The mycobacterial Eis proteins are aminoglycoside acetyltransferases. Here, the crystal structure of M. smegmatis Eis in the paromomycin-bound form is reported at 3.3 Å resolution.
Keywords: tuberculosis, aminoglycoside, paromomycin, tobramycin, drug resistance, Mycobacterium smegmatis, Mycobacterium tuberculosis
Abstract
The Rv2416c gene of Mycobacterium tuberculosis (Mtb) encodes the enhanced intracellular survival (Eis) protein that enhances intracellular survival of the pathogen in host macrophages during infection. The Mtb Eis protein is released into the cytoplasm of the phagocyte during intracellular infection and modulates the host immune response. It also contributes to drug resistance by acetylating multiple amine groups of aminoglycosides. Interestingly, the nonpathogenic M. smegmatis (Msm) contains a homologous eis gene (MSMEG_3513). The overall structures of Mtb Eis and Msm Eis are highly similar to each other, reflecting the high level (58%) of amino-acid sequence identity between them. Both Mtb Eis and Msm Eis are active as aminoglycoside acetyltransferases, while only Mtb Eis functions as an N ∊-acetyltransferase to acetylate Lys55 of dual-specificity protein phosphatase 16 (DUSP16)/mitogen-activated protein kinase phosphatase 7 (MKP-7), leading to the suppression of host immune responses. Here, the crystal structure of Msm Eis in the paromomycin-bound form is reported, revealing detailed interactions between an aminoglycoside antibiotic and Msm Eis. The crystal structure of Msm Eis in the paromomycin-bound form has been determined at 3.3 Å resolution. This work provides potentially useful information for structure-guided discovery of Eis inhibitors as a novel antituberculosis drug against drug-resistant Mtb.
1. Introduction
Tuberculosis remains one of the world’s most destructive bacterial infectious diseases. Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, is a highly successful human pathogen. It infects one third of the global population and claims nearly two million lives every year (Dye & Williams, 2010 ▶). It has the ability to persist in the form of a long-term asymptomatic infection, referred to as latent tuberculosis (Lin & Flynn, 2010 ▶). Latent tuberculosis becomes activated when the body’s immune system is weakened. As a result, tuberculosis is the major cause of death among immunocompromised AIDS patients (Getahun et al., 2010 ▶). Despite efforts to ensure proper drug dosages and patient compliance with drug regimens, multidrug-resistant and extensively drug-resistant strains of Mtb have emerged (Chiang et al., 2010 ▶). This makes the search for targets of new antituberculosis drugs urgent. Mtb has evolved a number of very effective intracellular survival strategies (Meena & Rajni, 2010 ▶) and is able to survive and multiply within human macrophages (Dahl et al., 2001 ▶). Since intracellular survival plays a central role in the pathogenesis of Mtb (Wei et al., 2000 ▶), the mycobacterial proteins responsible for intracellular survival remain a priority for the development of new antituberculosis drugs.
The Rv2416c gene in Mtb H37Rv, designated eis (enhanced intracellular survival), was found to enhance intracellular survival of M. smegmatis (Msm) in the human macrophage-like cell line U-937 (Wei et al., 2000 ▶). The Mtb Eis protein is produced during human tuberculosis infection and is released into the culture medium (Dahl et al., 2001 ▶). It has been shown that the sigma factor SigA binds the eis promoter in the W-Beijing strain of Mtb and that activation of the eis gene correlates with increased SigA levels and enhanced intracellular survival (Wu et al., 2009 ▶). Treatment of T cells with Eis modulates extracellular signal-regulated kinase 1/2 (ERK1/2), the JAK pathway and the production of tumour necrosis factor α (TNF-α) and interleukin 4 (IL-4) (Lella & Sharma, 2007 ▶). Mtb Eis has been shown to suppress host innate immune defences by negatively modulating inflammation, autophagy and cell death in a redox-dependent manner (Shin et al., 2010 ▶). It also contributes to drug resistance by acetylating multiple amine groups of aminoglycosides (Chen et al., 2011 ▶). Interestingly, the nonpathogenic Msm contains a homologous eis gene (MSMEG_3513). A high level (58%) of amino-acid sequence identity exists between the Eis proteins from Mtb and Msm. As expected, their overall structures are highly similar to each other (Kim et al., 2012 ▶). We showed that both the Mtb Eis and Msm Eis proteins are active as aminoglycoside acetyltransferases (Kim et al., 2012 ▶). However, they differ in the aminoglycoside substrate preference and in the number of acetylated amine groups per aminoglycoside (Chen et al., 2012 ▶). We also showed that only Mtb Eis functions as an N ∊-acetyltransferase to acetylate Lys55 of dual-specificity protein phosphatase 16 (DUSP16)/mitogen-activated protein kinase phosphatase 7 (MKP-7), a JNK-specific phosphatase, leading to the suppression of host immune responses (Kim et al., 2012 ▶).
