Abstract
Mobilization of the staphylococcal plasmid pC221 requires at least one plasmid-encoded protein, MobA, in order to form a relaxosome. pC221 and closely related plasmids also possess an overlapping reading frame encoding a protein of 15 kDa, termed MobC. By completing the nucleotide sequence of plasmid pC223, we have found a further example of this small protein, and gene knockouts have shown that MobC is essential for relaxosome formation and plasmid mobilization in both pC221 and pC223. Primer extension analysis has been used to identify the nic site in both of these plasmids, located upstream of the mobC gene in the sense strand. Although the sequence surrounding the nic site is highly conserved between pC221 and pC223, exchange of the oriT sequence between plasmids significantly reduces the extent of relaxation complex formation, suggesting that the Mob proteins are selective for their cognate plasmids in vivo.
The horizontal transfer of plasmid DNA by conjugative transfer requires two distinct processes: the formation of a mating pair between donor and recipient bacteria, and a series of DNA processing reactions to prepare the plasmid for transfer (45, 65, 66). The latter requires the formation of a protein-DNA complex at the origin of transfer (oriT), termed the relaxosome (10, 31), requiring a plasmid-encoded tyrosine transesterase (also referred to as a relaxase, or Mob protein) capable of introducing a single-stranded nick within oriT. This nick permits the transfer of single-stranded DNA into the recipient and may be captured by sodium dodecyl sulfate (SDS), alkali, heat, or protease treatment of the complex in vitro.
Relaxases typically carry three conserved amino acid sequence motifs (23, 27) related to those required for rolling circle replication initiator (Rep) proteins: these contain (motif I) the sequence surrounding the active site tyrosine, which nicks the DNA and typically forms a phosphodiester linkage with the nucleotide at the 5′ side of the nick; (motif II) a broadly hydrophobic region implicated in maintaining the stability of the trapped nick by the relaxosome (46); and (motif III) the histidine-hydrophobic-histidine motif responsible for binding the divalent metal cation essential for the cleavage process (22). Relaxases may be further classified into families based on both the amino acid sequence within these motifs and the nucleotide sequence surrounding the nick site: these include those related to the IncP plasmids such as TraI of RP4 (68) and VirD2 of the agrobacterial Ti plasmids (67); the IncQ plasmids such as MobA of RSF1010 and R1162 (54) and the Nes protein of staphylococcal pGO1 (13); the IncF/IncW family, including TraI of R1 and F (17) and TrwC of R388 (32); and relatives of the MobM protein of streptococcal plasmid pMV158 (48). Further characteristics are associated within these family groupings: for example, the IncQ-family MobA polypeptides include a primase activity which may be separated from the relaxase activity (7); and the IncF/IncW family TraI/TrwC polypeptides include a distinct helicase domain (11, 34, 60).
In addition to the relaxases, formation of the relaxosome typically requires one or more additional accessory proteins, usually encoded in cis with the relaxase. Thus, RP4 requires both TraI and the accessory TraJ proteins (18), the Ti plasmids require VirD2 and VirD1 (16), and RSF1010 utilizes MobA with MobC (58). No such partner has yet been identified for MobM of pMV158, but while the F plasmid TraI and R388 TrwC have both been observed to cleave plasmid DNA without accessory proteins in vitro (33, 36) the presence of accessory proteins TraY, TraM, and TrwA and chromosomally encoded IHF are required for optimal activity in vivo (25, 30, 37).
Compared to the gram-negatives, there are relatively few examples of well-characterized self-transmissible or mobilizable plasmids from the gram-positives (20), which includes pGO1 and pMV158 described above. The subject of the present study, pC221, is a 4.6-kb staphylococcal plasmid encoding resistance to chloramphenicol (43) that is mobilized in the presence of pGO1 (49). pC221 is a class I plasmid of the pT181 family (42, 53); along with the related pC223 and the streptomycin resistance plasmid pS194 (52), it has been observed to form relaxation complexes (41). The origin of transfer of pC221 has been localized to a 692-bp AluI fragment, and two overlapping open reading frames (MobA and MobB) identified as essential for mobilization, of which MobA is required for relaxation complex formation (49). Related reading frames are also present in pS194 (52) and pC223 (14). MobA has the three motifs common to relaxases and has been aligned with those of the IncP family (23, 27, 44).
