2011 ▶).
Hyaluronan (HA) serves as a primary substrate for CD44 and modulates cell adhesion and proliferation (Ponta et al., 2003 ▶), which is important in inflammatory diseases (Krettek & Sjöberg, 2009 ▶) and in the progression of many cancers (Toole, 2004 ▶, 2009 ▶). CD44 binds HA through a disulfide-stabilized HA-binding domain (HABD). It includes a N-terminal lectin-like fold of about 100 residues called the ‘link module’ and additional residues that are essential to bind HA and stabilize the fold (Banerji et al., 1998 ▶). While variant isoforms of the membrane-proximal region of the ectodomain arise from alternative splicing of variable exons and by post-translational modifications (Toole, 2004 ▶), the HABD is present and conserved in most CD44 splice variants and is known to be sufficient for the binding of hyaluronan (Banerji et al., 1998 ▶). It therefore constitutes an attractive target for the development of small-molecule inhibitors of HA binding (Liu & Finzel, 2014 ▶).
Crystallographic and solution structural studies have revealed much about the structure of the CD44 HABD. A crystal structure of human HABD (residues 20–178; hHABD20–178) has been reported at 2.20 Å resolution (Teriete et al., 2004 ▶) and solution studies conducted with NMR have revealed details of protein dynamics related to HA binding (Teriete et al., 2004 ▶; Takeda et al., 2006 ▶). Other studies with murine HABD (mHABD), which shares 86% sequence identity with the human protein, have produced high-resolution structures of mHABD with and without bound HA oligosaccharides (Banerji et al., 2007 ▶; Liu & Finzel, 2014 ▶). We have recently used crystals of the murine domain to characterize the binding of small-molecule inhibitors of HA binding (Liu & Finzel, 2014 ▶).
Motivated by a desire to obtain a crystal form of human CD44 HABD suitable for use in binding studies with small-molecule inhibitors, we have undertaken a search for alternate crystal forms. We sought to improve upon the resolution of diffraction and to obtain alternate crystal forms with a small asymmetric unit and an accessible HA-binding groove for use in soaking experiments with HA oligosaccharides and other ligands. To obtain the previously known hHABD orthorhombic crystal form (PDB entry 1uuh), Jackson and coworkers expressed and purified hHABD20–178 (Teriete et al., 2004 ▶). Using a slightly different construct, hHABD18–178, we have identified two alternate crystal forms, one hexagonal and one monoclinic, that diffract to high resolution. An unidentified peptide, surreptitiously co-purified along with CD44 from the bacterial expression host, is found to occupy a binding site in the monoclinic form far from the HA-binding groove. We speculate that this may be a site for interaction with one of the endogenous signaling partners of CD44.
2. Materials and methods
2.1. Macromolecule production
The CD44 protein constructs hHABD20–178, hHABD18–178 and hHABD20–168 (Table 1 ▶) were expressed in Escherichia coli, extracted, refolded and purified from insoluble inclusion bodies following the procedures described for the preparation of hCD4420–178 (Banerji et al., 1998 ▶) and as adapted previously (Liu & Finzel, 2014 ▶). The variant construct CD4418–178 was amplified using the forward primer 5′-CCTCGTGCATATGG50CTATGGCGCAGATCGATTTGAATATAACCTGCCG-3′ and the reverse primer 5′-AGGACTCGAGCTAC530ACGTCATCATCAGTAGGGTTGCTG-3′. An error in primer design resulted in the expression of the construct CD4418–178 with two unintended residues (Ala18-Met19) at the N-terminus, but this proved fortuitous. Amplified segments of the complementary DNA for each construct were prepared from human CD44 cDNA generously provided by James B. McCarthy at the University of Minnesota (Minneapolis, Minnesota, USA) using appropriate primers incorporating recognition sites (NdeI and XhoI, respectively; shown in bold) for ligation into expression vector pMCSG7 (Stols et al., 2002 ▶). Vector sequences were confirmed by DNA sequencing and the resulting plasmids were transformed into E. coli strain BL21(DE3)pLysS for bacterial expression. Inclusion bodies were isolated from the cell lysate, washed and the protein was extracted with 8 M denaturant urea. The soluble, denatured fraction (typically 10 ml from 1 l culture) is mainly recombinant HABD as assessed by SDS–PAGE analysis. The protein was chemically refolded in 2 l buffer consisting of 250 mM l-arginine and 2 mM reduced and 1 mM oxidized glutathione. The solution was slowly stirred at 4°C for 24 h followed by ultrafiltration through Amicon PM10 membranes. Monomeric HABDs were separated from the aggregates by a single pass through Sephacryl S-100 and were analyzed on SDS–PAGE as described previously for other constructs (Liu & Finzel, 2014 ▶). This resulted in yields of more than 20 mg recombinant HABDs from 1 l culture. The sequence and the formation of three disulfides in the construct hHABD18–178 were confirmed by mass spectrometry and crystallography. The molecular mass of the refolded protein was 17 783.1 Da, consistent with that of 17 788.8 Da expected for the fully reduced protein.