Paromomycin is an aminoglycoside antibiotic; it inhibits bacterial protein synthesis by binding to 16S ribosomal RNA. Many aminoglycosides contain a 2-deoxystreptamine part substituted at different positions by amino sugars. Paromomycin is an example of the 4,5-disubstituted subclass, while tobramycin belongs to the 4,6-disubstituted subclass (Fig. 1 ▶). Previously, we showed that the acetyltransferase activity of Msm Eis towards paromomycin is higher than that of Mtb Eis (Kim et al., 2012 ▶). Here, we have determined the crystal structure of Msm Eis in complex with paromomycin to reveal the detailed interactions between Msm Eis and paromomycin and to allow a comparison of its binding mode with that of tobramycin by Mtb Eis (Houghton et al., 2013 ▶). This study provides potentially useful information for structure-guided discovery of Eis inhibitors as novel antituberculosis drugs against drug-resistant Mtb.
Figure 1.
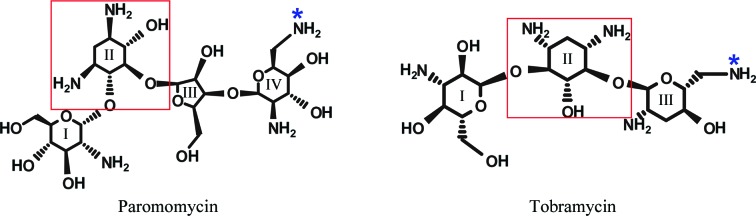
Chemical structures of paromomycin and tobramycin. The 2-deoxystreptamine group is enclosed in a red box. The N64 atom of paromomycin and the N61 atom of tobramycin are marked with asterisks.
2. Materials and methods
2.1. Protein expression, purification and crystallization
Msm Eis was overexpressed and purified as described previously (Kim et al., 2012 ▶). The purified Msm Eis protein was concentrated to 48.0 mg ml−1 in 20 mM Tris–HCl pH 8.5, 0.1 mM TCEP, 150 mM NaCl (1.0 mM monomer concentration) for crystallization using an YM10 ultrafiltration membrane (Amicon). Crystals were grown by the sitting-drop vapour-diffusion method at 297 K by mixing 1 µl protein solution and 1 µl reservoir solution. Msm Eis was pre-incubated with acetyl-CoA (100 mM) for 30 min and was then further incubated with paromomycin (50 mM) for a further 30 min in 20 mM Tris–HCl pH 8.5, 0.1 mM TCEP, 150 mM NaCl. Thick plate-shaped crystals were obtained with a reservoir solution consisting of 1.26 M ammonium sulfate, 100 mM MES pH 6.0. They grew to approximate dimensions of 0.2 × 0.2 × 0.1 mm within 2–3 d.