The nucleotide sequence of pC221 has been obtained in two independent laboratories (9, 51). These sequences differ in three places: two fall within the AluI oriT fragment, and the third alters the reading frame of MobA resulting in products of either 227 amino acids (aa) or 315 aa. Both pC221 and pS194 also encode a putative MobC open reading frame preceding (and overlapping) that of MobA, which is also represented in the trimethoprim resistance plasmid pSK639 of S. epidermidis (2) among others. However, the available sequence for pC223 does not indicate such a product. Nor does the sequence for pC223 cover the oriT region of pC221; comparison of the latter with consensus sequences of the nick site from the IncP family does not reveal any obvious homology.
The purpose of the present study was thus to address these issues: by reexamining the sequence of pC221 and pC223, we sought to define the limits of the MobA and MobC reading frames and, by a series of knockout mutations, demonstrate their requirement as relaxase and accessory protein in the formation of the plasmid relaxation complexes. In addition, characterization of the nick site in these plasmids shows that it is distantly related to others of the IncP family, suggesting a modification to the consensus sequence. Although further staphylococcal plasmids share the nick site sequence, especially pC221 and pC223, it appears that the MobA and MobC proteins of these plasmids are able to discriminate between such closely related oriT sequences in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, purification, and manipulation of DNA.
Staphylococcus aureus strain RN4220 (29, 38) was used as a host for plasmid maintenance and as the donor in all filter-mating experiments; RN2677 (resistant to novobiocin and rifampin) (28) was used as the recipient in filter-mating experiments. Plasmids studied are listed in Table 1. Bacterial cultures were grown at 37°C in Luria-Bertani or 2YT broth containing chloramphenicol at a concentration of 10 μg/ml, gentamicin at 5 μg/ml, novobiocin at 5 μg/ml, or rifampin at 5 μg/ml as required. To purify plasmid DNA, staphylococcal cell cultures were harvested by centrifugation and washed with 10 mM EDTA (pH 8.0). After resuspension and incubation with lysostaphin (Sigma) at 15 μg/ml for 30 to 60 min at 37°C, negatively supercoiled DNA was purified by the alkaline-SDS method of Birnboim and Doly (8), followed by small-scale purification by using silica filters (Qiagen, Crawley, United Kingdom) or large-scale isolation of negatively supercoiled DNA by cesium chloride-ethidium bromide density gradient centrifugation (55). Alternatively, whole-cell lysates were prepared essentially as described by Projan et al. (50) by SDS-induced lysis, freeze-thawing, and protease digestion, but with pronase (Sigma) instead of proteinase K and with increased volumes to reduce viscosity. Plasmid manipulations followed standard procedures (55); recombinant plasmids were transformed into S. aureus RN4220 by electroporation (5, 57) with pulse voltages of 1.8 to 2.5 kV. All recombinants were screened for loss of restriction site(s) as appropriate, and mutations were confirmed by DNA sequence analysis. Relative measurements of open circular and negatively supercoiled plasmid forms were made following agarose gel electrophoresis of whole-cell lysates, followed by image capture of ethidium bromide-stained gels and analysis of digitized images by using a GDS-8000 gel documentation system (Ultra-Violet Products, Cambridge, United Kingdom).
TABLE 1.
Plasmids described in the present study
| Plasmid | Relevant characteristics | OC (%)a | Mobilization efficiencyb | Reference or source |
|---|---|---|---|---|
| pC221 | pC221 wild type (inc4, chloramphenicol resistance) | 43 | 0.460 | 43 |
| pC221mobA1 | pC221 with 75-bp deletion between the MwoI sites | <5 | 0.038 | This study |
| pC221mobA4 | pC221 with extra 1 bp at the ScrFI site | <5 | 0 | This study |
| pC221mobAB2 | pC221 with extra 4 bp at the EcoRI site | 19 | 0 | This study |
| pC221mobC1 | pC221 with extra 2 bp at the ClaI site | <5 | 0 | This study |
| pC221cop903 | pC221cop903 high-copy-number mutant of pC221 | 75 | NT | 51 |
| pC221mobA2 | pC221cop903 with extra 5 bp at the SexAI site | <5 | NT | This study |
| pC221mobB1 | pC221cop903 with extra 4 bp at the HindIII site | 52 | NT | This study |
| pC221mobC3 | pC221cop903 with extra 4 bp at the XbaI site | 74 | NT | This study |
| pC223 | pC223 wild type (inc10, chloramphenicol resistance) | 32 | 0.150 | 41 |
| pC223mobA3 | pC223 with 461-bp deletion between the BsgI and BbvI sites | <5 | 0 | This study |
| pC223mobAB1 | pC223 with 4-bp deletion at the BanII site | 27 | 0 | This study |
| pC223mobC4 | pC223 with fortuitous 1-bp deletion at the ClaI site | <5 | 0 | This study |
| pC223mobC5 | pC223 with a BclI site engineered adjacent to the ClaI site | <5 | 0 | This study |
| pC223oriT1 | pC223 with replacement of the 374-bp BstEII-BstBI fragment from pC221 | <5 | 0 | This study |
| pC223oriT2 | pC223 with replacement of the 374-bp BstEII-BstBI fragment from pC221cop903 | -c | -c | This study |
| pC221oriT3 | pC221 with replacement of the 360 bp BstEII-BstBI fragment from pC223 | <5 | 0.023 | This study |
| pC221oriT4 | pC221cop903 with replacement of the 360-bp BstEII-BstBI fragment from pC223 | 14 | NT | This study |
| pGO1 | Self-transmissible, gentamicin resistance | 3 |
Relative percentage of total plasmid DNA measured in the relaxed, open circular form (OC) after agarose gel electrophoresis of whole-cell lysates (results of three to six separate determinations).
