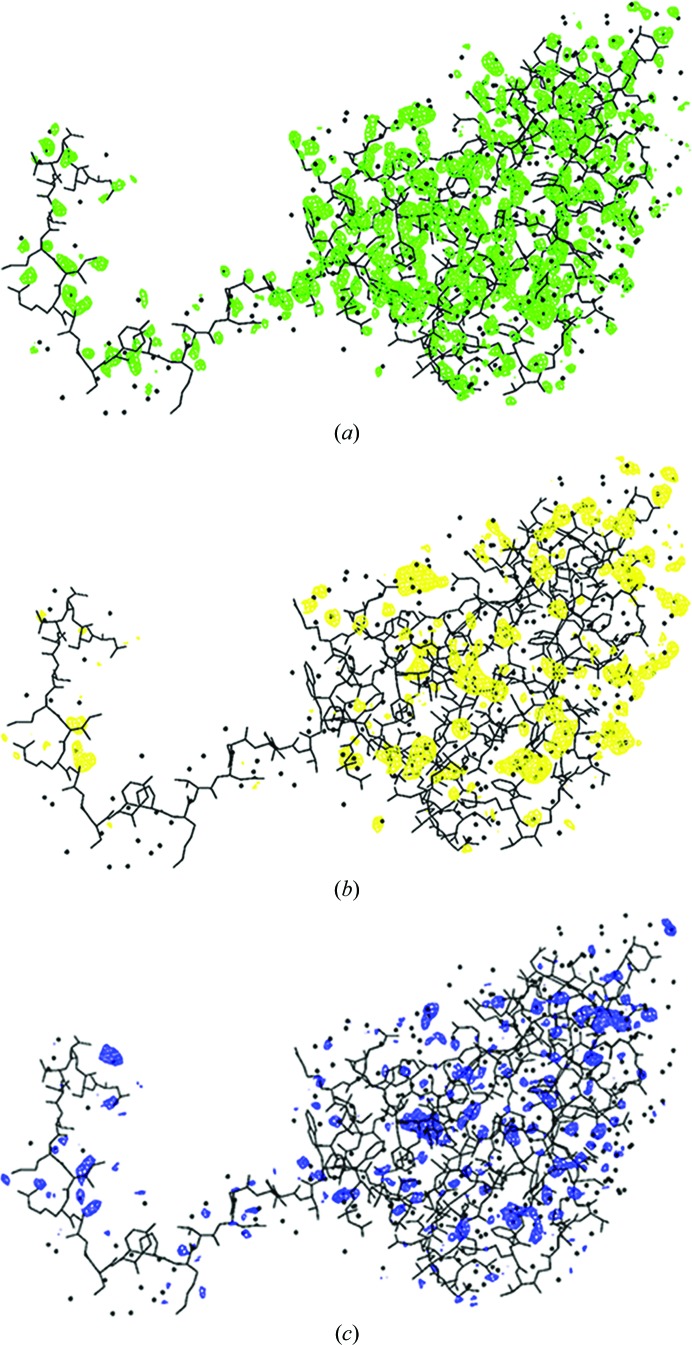
The crystal structure of Satellite tobacco mosaic virus (STMV) at 1.4 Å resolution represents the highest resolution and most precise native virus structure, utilizing more than twice as many reflections in refinement as were used for the previous STMV model. New details regarding the multiple ion arrangement on fivefold axes, the disposition of the ‘free’ nucleotide, alternate conformations of amino-acid side chains and the distortion of the capsid from strict icosahedral symmetry have emerged.
Keywords: virus structure, ion pores, nucleic acid, icosahedral symmetry
Abstract
Satellite tobacco mosaic virus (STMV) is among the smallest viruses, having 60 identical subunits arranged with T = 1 icosahedral symmetry. Its crystal structure was solved at 290 K and was refined using, in part, crystals grown in microgravity. Electron-density maps revealed nearly 57% of the genomic ssRNA. Using six flash-cooled crystals, diffraction data were recorded to 1.4 Å resolution and independent refinements of the STMV model were carried out versus the previous 1.8 Å resolution data representing merged data from 21 crystals (271 689 unique reflections), data consisting of corresponding reflections to 1.8 Å resolution from the cooled crystals and 1.4 Å resolution data from the cooled crystals comprised of 570 721 unique reflections. Models were independently refined with full NCS constraints using the program CNS and in restrained mode using the programs CNS, REFMAC5 and SHELX-97, with the latter two procedures including anisotropic temperature factors. Significant additional structural detail emerged from the analyses, including a unique cation and anion arrangement on fivefold axes and a precise assessment of icosahedral symmetry exactness in the crystal lattice. STMV represents the highest resolution native virus structure currently known by a substantial margin, and it permits the evaluation of a precise atomic model of a spherical virus at near-atomic resolution for the first time.
1. Introduction
1.1. STMV properties and prior structural studies
Satellite tobacco mosaic virus (STMV) is a spherical satellite virus to the helical rod-shaped Tobacco mosaic virus (Valverde & Dodds, 1986 ▶, 1987 ▶; Valverde et al., 1991 ▶; Ban et al., 1995 ▶). It is the second smallest virus currently known (Ban et al., 1995 ▶) and is a T = 1 icosahedron (Caspar & Klug, 1962 ▶) composed of 60 identical protein subunits of 159 amino acids with M r = 17 500 Da. The capsid proteins exhibit the canonical eight-stranded Swiss-roll β-barrel structure with a long 37-amino-acid extended tail that is responsible, in part, for protein dimer formation and for interaction with a genomic positive-sense ssRNA molecule of 1058 nucleotides (Mirkov et al., 1989 ▶; see Fig. 1 ▶). The entire virus particle has a molecular weight of about 1.4 MDa, about a quarter of which is RNA (Mirkov et al., 1989 ▶; Ban et al., 1995 ▶).
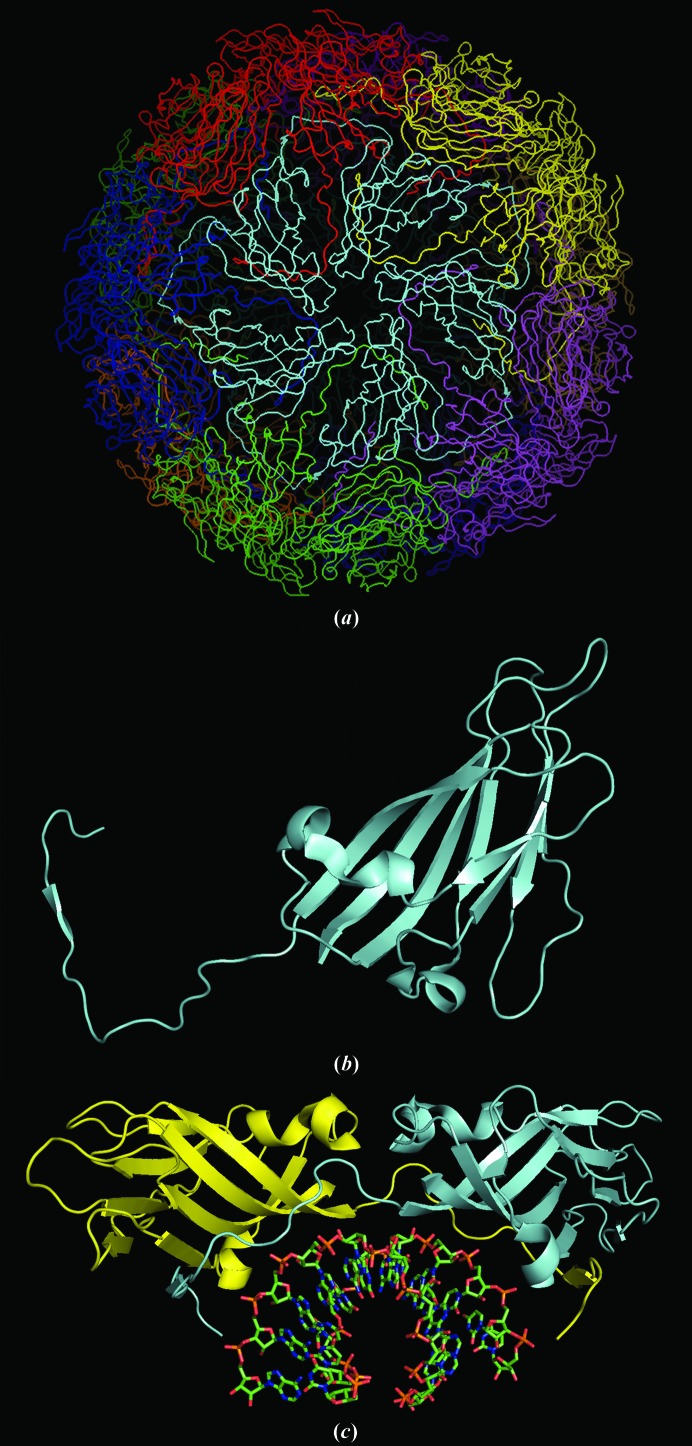
Figure 1.
STMV structure. In (a), the full capsid structure is illustrated. In (b), a monomer is shown with the extended N-terminal arm that wraps around its dimer partner. In (c), a dimer with its associated RNA double helix is displayed.
The structure of STMV was solved previously by isomorphous replacement and phase extension (Larson et al., 1993a ▶,b ▶). A noteworthy feature of that analysis was the appearance in the electron-density map of double-stranded helical segments of RNA at the 30 icosahedral dyad axes of the virus, constituting nearly 57% of the entire genome. Using, in part, unusually large crystals grown in microgravity aboard the US Space Shuttle (Day & McPherson, 1992 ▶), the virus particle, both protein and visible RNA, was refined using the program X-PLOR (Brünger, 1991 ▶; Brünger et al., 1987 ▶) to a resolution of 1.8 Å (Larson et al., 1998 ▶). Data were recorded from 21 different crystals at room temperature using a conventional rotating-anode source and a multiwire area detector (San Diego Multiwire Systems, San Diego, California, USA). The final R factor and R free for the model deposited in the Protein Data Bank (PDB; Berman et al., 2000 ▶; PDB entry 1a34) were 0.179 and 0.184, respectively.
