The N-acetyltransferase PseH from the H. pylori pseudaminic acid biosynthesis pathway has been purified and co-crystallized with acetyl-CoA by the hanging-drop vapour-diffusion method. A diffraction data set has been collected to 2.5 Å resolution.
Keywords: Helicobacter pylori, N-acetyltransferase, acetyl-CoA, pseudaminic acid biosynthesis
Abstract
Helicobacter pylori infection is the common cause of gastritis and duodenal and stomach ulcers, which have been linked to a higher risk of the development of gastric cancer. The motility that facilitates persistent infection requires functional flagella that are heavily glycosylated with 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-nonulosonic acid (pseudaminic acid). Pseudaminic acid biosynthesis protein H (PseH) catalyzes the third step in its biosynthetic pathway, producing UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose. Crystals of H. pylori PseH have been grown by the hanging-drop vapour-diffusion method using diammonium tartrate as a precipitating agent. The crystals belonged to space group I222 or I212121, with unit-cell parameters a = 107.8, b = 145.4, c = 166.3 Å. A complete X-ray diffraction data set has been collected to 2.5 Å resolution using cryocooling conditions and synchrotron radiation.
1. Introduction
Infection with Helicobacter pylori can lead to a variety of gastroduodenal disorders such as gastritis, gastric and duodenal ulcers and malignant tumours, including gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (Suerbaum & Michetti, 2002 ▶; Bauer & Meyer, 2011 ▶). H. pylori was the first bacterium to be classified as a class I, or definite, human carcinogen by the IACR (International Agency for Research on Cancer, 1994 ▶), since H. pylori infection was shown to increase the risk of gastric cancer approximately tenfold. H. pylori has a bundle of four to six flagella at one end of the cell body that help it drill into the mucus layer of the stomach for successful initial colonization of the gastric epithelium (Yoshiyama & Nakazawa, 2000 ▶; Kavermann et al., 2003 ▶; Baldwin et al., 2007 ▶). Motility by the flagellar motor is also required for attaining full infection levels and persistence of H. pylori in the high-flow environment of the stomach under conditions of constant and rapid turnover of gastric mucosa (Ottemann & Lowenthal, 2002 ▶).
The H. pylori flagellar filaments are polymers of two homologous flagellins FlaA and FlaB, both of which are O-glycosylated on serines and threonines with the unusual nine-carbon sugar pseudaminic acid (Pse), which is unique to bacteria (Schirm et al., 2003 ▶). Flagellin glycosylation is essential for flagellar assembly and bacterial motility (Josenhans et al., 2002 ▶; Schirm et al., 2005 ▶). Therefore, the Pse biosynthesis pathway can potentially be exploited in the design of novel therapeutics targeting H. pylori motility. Schoenhofen and coworkers have recently identified all six enzymatic components and reconstituted the complete five-step Pse biosynthesis pathway of H. pylori (Schoenhofen, McNally et al., 2006 ▶). Whilst the crystal structures of the first two enzymes of this pathway in H. pylori, PseB (FlaA1) and PseC, have been elucidated (Ishiyama et al., 2006 ▶; Schoenhofen, Lunin et al., 2006 ▶), the structures of the remaining components are not yet known. The third step of this pathway, N-4 acetylation of UDP-4-amino-4,6-dideoxy-N-acetyl-β-l-altrosamine to UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose, is catalyzed by a 21.1 kDa Pse biosynthesis protein H (PseH), also known as flagellin modification protein H (FlmH) or flagellar protein G (FlaG1). This enzyme utilizes acetyl-coenzyme A (acetyl-CoA) as the donor of the acetyl group. Mutation of the pseH gene in the closely related species Campylobacter jejuni resulted in a nonmotile phenotype that lacked flagella filaments and hook structures (Guerry et al., 2006 ▶), suggesting that PseH plays an essential role in flagella assembly.