Table 1. Sequence comparison of the constructs used to generate different crystal forms.
Non-native residues are shown underlined.
2.2. Crystallization
Hexagonal and monoclinic crystals of hCD4418–178 were obtained by hanging-drop vapor diffusion using the same protein concentrated to 8 mg ml−1 in a buffer consisting of 20 mM Tris–HCl, 150 mM NaCl. To obtain hexagonal crystals, this protein was mixed in a 1:1 ratio with well solution consisting of 30% MME 5000, 200 mM Na2SO4, 100 mM MES pH 6.5 and suspended over the well solution in 2 µl drops. Crystals with symmetry consistent with space group P6122 grew to 30 × 30 × 100 µm over a period of two weeks at 4°C with or without 5% DMSO. Crystal growth could be accelerated to one week by introducing 0.3 µl of the well solution containing micro-crystals from previous experiments into the crystallization drops. Monoclinic crystals with C2 space-group symmetry grew in several weeks at 4°C over a well solution comprising 25% PEG 3350, 100 mM HEPES pH 7.5, 100 mM NaCl, 5% DMSO. The monoclinic crystals were less numerous, grow more slowly and became larger (up to 100 × 100 × 150 µm).
2.3. Data collection and processing
Diffraction data were collected from single crystals at IMCA-CAT station 17-ID-B or GMCA-CAT station 23-ID-D at the Advanced Photon Source (APS), Argonne, Illinois, USA. Data were processed using XDS (Kabsch, 2010 ▶) and scaled with SCALA (Evans, 2006 ▶). A summary of the scaling statistics is provided in Table 2 ▶. Structures of the hexagonal P6122 or monoclinic C2 crystal forms were determined by molecular replacement with Phaser (McCoy et al., 2007 ▶) using monomer A of PDB entry 1uuh (Teriete et al., 2004 ▶) as a search model. Both crystal forms have two protein molecules in the asymmetric unit. Iterative rounds of model building and restrained refinement were carried out with Coot (Emsley et al., 2010 ▶) and REFMAC5 (Murshudov et al., 2011 ▶). Discreet alternate conformations were modelled only when it appeared that two states with roughly equal occupancy could best explain the electron density. The occupancies of atoms in alternate conformations were constrained to sum to 1.0 and were refined as a group using REFMAC5. Refined structures were validated with MolProbity (Chen et al., 2010 ▶). Atomic coordinates and reflection data have been deposited in the Protein Data Bank (Berman et al., 2000 ▶; PDB entries 4pz3 and 4pz4).
Table 2. Crystallographic data and refinement statistics for hHABD18–178 .
Values in parentheses are for the highest resolution shell.