2.2. X-ray data collection and phasing
The crystals were flash-cooled using a cryoprotectant solution consisting of 1.26 M ammonium sulfate, 100 mM MES pH 6.0, 25%(v/v) glycerol. The crystals were soaked in 5 µl cryoprotectant solution for 10 s before being flash-cooled in liquid nitrogen. X-ray diffraction data from a crystal of paromomycin-complexed Msm Eis were collected at 100 K using an ADSC Quantum 315r CCD detector system (Area Detector Systems Corporation, Poway, California, USA) on BL-5C at Pohang Light Source, Republic of Korea. The crystals of paromomycin-complexed Msm Eis belonged to space group P212121, with unit-cell parameters a = 107.27, b = 126.54, c = 236.64 Å. Six monomers are present in the asymmetric unit, giving a Matthews coefficient and solvent fraction of 2.70 Å3 Da−1 and 54.5%, respectively. Data-collection and phasing statistics are summarized in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data-collection statistics | |
| Protein | M. smegmatis Eis |
| Data set | Paromomycin complex |
| Resolution range () | 30.03.30 (3.363.30) |
| X-ray source | BL-5C, Pohang Light Source |
| X-ray wavelength () | 1.0000 |
| Space group | P212121 |
| a, b, c () | 107.27, 126.54, 236.64 |
| , , () | 90, 90, 90 |
| Total/unique reflections | 89672/49506 |
| Completeness (%) | 95.4 (92.0) |
| Mean I/(I) | 22.1 (6.98) |
| R merge † (%) | 17.7 (55.9) |
| Refinement statistics | |
| Resolution range () | 15.03.30 |
| R work/R free ‡ (%) | 15.1/23.4 |
| No. of protein residues | 2412 |
| No. of sulfates | 6 |
| No. of paromomycins | 6 |
| Mean B factor (2) | |
| Protein | 48.9 |
| Sulfate | 81.9 |
| Paromomycin | 53.0 |
| R.m.s. deviations from ideal geometry | |
| Bond lengths () | 0.014 |
| Bond angles () | 1.69 |
| Ramachandran plot | |
| Most favoured (%) | 91.0 |
| Allowed (%) | 9.0 |
| Generously allowed (%) | 0 |
R
merge = 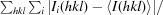
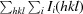 , where I(hkl) is the intensity of reflection hkl,
, where I(hkl) is the intensity of reflection hkl, 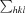 is the sum over all reflections and
is the sum over all reflections and 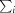 is the sum over i measurements of reflection hkl.
is the sum over i measurements of reflection hkl.
R = 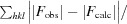
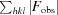 , where R
free is calculated for a randomly chosen 10% of reflections, which were not used for structure refinement, and R
work is calculated for the remaining reflections.
, where R
free is calculated for a randomly chosen 10% of reflections, which were not used for structure refinement, and R
work is calculated for the remaining reflections.
2.3. Model building and refinement
The structure was solved by molecular replacement using Phaser (McCoy et al., 2007 ▶) in the CCP4 program suite (Winn et al., 2011 ▶). We used the hexameric model of CoA-bound Msm Eis (PDB entry 3sxn; Kim et al., 2012 ▶) as the search model, with the ligands and waters removed. Subsequent manual model rebuilding was carried out using Coot (Emsley & Cowtan, 2004 ▶) interspersed with rounds of automatic refinement by REFMAC5 (Murshudov et al., 2011 ▶) and PHENIX (Adams et al., 2010 ▶). The geometry of the final refined model was checked with MolProbity (Chen et al., 2010 ▶). Refinement statistics are summarized in Table 1 ▶.
2.4. Accession code
The coordinates and structure factors for paromomycin-complexed Msm Eis have been deposited in the Protein Data Bank with accession code 4qb9.