1.2. Objectives
We subsequently devised conditions for cryo-preservation of large STMV crystals and collected X-ray diffraction intensities to 1.4 Å resolution from six flash-cooled crystals. An image of one frame of data can be seen in Fig. 2 ▶. With the acquisition of cryogenic data, we developed five principal objectives: (i) to increase the precision of the refined model, (ii) to determine how closely virus particles adhere to strict icosahedral symmetry, (iii) to obtain additional structural details of the protein capsid, (iv) to further elucidate the structure of the RNA within the capsid and (v) to compare the cryogenic crystal structure with the room-temperature structure. We re-refined the model against the original room-temperature data and against the cryogenic crystal data with a resolution cutoff of 1.8 Å and refined the model against the 1.4 Å resolution data, with the latter consisting of 570 721 independent reflections compiled from 4 434 593 intensity observations.
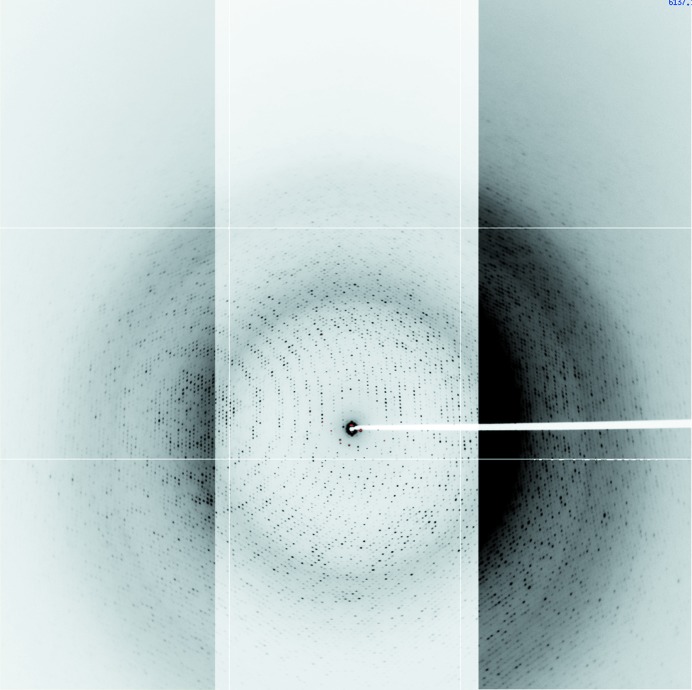
Figure 2.
Diffraction image from a crystal of STMV cooled to 100 K obtained on ALS beamline 8.2.2 using an ADSC 3 × 3 CCD detector at a 2Θ tilt of 10°. The image is a composite of three contrast levels in order to illustrate the resolution obtained near the edges of the detector. If the contrast level that was used for the right side of the image was used for the entire image, the center would be too dark to see any spots, as seen on the inside portion of the right side of the image. The maximum resolutions at the top and bottom corners are 1.25 and 1.44 Å, respectively; the resolutions at the upper, lower and side edges are 1.44, 2.05 and 1.66 Å, respectively.
Refinement was undertaken in the orthorhombic I222 crystal unit cell using four different procedures. The first utilized the program CNS with full NCS icosahedral constraints on the 15 protein subunits and associated RNA making up a crystallographic asymmetric unit. The final model, constrained to exact icosahedral symmetry, served as a baseline and provided common starting models for restrained refinement by each of the other approaches. In the fully constrained model every atom of every subunit in the asymmetric unit was obligated to exhibit icosahedrally symmetric identity in position and isotropic temperature-factor parameters. An important feature of the constrained refinement at 1.4 Å resolution is that for the 13 624 refined atomic parameters (including H atoms) there were 570 721 unique reflections, thereby yielding an X-ray observation:parameter ratio of almost 41.9.
The congruence of the four asymmetric units of the STMV particle is unequivocal as it is a consequence of crystallographic symmetry. The overall icosahedral symmetry of the particle, however, is imposed by biochemical factors, thus it need not be physically and mathematically exact. In the crystal, each protein subunit in the crystallographic asymmetric unit is exposed to a different chemical and physical environment, i.e. some subunits make direct lattice contacts with the protein shell of neighboring particles, while others are entirely exposed to solvent.
We believed it justified, therefore, to further refine STMV in a manner that allowed the atomic parameters within an asymmetric unit to vary among subunits. We present evidence below that this approach was proper. In restrained refinements strict icosahedral constraints were replaced by NCS restraints. Using the fully constrained model as a starting point, independent restrained refinements were carried out using CNS v.1.2 (Brünger et al., 1998 ▶), REFMAC5 v.5.5.0109 (Murshudov et al., 2011 ▶) and SHELX-97 (Sheldrick & Schneider, 1997 ▶), the first two of which employ maximum-likelihood target functions. The three procedures were carried out with each of the three data sets.
2. Materials and methods
2.1. Crystallization and data collection
STMV was purified from leaves of infected tobacco plants (Nicotiana tabacum) as described previously (Koszelak et al., 1989 ▶; Larson et al., 1993a ▶,b ▶) and was recrystallized four times by the addition of ammonium sulfate to 15% saturation in the presence of 0.05 M sodium phosphate pH 6.5. The X-ray diffraction data to 1.8 Å resolution used in the earlier study (Larson et al., 1993a ▶,b ▶, 1998 ▶) were recorded from STMV crystals at 290 K. These crystals were grown on earth by vapor diffusion in sitting drops (McPherson, 1999 ▶) and in microgravity (International Microgravity Laboratory I, January 1992) by liquid–liquid diffusion (Day & McPherson, 1992 ▶; Larson et al., 1998 ▶). High-resolution data collected from nine microgravity-grown crystals were combined with data from 12 earth-grown crystals (Larson et al., 1993a ▶,b ▶). The crystal data are summarized in Table 1 ▶. Crystals were mounted in quartz capillaries by conventional methods and data were collected on a San Diego Multiwire Systems (SDMS) area detector (Xuong et al., 1985 ▶). A Rigaku RU-200 rotating-anode generator fitted with a Supper graphite monochromator and operating at 45 kV and 40 mA provided Cu Kα radiation. The data were processed with the SDMS software.
Table 1. Summary of crystallization, data collection and processing for room-temperature and low-temperature data.
| Room-temperature data | Low-temperature data | ||
|---|---|---|---|
| Crystallization conditions | |||
| Temperature (K) | 296 | 296 | 296 |
| Crystallization conditions | Earth-grown by vapor diffusion in sitting drops | Microgravity-grown by liquid–liquid diffusion in cryostat | Earth-grown by vapor diffusion in sitting drops |
| Drop | 8% ammonium sulfate | 20% ammonium sulfate | 15% ammonium sulfate |
| Reservoir | 14–16% ammonium sulfate | None | 15% ammonium sulfate |
| pH | 6.5 | 6.5 | 6.5 |
| Growth period (d) | 14 | 14 | 14 |
| Data-collection parameters | |||
| Radiation source | Rigaku RU-200 Cu rotating anode, 45 kV, 40 mA | Rigaku RU-200 Cu rotating anode, 45 kV, 40 mA | Beamline 8.2.2, ALS synchrotron, λ = 1.06 Å |
| Detector | SDMS 2-detector system; distances of 930 and 985 mm | SDMS 2-detector system; distances of 940 and 1000 mm | ADSC Q315 3 × 3 CCD detector; distance of 205 mm |
| Monochromator | Supper graphite | Supper graphite | Double crystal, Si(111) |
| Scans | 45° ω scans in 0.10–0.12° increments | 50° ω scans in 0.10° increments | 0.333° ϕ scans |
| Frame collection times (s) | 120–200 | 300–360 | 45 |
| No. of crystals | 12 | 9 | 6 |
| Temperature (K) | 290 | 290 | 100 |
| Mounting | Quartz capillaries | Quartz capillaries | Cryoloops |
| Data-processing statistics | |||
| Resolution (Å) | 30.0–1.82 | 48.35–1.40 (1.45–1.40) | |
| R sym or R merge | 0.091 | 0.100 (0.354) | |
| 〈I/σ(I)〉 | 13.0 | 11.6 (2.9) | |
| Completeness (%) | 89.0 | 95.4 (67.3) | |
| Multiplicity | 8.2 | 7.23 (2.17) | |
| No. of observations | 2239616 | 4434593 | |
| Unique reflections | 271689 | 613720 | |
| Crystal data | |||
| Space group | I222 | I222 | |
| Unit-cell parameters (Å) | a = 174.27, b = 191.77, c = 202.50 | a = 172.69, b = 190.30, c = 201.69 | |
| Z (particles per cell) | 2 | 2 | |
For the collection of low-temperature data, STMV was purified as described above and crystallized by the sitting-drop vapor-diffusion method (McPherson, 1999 ▶) with 15% saturated ammonium sulfate as precipitant. Crystals were mounted in CryoLoops (Hampton Research, Aliso Viejo, California, USA) and passed briefly through a cryoprotectant composed of 30% MPD, 20% PEG 3350 and 15% saturated ammonium sulfate in water. The crystals were immediately flash-cooled in the nitrogen cold stream at the goniometer. Data were collected on beamline 8.2.2 at the Advanced Light Source, Berkeley, California, USA at 100 K using a 3 × 3 CCD detector (Advanced Detector Systems Corp., San Diego, California, USA) at a wavelength of 1.06 Å. The crystal-to-detector distance was 204.8 mm and the images were obtained as 0.333° ω scans.
A total of 848 images from six crystals were processed with d*TREK (Pflugrath, 1999 ▶), yielding 4 434 593 observations and 613 720 unique reflections to a resolution of 1.4 Å. With a variation in unit-cell parameters of less than 1%, the images from each crystal were corrected for absorption and scaled as a group but were not merged. The six groups were then scaled and merged as though they came from a single crystal. The R merge was 0.100 with an 〈I/σ(I)〉 of 11.6. Additional data statistics are presented in Table 1 ▶.