Ratio of chloramphenicol-resistant to gentamicin-resistant recipients after filter mating (result of at least three separate experiments; representative data are shown). NT, not tested.
Plasmids pC223oriT1 and pC223oriT2 are identical.
DNA sequence analysis.
Nucleotide sequencing was performed by using the Prism Ready Reaction DyeDeoxy terminator cycle sequencing kit and an ABI 373 DNA sequencer (Applied Biosystems, Foster City, Calif.). Oligonucleotide sequencing primers for pC223 were initially based on available data (GenBank accession nos. X07371 and X12831), after which novel data were obtained by using a genomic-walking strategy.
Generation of knockout mutations.
Plasmids were digested with the appropriate restriction enzyme and then treated with Klenow polymerase (Pharmacia) to trim back 3′ overhangs or fill-in 5′ overhangs. Pfu polymerase (Stratagene) was used for the same purpose following digestion of pC223 with BsgI and BbvI. The resultant blunt-ended DNA was self-ligated and transformed into S. aureus RN4220 by electroporation (5, 57). In the case of pC223mobC5, a product encompassing the 608-bp BstEII-ClaI fragment was generated by PCR with the primers ORI(+) (5′-CCTAAAAAACCGATACCTGAAAACAC-3′) and BCL(−) (5′-TCTTTAGCAATCGATGATCACGTCTCTTTATCTAATTTTGGCGC-3′), the latter incorporating an additional BclI site immediately adjacent to that for ClaI (-GTGATCATCGATT-), which also introduces a termination codon into the reading frame for MobC (underlined). After digestion with BstEII and ClaI, this fragment was ligated to the reciprocal part of pC223 yielding the mutant plasmid.
Generation of oriT exchange plasmids.
Products encompassing the 360- to 374-bp BstEII-BstBI fragments of pC221, pC221cop903, and pC223 were amplified by PCR with primers ORI(+) (as described above) and ORI(−) (5′-GGCACACTCATATTCAAAGTTTCGGC-3′). After digestion with these enzymes, the fragments were religated with the reciprocal parts of the noncognate replicons as indicated in Table 1, and transformed into S. aureus RN4220.
Filter-mating experiments.
Mobilization of chloramphenicol resistance plasmids by pGO1 (3) was determined by filter mating based on the method of Schaberg et al. (56). Cultures of RN4220 (containing pGO1 and pC221/pC223 variants) and RN2677 were grown to mid-exponential phase in 2YT broth, and volumes containing 3 × 108 cells were mixed and filtered through 13-mm-diameter, 0.2-μm-pore size nitrocellulose filters (Whatman). After growth on brain heart infusion agar overnight, cells were resuspended by vortexing them in 2YT broth and plated out with appropriate selection for donors, recipients, or transconjugants. Conjugation frequency was calculated from the number of gentamicin-resistant transconjugants obtained per recipient cell; relative mobilization efficiency is described by the ratio of chloramphenicol-resistant to gentamicin-resistant transconjugants.
Radiolabeling of restriction fragments and primer extension products.