3. Results and discussion
3.1. Overall quality of the structure
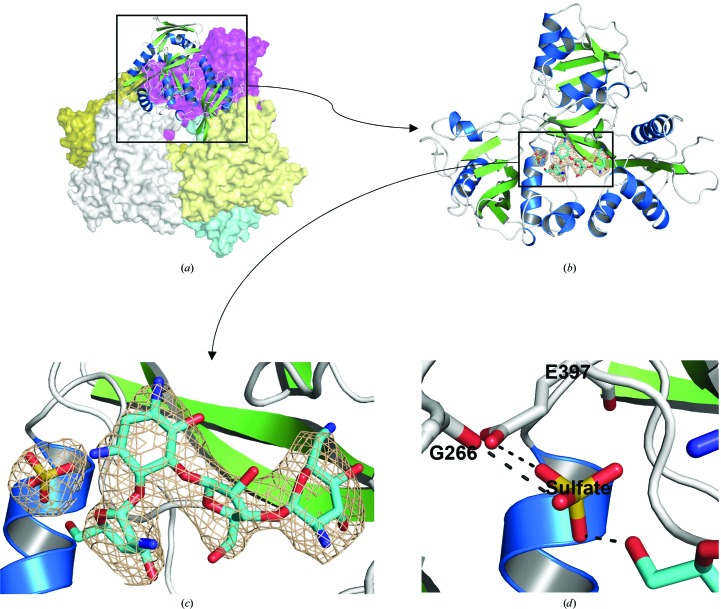
We have determined the crystal structure of the Msm Eis protein in complex with paromomycin at 3.3 Å resolution (Table 1 ▶). The refined model includes 2412 residues in six independent Eis monomers (residues 1–402 for chains A–F) and six molecules of paromomycin in the asymmetric unit. The N-terminal fusion tag is disordered in the crystal and is not visible in the electron-density map for all six monomers. The six monomers of paromomycin-bound Msm Eis in the asymmetric unit form a hexameric molecule in the crystal (Fig. 2 ▶). They are highly similar to each other, as indicated by the small r.m.s. deviations of 0.36–0.47 Å for pairwise comparisons of chain A against all others over 402 Cα atoms. One molecule of paromomycin is noncovalently bound to the active site of each monomer and all six paromomycin molecules in the asymmetric unit are well defined by the electron density (Fig. 2 ▶ and Supplementary Fig. S11). An extra electron density was observed adjacent to paromomycin and was modelled as a sulfate ion because the crystallization condition contained a high concentration of sulfate ions (Fig. 2 ▶ and Supplementary Fig. S1). The sulfate ion interacts with the main-chain O atoms of Gly266 (O–O distance of 3.1 Å) and Glu397 (O–O distance of 3.0 Å) and with O61 of paromomycin (O–O distance of 2.7 Å) (Fig. 2 ▶ d). Although acetyl-CoA was present at a 100-fold excess over the Msm Eis monomer in the crystallization conditions, its electron density was lacking at the expected binding site.
Figure 2.
The crystal structure of Msm Eis in complex with paromomycin. (a) Cartoon and surface representation of the Msm Eis hexameric structure. Five monomers of the hexamer are shown in surface mode (coloured magenta, slate, aquamarine, pale yellow and olive) and one is shown in cartoon mode. (b) Cartoon representation of the monomeric structure. (c) The electron density of paromomycin and a sulfate ion bound to chain A. The wheat-coloured mesh is the simulated-annealing OMIT F o − F c electron-density map calculated with paromomycin and the sulfate ion removed from the model (contoured at 3.0σ). (d) Cartoon representation of the sulfate-binding region in Msm Eis. Amino-acid residues and paromomycin around the sulfate ion are shown as stick models. N and O atoms are coloured blue and red, respectively. Black dotted lines denote hydrogen bonds.