To obtain diffraction intensities from virus crystals, as a consequence of their large unit-cell parameters of generally hundreds of angstroms, virus crystals must be considerably larger in physical size than those of more conventional samples. This, we have found, explains the difficulty that investigators have had in cooling virus crystals, not the unique virus composition. Small virus crystals are no more difficult to freeze than are other macromolecular crystals. Our good fortune, which provided a major advance regarding STMV, was to identify conditions that allowed even large virus crystals to be flash-cooled. Using flash-cooled crystals reduced the multiplicity of scaling together data from a large number of crystals, as was necessary with the previous 290 K data collection. There, because of radiation-induced decay, data from 21 separate crystals were merged. Use of flash-cooled crystals also allowed us to utilize synchrotron X-rays with a minimum number of crystals as opposed to the conventional radiation source used previously.
In this investigation, the resolution of the diffraction data used was extended from the previous 1.8 to 1.4 Å, an increase of 0.4 Å. Because of the spherical nature of the diffraction pattern, this increment, in this case, led to a doubling of the number of unique diffraction intensities to 570 721. Furthermore, statistical measures (see Table 1 ▶) indicate that the additional data were comparable in quality in almost all ways to the data lying between the low-resolution limit and 1.8 Å. Although data were processed to 1.40 Å resolution, an initial phase extension from 10 to 1.4 Å showed a marked jump in the R factor from R ≃ 0.3 at 1.45 Å to R ≃ 0.7 at 1.44 Å. Therefore, data to 1.45 Å resolution were used for refinement purposes.
The test set for the room-temperature data set was the same as the set employed in the original structure refinement, which was randomly generated. Similarly, we used a randomly generated test set for the low-temperature data set as well. While this method may endow some bias to the test set owing to the high NCS present in the crystal, we believe that the 1.4 Å resolution mitigates to some extent the overfitting that some might fear. Furthermore, we use difference maps as justification for our conclusions, not decreases in R and/or R free.
2.2. Initial models
The starting model for constrained refinement was the previously described (Larson et al., 1998 ▶) 1.8 Å resolution model (PDB entry 1a34) with several modifications. In the capsid protein, we removed the first three amino-terminal residues because they were modeled based on very weak density. In the RNA, we also removed two nucleotides from each end of each strand for the same reason, leaving only six nucleotides per strand. Furthermore, the 1a34 model contained a sulfate ion on the icosahedral fivefold axis. We had reasonably assumed that this ion was sulfate in the earlier study because the crystals were grown from a 15% saturated ammonium sulfate solution. However, we subsequently grew crystals of STMV with a variety of salt precipitants and buffers that were devoid of sulfate and the electron density on the fivefold axis attributed to the anion did not appear to change. This ion is almost certainly phosphate, either from the plant source or from the extraction and purification steps, which utilized phosphate buffers throughout. For this analysis we therefore replaced the sulfate with a phosphate ion. Lastly, we removed from the starting model the putative ‘free nucleotide’ (Larson et al., 1998 ▶) that was not contiguous with any of the RNA helical segments. We added H atoms to the protein and RNA in ideal positions. When the constrained model was expanded to the 15-subunit crystallographic asymmetric unit, the singular subunit became subunit A and the 14 that were generated from it became subunits B through O, with subunits A–E, subunits F–J and subunits K–O comprising pentamers of the three-pentamer crystallographic asymmetric unit.
2.3. Comparison of refinement programs and procedures
The starting model was initially refined using CNS (Brünger et al., 1998 ▶) with rigorous icosahedral constraints; that is, the model was composed of a single capsid protein and its associated RNA strands, as well as the ions and solvent that make up the icosahedral asymmetric unit (1/60th of the virus particle). The model was refined against the room-temperature data at 1.8 Å resolution, as well as against the low-temperature data at both 1.8 and 1.4 Å resolution. During refinement, models were manually modified by the addition, deletion or adjustment of disordered residues, water molecules and ions. Initially, all water molecules were harmonically restrained to remain close to their initial positions as were the ions on the fivefold icosahedral axes. Icosahedrally averaged σA-weighted 2F o − F c and F o − F c maps were used to evaluate and rebuild models.
Upon convergence of the constrained refinements, the models were expanded to a crystallographic asymmetric unit consisting of 15 capsid proteins and 30 segments of RNA and associated water molecules and ions. Fastidious evaluation of the water molecules was required to eliminate duplicate and overlapped molecules that were inadvertently generated or that were found not to be in density in σA-weighted 2F o − F c maps. The expanded models for each data set were then refined with CNS using harmonic restraints for the water molecules and NCS restraints to force equivalent molecular entities (protein or RNA backbone) to be similar and with REFMAC5 (Murshudov et al., 2011 ▶) and SHELX-97 (Sheldrick & Schneider, 1997 ▶). No further rebuilding or any other manual intervention was imposed on the models in order to allow evaluation of the three refinement programs on their merits alone. Each has unique capabilities, some of which are presented in Table 2 ▶. Models were generated by an approach that matched the corresponding capabilities of the refinement programs so that comparisons could eventually be made between final models that were otherwise treated in similar ways.
Table 2. Programs used for refinement of STMV models and their capabilities.
| CNS | REFMAC5 | SHELXL-97 | ||||
|---|---|---|---|---|---|---|
| Features | Available | Utilized | Available | Utilized | Available | Utilized |
| Simulated annealing | Yes | No | No | — | No | — |
| Conjugate-gradient minimization | Yes | Yes | Yes | Yes | Yes | Yes |
| Full least squares | No | — | No | — | Yes | No |
| NCS constraints | Yes | Yes | No | — | No | — |
| NCS restraints | Yes | Yes | Yes | Yes | Yes | Yes |
| Simultaneous refinement of positions and B factors | No | — | Yes | Yes | Yes | Yes |
| Positional restraints | Yes (harmonic) | Yes | No | — | Yes (DAMP) | Yes |
| Fixed atomic positions | Yes | Sometimes | No | — | Yes | Sometimes |
| Bulk-solvent correction | Yes | Yes | Yes | Yes | Yes | Yes |
| TLS refinement | No | — | Yes | Yes | No | — |
| Atomic anisotropic B/U-factor refinement | No | — | Yes | Yes | Yes | Yes |
| Blocked parameter refinement | Yes | Yes | No | — | Yes | Yes |
| Refinement against F or F 2 | Both | Both | F | Yes | F 2 | Yes |
| Maximum-likelihood target | Yes | Yes | Yes | Yes | No | — |
| Direct Fourier summation or fast Fourier (FFT) | Both | FFT | FFT | FFT | Direct | Direct |
In order to evaluate the refinement programs and procedures solely on their performance, the starting models were the same (subject to the NCS treatment) and the deviations from ideality were roughly the same to ensure that the R factors were not affected in one program vis-à-vis the other programs owing to relaxed geometric restraints. To this end, for each model, r.m.s. deviations in bond lengths and angles were computed by the respective refinement program, by CNS (using the parameter files protein-allhdg.param and dna-rna-allatom.param) and by the validation server of the PDB (Berman et al., 2000 ▶). Automatic weighting in CNS resulted in r.m.s. deviations in bond lengths similar to those obtained in SHELX-97, and the weighting term in REFMAC5 was adjusted in order to obtain similar bond and angle r.m.s. deviations. By so doing, the stereochemical criteria applied to each of the models were comparable and therefore did not introduce ambiguity in comparing residuals produced by the various programs. In this way, the main criteria for program evaluation are the R and R free values under conditions in which the contents of the models are the same and the agreement of the stereochemistry to ideality is similar. The assessment of the program for efficiency was evaluated by time per cycle, however a cycle is defined for the program.
At the end of all refinements by all of the procedures, based on R free, we selected the best NCS-restrained model produced from the room-temperature data and the best constrained and restrained models emerging from the 1.4 Å resolution low-temperature data for further refinement. In the extended refinements of these three models (using REFMAC5, CNS and REFMAC5, respectively), we restored as many nucleotides, putative nucleotide fragments and additional solvent molecules as we could deduce from σA-weighted difference electron-density maps. These three final models were used for model analyses and to produce figures and have been deposited in the PDB (Berman et al., 2000 ▶).
2.4. The refinement programs
CNS allows refinement with both strict icosahedral NCS constraints and NCS restraints. In going from NCS constraints to NCS restraints, the number of parameters increases by approximately 15-fold as there are 15 subunits in the crystallographic asymmetric unit. For the 1.8 Å resolution refinements using REFMAC5, TLS refinement (Schomaker & Trueblood, 1968 ▶) allowed bulk anisotropic thermal motion modeling. TLS treatment by itself, applied to both the capsid proteins and the individual RNA strands, increased the number of refined parameters by 900.
For refinements using 1.4 Å resolution data, one objective was to investigate the value of treating model atoms as anisotropically vibrating scattering centers. At the time that we began these experiments, there were two programs from which to choose: REFMAC5 and SHELX-97 (others now exist). We used both programs so that we could evaluate which might produce the best results under conditions of high NCS. Anisotropic thermal factors applied to all atoms increased the number of parameters by a factor of 2.25 and reduced the X-ray observation-to-parameter ratio by about 50%.
The three programs emerged from different concepts. SHELX-97 is an enhancement of the small-molecule refinement program SHELX-76 (Sheldrick, 1976 ▶). It employs conventional Fourier summation and has full-matrix least-squares capability, although conjugate-gradient minimization has been implemented and, from an execution-time perspective, is the preferred mode for most routine macromolecular refinements. CNS was developed on the concept of simulated annealing, providing an increased radius of convergence that could overcome problems of models failing to escape local minima. REFMAC5 was created based on the idea that maximum-likelihood refinement would be more effective at dealing with errors than least-squares refinement, upon which most other macromolecular refinement packages were based.