| Monoclinic form | Hexagonal form | |
|---|---|---|
| PDB code | 4pz3 | 4pz4 |
| Data collection and processing | ||
| Diffraction source | APS 17-ID-B | APS 23-ID-D |
| Space group | C121 | P6122 |
| a, b, c (Å) | 72.50, 60.29, 75.13 | 70.27, 70.27, 286.46 |
| α, β, γ (°) | 90, 113.40, 90 | 90, 90, 120 |
| Resolution range (Å) | 44.67–1.08 (1.14–1.08) | 286.46–1.598 (1.604–1.598) |
| Total No. of reflections | 376553 (34299) | 720488 (5519) |
| No. of unique reflections | 122395 (15623) | 56356 (455) |
| Completeness (%) | 97.3 (85.5) | 99.3 (80.4) |
| Multiplicity | 3.1 (2.2) | 12.8 (12.1) |
| 〈I/σ(I)〉 | 21.2 (6.7) | 26.4 (5.3) |
| R merge | 0.031 (0.154) | 0.063 (0.490) |
| R r.i.m. † | 0.038 (0.208) | 0.066 (0.544) |
| Wilson B factor (Å2) | 12 | 28 |
| Structure solution and refinement | ||
| Resolution range (Å) | 68.96–1.08 (1.11–1.08) | 60.860–1.600 (1.640–1.598) |
| No. of reflections, working set | 116222 (6803) | 53422 (3738) |
| No. of reflections, test set | 6171 (339) | 2864 (180) |
| Final R cryst | 0.166 (0.187) | 0.197 (0.260) |
| Final R free | 0.177 (0.190) | 0.229 (0.284) |
| Cruickshank DPI (Å) | 0.03 | 0.08 |
| No. of non-H atoms | 2596 | 2772 |
| No. of solvent waters | 372 | 199 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.012 | 0.013 |
| Angles (°) | 1.5 | 1.4 |
| Average B factor (Å2) | 11.6 | 18.4 |
| Ramachandran plot | ||
| Most favored (%) | 94.0 | 95.8 |
| Allowed (%) | 5.2 | 3.8 |
| Outliers | 2‡ | 1§ |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
Glu126 of both chains A and B.
Tyr114 of chain B.
3. Results and discussion
3.1. Construct evaluation
To begin a search for alternate crystal forms, three different constructs of human HABD were expressed, purified and evaluated in crystallization trials. Broad screening with commercially available incomplete factorial crystallization screens did not result in the discovery of new conditions. Instead, targeted screening guided by the known conditions for successful crystallization of murine CD44 constructs (Banerji et al., 2007 ▶) resulted in the identification of two new crystal forms. Hexagonal crystals of hHABD18–178 arise from conditions using MME 5000 as a precipitant, similar to those used for the crystallization of apo murine HABD. Monoclinic crystals result from the same construct under conditions similar to those used in the crystallization of the murine CD44–hyaluranon oligosaccharide complex. Both crystal forms were obtained using a construct (hHABD18–178) that extends the constructs used in earlier work by two residues at the N-terminal end and substitutes the naturally occurring Ser18-Leu19 of CD44 with Ala18-Met19. A shorter construct (hHABD20–168) was engineered to resemble the molecular extent of the mHABD24–174 construct used extensively in our laboratory, while the construct hHABD20–178 replicates the exact construct successfully crystallized by Teriete et al. (2004 ▶). No usable crystals of either hHABD20–178 or hHABD20–168 were obtained.
Throughout this discussion, protein constructs are identified by species and residue range (e.g. hHABD20–178 or mHABD24–174), while crystal structures are identified unambiguously by the four-letter PDB code (e.g. 1uuh, 4pz3 or 4pz4). Individual protein monomers within a crystal structure are identified by the PDB code and chain identifier (e.g. 1uuh-A or 4pz3-B).
3.2. New crystal forms
Structures of both the hexagonal and the monoclinic forms were determined by molecular replacement and were refined against high-resolution diffraction data (1.60 and 1.08 Å, respectively). A summary of the refinement statistics for 4pz4 and 4pz3 is provided in Table 2 ▶. Both structures include two protein chains in the crystallographic asymmetric unit. The higher resolution of either structure significantly improves upon that achieved with the previously reported orthorhombic crystal form (2.20 Å resolution; PDB entry 1uuh; Teriete et al., 2004 ▶). The structures recapitulate earlier findings of overall fold and secondary structure reported for both the human (Teriete et al., 2004 ▶) and murine (Banerji et al., 2007 ▶) CD44 HA-binding domains (Fig. 1 ▶ a). All four chains are well ordered from Ala20 to Tyr169. The two additional residues at the N-terminus (Ala18-Met19) are disordered in all but one chain of the hexagonal form (chain A). It is not at all clear how these residues might alter the crystallization properties as they are not involved in specific intermolecular contacts in either of the new crystal forms (4pz3 or 4pz4) or the original orthorhombic form (1uuh). The C-terminal residues are disordered beyond Ser171 in all structures. In the hexagonal form, 13 residues are modeled with side chains in two discrete conformations. In the higher resolution monoclinic form, 24 residues are so modeled.