3.2. Comparisons of paromomycin-bound Msm Eis with other Eis structures
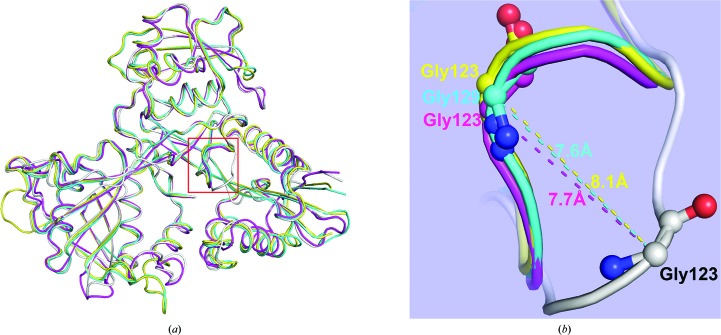
To assess possible structural changes upon binding paromomycin, we superimposed paromomycin-bound Msm Eis (chain A) with other Eis structures (Figs. 3 ▶ and 4 ▶). Against CoA-bound Msm Eis (PDB entry 3sxn; chain A; Kim et al., 2012 ▶), the r.m.s. deviations were 0.60–0.83 Å for monomer–monomer comparisons over 402 Cα atoms and 1.07 Å for a hexamer–hexamer comparison over 2412 Cα atoms. Notable structural differences are observed around the CoA-binding site. That is, part of the β5–α4 loop (Ser121–Tyr126) shows a relatively large conformational difference, with a maximum Cα deviation of 7.7 Å at Gly123 of Msm Eis (Fig. 3 ▶ b). Tyr126 is one of the key catalytic residues. Superposition of paromomycin-bound Msm Eis and acetyl-CoA-bound Mtb Eis (PDB entry 3ryo; chain A; Kim et al., 2012 ▶) gives r.m.s. deviations of 1.4–1.7 Å for monomer–monomer comparisons over 396 Cα atoms and an r.m.s. deviation of 1.5–1.8 Å for a hexamer–hexamer comparison over 2376 Cα atoms. Ser121–Tyr126 in Msm Eis (corresponding to Ser127–Tyr132 in Mtb Eis) show a relatively large conformational difference, with a maximum Cα deviation of 7.6 Å at Msm Eis Gly123 (and Mtb Eis Gly129) (Fig. 3 ▶ b). The same region shows a similarly large difference between paramomycin-complexed Msm Eis and CoA/tobramycin-complexed Mtb Eis (Houghton et al., 2013 ▶), with a maximum Cα deviation of 8.1 Å at Eis Gly123 (Fig. 3 ▶ b). The observed large structural differences are most likely owing to the absence of bound CoA or acetyl-CoA in our paromomycin-complexed Msm Eis, because the region encompassing Ser121–Tyr126 plays an important role in the interaction with CoA or acetyl-CoA (Supplementary Fig. S2; Kim et al., 2012 ▶). One significant structural difference between Msm Eis and Mtb Eis is the 310-helix α6′. It is present in both the paromomycin-bound and CoA-bound Msm Eis structures, but not in the apo and acetyl-CoA-complexed Mtb Eis structures (Supplementary Fig. S2).
Figure 3.
Structural comparison of paromomycin-complexed Msm Eis with other Msm Eis and Mtb Eis structures. (a) Monomers of paromomycin-bound Msm Eis (grey; chain A), CoA/tobramycin-bound Mtb Eis (yellow; PDB entry 4jd6, chain A), CoA-bound Msm Eis (magenta; PDB entry 3sxn, chain A) and acetyl-CoA-bound Mtb Eis (cyan; PDB entry 3ryo, chain A) are superimposed. (b) Close-up view of the region encompassing residues Ser121–Tyr126, which are enclosed in a red box in (a), which displays a large structural difference. Amino-acid residues around this region are shown as stick models. Dotted lines denote the distance between Cα atoms.
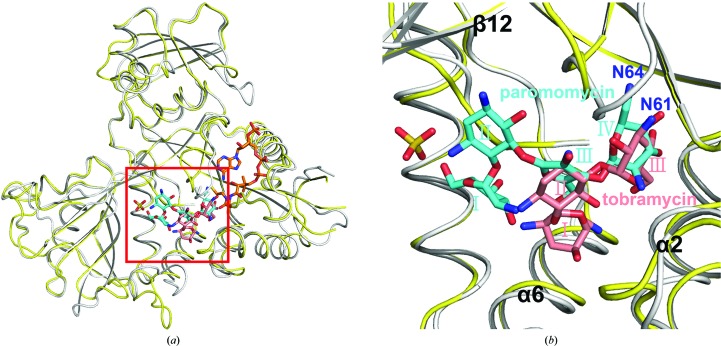
Figure 4.
