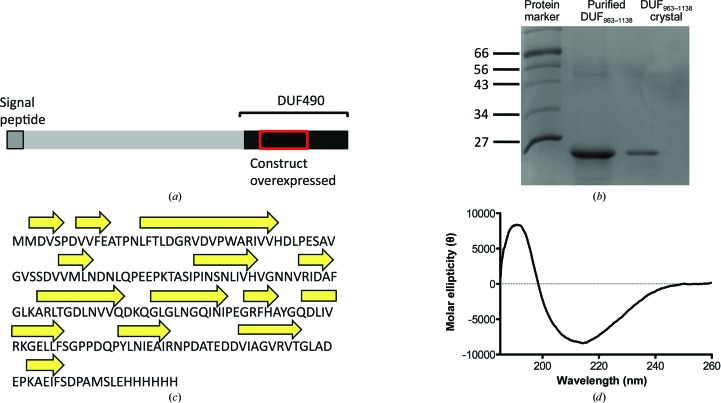
The C-terminal DUF490963–1138 domain of TamB (previously YtfN) from E. coli strain K-12 has been overexpressed, purifed and crystallized. Preliminary diffraction data have been collected to 2.1 Å resolution.
Keywords: DUF490, TamB, autotransporter, Escherichia coli
Abstract
TamB is a recently described inner membrane protein that, together with its partner protein TamA, is required for the efficient secretion of a subset of autotransporter proteins in Gram-negative bacteria. In this study, the C-terminal DUF490963–1138 domain of TamB was overexpressed in Escherichia coli K-12, purified and crystallized using the sitting-drop vapour-diffusion method. The crystals belonged to the primitive trigonal space group P3121, with unit-cell parameters a = b = 57.34, c = 220.74 Å, and diffracted to 2.1 Å resolution. Preliminary secondary-structure and X-ray diffraction analyses are reported. Two molecules are predicted to be present in the asymmetric unit. Experimental phasing using selenomethionine-labelled protein will be undertaken in the future.
1. Introduction
Proteobacterial genomes are littered with proteins devoid of functional annotation. A function for TamB (previously YtfN) and its partner protein TamA (previously YtfM) has recently been elucidated (Selkrig et al., 2012 ▶). Both proteins constitute the translocation and assembly module (TAM), a cell envelope-spanning protein complex found in most proteobacteria that contributes to the efficient secretion of autotransporter (AT) proteins (Selkrig et al., 2012 ▶). ATs are secreted outer membrane proteins that possess a C-terminal outer membrane (OM)-bound 12-stranded β-barrel domain and an N-terminal passenger domain consisting of an extended β-helical structure (Nishimura et al., 2010 ▶). The insertion of the β-domain into the OM is dependent on the β-barrel assembly machinery (BAM) complex, whereas the secretion of the passenger domain seemingly relies on the TAM complex for efficient OM translocation (Selkrig et al., 2012 ▶; Sauri et al., 2009 ▶). TamB is a large (137 kDa) multi-domain inner membrane protein for which little structural information is available. Amino-acid sequence analysis of TamB reveals the presence of a C-terminal conserved domain of unknown function DUF490 (Fig. 1 ▶ a), which has previously been shown to associate with its partner protein TamA (Selkrig et al., 2012 ▶). Bioinformatic analyses of the DUF490 domain reveal the widespread occurrence of this domain, and some examples of other proteins with this domain, as well as sequence alignments, are presented in Supplementary Fig. S11. Structural characterization of the DUF490 domain is therefore of interest since it might aid in shedding light on the biological functions of TamB and the numerous other proteins of unknown function that contain this domain. Here, we report the cloning, expression, purification, crystallization and collection of X-ray crystallographic data for DUF490963–1138 from the Escherichia coli K-12 TamB protein.
Figure 1.
Purification and characterization of DUF490963–1138 from TamB of E. coli. (a) Domain representation of TamB highlighting the C-terminal DUF490 domain and an N-terminal signal peptide that also serves as a membrane anchor. (b) 15% SDS–PAGE of purified DUF490963–1138 shows a single band migrating at the approximate molecular weight expected for the construct (20 kDa). Protein markers are labelled in kDa. Additionally, several crystals were washed with 35% PEG 400 solution three times and then dissolved in pure H2O and loaded onto the gel. (c) Secondary-structure prediction of the DUF490963–1138 fold using PSIPRED suggests that the protein consists predominantly of β-strands and random coil. (d) The far-UV CD spectrum of the construct confirms that the protein consists predominantly of β-strands (30%), turns (25%) and disordered polypeptide (33%) with minor α-helical content (12%). Values were calculated based on the average of all matching solutions.