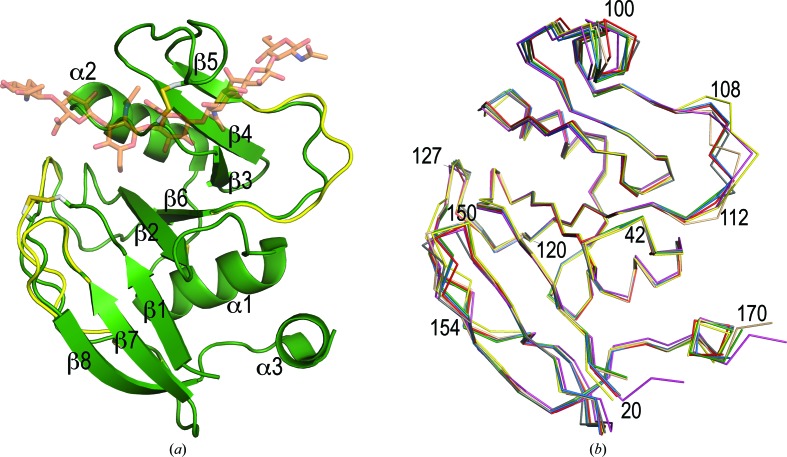
Figure 1.
(a) The structure of the hHABD18–178 monomer (green; 4pz3-A) showing secondary-structure assignments and disulfide-bond locations. Apo mHABD24–174 (2jcp; yellow) is shown for comparison, but regions of the backbone that overlay precisely with 4pz3-A are omitted for clarity. The HA-binding position inferred from the complex of mHABD24–174 with an oligosaccharide octamer (2jcp; orange) is shown for positional reference only. (b) Comparison of the Cα backbone geometry in six crystallographically independent hHABD crystal structures: hHABD20–178 (1uuh-A, gray; 1uuh-B, red) and hHABD18–178 (4pz3-A, blue; 4pz3-B, green; 4pz4-A, magenta; 4pz4-B, wheat). Also shown is the backbone from the mHABD24–174 apo structure (2jcp; yellow).
3.3. Comparison of hHABD crystal structures
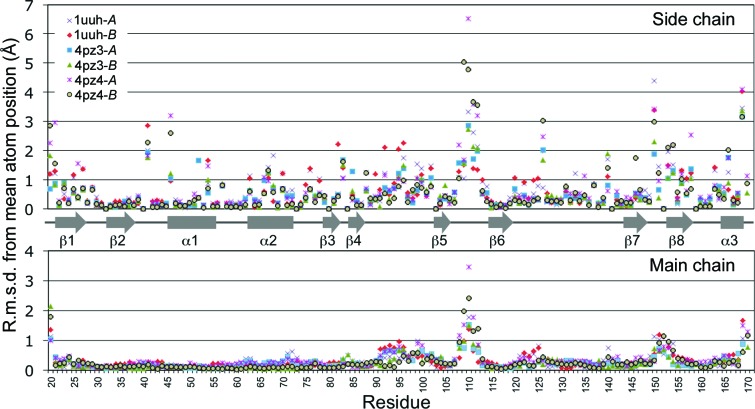
The four crystallographically independent snapshots of hHABD18–178 revealed in these crystal structures (4pz3-A, 4pz3-B, 4pz4-A and 4pz4-B) can be pooled with the other two conformations available from the orthorhombic hHABD20–178 crystals (1uuh-A and 1uuh-B) to conduct an analysis of conformational flexibility in these domains. To look for and quantitate differences in the six available structures, we overlaid them on a common positional framework (1uuh-A; Fig. 1 ▶ b), computed a mean position for each atom from all six structures and then calculated root-mean-square deviations (r.m.s.d.s) from this mean position by residue. Results are presented graphically in Fig. 2 ▶. The mean r.m.s.d. for all atoms was 0.53 Å.
Figure 2.
R.m.s.d.s between hHABD structures and a consensus mean structure. Differences by residue are separated to display main-chain atoms (N, Cα, C, O) and side-chain atoms separately. The consensus was obtained by averaging atomic positions for each atom after co-superposition on 1uuh-A.
Large differences (greater than three times the mean r.m.s.d.) in the backbone conformation occur only at the N- and C-termini and in residues 108–112 of the β5–β6 loop (Fig. 1 ▶ b). Apart from some side chains along the C-terminal helix, large differences are restricted to the side chains of Arg41 and Arg150 and the loop segment with diverse backbones (108–112). While these are all unliganded apo structures (besides chain B of 4pz3, which will be discussed below), these are side chains that are known to shift in response to HA or inhibitor binding (Takeda et al., 2006 ▶; Banerji et al., 2007 ▶; Liu & Finzel, 2014 ▶). Some contiguous portions of the backbone (e.g. residues 94–104, 122–127 and 150–154) have moderately large r.m.s.d.s (elevated but less than three times the mean). These differences are not the result of any specific backbone torsional changes, but instead reflect small but concerted shifts in positions of each segment.