Structural comparison of Msm Eis complexed with paromomycin only and Mtb Eis complexed with both CoA and tobramycin (binding mode A; Houghton et al., 2013 ▶). (a) Monomers of paromomycin-bound Msm Eis (gray) and CoA/tobramycin-bound Mtb Eis (yellow) are superimposed. (b) Close-up view of the aminoglycoside-binding pocket, as indicated by a red rectangle in (a). Paromomycin (cyan) and tobramycin (salmon) are shown as stick models.
In our current structure, acetyl-CoA did not bind to Msm Eis despite pre-incubation of the enzyme with a 100-fold excess of acetyl-CoA over the Eis monomer. The acetyl-CoA-binding site in our structure is vacant and is not even partly occupied by bound paromomycin. The absence of acetyl-CoA in the crystal may be owing to the presence of 1.26 M ammonium sulfate in the reservoir solution and/or to the absence of acetyl-CoA in the cryoprotectant solution.
3.3. Paromomycin binding to the Msm Eis active site
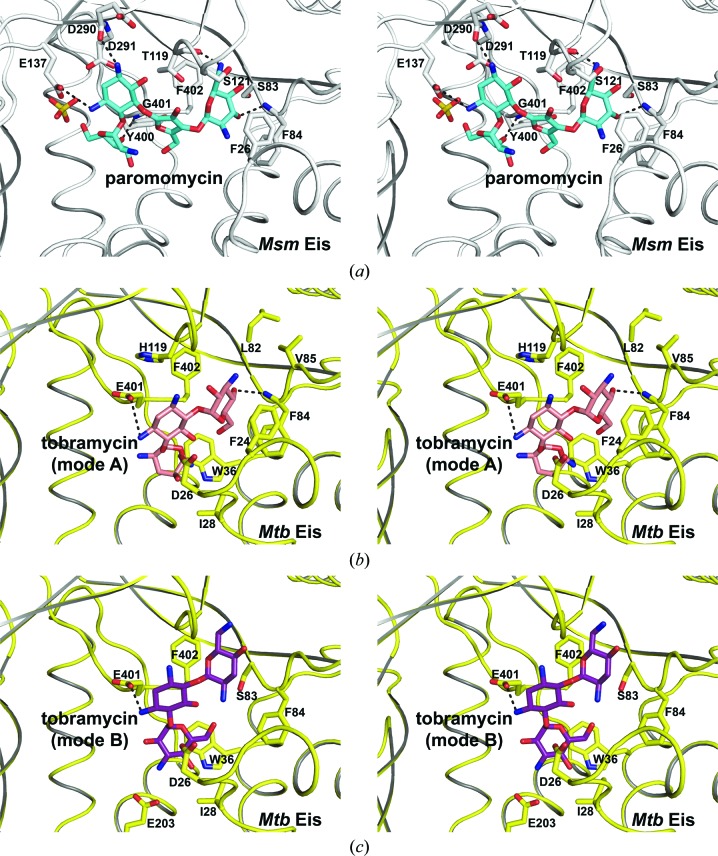
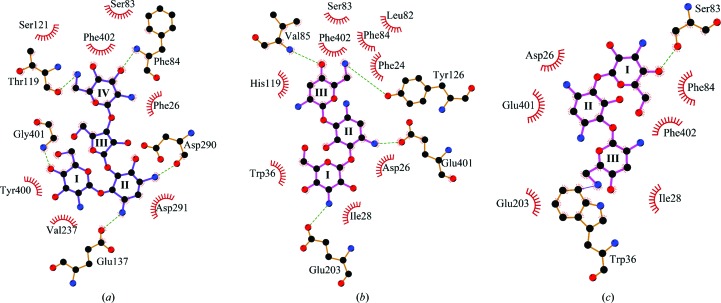
The six paromomycin molecules bound to the active sites of the Msm Eis hexamer in the asymmetric unit adopt essentially a single conformation in the crystal (Fig. 2 ▶ and Supplementary Fig. S1). Msm Eis was shown to triacetylate paromomycin (Chen et al., 2012 ▶). It is not clear why the observed binding mode is preferred over other possible binding modes. In the observed binding mode, the side chain of Glu137 and the carbonyl group of Asp290 are hydrogen-bonded to two amines of paromomycin ring II, while the amino group of Gly401 interacts with the hydroxyl groups of ring I and ring III (Figs. 5 ▶ a and 6 ▶ a). Paromomycin ring IV is oriented by contacts with the backbone amino group of Phe84, the carbonyl group of Thr119 and the side chains of Ser121 and Phe402. Most residues of Msm Eis that interact with paromomycin are well conserved in Mtb Eis, except for Thr119, Glu137 and Gly401 of Msm Eis, which are replaced by His125, Leu143 and Glu407 in Mtb Eis (Supplementary Fig. S2).