Restriction fragments were 5′ end labeled by using T4 polynucleotide kinase (Pharmacia) and [γ-32P]ATP (ICN Biochemicals) prior to separation on denaturing polyacrylamide gels (6% [wt/vol] polyacrylamide, 8 M urea, 1× Tris-borate-EDTA buffer) (55). Primer extension products were generated by using primers designed to anneal 150 nucleotides (nt) away from the predicted nic site: the sequences were 5′-GAACGTATAGCAACCAC-3′ for the positive strand and 5′-CACTCATTCAATCCCACC-3′ for the (−) strand. Template plasmid DNA (negatively supercoiled, predigested with HpaII, or derived from whole-cell lysates; 5 μg per reaction) was chemically denatured and recovered by ethanol precipitation prior to use. After an annealing step, primers (10 ng per reaction) were extended by using T7 Sequenase (Amersham Pharmacia) in the presence of [α-32P]dATP (ICN Biochemicals) and 80 μM (each) dATP, dCTP, dGTP, and dTTP at 37°C for 5 min prior to separation with denaturing gels as described above. Sequencing ladders were produced with the T7 Sequenase kit (Amersham Pharmacia) with the same primer-template combinations. All radiolabeled gels were imaged by using Fuji BAS plates and a BAS-1000 phosphorimager.
Sequence availability for pC223.
The complete pC223 sequence has been deposited in GenBank (accession no. AY355285).
RESULTS
DNA sequence analysis.
Plasmid pC221 has previously been sequenced independently by two different groups, and the data were deposited in GenBank under accession numbers X02166 (9) and X02529 (51). These sequences differ in three places: (i) an additional A next to nucleotide 2966 in X02529; (ii) A3143 of X02166 is replaced by G in X02529; and (iii) an additional C next to nucleotide at 4263 of X02529. Our laboratory stock of plasmid pC221 was derived from that used by Brenner and Shaw (9); pC221cop903 was from the study of Projan et al. (51). Upon sequencing each of these regions in both plasmids we found G at nucleotide 3143, but otherwise confirmed the sequences to be as for X02166. Crucially, this leads to a MobA translational product of 315 amino acids (aa) rather than a 227-aa product resulting from the presence of an extra base, which would terminate prior to the unique EcoRI site. It should be noted pC221cop903 differs from the wild-type plasmids by a deletion within the copy control region (51), resulting in an increase in copy number; the oriT/mob regions of the two plasmids are identical.
Two overlapping sequences of pC223 were available via GenBank prior to the present study: X07371 (15) and X12831 (14). In addition, that of the replication initiator (here referred to as RepJ) has also been deposited independently as M21928 (53). However, although X12831 indicated the presence of a MobA product analogous to that of pC221, it did not support an overlapping MobC reading frame, nor did it extend significantly into the predicted oriT region. We therefore sequenced the region of pC223 corresponding to X12831 and also completed the sequence of the plasmid between the repJ and mobA genes.
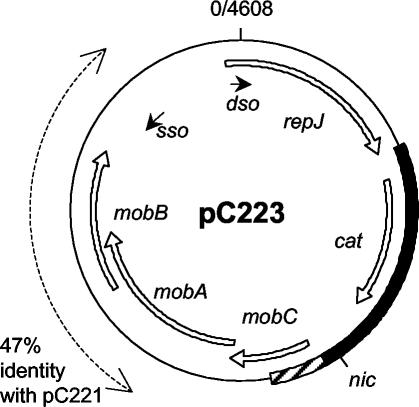
The resultant plasmid map is displayed in Fig. 1. The plasmid is 4,608 bp in size (compared to 4,555 bp for pC221); the organization of replication features and reading frames is the same as for pC221, and the two plasmids are closely related at the nucleotide sequence level (82% identity for most of the plasmid, falling to 47% over a region covering the mobB gene). Our novel sequence includes a cat gene most closely related to GenBank accession no. M58516 (59), and we find trivial differences within the region encoding the C terminus of RepJ, leading to translation of aa 293 as lysine instead of threonine. More significantly, several amendments to X12831 result in the identification of a potential pC223 mobC gene: this translates as a product of 15.2 kDa (132 aa) compared to 14.6 kDa (127 aa) for that of pC221, with 73% amino acid sequence identity between the two proteins. In both plasmids the MobC and MobA reading frames overlap to the same extent (16 nucleotides). The overlap between MobA and MobB reading frames differs due to the lower sequence conservation surrounding that for MobB: in pC221 MobA (315 aa) overlaps MobB (230 aa) by 266 nucleotides and in pC223 MobA (330 aa) overlaps MobB (236 aa) by 281 nucleotides.
FIG. 1.