Others have characterized residues 120–127 as ‘potentially conformationally dynamic’ (Piotrowicz et al., 2011 ▶), but the structural comparison does not support this. Moreover, the bracketing disulfide bonds on either side of this peptide leave little opportunity for flexibility. In general, the disulfide-stabilized link domain is very rigid and adopts the same conformation when different ligands are bound or in different environments (Fig. 3 ▶ a).
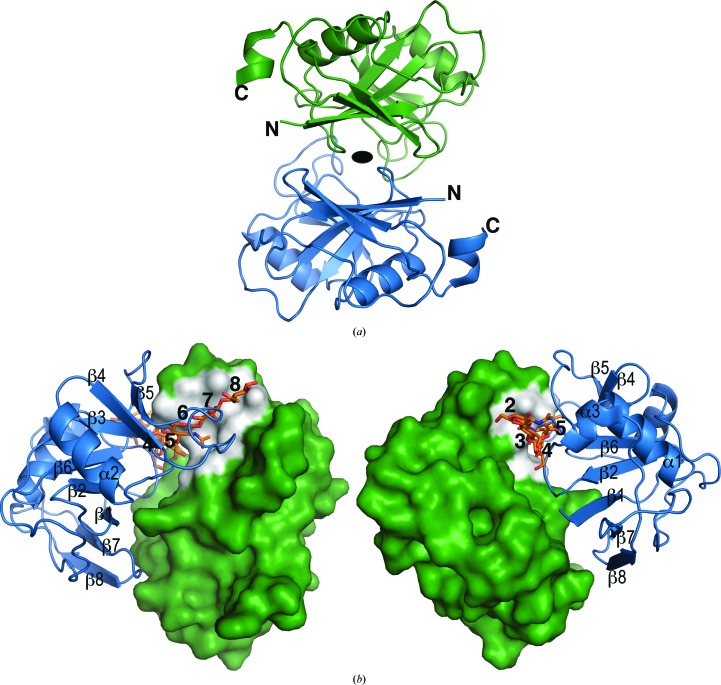
Figure 3.
(a) A similar noncrystallographic twofold axis of symmetry relates two molecules in the asymmetric unit of both the monoclinic and hexagonal crystal forms of hHABD18–178. 4pz3-A (green) and 4pz3-B (blue) are shown as illustrative of the dimer in both forms. (b) Two different views of the HA-binding groove show how intermolecular contacts made within the dimer prevent HA oligosaccharides and small molecules from binding. HA (orange) is included for positional reference only using its location upon superposition of the mHABD24–174 structure with bound HA (2jcr) onto 4pz3-A (green). HA saccharide subsites (white) are numbered for reference in the text.
Fig. 1 ▶(a) also includes a comparison with the murine mHABD24–174 structure (2jcp; Banerji et al., 2007 ▶). Backbone differences are limited to portions of the loops joining β5 to β6 (particularly residues 108–112, human numbering), β6 to β7 and β7 to β8. These are the same regions that show conformational diversity in different human structures, suggesting that these regions have intrinsic flexibility even when the primary sequence is unchanged. The difference between the murine and human CD44 conformation in the vicinity of 108–112 has been noted and attributed to the insertion of Val112 (murine numbering) in the murine protein (as shown by Banerji et al., 2007 ▶). It is worth noting that there is no difference in the backbone conformation in residues 40–42, which undergo a significant change upon ligand binding (Takeda et al., 2006 ▶; Ogino et al., 2010 ▶). We see no evidence that this is an intrinsically flexible loop when no HA is bound.