Figure 5.
Stereoview of aminoglycoside binding to Msm Eis and Mtb Eis. Amino-acid residues around paromomycin or tobramycin are shown as stick models. N and O atoms are coloured blue and red, respectively. Black dotted lines denote hydrogen bonds. (a) Paromomycin bound to Msm Eis monomer (chain A). (b) Tobramycin bound to Mtb Eis monomer (chain A; mode A; Houghton et al., 2013 ▶). (c) Tobramycin bound to Mtb Eis monomer (chain A; mode B).
Figure 6.
Interactions of aminoglycoside drugs with Msm Eis and Mtb Eis. (a) Paromomycin bound to Msm Eis (chain A). (b) Tobramycin bound to Mtb Eis (binding mode A; Houghton et al., 2013 ▶). (c) Tobramycin bound to Mtb Eis (binding mode B; Houghton et al., 2013 ▶). C, N and O atoms are coloured black, blue and red, respectively. The figure was prepared with LigPlot+ (Laskowski & Swindells, 2011 ▶).
Recently, a crystal structure of the Mtb Eis C204A mutant bound to tobramycin and CoA was reported at 3.5 Å resolution (Houghton et al., 2013 ▶). Strong and continuous electron density for tobramycin was observed in two of the six aminoglycoside-binding pockets of the Mtb Eis hexamer in the asymmetric unit, and two possible binding modes (A and B) were modelled (Figs. 5 ▶ b, 5 ▶ c, 6 ▶ b and 6 ▶ c). This is in agreement with the finding that Mtb Eis (as well as Msm Eis) can acetylate multiple amines of several aminoglycosides (Chen et al., 2012 ▶). The two binding modes are approximately related by a 180° rotation, a shift and a minor conformational change (Houghton et al., 2013 ▶). Binding mode A is consistent with 6′-acetylation of tobramycin by Mtb Eis, while binding mode B is consistent with 3′′-acetylation (Houghton et al., 2013 ▶). The mode of paromomycin binding to Msm Eis is more similar to mode A of tobramycin binding to Mtb Eis. Rings III and IV of paromomycin and rings II and III of tobramycin are located at similar positions in the active sites of Msm Eis and Mtb Eis, respectively. As a consequence, the potential acetylation sites of paromomycin (the N64 atom) and tobramycin (the N61 atom) are located in highly similar positions (Fig. 4 ▶ b). Rings III and IV of paromomycin are recognized by the active-site residues of Msm Eis in a manner somewhat similar to the recognition of rings II and III of tobramycin by the active-site residues of Mtb Eis (Figs. 5 ▶ and 6 ▶). Paromomycin rings III and IV are recognized by Phe26, Ser83, Phe84, Thr119, Ser121 and Phe402 of Msm Eis (Figs. 5 ▶ a and 6 ▶ a). Tobramycin rings II and III are recognized by Phe24, Leu82, Phe84, Val85, His119 and Phe402 of Mtb Eis in binding mode A (Figs. 5 ▶ b and 6 ▶ b). In mode B (Houghton et al., 2013 ▶), tobramycin is recognized by Asp26, Ile28, Trp36, Ser83, Phe84, Glu203, Glu401 and Phe402 of Mtb Eis (Figs. 5 ▶ c and 6 ▶ c). It is hoped that our structural data on paromomycin-bound Msm Eis will be useful in the design of Mtb Eis inhibitors as novel antituberculosis antibiotics.