For our purposes, we wanted not only to obtain the best model for the virus but also to determine which program provided the greatest efficiency and easiest application for virus structure refinement where high, but well defined, NCS was present. From this perspective, we considered how coordinates were handled, the speed of the refinement process and the capability of incorporating unique features of the virus structure. For STMV these included ions located on fivefold axes and RNA strands disordered about both crystallographic twofold axes and noncrystallographic icosahedral twofold axes. The latter were particularly troublesome because symmetry, both crystallographic and NCS, could cause nucleic acid strands to superimpose on one another. To carry out the refinements, array sizes were necessarily increased in both CNS and SHELX-97, but no modifications to REFMAC5 were necessary.
Table 2 ▶ presents additional features of the three programs and notes which of those features we invoked during refinement. The two internal procedures that most impact the speed of the programs are the Fourier calculations and the inversion of the normal matrix. As can be seen, direct summation is the only method of Fourier calculation available in SHELX-97, whereas CNS and REFMAC5 utilize fast Fourier transforms (Ten Eyck, 1977 ▶). Through conjugate-gradient minimization, all of the programs avoid calculating and inverting the full normal matrix, greatly increasing the efficiency. In addition, REFMAC5 uses a ‘sparse’ matrix to reduce the number of matrix elements calculated, which additionally speeds up refinement (see Table 3 ▶).
Table 3. Refinement results for the best models from each program and for each data set.
R.m.s. deviations are from the PDB validation server.
| Program | CNS | CNS | REFMAC5 | SHELX-97 |
|---|---|---|---|---|
| Refinement type | Constrained | Restrained | Restrained | Restrained |
| Room-temperature data to 1.8 Å resolution | ||||
| R | 0.172 | 0.145 | 0.136 | 0.148 |
| R free | 0.174 | 0.172 | 0.163 | 0.181 |
| R.m.s.d., bonds (Å) | 0.009 | 0.010 | 0.010 | 0.009 |
| R.m.s.d., angles (°) | 1.2 | 1.2 | 1.2 | 2.0 |
| Cryo data to 1.8 Å resolution | ||||
| R | 0.253 | 0.208 | 0.180 | 0.207 |
| R free | 0.255 | 0.241 | 0.216 | 0.237 |
| R.m.s.d., bonds (Å) | 0.010 | 0.010 | 0.010 | 0.008 |
| R.m.s.d., angles (°) | 1.3 | 1.3 | 1.2 | 1.8 |
| Cryo data to 1.4 Å resolution | ||||
| R | 0.278 | 0.238 | 0.229† | 0.258‡ |
| R free | 0.280 | 0.261 | 0.255 | 0.282 |
| R.m.s.d., bonds (Å) | 0.009 | 0.010 | 0.010 | 0.009 |
| R.m.s.d., angles (°) | 1.3 | 1.3 | 1.3 | 1.8 |
| Time per cycle (h) | 2.8 | 3.9 | 0.1 | 18.3 |
R = 0.215 and R free = 0.253 with anisotropic refinement.
R = 0.210 and R free = 0.278 with anisotropic refinement.
With all models, H atoms were removed before submitting coordinates to the validation server. This, in and of itself, could increase the r.m.s. deviations of bond lengths calculated by that server. For all programs the range of r.m.s. deviations in bond lengths was 0.007–0.008 Å. However, CNS, in calculating the r.m.s. deviations in bond lengths, produced a range of 0.007–0.010 Å; the validation server produced a range of 0.008–0.010 Å. The differences in the ranges are probably not significant. A similar observation also pertained when discrepancies in r.m.s. deviations in angles were compared. SHELX-97 parameterizes bond angles as distances between the non-central atoms of a bond angle rather than as the angle between the bonds. Therefore, r.m.s. deviations in angles are in effect r.m.s. deviations in distances. Hence, it was necessary to calculate the r.m.s. deviations in angles using either CNS or the validation server to compare SHELX-97-generated models with models from the other programs. It is apparent that the angle restraints in SHELX-97 are considerably more lax than in the other two programs. The SHELX-97 models had r.m.s. deviations for angles in the range 1.82–1.94° when calculated with CNS and 1.8–2.0° when calculated with the validation server. The CNS-refined models had r.m.s. deviations in the range 1.13–1.30°, which were very comparable to the values produced by the validation server (1.2–1.3°). The REFMAC5 values were also comparable to the values from the validation server, with a range of 1.06–1.18°. The supporting information in Supplementary Tables S1, S2 and S31 allows comparison of the thermal factors, r.m.s. deviations and R and R free values between the various refined models, respectively.
In addition to the refinement programs discussed above, we used Coot (Emsley & Cowtan, 2004 ▶) for model building and we created all figures except for Fig. 2 with PyMOL v.1.0 (DeLano, 2002 ▶). For model building, both averaged and non-averaged σA-weighted 2F o − F c and F o − F c maps were employed, with greater emphasis on the non-averaged maps for the restrained refinements.
2.5. Modeling of sulfate and magnesium ions on the fivefold axis
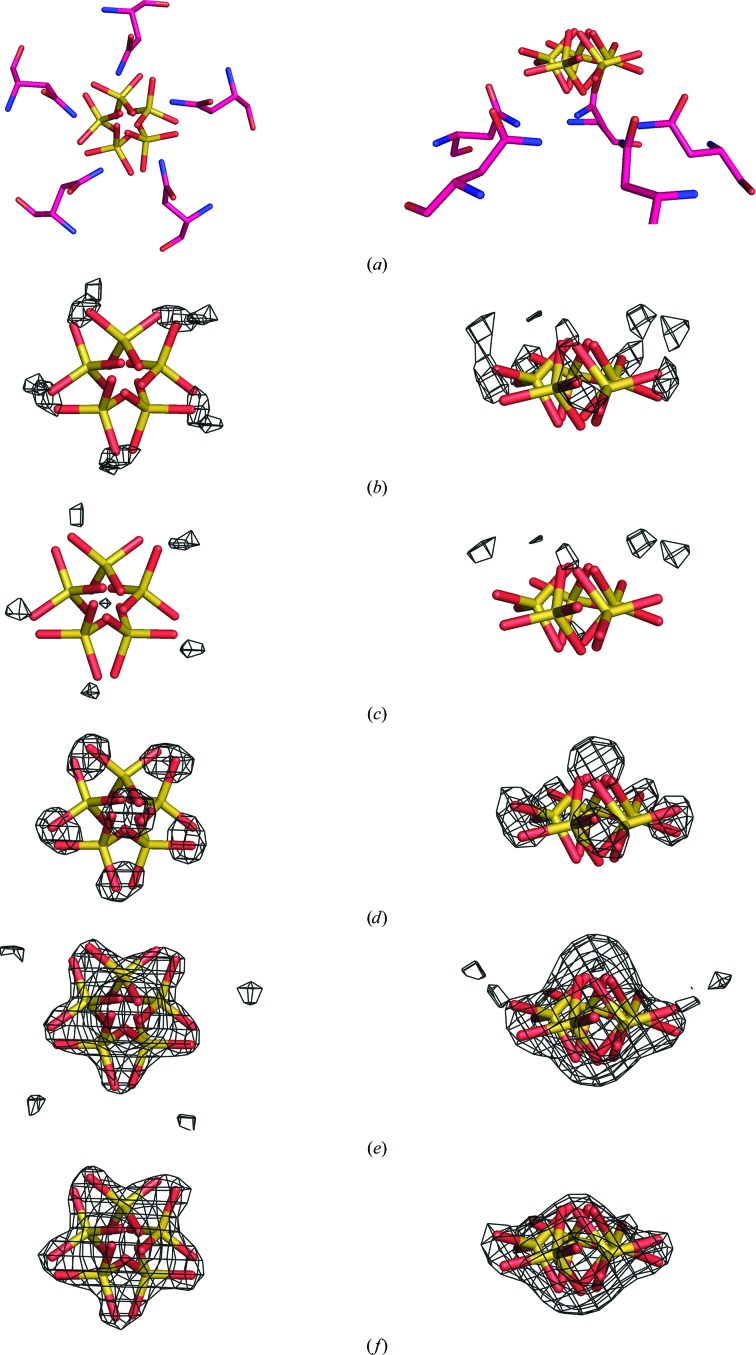
After the refinements of the initial models using the three programs, σA-weighted 2F o − F c and F o − F c maps phased by the best constrained and restrained models for the room-temperature and low-temperature data sets were analyzed for additional features. The maps for the room-temperature restrained model using REFMAC5 showed no appreciable density on the fivefold axis outside of the ring of Asn117 side chains. However, there was significant density at this position in the low-temperature maps. Owing to the crystallization conditions, this density was modeled as a sulfate ion with the S atom on the fivefold axis. However, the tendency was for the S atom to move off the fivefold axis during refinement. To investigate this behavior, we ran a series of refinements with both the constrained and restrained models, varying the entity at the sulfate ion position. Although the results were similar for both constrained and restrained treatments, we describe the procedure for the constrained case only. The entities were assigned occupancies of 0.2 and antibumping restraints between the NCS equivalents were turned off. The averaged difference maps from the constrained model are displayed in Fig. 3 ▶. In the series we had (i) the sulfate ion with the S atom fixed on the fivefold axis, (ii) the sulfate ion free to move off the axis, (iii) the sulfate ion replaced by a chloride ion, (iv) the sulfate ion replaced by a water molecule and (v) nothing at the sulfate position. As can be seen in Fig. 3 ▶(b), a fixed S atom located on the fivefold axis results in five density peaks in the vicinity of the O atoms of the final model. These peaks diminish in Fig. 3 ▶(c) when the S atom is allowed to move off the axis, allowing the O atoms to move into place. The full weight of the O atoms appears as strong density peaks in Fig. 3 ▶(d) when the O atoms are removed by replacing the sulfate ion with a chloride ion. When water or nothing occupies this site, as shown in Figs. 3 ▶(e) and 3 ▶(f), respectively, a single difference peak having the shape of a fivefold-disordered sulfate ion is obtained. We therefore feel confident that the off-axis sulfate ion model is the best representation for this position.