Limited structural data are available on bacterial enzymes that are functionally homologous to PseH. Transfer of an acetyl group from acetyl-CoA to the 4-amino moiety of the nucleotide-linked sugar substrate in a different biosynthetic pathway leading to legionaminic acid is catalyzed by the trimeric N-acetyltransferase PglD, which has a left-handed β-helix (LbH) fold (Olivier & Imperiali, 2008 ▶). PseH shows no detectable sequence similarity to PglD. A different characterized example of a bacterial nucleotide-sugar N-acetyltransferase, the Escherichia coli dTDP-fucosamine acetyltransferase WecD (Hung et al., 2006 ▶), belongs to a large GCN5-related N-acetyltransferase (GNAT) superfamily (Vetting et al., 2005 ▶), members of which are present in all kingdoms of life and utilize acetyl-CoAs to acylate their cognate substrates. The crystal structure of the WecD–acetyl-CoA complex (Hung et al., 2006 ▶) revealed that WecD forms homodimers in which each monomer contains a funnel-like acetyl-CoA binding pocket. However, to the best of our knowledge, no crystal structure of WecD in complex with the nucleotide-linked sugar substrate is available and the mode of substrate binding remains unknown. Although PseH shares only 15% sequence identity with WecD, it is predicted to be a member of the same GNAT superfamily based on sequence analysis using the SUPERFAMILY hidden Markov models (http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY; Gough et al., 2001 ▶). In this paper, we report the crystallization and preliminary X-ray analysis of recombinant H. pylori PseH. Analysis of the crystal structure of this enzyme and its comparison with the structures of WecD and other GNAT superfamily N-acetyltransferases would be an important step towards our understanding of how the structural variations within this class of enzymes are related to substrate and reaction specificity.
2. Materials and methods
2.1. Vector construction, protein expression and purification
The coding sequence for PseH (HP0327, UniProtKB O25094; 180 amino-acid residues) was amplified by PCR from H. pylori genomic DNA (ATCC 26695) using OneTaq Hot Start DNA Polymerase (New England Biolabs) and the primers CACCAAAAAAAATTATTCTTATAAAAATATC (forward) and CTAAAGTTTTAGAAGAGATTGATCATTATATC (reverse). The amplified fragment was ligated into the pET151/D-TOPO vector using the TOPO cloning kit (Invitrogen) to produce an expression vector that contains an N-terminal His6 tag followed by six amino-acid residues GIDPFT that comprise the Tobacco etch virus (TEV) protease cleavage site. The expression construct was verified by DNA sequencing (see Table 1 ▶). The vector was introduced into E. coli strain BL21-CodonPlus (DE3)-RIPL (Stratagene). The cells were grown in Luria–Bertani medium supplemented with 50 mg l−1 ampicillin and 34 mg l−1 chloramphenicol at 310 K to an optical density OD600 of 0.6, at which point protein overexpression was induced by adding 0.5 mM isopropyl β-d-1-thiogalactopyranoside. The cells were grown for a further 3 h, harvested by centrifugation, resuspended in a buffer consisting of 20 mM sodium phosphate pH 7.4, 200 mM NaCl, 1 mM PMSF and lysed using an EmulsiFlex-C5 high-pressure homogenizer (Avestin). The cell lysate was clarified by centrifugation at 12 000g for 30 min at 277 K. NaCl and imidazole were then added to the supernatant to final concentrations of 500 and 15 mM, respectively, before loading it onto a 5 ml HiTrap Chelating HP column (GE Healthcare) pre-washed with buffer A (20 mM sodium phosphate pH 7.4, 500 mM NaCl, 15 mM imidazole). The column was washed with 20 column volumes of buffer A containing 60 mM imidazole and protein was eluted with buffer A containing 500 mM imidazole. The N-terminal tag was removed with His6-TEV protease (Invitrogen, 300 U per milligram of protein) overnight at 277 K whilst dialyzing the sample against buffer B [50 mM Tris–HCl pH 8.0, 2 mM dithiothreitol, 200 mM NaCl, 1%(v/v) glycerol]. NaCl and imidazole were then added to the sample to final concentrations of 500 and 15 mM, respectively, and the TEV protease and the uncleaved protein were removed over a HiTrap Chelating HP column. The flowthrough was concentrated to 2 ml in a VivaSpin 10 000 Da cutoff concentrator and purified further by passing it through a Superdex 200 HiLoad 26/60 gel-filtration column (GE Healthcare) equilibrated with buffer C (50 mM Tris–HCl pH 8.0, 200 mM NaCl). The protein purity was estimated to be greater than 95% by SDS–PAGE. Protein concentration was determined using the Bradford assay (Bradford, 1976 ▶).