In both new crystal forms the two protein chains are positioned face to face in the crystallographic asymmetric unit about a noncrystallographic dyad axis (Fig. 3 ▶ a) to form almost identical dimers. In this packing arrangement, access to the HA-binding groove of each monomer is blocked by the presence of the opposing monomer. Residues Cys77, Arg78 and Tyr79 sit squarely in the HA5 and HA6 subsites, and Ile96–Ala99 protrude from the long loop joining β4 and β5 to obstruct the HA6 subsite (Fig. 3 ▶ b, right). Gly40-Arg41 of loop β2–α1 occupy the HA4 subsite (Fig. 3 ▶ b, left) and are likely to prevent the rearrangement of the inducible binding site accessible to small-molecule inhibitors of HA binding in murine HABD crystals (Liu & Finzel, 2014 ▶). We have been unable to form co-crystals with small-molecule inhibitors that bind to the same site by either co-crystallization or soaking using either of these crystal forms, which is likely to be because access to the groove is occluded by intermolecular packing. No similar dimer exists in the orthorhombic crystals (1uuh); in these crystals the HA-binding site is also blocked by packing, but differently. The search for a crystal form suitable for use in HA-binding studies must continue.
3.4. Peptide-binding site
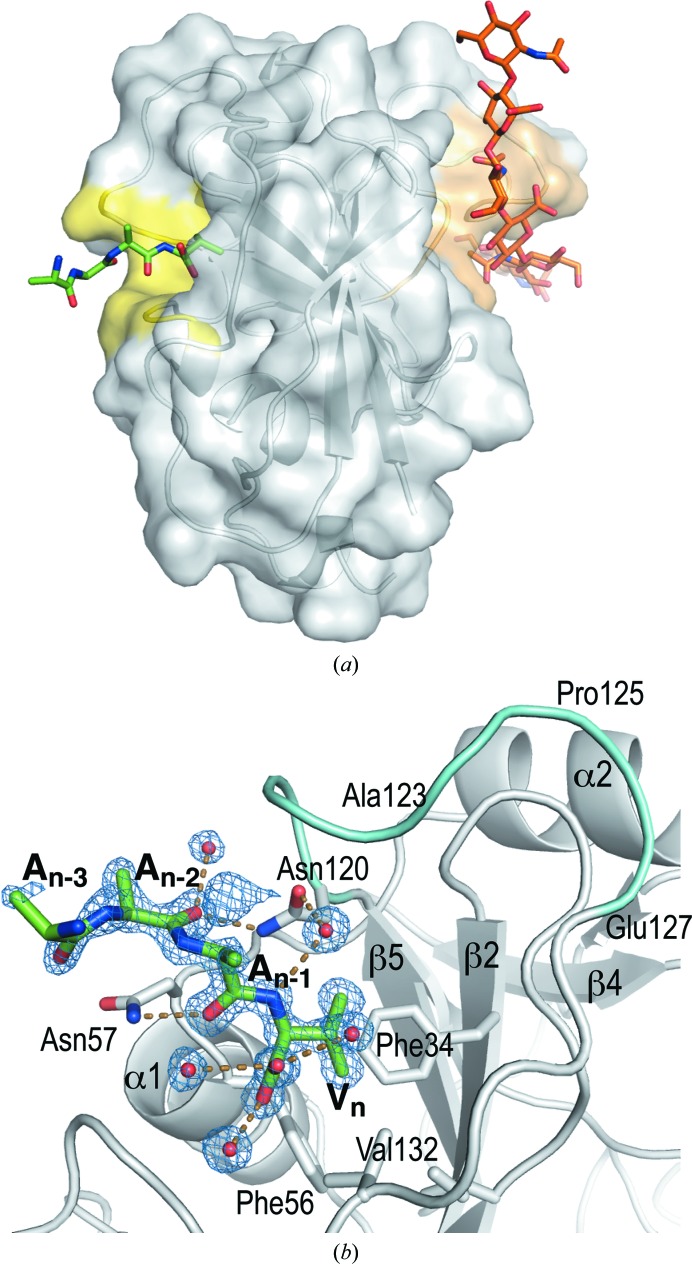
Quite unexpectedly, an unidentified peptide (or likely a mixture of short peptides) lies in a hydrophobic pocket of one molecule (chain B) in the monoclinic crystal form of hHABD18–178. This binding site is on the opposite side of the HABD from the HA-binding groove (Fig. 4 ▶ a). The peptide has been present in every monoclinic crystal we have examined, but never associated with chain A, where the pocket geometry is unchanged. The peptide is likely to have co-purified along with the protein from the expression system and has been retained throughout refolding, gel filtration and crystallization, but we have been unable to separate it for unambiguous identification or to replace the peptide with synthetic peptides introduced as potential surrogates.
Figure 4.