Figure 3.
Modeling of the sulfate ion on the fivefold axis. In all panels the sulfate ion on the fivefold axis from the final constrained model is shown. In (a), the surrounding Asn117 residues are also shown to illustrate the hydrogen bonding between the amide N atoms and the equatorial O atoms of the sulfate ion. On the left the view is down the fivefold axis and on the right it is perpendicular to the axis. In (b)–(f), σA-weighted F o − F c maps (contoured at 2.5σ) are superposed on the model, each phased by the constrained model in which the sulfate ion position was treated differently and then icosahedrally averaged. In (b), the map was phased by a model in which the S atom of the sulfate ion was fixed on the fivefold axis. In (c), the map was phased by a model in which the sulfate ion was not fixed on the fivefold axis. It is seen that the density around the equatorial O atoms decreases from (b) to (c). In (d), the map was phased by a model in which the sulfate ion was replaced by a chloride ion. The increased density around the equatorial O atoms reflects their absence in the model. In (e), the map was phased by a model in which the sulfate ion was replaced by a water molecule. In (f), the map was phased by a model in which nothing occupied the sulfate ion position. The density in both (e) and (f) suggests a sulfate ion with the S atom off the fivefold axis and the equatorial O atoms strategically located between the asparagine side chains to minimize steric clashes.
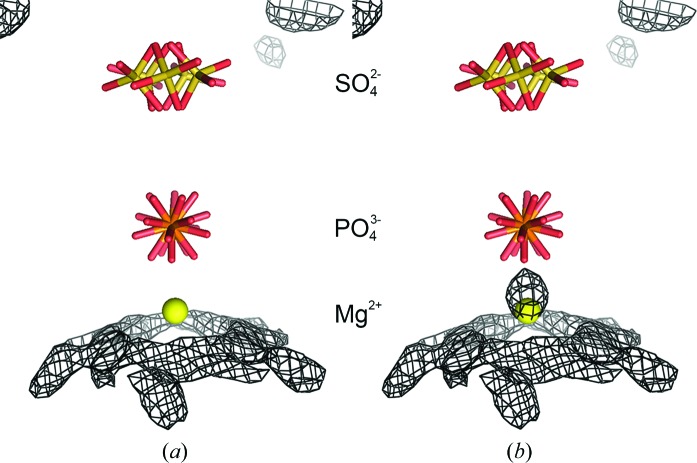
In the deposited model from which the initial models for this study were derived, there was a water molecule on the fivefold axis on the interior side of the phosphate ion. When this molecule was allowed to refine freely, it moved off the fivefold axis. When it was fixed on the fivefold, it refined to a position that was too close to the phosphate ion. Similar to the sulfate ion treatment described above, we tested various models for this site using water and magnesium ion and leaving the site vacant. In the constrained treatment, the water or magnesium ion had an occupancy of 0.2 and the antibumping restraints between their NCS equivalents were turned off. In the restrained treatment, the occupancies were set to 1.0 and there were no antibumping restraints to consider. Fig. 4 ▶(a) displays the averaged difference map resulting from the freely refined magnesium ion and shows that there is no density around the final magnesium position. On the other hand, Fig. 4 ▶(b) displays the averaged difference map resulting from the freely refined water molecule which moves off the fivefold and away from the phosphate ion. The residual density, as expected, lies between the final magnesium position and the phosphate ion, with the water molecule accounting for some of the density of the missing magnesium ion. Other scenarios that were tested gave residual difference density as expected for each scenario under the assumption that the site is occupied by a magnesium ion.
Figure 4.
Modeling of the magnesium ion on the fivefold axis. (a) and (b) show the final constrained model along the fivefold axis. The maps are averaged σA-weighted F o − F c maps contoured at 2.5σ. The map in (a) was phased by a model in which the magnesium ion was free to move along or off the fivefold axis. There is no negative density at the magnesium position nor is there positive density between the Mg2+ and the phosphate ions, suggesting that the entity at this position is modeled well by a Mg2+ ion. The map in (b) was phased by a model in which a water molecule was free to move along or off the fivefold axis. While the Mg2+ in (a) stayed on the fivefold axis, the water molecule moved off the axis by almost 0.5 and 0.71 Å from the Mg2+ position. As seen here, there was a large positive density peak at the site extending towards the phosphate ion, all of which suggests that water inadequately fits the data and that there is some entity here, such as an Mg2+ ion, that should be closer to the phosphate than water. The larger van der Waals contact distance for water accounts for the manner in which the water molecule refines away from the fivefold axis while producing difference density on the fivefold that is closer to the phosphate than a water can be.
2.6. PDB accession codes
Atomic coordinates and structure factors of the final models have been deposited in the PDB as the following entries: the model of STMV rigorously constrained to exact icosahedral symmetry using 1.4 Å resolution X-ray data from the cooled crystals as entry 4oq8, the model produced by NCS-restrained refinement using the previous 1.8 Å room-temperature data as entry 4nia and the 1.4 Å resolution model produced by restrained refinement using data from the flash-cooled crystals as entry 4oq9.
3. Results
3.1. Statistical comparison of refined models from different programs
The results of the individual refinements are presented in Table 3 ▶. For icosahedrally constrained refinement, the models produced by re-refinement here with CNS, using the identical room-temperature X-ray data to 1.8 Å resolution, can be compared with the previous refinement results using X-PLOR, which were also based on rigorous constraints, and those results with comparable CNS constrained refinement to the same resolution using the cryogenic crystal data. Neither REFMAC5 nor SHELX-97 incorporate completely constrained refinement and could not be used for this purpose. At 1.8 Å resolution, with all programs, both constrained and restrained refinements were restricted to positional parameters and isotropic B factors (see Supplementary Table S1), except for REFMAC5, in which TLS refinement was also utilized.
For the previous model refined with X-PLOR (Larson et al., 1998 ▶), the R and R free were 0.179 and 0.184, respectively. CNS constrained refinement against the same data produced a model with R = 0.172 and R free = 0.174. Despite the abbreviated model, the lower residuals may reflect an improved treatment of the bulk solvent. Constrained refinement using CNS at 1.8 Å resolution with data from cooled crystals yielded residuals that were significantly poorer than those for the room-temperature data, both from previous refinement with X-PLOR or current re-refinement with CNS. R and R free were 0.253 and 0.255, respectively, with comparable r.m.s. deviations in bond lengths and angles. Restrained refinement in CNS using 1.8 Å data produced uniformly improved residuals, both R and R free, for the 290 K data and the 100 K data. Comparison of results using the data from the two different temperatures, however, showed the same marked difference. For room-temperature data R = 0.145 and R free = 0.172 and for the cryogenic crystal data R = 0.208 and R free = 0.241.
With REFMAC5, even more significant improvements were realised using restrained refinement at 1.8 Å resolution at both temperatures. However, SHELX-97 produced results similar to CNS. As seen in Table 3 ▶, REFMAC5 yielded residuals of R = 0.136 and R free = 0.163 for the 290 K data and R = 0.180 and R free = 0.216 for the 100 K data, while SHELX-97 produced R = 0.148 and R free = 0.181 for the room-temperature crystals and R = 0.207 and R free = 0.237 for the cryogenically cooled crystals, respectively. It should be noted that throughout all of the refinements, superior residuals were consistently obtained with the old room-temperature data.
The noteworthy reductions in residuals, both R and R free, when constraints were liberalized and restrained refinement instituted, in all of the programs, indicated that exact icosahedral symmetry may not prevail within STMV particles, further suggesting that parameter variation among subunit polypeptides was appropriate. The particularly large improvement exhibited by REFMAC5 may also have been owing in part to its incorporation of TLS treatment.
No room-temperature data exist to 1.4 Å resolution, thus, with one exception, the only comparisons that can be made are the refinements using the three different procedures with restrained refinement against the data from the cryogenic crystals. The one exception is between the constrained and restrained refinements using CNS with 1.4 Å resolution cryogenic crystal data. In this comparison it can be seen in Table 3 ▶ that restrained refinement again produced improved residuals over those for the constrained case, an R of 0.238 versus an R of 0.278 and an R free of 0.261 versus an R free of 0.280. It is noteworthy that the residuals obtained with 1.4 Å resolution data versus 1.8 Å resolution data for the icosahedrally constrained case were not very different. This further indicates that the quality of the cryogenic measurements between 1.4 and 1.8 Å resolution were comparable to those between the low-resolution limit and 1.8 Å.
With restrained isotropic refinement and 1.4 Å resolution X-ray data, CNS, REFMAC5 (including TLS parameters) and SHELX-97 working R factors converged at 0.238, 0.229 and 0.258, respectively, and the corresponding R free values were 0.261, 0.255 and 0.282, respectively. From the residuals, and given comparable r.m.s. deviations for bond lengths and angles, REFMAC5 proved to be the most effective procedure. The elevated residuals for SHELX-97 may result from the less adequate angle parameterizations that produce r.m.s. deviations in angles that are at least 0.5° greater than those obtained with the other programs. Consideration of the cycle times (Table 3 ▶) required by each program further showed that the speed of REFMAC5 is far greater than that of either CNS or SHELX-97. It must be emphasized, however, that these results were obtained on a refinement problem exhibiting high NCS, and the outcomes could be different for more conventional crystallographic applications.