Table 1. Macromolecule-production information.
| Source organism | H. pylori strain 26695 |
| DNA source | Genomic DNA |
| Forward primer | CACCAAAAAAAATTATTCTTATAAAAATATC |
| Reverse primer | CTAAAGTTTTAGAAGAGATTGATCATTATATC |
| Cloning vector | pET151/D-TOPO |
| Expression vector | pET151/D-TOPO |
| Expression host | E. coli strain BL21-CodonPlus (DE3)-RIPL |
| Complete amino-acid sequence of the construct produced | GIDFPTKKNYSYKNIQAIDFTNLNDGEKLLVLEFRNHPNTALWMYSTFISLKTHLQFIEDLKNSPNHRYFLFKEEGVYLGVGSITKINFFHKHGYLGIYKNPFLKNGGETILKALEFIAFEEFQLHSLHLEVMENNFKAIAFYEKNHYELEGRLKGFISKDKEFIDVLLYYKDKKGYNDQSLLKL |
2.2. Crystallization
Prior to crystallization, protein was concentrated to 10.5 mg ml−1 using a 10 000 Da cutoff concentrator, mixed with acetyl-CoA (final concentration 1.5 mM) and centrifuged for 20 min at 13 000g to clarify the solution. Initial screening of crystallization conditions was performed by the vapour-diffusion method in a hanging-drop format using a Phoenix crystallization robot (Art Robbins Instruments) and Crystal Screen HT and PEG/Ion Screen HT (Hampton Research). The initial crystallization droplets consisted of 100 nl protein solution mixed with 100 nl reservoir solution and were equilibrated against 50 µl reservoir solution in a 96-well Art Robbins CrystalMation Intelli-Plate (Hampton Research). Crystals appeared after 1 d from condition No. 34 of Crystal Screen HT (Fig. 1 ▶), which consisted of 1.1 M diammonium tartrate, 100 mM sodium acetate trihydrate pH 4.6. This condition was refined to improve the crystal quality, yielding an optimal crystallization reservoir solution of 0.9 M diammonium tartrate, 80 mM sodium acetate trihydrate pH 4.2 and acetyl-CoA at a concentration of 3 mM.
Figure 1.
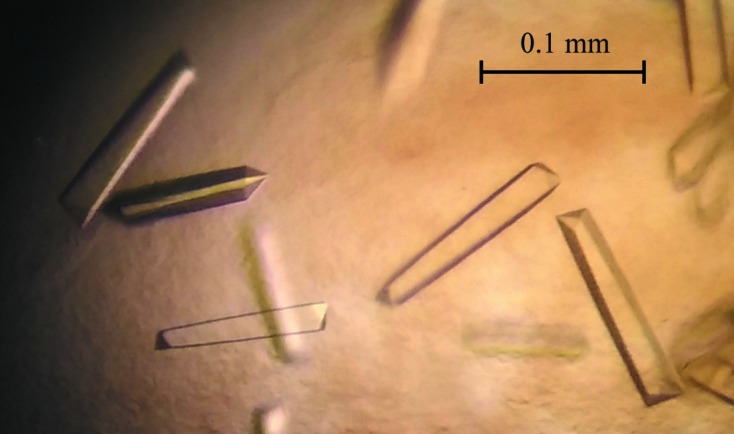
Crystals of a putative H. pylori PseH–acetyl-CoA complex.