The unexpected binding site for a peptide on the back side of chain B in the monoclinic hHABD18–178 structure (4pz3-B). (a) The binding site (yellow) in relation to the known HA-binding site (orange). (The HA position is modeled by superposition of the murine HABD structure 2jcr.) (b) Detail of hydrogen-bonding interactions (mostly water-mediated) and the 3σ OMIT (F o − F c) electron density (blue) validating the peptide geometry and model integrity. Backbone geometry for the three C-terminal residues (An−2, An−1 and Vn) and the side chain of the valine is very well defined. The A6 peptide homolog on CD44 (residues 120–127) is colored cyan.
The peptide-binding site is a pocket inside the concave curl of the extended HABD β-sheet and at the C-terminal end of helix α1. It is rimmed by portions of the long meandering loop joining β6 to β7 (residues 120–136). Our comparison of hHABD structures shows this to be one of the more rigid portions of the structure (Fig. 2 ▶). Ile26, Arg29, Phe34, Phe56 and Val132 surround a hydrophobic subsite for the C-terminal residue of the peptide, which we confidently identify as a valine based on high-resolution electron density (Fig. 4 ▶ b). Threonine is an unlikely candidate as there is no possible partner for a side-chain hydroxyl group in the hydrophobic subsite. The fact that this is the C-terminus of the peptide is also convincingly shown by the presence of well resolved water molecules bound to the carboxylate. Most hydrogen bonds stabilizing the peptide binding are water-mediated, although hydrogen bonds from peptide backbone carbonyl O atoms to the side-chain amides of Asn57 and Asn120 also exist. Other amino acids in the peptide cannot be identified with certainty based on electron density alone and we cannot exclude the possibility that more than one species is bound; the peptide has been modeled as An−3-A-A-Vn, with all peptide bonds in a trans conformation. Longer side chains than alanine might fit the broken density, particularly in place of An−1 (Fig. 4 ▶ b).
This pocket and all residues bordering it are also conserved in murine CD44 HABD. A small molecule identified by fragment screening has been confirmed to bind in this location by co-crystallization with mHABD24–174 (PDB entry 4mrh; Liu & Finzel, 2014 ▶), but binding by peptides has not been observed in any other crystal form, including the hexagonal form prepared from the same protein preparation. Peptide binding does not seem to be prohibited by crystal packing in any of these forms, so we cannot offer any reasoned explanation for the lack of binding at other essentially equivalent sites.
The newly identified pocket may represent a binding site for some yet-to-be-identified proteinaceous partner of CD44, which is thought to be involved in several different signaling pathways and regulatory protein assemblies (for a review, see Zöller, 2011 ▶). A number of studies have been conducted to identify peptides that specifically interfere with CD44-mediated cell motility or tumor migration (Hibino et al., 2004 ▶; Piotrowicz et al., 2011 ▶; Ugarte-Berzal et al., 2014 ▶), but the site of action of these peptides is not known. Peptide A6 (acetyl-KPSSPPEE-NH2) has antitumor activity (Mishima et al., 2000 ▶; Ghamande et al., 2008 ▶) and inhibits CD44-medited migration and metastasis (Piotrowicz et al., 2011 ▶). It has been suggested that A6 may bind to CD44 and facilitate dissociation of CD44 homodimers that repress biological activity and HA recognition (Piotrowicz et al., 2011 ▶). We looked for binding of A6 to CD44 using a sensitive SPR binding assay (Liu & Finzel, 2014 ▶), but could detect no direct binding against hHABD20–178 (data not shown). This does not rule out the possibility that binding occurs elsewhere. A6 shares sequence homology with CD44 residues 120–127 (NASAPPEE), so it has also been proposed that it may act as a decoy to antagonize the association of CD44 with another protein whose identity is not yet known (Piotrowicz et al., 2011 ▶). These residues surround half of the outer rim of the binding site of the peptide that we have detected crystallographically (Fig. 4 ▶ b), lending support to a hypothesis that this region of the HABD surface is the site for a biologically significant protein–protein interaction. The subsite surrounding the peptide found in the monoclinic crystal form may be the epicenter of this interaction.