3.2. Structural features of the refined models
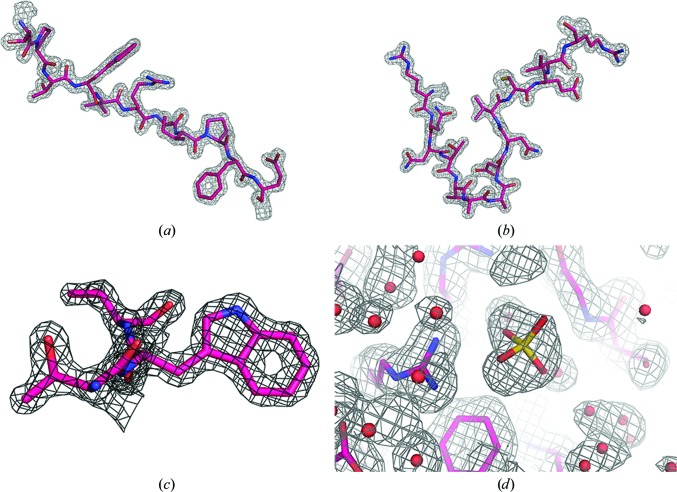
A salient question is whether new and useful structural details emerged as a consequence of the increase in resolution, or whether there was simply an enhancement of precision. Indeed, new and valuable structural information did appear, in addition to expected features such as greater definition of alternate side-chain conformations and a more complete and assured water structure. There were indeed numerous explicit examples of increased structural clarity or novelty. The following results are derived from the three final models, the refinement and model statistics of which are found in Table 4 ▶. Fig. 5 ▶ illustrates the general quality of the electron-density maps phased by the low-temperature restrained model.
Table 4. Model statistics for the three structures deposited in the Protein Data Bank.
H atoms were included in the models as riding atoms in REFMAC5 and refined with tight stereochemical restraints in CNS.
| Room temperature | Low temperature | ||
|---|---|---|---|
| Refinement program | REFMAC5 | CNS | REFMAC5 |
| Refinement mode | NCS restrained | NCS constrained | NCS restrained |
| Resolution (Å) | 87.04–1.82 | 48.35–1.45 | 48.35–1.45 |
| No. of reflections (work/test) | 264332/3984 | 542362/28359 | 542362/28359 |
| R work/R free | 0.123/0.156 | 0.278/0.279 | 0.217/0.250 |
| No. of refined atoms | |||
| Protein (non-H/H) | 18231/18751 | 1205/1241 | 18067/18670 |
| RNA (non-H/H) | 6720/3300 | 448/219 | 6720/3285 |
| Ions | 71 | 16 | 233 |
| Water | 3381 | 277 | 4276 |
| B factors 〈B〉 (Å2) | |||
| Protein | 22.0 | 25.4 | 27.4 |
| RNA | 108.2 | 72.8 | 112.6 |
| Ions | 25.7 | 42.0 | 61.4 |
| Water | 41.8 | 45.1 | 42.7 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.009 | 0.007 | 0.009 |
| Bond angles (°) | 1.26 | 1.28 | 1.25 |
Figure 5.
Examples of the quality of the electron-density maps. The maps in (a), (b) and (c) are F o maps phased by the low-temperature restrained model. The fragment in (a) is residues 34–44 and the map is contoured at 2.2σ, in (b) the fragment contains residues 131–144 and the map is contoured at 2.0σ, and (c) displays residues 36–38 with the map contoured at 2.5σ. In (d), a sulfate ion bound to Arg95 in the restrained low-temperature model is shown superposed on the density of the non-averaged σA-weighted 2F o − F c map contoured at 1.0σ.
3.3. The free nucleotide
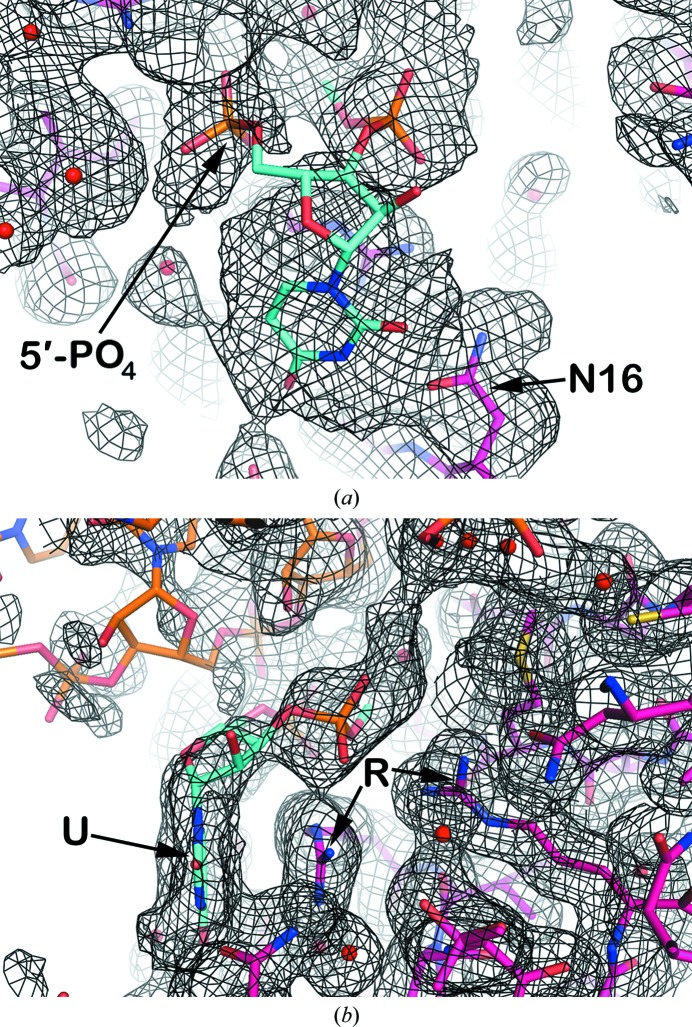
In earlier reports (Larson et al., 1998 ▶; Larson & McPherson, 2001 ▶), we described a singular nucleotide, which we termed the ‘free nucleotide’, that was not contiguous with any helical segment of RNA. The nucleotide was particularly significant because it suggested the course of the single-stranded loop that closed the end of a helix (Larson & McPherson, 2001 ▶). In the earlier analysis, only the ribose and the 5′-phosphate group of the nucleotide, pinned by the side chains of two arginine residues, was visible in electron-density maps. The continued course of the polynucleotide strand in both directions away from this nucleotide was at best ambiguous.
The current results, illustrated by Fig. 6 ▶, makes visible the entire ‘free nucleotide’ including the base, and brings increased clarity to the path of the single-stranded polynucleotide through the ‘free nucleotide’. It can now be seen that the base is stacked upon the guanidinium group of Arg125. In addition, the side chain of Asn16 is nearly coplanar with and approaches the edge of the base, where it could be oriented to form hydrogen bonds with appropriate atoms on the nucleotide base. The latter feature, as proposed by Seeman et al. (1976 ▶), could thereby provide some degree of base specificity at the ‘free nucleotide’ position.
Figure 6.
The free nucleotide fragment. Displayed is the low-temperature restrained model superposed on averaged σA-weighted 2F o − F c density phased by a model which did not contain the free nucleotide. The density for both panels is contoured at 0.2σ. The free nucleotide is shown in cyan, the protein is shown in magenta, the RNA is shown in orange and water molecules are shown as red spheres. The free nucleotide is modeled with a uridine base, but the density could conceivably accommodate any base, and both the 5′-phosphate and 3′-phosphate groups are included. In (a), Asn16 is identified by ‘N16’ and can possibly interact with the uridine base through its side chain. In (b), the uridine base, identified by the symbol ‘U’, is stacked against the guanidinium group of Arg125. Arg125 and Arg131, identified by the symbol ‘R’, are hydrogen-bonded to the 3′-phosphate of the ‘free nucleotide’.
3.4. The sulfate, magnesium and phosphate ions
Two anions appeared in the high-resolution electron-density maps that were not apparent in previous room-temperature maps. One of these is of rather little consequence and is shown in Fig. 5 ▶(d) to be bound by the guanidinium group of Arg95 on the exterior of the molecule. It is not likely to have any functional significance. The second anion, however, appears to represent a general anion-binding site, as it can be replaced by halides and other anions when different mother liquors are utilized. This site lies on an icosahedral fivefold axis and may be functionally important, as it completes a rather intricate arrangement of ions that extends from the interior of the particle, through a pore in the protein shell, to the exterior.
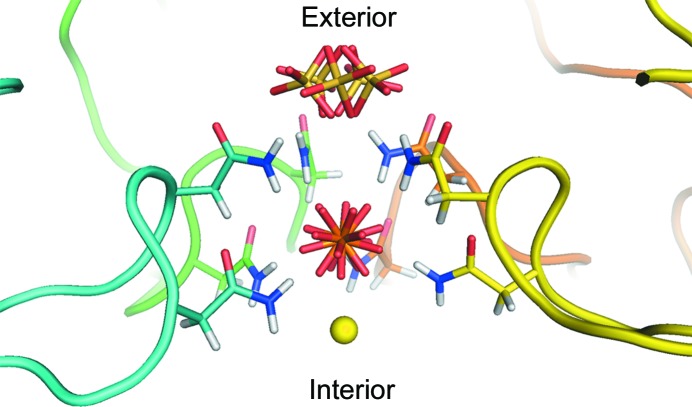
Fig. 7 ▶ shows this pore and the associated ions, which are presented in a manner reflecting their modes of disorder as deduced from difference Fourier syntheses (see §2.5). At the most interior is a magnesium ion on the fivefold axis, the coordination of which is not clear. As seen in Fig. 4 ▶, there is a ring of smeared density just below the ion, which has peaks corresponding to water molecules coordinated to the amide N atoms of the Asn115 residues and the hydroxyls of Thr118 residues, both of which cannot be occupied simultaneously; hence, the density is smeared in a ring around the fivefold axis near these ten residues. The coordinating waters around the magnesium ion must be so severely disordered that no density is seen at all. Above the magnesium ion is the phosphate ion, which was previously thought to be sulfate but is almost certainly phosphate. The P atom lies on the fivefold axis and is surrounded by a ring of five asparagine (Asn115) side groups below it and another ring of five asparagine (Asn117) side chains above. The four O atoms of the phosphate are hydrogen-bonded variously to the amide N atoms of the ten asparagine residues. Hence, the phosphate has a multitude of rotational dispositions. Whether these are dynamic in character or static or both we cannot determine.
Figure 7.