2.3. Data collection and processing
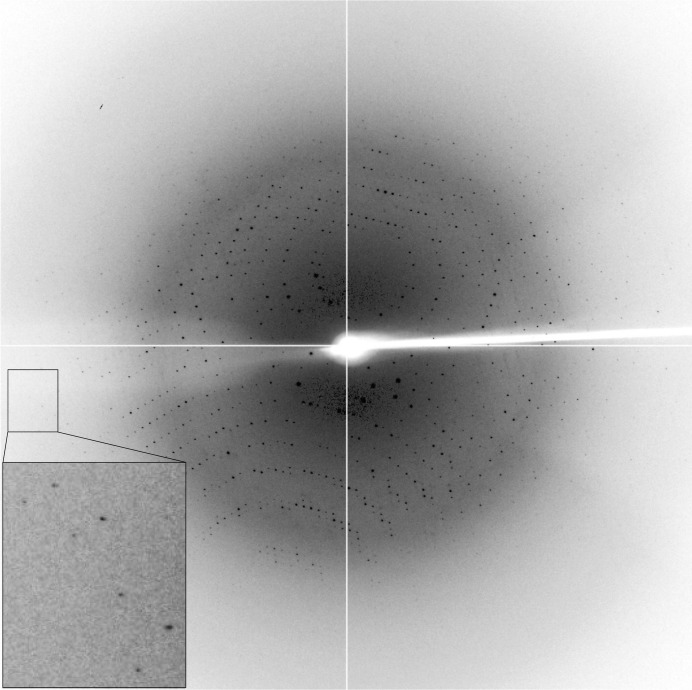
The PseH crystal was soaked in a cryo-stabilizing solution consisting of 0.9 M diammonium tartrate, 80 mM sodium acetate trihydrate pH 4.2, 20% glycerol, 3.3 mM acetyl-CoA and was flash-cooled by plunging it into liquid nitrogen prior to data collection. X-ray data were collected to 2.5 Å resolution on the MX1 beamline of the Australian Synchrotron (Fig. 2 ▶). A total of 360 images were collected using a 0.5° oscillation width. The data were processed using iMosflm (Battye et al., 2011 ▶) and SCALA (Evans, 2006 ▶) from the CCP4 suite (Winn et al., 2011 ▶). Calculation of the self-rotation function was performed using POLARRFN (Winn et al., 2011 ▶).
Figure 2.
A representative 0.5° oscillation image of the data collected using an ADSC Quantum 210r CCD detector at station MX1, Australian Synchrotron, Victoria, Australia. A magnified rectangle shows diffraction spots beyond 2.5 Å resolution.
3. Results and discussion
Recombinant H. pylori PseH was expressed in E. coli strain BL21-CodonPlus (DE3)-RIPL from the pET151/D-TOPO plasmid upon induction by T7 polymerase. The enzyme was purified to >95% electrophoretic homogeneity based on Coomassie Blue staining of SDS–PAGE gels. The protein migrated on SDS–PAGE with an apparent molecular weight of 21–22 kDa, which is close to the value calculated from the amino-acid sequence (21.4 kDa).
Crystals of the H. pylori PseH–acetyl-CoA complex were obtained using a sparse-matrix crystallization approach. Analysis of the X-ray diffraction data by the autoindexing routine in iMosflm is consistent with a body-centred orthorhombic crystal system (I222 or I212121), with unit-cell parameters a = 107.8, b = 145.4, c = 166.3 Å. The average I/σ(I) value is 17.6 for all reflections (resolution range 39.0–2.5 Å) and 6.0 in the highest resolution shell (2.64–2.50 Å). A total of 337 091 measurements were made of 45 481 independent reflections. Data processing gave an R merge of 0.082 for intensities (0.362 in the resolution shell 2.64–2.50 Å) and these data were 100% complete (100% completeness in the highest resolution shell) (see Table 2 ▶).
Table 2. Data collection and processing.
| Diffraction source | MX1 beamline, Australian Synchrotron |
| Wavelength (Å) | 1.0 |
| Temperature (K) | 100 |
| Detector | ADSC Quantum 210r CCD |
| Crystal-to-detector distance (mm) | ? |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 1 |
| Space group | I222 or I212121 |
| a, b, c (Å) | 107.8, 145.4, 166.3 |
| α, β, γ (°) | 90, 90, 90 |
| Mosaicity (°) | 0.35 |
| Resolution range (Å) | 39–2.5 |
| Total No. of reflections | 337091 |
| No. of unique reflections | 45481 |
| Completeness (%) | 100 |
| Multiplicity | 7.4 |
| 〈I/σ(I)〉 | 17.6 |
| R r.i.m. | 0.088 |
| Overall B factor from Wilson plot (Å2) | 31 |
Calculations of the Matthews coefficient for four, five or six molecules in the asymmetric unit give values of 3.8, 3.1 and 2.6 Å3 Da−1, respectively, all of which lie in the range observed for protein crystals (Matthews, 1977 ▶). For the self-rotation function calculated using data in the resolution range 10–6 Å with an integration radius of 25 Å, no dominant features that can be confidently assigned to noncrystallographic axes were found in the κ = 90, 120 or 180° sections. Thus, we are currently unable to determine the protein contents of the asymmetric unit. Our efforts are currently being directed towards a search for heavy-atom derivatives and the solution of the structure using multiple isomorphous replacement and/or multi-wavelength anomalous dispersion methods.