Supplementary Material
PDB reference: human CD44 hyaluronan-binding domain, in new space group, 4pz4
PDB reference: complex with undefined peptides, 4pz3
Acknowledgments
We thank Dr James B. McCarthy for the generous gift of plasmids. We also gratefully acknowledge the University of Minnesota Supercomputing Institute for access to software and computational resources. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman–Woodward Medical Research Institute. GM/CA @ APS has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science under contract No. W-31-109-ENG-38. This work was supported by a grant from the Minnesota Department of Employment and Economic Development (#SPAP-06-0014-P-FY07) to BCF, an American Heart Association Predoctoral Fellowship (#14PRE17890008) to L-KL and a grant from the Masonic Cancer Center of the University of Minnesota.
References
- Banerji, S., Day, A. J., Kahmann, J. D. & Jackson, D. G. (1998). Protein Expr. Purif. 14, 371–381. [DOI] [PubMed]
- Banerji, S., Wright, A. J., Noble, M., Mahoney, D. J., Campbell, I. D., Day, A. J. & Jackson, D. G. (2007). Nature Struct. Mol. Biol. 14, 234–239. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Ghamande, S. A., Silverman, M. H., Huh, W., Behbakht, K., Ball, G., Cuasay, L., Würtz, S. O., Brunner, N. & Gold, M. A. (2008). Gynecol. Oncol. 111, 89–94. [DOI] [PubMed]
- Hibino, S., Shibuya, M., Engbring, J. A., Mochizuki, M., Nomizu, M. & Kleinman, H. K. (2004). Cancer Res. 64, 4810–4816. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Krettek, A. & Sjöberg, S. (2009). Cardiovasc. Hematol. Disord. Drug Targets, 9, 293–302. [DOI] [PubMed]
- Liu, L.-K. & Finzel, B. C. (2014). J. Med. Chem. 57, 2714–2725. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Mishima, K., Mazar, A. P., Gown, A., Skelly, M., Ji, X.-D., Wang, X.-D., Jones, T. R., Cavenee, W. K. & Huang, H.-J. S. (2000). Proc. Natl Acad. Sci. USA, 97, 8484–8489. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ogino, S., Nishida, N., Umemoto, R., Suzuki, M., Takeda, M., Terasawa, H., Kitayama, J., Matsumoto, M., Hayasaka, H., Miyasaka, M. & Shimada, I. (2010). Structure, 18, 649–656. [DOI] [PubMed]
- Park, H.-Y., Lee, K.-J., Lee, S. -J. & Yoon, M.-Y. (2012). Mol. Biotechnol. 51, 212–220. [DOI] [PubMed]
- Piotrowicz, R. S., Damaj, B. B., Hachicha, M., Incardona, F., Howell, S. B. & Finlayson, M. (2011). Mol. Cancer Ther. 10, 2072–2082. [DOI] [PubMed]
- Ponta, H., Sherman, L. & Herrlich, P. A. (2003). Nature Rev. Mol. Cell Biol. 4, 33–45. [DOI] [PubMed]
- Stols, L., Gu, M., Dieckman, L., Raffen, R., Collart, F. R. & Donnelly, M. I. (2002). Protein Expr. Purif. 25, 8–15. [DOI] [PubMed]
- Takeda, M., Ogino, S., Umemoto, R., Sakakura, M., Kajiwara, M., Sugahara, K. N., Hayasaka, H., Miyasaka, M., Terasawa, H. & Shimada, I. (2006). J. Biol. Chem. 281, 40089–40095. [DOI] [PubMed]
- Teriete, P., Banerji, S., Noble, M., Blundell, C. D., Wright, A. J., Pickford, A. R., Lowe, E., Mahoney, D. J., Tammi, M. I., Kahmann, J. D., Campbell, I. D., Day, A. J. & Jackson, D. G. (2004). Mol. Cell, 13, 483–496. [DOI] [PubMed]
- Toole, B. P. (2004). Nature Rev. Cancer, 4, 528–539. [DOI] [PubMed]
- Toole, B. P. (2009). Clin. Cancer Res. 15, 7462–7468. [DOI] [PMC free article] [PubMed]
- Ugarte-Berzal, E., Bailón, E., Amigo-Jiménez, I., Albar, J. P., García-Marco, J. A. & García-Pardo, A. (2014). J. Biol. Chem. 289, 15340–15349. [DOI] [PMC free article] [PubMed]
- Zöller, M. (2011). Nature Rev. Cancer, 11, 254–267. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human CD44 hyaluronan-binding domain, in new space group, 4pz4
PDB reference: complex with undefined peptides, 4pz3