The pore along the fivefold axis. The pore is viewed perpendicular to the axis. The forward-most subunit of the pentamer forming the pore has been removed for clarity. The backbone of each subunit is shown in a different color and only the side chains of the asparagine residues are shown. The Asn115 (lower residues) and Asn117 (upper residues) side chains form a cage around the phosphate ion. This ion appears to be tightly bound through ten hydrogen bonds formed by the amide N atoms of the asparagine residues. As far as we can tell, despite the disorder of the O atoms, the P atom lies on the fivefold axis. Although we originally modeled this position as a sulfate owing to the fact that the crystals were grown from ammonium sulfate, we believe that the position is probably occupied even within the plant and most probably by phosphate, which is abundant in plants. Furthermore, virus preparation involves the use of phosphate buffers that would also be a source of phosphate. Crystallographically, phosphate and sulfate are indistinguishable if both are present. At the outward side of the pore is an ion modeled as a positionally disordered sulfate because we believe that it is easily exchanged and thus is an artifact of the crystallization conditions. On the inward side of the pore we modeled a magnesium ion bound to the phosphate.
Above the upper ring of asparagine (Asn117) residues lies a newly observed sulfate anion and it is fixed there by hydrogen-bonding interactions with the five Asn117 side chains that form a crown slightly below it. The distances are about 3.34 Å from the nearest discrete water of the water ring to the magnesium ion, 3.35 Å from the magnesium to the P atom and 5.72 Å from the P atom to the average S atom position. The sulfate ion cannot simultaneous hydrogen-bond to all five of the Asn117 side chains, and like the phosphate, has rotational freedom. A meticulous analysis of the sulfate disposition using difference Fourier syntheses (see §2.5) was carried out, and this showed unambiguously that the S atom does not, in fact, lie exactly on the fivefold axis, but lies at five equivalent off-axis positions in order to form its hydrogen bonds. In Fig. 7 ▶, therefore, this disorder is shown by the five equivalent sulfate positions. Similarly, assignment of the cation on the fivefold axis, which might otherwise have been assumed to be a water molecule, was also a consequence of another series of difference Fourier maps (see §2.5). The difference Fourier analyses that identified these ions and their dispositions were only possible because of the extended resolution of the X-ray data.
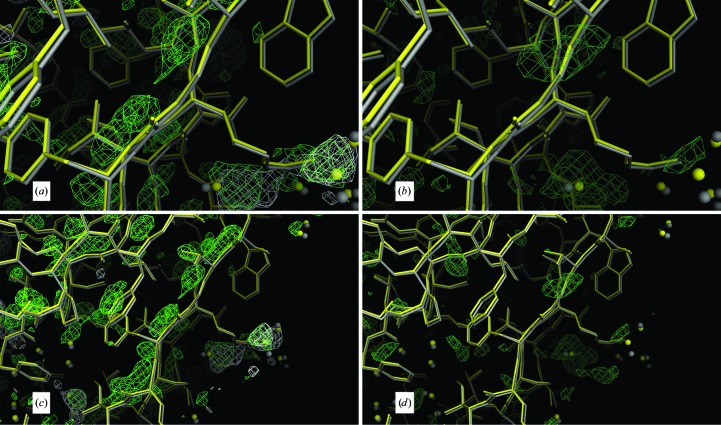
3.5. Distortion from strict icosahedral symmetry
The question of how closely the virus particle obeys strict icosahedral symmetry when embedded in the crystal lattice was also answered by the refinement results in combination with σA-weighted difference Fourier syntheses. These maps used phases based on the NCS-constrained model or the restrained model. Direct comparison of the restrained model produced by REFMAC5 with the constrained structure showed that overall there was little change in the restrained model from ideal symmetry. The r.m.s. deviations in Cα positions between corresponding subunits of the two models ranged from 0.08 Å to only 0.208 Å. Centroid differences of the 15 subunits in the asymmetric unit of the restrained model from strict icosahedral symmetry varied from 0.058 to 0.199 Å (see Supplementary Table S4).
In the crystal, there are only two significant lattice contacts; these are between subunits K and D (see §2.2) and between subunits N and F. The exteriors of all other subunits are mostly exposed to solvent. This means that any one particle in the lattice has a total of 16 subunits involved in interparticle contacts. Although the centroids of the contacting subunits show no exceptional movement from ideality, the largest deviations from the constrained model of individual Cα atoms, and side-chain atoms, are almost all in residues involved in interparticle contacts. In these contact areas, maximum Cα deviations range from 0.5 to 0.6 Å, whereas side-chain atoms have deviations as large as 2.8 Å (Supplementary Table S4).
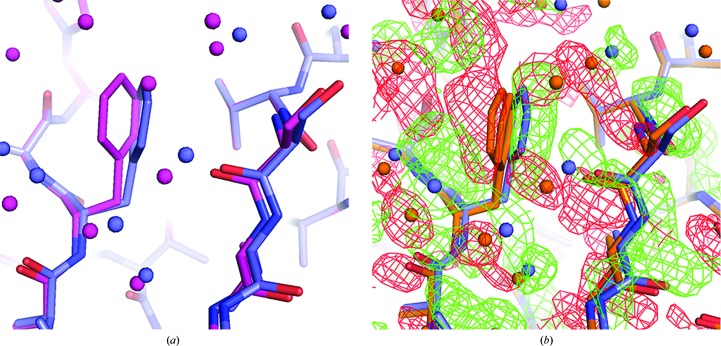
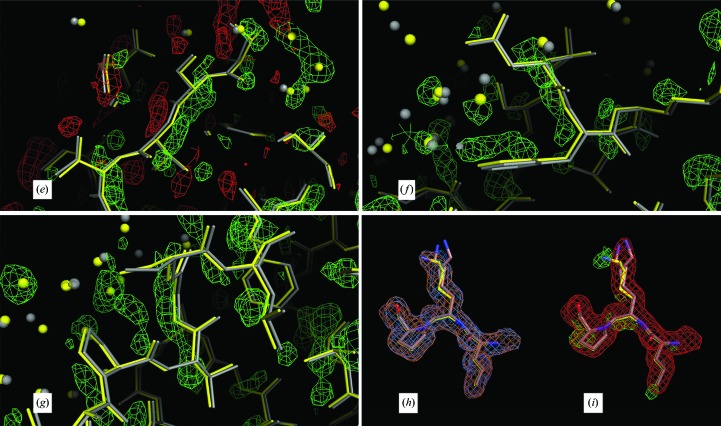
An example of the differences from ideality revealed by the restrained refinements is presented in Figs. 8 ▶(a) and 8 ▶(b), which involves a phenylalanine side chain (Phe53) and main-chain atoms in the contact area between subunits D and K. Difference Fourier syntheses with phases based on the constrained model clearly show the shift in positions away from perfect symmetry. In Fig. 9 ▶(a), a σA-weighted F o − F c Fourier map using phases from the constrained model shows the map to be cluttered with positive difference density. By icosahedrally averaging the difference map in Fig. 9 ▶(a) these differences are largely reduced (Fig. 9 ▶ b), and they are almost eliminated (Fig. 9 ▶ c) when the Fourier map is produced from the restrained model. Further examples of icosahedral symmetry distortion are presented in Fig. 10 ▶, in which positive difference density parallel to the main chain or lengthy side chains indicate an inadequate fit of the constrained model to the difference map (Figs. 10 ▶ a–10 ▶ g). Figs. 10 ▶(h) and 10 ▶(i) illustrate the difference between a side-chain conformation of the restrained model and the average conformation of the constrained model. Although it might be argued that the restrained model overfits the data and that the significant decrease in R and R free are a consequence of overfitting, the significant improvement in difference maps are our main justification for restrained refinement, and a significant portion of the decrease in the R factors results from real improvement in the model that reduces or eliminates the difference density seen in the difference maps generated from the constrained model.
Figure 8.
Particle packing-related disruption of NCS symmetry. The deviation from strict icosahedral symmetry owing to particle packing is illustrated. (a) Superposition of subunit K (blue) of the restrained low-temperature model using selected Cα atoms onto subunit A (magenta) of the same model showing the difference in the phenyl ring orientation of Phe53. The water molecules are colored according to the color of the chain with which they are associated. Subunit K makes major contacts with a neighboring particle in the crystal lattice, whereas subunit A makes only minor contacts. (b) The volume around the Phe53 of subunit K with density from the non-averaged σA-weighted F o − F c map phased by the low-temperature constrained model shown in orange. Subunit K of the low-temperature restrained model is shown in blue. Phe53 of the constrained model was refined as disordered to compensate for the differences between the subunits that interact with adjacent particles and those that do not. However, it can be seen that the disordered model does not adequately fit the restrained model. The difference density, with green being positive density and red being negative density, shows that while the disordered model may be better than a single phenyl ring position, it still does not adequately account for the density around the Phe53 side chain: the model places too much density on the left side of the phenyl ring (red density) and not enough on the right side (green density).
Figure 9.
General breakdown of strict icosahedral symmetry. The general breakdown of the icosahedral symmetry at 1.4 Å resolution is illustrated by subunit M, which exhibits the greatest r.m.s. deviation between the Cα positions of corresponding subunits of the expanded constrained model and the restrained model (see Supplementary Table S4). In (a), the non-averaged σA-weighted difference map using the low-temperature constrained model phases is displayed. In (b), the map in (a) was averaged and demonstrates that much of the error in the model has been averaged away. In (c), the σA-weighted difference map, phased by the low-temperature restrained model, exhibits the least amount of density and hence suggests that the model is a better model. Density in all panels is contoured at 2.5σ.
Figure 10.