Organization of plasmid pC223 (4,608 bp). The map is oriented with the nick site of the double-stranded replication origin (dso) at the +1 position. Novel sequence data obtained in the present study are indicated by the solid bar on the map; amendments to previously available data are concentrated in the shaded region. The principal reading frames are indicated by the open arrows. sso indicates the site and orientation of the single-stranded (−) replication origin. The nick site within the origin of transfer determined in the present study is given as nic. The layout of pC221 is similar, although the two plasmids have considerably reduced sequence identity in the region shown by the dashed line.
MobA and MobC are both required for the formation of relaxation complexes.
In the previous study by Projan and Archer (49), a copy mutant of pC221, pC221cop905, was used to investigate the effects of interruption of Mob reading frames on formation of the relaxation complex. Our first objective was to verify such data for the wild-type copy number plasmid. In addition, we wanted to investigate the effects of interruption to the MobC reading frame alone. We adopted a similar strategy of restriction digestion and end modification prior to religation in order to disrupt reading frames at defined locations.
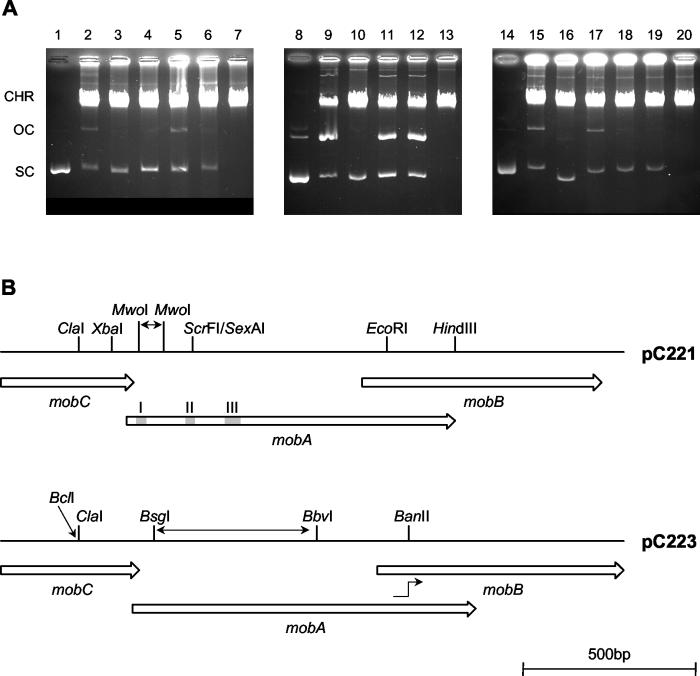
The results are shown in Fig. 2 and summarized in Table 1. Isolation of pC221 from whole-cell lysates typically resulted in roughly half the material in the nicked, open circular form (form II) indicative of relaxation complex formation. Deletion of 75 bp between the MwoI sites produced plasmid pC221mobA1, with an in-frame deletion of residues 14 to 38 of MobA. This includes the active site tyrosine at position 18 within conserved motif I: without this nucleophile, the relaxase is inactive and no significant relaxation complex formation is seen. Disruption at the ScrFI site in pC221mobA4 retains motifs I and II but interrupts MobA after 64 aa, again abolishing relaxase activity. In the context of our sequence of pC221, disruption via the EcoRI site in pC221mobAB2 interrupts MobA after 251 aa, leaving the three motifs intact, and also truncates MobB after only 25 aa; as observed previously (49), relaxation complex formation is still evident. Blunting of the ends after digestion with ClaI to produce pC221mobC1 interrupts MobC after 76 aa: no relaxation complex was detected, indicating that MobC is also essential for relaxation complex formation in vivo.
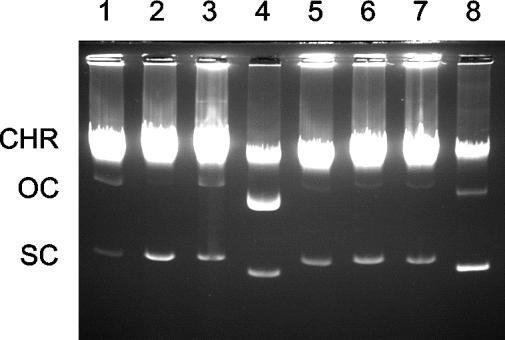
FIG. 2.