2. Materials and methods
2.1. Macromolecule production
DUF490963–1138 was cloned from a TamBDUF490 construct previously produced from E. coli strain K-12 (Selkrig et al., 2012 ▶). The amplified PCR product was digested with NdeI and XhoI and ligated into pET-21a to produce an expression plasmid which encodes DUF490963–1138 with a C-terminal His6 tag.
E. coli BL21 (DE3) cells transformed with the DUF490963–1138 expression plasmid were grown in LB medium supplemented with 3%(v/v) glycerol and 100 mg ml−1 ampicillin at 37°C to an OD600 of 0.4–0.6; isopropyl β-d-1-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.5 mM to induce protein overexpression and the cells were grown for a further 15 h at 25°C. The cell pellet was collected by centrifugation at 4400g and resuspended in buffer A [20 mM Tris–HCl, 10 mM imidazole, 0.5 M NaCl, 5%(v/v) glycerol, 0.05% LDAO pH 7.5] supplemented with cOmplete protease-inhibitor cocktail (Roche) plus lysozyme (2 mg ml−1) and lysed by sonication. Cell debris were cleared by additional centrifugation at 46 000g and the supernatant was passed through a nickel-charged HisTrap HP column (GE Healthcare). The bound fractions were collected after elution with buffer B [20 mM Tris–HCl, 350 mM imidazole, 0.5 M NaCl, 5%(v/v) glycerol, 0.05% LDAO pH 7.5]. Fractions containing the protein of interest were pooled, dialysed in 50 mM Tris, 200 mM NaCl, 3%(v/v) glycerol, 0.05% LDAO pH 7.5 and run on a Superdex S200 gel-filtration column equilibrated with the same buffer. After SEC, DUF490963–1138 was dialysed into the same buffer omitting the 3% glycerol and stored at −80°C. Purified DUF490963–1138 was analysed by far-UV circular-dichroism (CD) spectroscopy. CD measurements were obtained using a protein concentration of 25 µM in a 0.02 cm quartz cuvette with a Jasco J-810 spectropolarimeter (Jasco UK Ltd). Secondary-structure estimates were obtained using the CONTIN procedure which was available from the DichroWeb server (Provencher & Glöckner, 1981 ▶; Whitmore & Wallace, 2008 ▶). Bioinformatic analysis of DUF490 was carried out using the Conserved Domain Architecture Retrieval Tool (CDART; Geer et al., 2002 ▶). Macromolecule production is summarized in Table 1 ▶.
Table 1. Macromolecule-production information.
| Source organism | E. coli strain K-12 |
| Forward primer | TCAGCATATGATGGATGTATCGCCAGATGTTGTA |
| Reverse primer | GGTACTCGAGCGACATCGCCGGGTCAGA |
| Cloning vector | pET-21a |
| Expression host | E. coli BL21 (DE3) |
| Complete amino-acid sequence of the construct produced | MMDVSPDVVFEATPNLFTLDGRVDVPWARIVVHDLPESAVGVSSDVVMLNDNLQPEEPKTASIPINSNLIVHVGNNVRIDAFGLKARLTGDLNVVQDKQGLGLNGQINIPEGRFHAYGQDLIVRKGELLFSGPPDQPYLNIEAIRNPDATEDDVIAGVRVTGLADEPKAEIFSDPAMSLEHHHHHH |
2.2. Crystallization
For crystallization, DUF490963–1138 was concentrated to 15 mg ml−1 using Vivaspin ultracentrifugation spin columns (MWCO 4000–6000) and filtered prior to dispensing into crystallization trays. Initial crystallization screens were set up in a 96-well MRC sitting-drop vapour-diffusion format (60 µl reservoir solution, 0.5 µl protein + 0.5 µl reservoir) using a Cartesian Honeybee 8+1 dispensing robot. Crystals appeared after several days in 0.1 M HEPES, 15%(v/v) PEG 400, 0.2 M CaCl2 pH 7.0 at 289 K. After optimization, diffraction-quality crystals were obtained in 0.1 M HEPES, 25%(v/v) PEG 400, 0.2 M CaCl2 pH 8.0 using 8 mg ml−1 protein and a protein:reservoir ratio of 1:1 at 289 K. The crystals were cryoprotected using 0.1 M HEPES, 30%(v/v) PEG 400, 0.2 M CaCl2 pH 8.0 by transferring the crystals using a LithoLoop into the cryosolution for 3 s and flash-cooling in a nitrogen-gas stream at 110 K. Crystallization is summarized in Table 2 ▶.