Further evidence in support of a restrained model. In (a)–(i), the low-temperature constrained (gray) and restrained (yellow) models are displayed. All maps are σA-weighted Fourier maps, with the 2F o − F c maps contoured at 1.0σ and the F o − F c maps contoured at 2.5σ. (a)–(g) show chain M and (h) and (i) show chain I. (a)–(d) show the segment V122-G-T124 in the foreground. In (a) and (c), the maps that are superposed on the models are the non-averaged F o − F c density (light green) and the averaged F o − F c density (in white) phased by the constrained model. In (b) and (d), the model is superposed on the non-averaged F o − F c density phased by the restrained model. In (e), the segment T71-S-V-T74 is superposed on the non-averaged F o − F c density phased by the constrained model, with negative density displayed in red and positive density in light green. In (f), the segment N64-F65 is shown superposed on the positive density of the non-averaged F o − F c map phased by the constrained model. In (g), the segment V57-G-S59 is superposed on the positive density of the F o − F c map phased by the constrained model. Chain M has the largest r.m.s. deviation of 0.208 Å between the restrained and constrained models. In (a), (c), (e), (f) and (g) the non-averaged difference map (light green) suggests that a deviation from strict icosahedral noncrystallographic symmetry exists because it always lies on the opposite side of the restrained model (yellow) from the constrained model (gray), suggesting that the constrained model should move toward the restrained model to accommodate the difference density. In (a) and (c) the density of the averaged difference map (white) has been averaged out around the chains, leaving only random difference peaks away from the chain. In (b) and (d) the restrained refinement has produced a cleaner difference map without the density around the chain as the chain moved towards the density seen in (a) and (c). In (h), the fragment L96-K97-P98 is shown superposed on the non-averaged constrained 2F o − F c (orange) and the non-averaged restrained 2F o − F c (blue) electron-density maps. Since these two maps match well, the density color appears almost white. In (i), the non-averaged constrained F o − F c map (green) and the averaged constrained 2F o − F c map (red) are shown. Clearly, from the difference density and the non-averaged 2F o − F c density, Lys97 on chain I does not have the same conformation as the refined constrained model, which was modeled with alternate conformations, neither of which coincide with the conformation found in chain I.
4. Discussion
4.1. Virus-particle distortion from strict icosahedral symmetry
Previous investigations have challenged the dogma that spherical viruses are exactly icosahedral at the molecular and atomic level even when in a crystal. Indeed, we have provided at least two examples where this was not the case (Larson et al., 2005 ▶; Makino et al., 2013 ▶). In light of the different subunit environments arising from packing interactions that can, in turn influence local protein conformation, the restrained refinements we carried out are, if anything, conservative. The deviations of general atomic positions were ultimately only marginally significant. Although the particles after restrained refinement are not perfect icosahedra, they remain nearly so, and what deviations there are from ideality are entirely explicable.
In the crystal, STMV virions make intimate contact along unit-cell body diagonals (111 directions). Residues at these interparticle interfaces do not have the same chemical and physical environments as do residues entirely exposed to solvent, as are found elsewhere on particle surfaces. Rigorously constrained refinement is therefore not adequate to produce a model of the virus as it exists in the crystal lattice. Other investigators have come to similar conclusions and for the same reasons. A Satellite tobacco necrosis virus-like particle produced in Escherichia coli, for example, was refined to high resolution using restraints (Lane et al., 2011 ▶).
4.2. Non-modeled RNA volume
At the end of refinements, σA-weighted 2F o − F c and F o − F c difference Fourier maps showed the interior space, separate from that occupied by the model, still to be cluttered with irregular patches and discontinuous density that were unrecognizable in chemical terms. Roughly 45% of the nucleotides unaccounted for in electron-density maps were statistically disordered, and these filled a substantial part of the interior of the virus. The resulting patchwork of density cannot be modeled by components of RNA because it is a molecular amalgam. A single bulk-solvent correction was only marginally adequate compensation because it assumed a uniform density, which the disordered RNA is not. We were therefore obligated to neglect the residual density. The missing RNA density, although 60-fold averaged, does however contribute to the X-ray scattering; thus, some fraction of the residuals is undoubtedly attributable to the spatially disordered RNA. Essentially the same is true for the disordered amino-terminal polypeptides. Although not lost owing to icosahedral averaging, the tails assume multiple conformations (Day et al., 2001 ▶) but still contribute to X-ray scattering. Their statistical spatial average is also too diffuse to be modeled.
4.3. Data quality
A curious feature of our results is that the 290 K data, collected in 1991–1992 and recorded with conventional X-ray sources and more primitive detector technology and scaled together from a large number of crystals, were consistently superior to the data collected using synchrotron radiation and a CCD detector. However, the two data sets were quite comparable in terms of redundancy, estimated errors and other statistical measures (see Table 1 ▶), so it is difficult to argue that the data from the cooled crystals were simply ‘poorer data’.
We believe that the higher R factors produced from the cooled crystals were attributable to perturbations introduced by the cooling process. Indeed, there is recent work indicating that room-temperature X-ray data have a higher information content than do data from cooled crystals (Fraser et al., 2011 ▶). It should not be neglected, however, that the high-resolution reflections for the room-temperature data set, and a substantial portion of lower resolution intensities as well, were recorded from STMV crystals that were grown in microgravity (Day & McPherson, 1992 ▶). Thus, the possibility cannot be ignored that the superiority of the room-temperature X-ray data arises not exclusively from the absence of cooling damage, but at least in part from positive effects of the absence of gravity on the inherent structure of the crystals. There is some evidence from previous microgravity crystallization experiments that this may be the case (Day & McPherson, 1992 ▶; McPherson, 1996 ▶).
Currently, the PDB-deposited models (§2.6) represent the most comprehensively refined and the highest resolution models of a naturally occurring virus. We believe the analysis presented here also serves as the most thorough comparison of current macromolecular crystallographic refinement procedures when applied to structures having high degrees of noncrystallographic symmetry.
Supplementary Material
Four tables that provide information to compare the different refinement procedures employed in this study. . DOI: 10.1107/S1399004714013789/mn5065sup1.pdf
PDB reference: Satellite tobacco mosaic virus, 4nia
PDB reference: 4oq8
PDB reference: 4oq9
Acknowledgments
This research was supported in part by a grant to AM from the NIH (GM080412).
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: MN5065).
References
- Ban, N., Larson, S. B. & McPherson, A. (1995). Virology, 214, 571–583. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Brünger, A. T. (1991). Annu. Rev. Phys. Chem. 42, 197–223.
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Brünger, A. T., Kuriyan, J. & Karplus, M. (1987). Science, 235, 458–460. [DOI] [PubMed]
- Caspar, D. L. & Klug, A. (1962). Cold Spring Harb. Symp. Quant. Biol. 27, 1–24. [DOI] [PubMed]
- Day, J., Kuznetsov, Y. G., Larson, S. B., Greenwood, A. & McPherson, A. (2001). Biophys. J. 80, 2364–2371. [DOI] [PMC free article] [PubMed]
- Day, J. & McPherson, A. (1992). Protein Sci. 1, 1254–1268. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fraser, J. S., van den Bedem, H., Samelson, A. J., Lang, P. T., Holton, J. M., Echols, N. & Alber, T. (2011). Proc. Natl Acad. Sci. USA, 108, 16247–16252. [DOI] [PMC free article] [PubMed]
- Koszelak, S., Dodds, J. A. & McPherson, A. (1989). J. Mol. Biol. 209, 323–325. [DOI] [PubMed]
- Lane, S. W., Dennis, C. A., Lane, C. L., Trinh, C. H., Rizkallah, P. J., Stockley, P. G. & Phillips, S. E. V. (2011). J. Mol. Biol. 413, 41–50. [DOI] [PubMed]
- Larson, S. B., Day, J., Greenwood, A. & McPherson, A. (1998). J. Mol. Biol. 277, 37–59. [DOI] [PubMed]
- Larson, S. B., Koszelak, S., Day, J., Greenwood, A., Dodds, J. A. & McPherson, A. (1993a). J. Mol. Biol. 231, 375–391. [DOI] [PubMed]
- Larson, S. B., Koszelak, S., Day, J., Greenwood, A., Dodds, J. A. & McPherson, A. (1993b). Nature (London), 361, 179–182. [DOI] [PubMed]
- Larson, S. B., Lucas, R. W. & McPherson, A. (2005). J. Mol. Biol. 346, 815–831. [DOI] [PubMed]
- Larson, S. B. & McPherson, A. (2001). Curr. Opin. Struct. Biol. 11, 59–65. [DOI] [PubMed]
- Makino, D. L., Larson, S. B. & McPherson, A. (2013). J. Struct. Biol. 181, 37–52. [DOI] [PMC free article] [PubMed]
- McPherson, A. (1996). Crystallogr. Rev. 6, 157–308.
- McPherson, A. (1999). Crystallization of Biological Macromolecules. Cold Spring Harbor Laboratory Press.
- Mirkov, T. E., Mathews, D. M., Du Plessis, D. H. & Dodds, J. A. (1989). Virology, 170, 139–146. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Schomaker, V. & Trueblood, K. N. (1968). Acta Cryst. B24, 63–76.
- Seeman, N. C., Rosenberg, J. M. & Rich, A. (1976). Proc. Natl Acad. Sci. USA, 73, 804–808. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (1976). SHELX-76. Program for Crystal Structure Determination. University of Cambridge.
- Sheldrick, G. M. & Schneider, T. R. (1997). Methods Enzymol. 277, 319–343. [PubMed]
- Ten Eyck, L. F. (1977). Acta Cryst. A33, 486–492.
- Valverde, R. A. & Dodds, J. A. (1986). J. Gen. Virol. 67, 1875–1884.
- Valverde, R. A. & Dodds, J. A. (1987). J. Gen. Virol. 68, 965–972.
- Valverde, R. A., Heick, J. A. & Dodds, J. A. (1991). Phytopathology, 81, 99–104.
- Xuong, N. H., Nielsen, C., Hamlin, R. & Anderson, D. (1985). J. Appl. Cryst. 18, 342–350.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Four tables that provide information to compare the different refinement procedures employed in this study. . DOI: 10.1107/S1399004714013789/mn5065sup1.pdf
PDB reference: Satellite tobacco mosaic virus, 4nia
PDB reference: 4oq8
PDB reference: 4oq9