Supplementary Material
PDB reference: Eis, complex with paromomycin, 4qb9
Supplementary figures. DOI: 10.1107/S2053230X14017385/tt5057sup1.pdf
Acknowledgments
We thank the beamline staff at BL-5C and BL-7A of Pohang Light Source, Republic of Korea for assistance during X-ray diffraction experiments. This work was supported by the Korea Ministry of Science, ICT and Future Planning, the National Research Foundation of Korea (NRF-2013R1A2A1A05067303), the Innovative Drug Research Center for Metabolic and Inflammatory Disease and the Korea Ministry of Health, Welfare and Family Affairs (Korea Healthcare Technology R&D Project A092006).
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: TT5057).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Chen, W., Biswas, T., Porter, V. R., Tsodikov, O. V. & Garneau-Tsodikova, S. (2011). Proc. Natl Acad. Sci. USA, 108, 9804–9808. [DOI] [PMC free article] [PubMed]
- Chen, W., Green, K. D., Tsodikov, O. V. & Garneau-Tsodikova, S. (2012). Biochemistry, 51, 4959–4967. [DOI] [PMC free article] [PubMed]
- Chiang, C.-Y., Centis, R. & Migliori, G. B. (2010). Respirology, 15, 413–432. [DOI] [PubMed]
- Dahl, J. L., Wei, J., Moulder, J. W., Laal, S. & Friedman, R. L. (2001). Infect. Immun. 69, 4295–4302. [DOI] [PMC free article] [PubMed]
- Dye, C. & Williams, B. G. (2010). Science, 328, 856–861. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Getahun, H., Gunneberg, C., Granich, R. & Nunn, P. (2010). Clin. Infect. Dis. 50, S201–S207. [DOI] [PubMed]
- Houghton, J. L., Biswas, T., Chen, W., Tsodikov, O. V. & Garneau-Tsodikova, S. (2013). Chembiochem, 14, 2127–2135. [DOI] [PMC free article] [PubMed]
- Kim, K. H. et al. (2012). Proc. Natl Acad. Sci. USA, 109, 7729–7734.
- Laskowski, R. A. & Swindells, M. B. (2011). J. Chem. Inf. Model. 51, 2778–2786. [DOI] [PubMed]
- Lella, R. K. & Sharma, C. (2007). J. Biol. Chem. 282, 18671–18675. [DOI] [PubMed]
- Lin, P. L. & Flynn, J. L. (2010). J. Immunol. 185, 15–22. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Meena, L. S. & Rajni (2010). FEBS J. 277, 2416–2427. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Shin, D.-M., Jeon, B.-Y., Lee, H.-M., Jin, H. S., Yuk, J.-M., Song, C.-H., Lee, S.-H., Lee, Z.-W., Cho, S.-N., Kim, J.-M., Friedman, R. L. & Jo, E.-K. (2010). PLoS Pathog. 6, e1001230. [DOI] [PMC free article] [PubMed]
- Wei, J., Dahl, J. L., Moulder, J. W., Roberts, E. A., O’Gaora, P., Young, D. B. & Friedman, R. L. (2000). J. Bacteriol. 182, 377–384. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wu, S., Barnes, P. F., Samten, B., Pang, X., Rodrigue, S., Ghanny, S., Soteropoulos, P., Gaudreau, L. & Howard, S. T. (2009). Microbiology, 155, 1272–1281. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Eis, complex with paromomycin, 4qb9
Supplementary figures. DOI: 10.1107/S2053230X14017385/tt5057sup1.pdf