Table 2. Crystallization.
| Method | Sitting-drop vapour diffusion |
| Plate type | 96-well MRC plate |
| Temperature (K) | 289 |
| Protein concentration (mg ml−1) | 15 |
| Buffer composition of protein solution | 50 mM Tris–HCl, 200 mM NaCl, 0.05% LDAO pH 7.5 |
| Composition of reservoir solution | 0.1 M HEPES, 15%(v/v) PEG 400, 0.2 M CaCl2 pH 7.0 |
| Volume and ratio of drop | 1 µl, 1:1 |
| Volume of reservoir (µl) | 60 |
2.3. Data collection and processing
X-ray diffraction data were collected on beamline I03 at the Diamond Light Source (DLS) synchrotron, Harwell, England. Data were collected using a PILATUS3 6M detector with an oscillation angle of 0.15° and 0.08 s exposure time. A total of 3600 frames were collected and indexed using iMosflm (Battye et al., 2011 ▶) and scaled and merged using AIMLESS (Evans & Murshudov, 2013 ▶) from the CCP4 program suite (Winn et al., 2011 ▶). Processed data are summarized in Table 3 ▶.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | I03, DLS |
| Wavelength (Å) | 0.9763 |
| Temperature (K) | 100 |
| Detector | PILATUS3 6M |
| Rotation range per image (°) | 0.15 |
| Exposure time per image (s) | 0.08 |
| Space group | P3121 or P3221 |
| a, b, c (Å) | 57.34, 57.34, 220.74 |
| α, β, γ (°) | 90, 90, 120 |
| Multiplicity | 9.6 (9.5) |
| Resolution range (Å) | 48.45–2.10 (2.17–2.10) |
| No. of unique reflections | 25570 (2303) |
| Completeness (%) | 99.7 (98.9) |
| 〈I/σ(I)〉 | 12.1 (3.2) |
| R p.i.m. † (%) | 3.2 (26.2) |
| R merge ‡ (%) | 9.3 (76.3) |
R
p.i.m. = 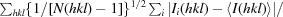
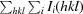 .
.
R
merge = 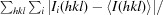
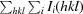 .
.
3. Results and discussion
DUF490963–1138 from TamB was produced recombinantly and purified to homogeneity. The protein migrates as a single band on an SDS–PAGE gel, with an estimated molecular weight of 20 kDa, which is close to the calculated molecular weight based on the amino-acid sequence of the His6-tagged protein (20.3 kDa; Fig. 1 ▶ b). We observed the presence of multimeric species following gel-filtration chromatography (Supplementary Fig. S2). Whether this oligomerization is physiologically relevant is not currently known. However, in the previous study describing the discovery of the TAM complex, TamB was shown to behave as a monomer on blue native PAGE (Selkrig et al., 2012 ▶). For crystallization purposes and further sample characterization we selected the gel-filtration peak that corresponded to the monomeric fraction. Far-UV circular-dichroism analysis of DUF490963–1138 suggests that the protein consists predominantly of β-strands, turns and random coil, with a small α-helical fraction (Fig. 1 ▶ d). This is in agreement with the analysis of the amino-acid sequence by the secondary-structure prediction program PSIPRED (McGuffin et al., 2000 ▶; Fig. 1 ▶ c).