Acknowledgments
We thank the staff at the Australian Synchrotron for assistance with data collection. We are also grateful to Dr Danuta Maksel and Dr Robyn Gray at the Monash University Protein Crystallography Unit for assistance with the robotic crystallization trials. AR is an Australian Research Council Research Fellow.
References
- Baldwin, D. N., Shepherd, B., Kraemer, P., Hall, M. K., Sycuro, L. K., Pinto-Santini, D. M. & Salama, N. R. (2007). Infect. Immun. 75, 1005–1016. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Bauer, B. & Meyer, T. F. (2011). Ulcers, 10.1155/2011/340157.
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Gough, J., Karplus, K., Hughey, R. & Chothia, C. (2001). J. Mol. Biol. 313, 903–919. [DOI] [PubMed]
- Guerry, P., Ewing, C. P., Schirm, M., Lorenzo, M., Kelly, J., Pattarini, D., Majam, G., Thibault, P. & Logan, S. (2006). Mol. Microbiol. 60, 299–311. [DOI] [PMC free article] [PubMed]
- Hung, M.-N., Rangarajan, E., Munger, C., Nadeau, G., Sulea, T. & Matte, A. (2006). J. Bacteriol. 188, 5606–5617. [DOI] [PMC free article] [PubMed]
- International Agency for Research on Cancer (1994). IARC Monogr. Eval. Carcinog. Risks Hum. 61, 177–240.
- Ishiyama, N., Creuzenet, C., Miller, W. L., Demendi, M., Anderson, E. M., Harauz, G., Lam, J. S. & Berghuis, A. M. (2006). J. Biol. Chem. 281, 24489–24495. [DOI] [PubMed]
- Josenhans, C., Vossebein, L., Friedrich, S. & Suerbaum, S. (2002). FEMS Microbiol. Lett. 210, 165–172. [DOI] [PubMed]
- Kavermann, H., Burns, B. P., Angermuller, K., Odenbreit, S., Fischer, W., Melchers, K. & Haas, R. (2003). J. Exp. Med. 197, 813–822. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1977). X-ray Structure of Proteins, Vol. 3, 3rd ed., edited by H. Neurath & R. L. Hill, pp. 468–477. New York: Academic Press.
- Olivier, N. B. & Imperiali, B. (2008). J. Biol. Chem. 283, 27937–27946. [DOI] [PMC free article] [PubMed]
- Ottemann, K. M. & Lowenthal, A. C. (2002). Infect. Immun. 70, 1984–1990. [DOI] [PMC free article] [PubMed]
- Schirm, M., Schoenhofen, I. C., Logan, S. M., Waldron, K. C. & Thibault, P. (2005). Anal. Chem. 77, 7774–7782. [DOI] [PubMed]
- Schirm, M., Soo, E. C., Aubry, A. J., Austin, J., Thibault, P. & Logan, S. M. (2003). Mol. Microbiol. 48, 1579–1592. [DOI] [PubMed]
- Schoenhofen, I. C., Lunin, V. V., Julien, J.-P., Li, Y., Ajamian, E., Matte, A., Cygler, M., Brisson, J.-R., Aubry, A. & Logan, S. M. (2006). J. Biol. Chem. 281, 8907–8916. [DOI] [PubMed]
- Schoenhofen, I. C., McNally, D. J., Brisson, J.-R. & Logan, S. M. (2006). Glycobiology, 16, 8C–14C. [DOI] [PubMed]
- Suerbaum, S. & Michetti, P. (2002). N. Engl. J. Med. 347, 1175–1186. [DOI] [PubMed]
- Vetting, M. W., de Carvalho, L. P. S., Yu, M., Hegde, S. S., Magnet, S., Roderick, S. L. & Blanchard, J. S. (2005). Arch. Biochem. Biophys. 433, 212–226. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yoshiyama, H. & Nakazawa, T. (2000). Microbes Infect. 2, 55–60. [DOI] [PubMed]