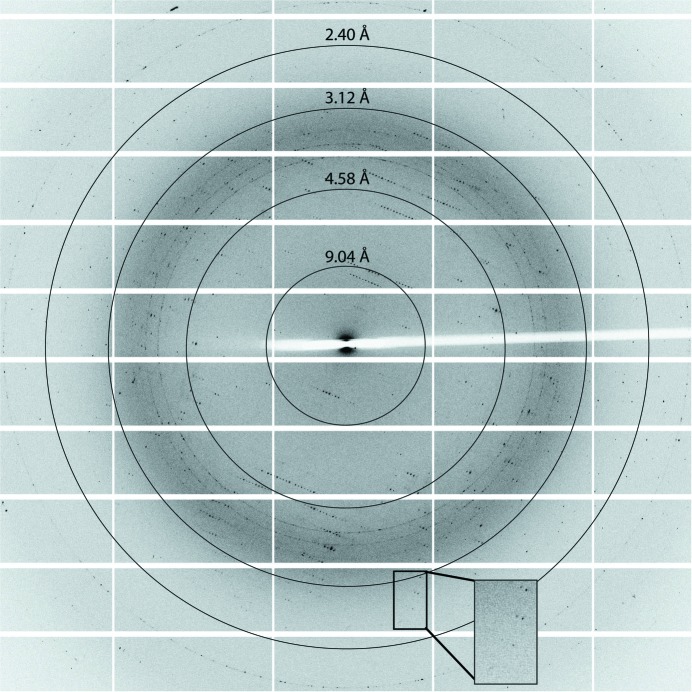
Initial crystal screening consisted of 384 crystallization conditions, of which only one produced a successful hit. Optimization of this crystallization condition was undertaken varying the levels of precipitant, pH and protein concentration (Fig. 2 ▶). The presence of DUF490963–1138 within crystals was confirmed by analysing washed crystals by SDS–PAGE (Fig. 1 ▶ b). X-ray diffraction data were collected to a resolution of 2.1 Å (Fig. 3 ▶). The crystals belonged to space group P3121 (or P3221). The unit-cell parameters were a = b = 57.34, c = 220.74 Å, α = β = 90, γ = 120°. Two monomers are predicted in the asymmetric unit, with a calculated Matthews coefficient of 2.90 Å3 Da−1 and a solvent content of 57.6%. Analysis of possible twinning was carried out using phenix.xtriage, which detected no twinning (Zwart et al., 2005 ▶). Owing to the absence of any structural models of DUF490963–1138 homologues, ab initio modelling of DUF490963–1138 was attempted using the I-TASSER, Phyre and SWISS-MODEL servers (Roy et al., 2010 ▶; Kelley & Sternberg, 2009 ▶; Biasini et al., 2014 ▶). However, the confidence in the models was very low and molecular replacement was unsuccessful. Initial experimental phasing with heavy-atom derivatives was also undertaken; however, heavy-metal soaks proved detrimental to the crystal quality and diffraction and co-crystallization trials failed to produce any crystals. Therefore, selenomethionine-labelled protein will be produced for experimental phasing (the hypothetical number of Met residues in the asymmetric unit is eight). The high-resolution structure of DUF490963–1138 from TamB will provide insight into the function of this domain and its possible contribution to autotransporter biogenesis.
Figure 2.

Optimized crystals of DUF490963–1138 grown using the sitting-drop vapour-diffusion method. The dimensions of the crystals were approximately 100 × 50 × 30 µm.
Figure 3.
Representative diffraction pattern of DUF490963–1138. The box highlights the high-resolution spots with an adjusted background.
Supplementary Material
Supporting Information.. DOI: 10.1107/S2053230X14017403/wd5236sup1.pdf
Acknowledgments
We would like to acknowledge the Diamond Light Source for access to I03 (proposal No. MX5689). IJ is supported by a four-year studentship from the Wellcome Trust (grant No. 093597/Z/10/Z). RG is supported by a Kelvin Smith Scholarship from the University of Glasgow.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: WD5236).
References
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., Kiefer, F., Cassarino, T. G., Bertoni, M. & Bordoli, L. (2014). Nucleic Acids Res. 42, W252–W258. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Geer, L. Y., Domrachev, M., Lipman, D. J. & Bryant, S. H. (2002). Genome Res. 12, 1619–1623. [DOI] [PMC free article] [PubMed]
- Kelley, L. A. & Sternberg, M. J. (2009). Nature Protoc. 4, 363–371. [DOI] [PubMed]
- McGuffin, L. J., Bryson, K. & Jones, D. T. (2000). Bioinformatics, 16, 404–405. [DOI] [PubMed]
- Nishimura, K., Tajima, N., Yoon, Y.-H., Park, S.-Y. & Tame, J. R. H. (2010). J. Mol. Med. 88, 451–458. [DOI] [PubMed]
- Provencher, S. W. & Glöckner, J. (1981). Biochemistry, 20, 33–37. [DOI] [PubMed]
- Roy, A., Kucukural, A. & Zhang, Y. (2010). Nature Protoc. 5, 725–738. [DOI] [PMC free article] [PubMed]
- Sauri, A., Soprova, Z., Wickström, D., de Gier, J.-W., Van der Schors, R. C., Smit, A. B., Jong, W. S. P. & Luirink, J. (2009). Microbiology, 155, 3982–3991. [DOI] [PubMed]
- Selkrig, J. et al. (2012). Nature Struct. Mol. Biol. 19, 506–510. [DOI] [PubMed]
- Whitmore, L. & Wallace, B. A. (2008). Biopolymers, 89, 392–400. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zwart, P., Grosse-Kunstleve, R. & Adams, P. (2005). CCP4 Newsl. Protein Crystallogr. 43, 27–35.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.. DOI: 10.1107/S2053230X14017403/wd5236sup1.pdf