Effects of plasmid mutations on relaxosome formation. (A) Agarose gel electrophoresis of plasmid variants. Control lanes show negatively supercoiled pC221 (lane 1), pC221cop903 (lane 8), and pC223 (lane 14). Other lanes show samples derived from whole-cell lysates of S. aureus RN4220 carrying different plasmids: lane 2, pC221; lane 3, pC221mobA1; lane 4, pC221mobA4; lane 5, pC221mobAB2; lane 6, pC221mobC1; lane 9, pC221cop903; lane 10, pC221mobA2; lane 11, pC221mobB1; lane 12, pC221mobC3; lane 15, pC223; lane 16, pC223mobA3; lane 17, pC223mobAB1; lane 18, pC223mobC4; and lane 19, pC223mobC5. Lysates from plasmid-free cells are shown in lanes 7, 13, and 20. CHR, chromosomal DNA; OC, SC, open circular and negatively supercoiled plasmid DNA. (B) Restriction sites used to generate mob plasmid mutations, and the relative locations of the mob genes involved. pC221 and pC221cop903 are identical in this region. Conserved motifs I, II, and III are indicated by the shaded regions within mobA. Note that pC221mobA1 results from deletion of 75 bp between the two MwoI sites encoding motif I, pC223mobA3 has lost 461 bp between the BsgI-BbvI sites deleting motifs II and III, and in pC223mobAB1 the mutation causes a frameshift fusing mobA to mobB at the position indicated.
The high-copy mutant pC221cop903 was also used, both to improve yields of plasmid DNA (through the higher copy number) and to increase the range of restriction sites unique to the mob region (due to the deletion in the copy control region). As stated above, the sequence within the mob region is identical with the wild-type plasmid, thus the corresponding restriction maps are the same (Fig. 2B). A higher proportion of plasmid isolated as the nicked form was observed in comparison to the wild-type replicon, but disruption of MobA via the SexAI site (coincident with the ScrFI site described above) in pC221mobA2 again abolished relaxase activity. Relaxation complex formation was still evident after either interruption of MobB (via HindIII, yielding pC221mobB1) or of MobC after 108 aa (via XbaI, yielding pC221mobC3), indicating that essential sequences of the latter product are not contained within the last 19 aa. Unfortunately, further studies based on variants of this replicon were hindered by instabilities, resulting in frequent deletions, rearrangements, or loss of the high-copy phenotype.
The differences in sequence between pC221 and pC223 precluded the use of identical interruptions within MobA of the latter plasmid. Nevertheless, a range of mutant plasmids was obtained (Fig. 2B). Deletion of the region between BsgI and BbvI interrupted MobA after 28 aa, leaving only motif I present in pC223mobA3: this abolished relaxase activity. Interruption of MobA via BanII led to a fusion between the first 266 aa of MobA and residues 33 to 236 of MobB in pC223mobAB1. Curiously, this fusion protein retains relaxase activity, suggesting that C-terminal sequences of MobA are dispensable, as observed for pC221mobAB2. Interruption of MobC via ClaI proved problematic, with the sole mutant recovered carrying a deletion of one nucleotide (rather than the addition of two) at the ClaI site of pC223mobC4. In a second strategy, the BstEII-ClaI fragment was replaced with a fragment generated by PCR to contain a BclI site with an in-frame termination codon adjacent to the ClaI site, truncating MobC at 73 aa in pC223mobC5. As expected, neither of these MobC mutants supported relaxase activity. We therefore conclude that MobC is essential for relaxosome formation in pC221 and pC223, as is a MobA protein encoding each of the conserved motifs I, II, and III at a minimum.
MobA, MobB, and MobC are all required for efficient mobilization.
MobA and MobB are already known to be required for mobilization of pC221 by the self-transmissible plasmid pGO1 (49). To establish the requirement for MobC in the context of pC221 and confirm the predicted mobilization requirements of pC223, each of the wild-type copy number variant plasmids described above was tested in filter-mating experiments. The conjugation frequency of pGO1 (measured as the proportion of recipients which had acquired gentamicin resistance) was routinely found to be within the range 10−6 to 10−5; the mobilization frequencies of pC221 and pC223 relative to pGO1 were typically 0.46 and 0.15, respectively (Table 1).
Disruption of each of the Mob proteins described above severely affected mobilization of the chloramphenicol resistance plasmids, by at least one or more orders of magnitude. Residual mobilization such as that observed for pC221mobA1 may be attributed to direct interaction between pGO1 and the pC221 origin of transfer as suggested previously (49), although it is to be noted that whole-cell lysate analysis of RN4220 carrying both pGO1, and each of the chloramphenicol resistance plasmids did not reveal any unusual perturbations in the degree of relaxation complex formation of the latter (data not shown). In addition to demonstrating the requirement for MobC, the results obtained with pC223mobAB1 also demonstrate the requirement for intact and distinct MobA and MobB proteins in mobilization.
Location of the nic site.
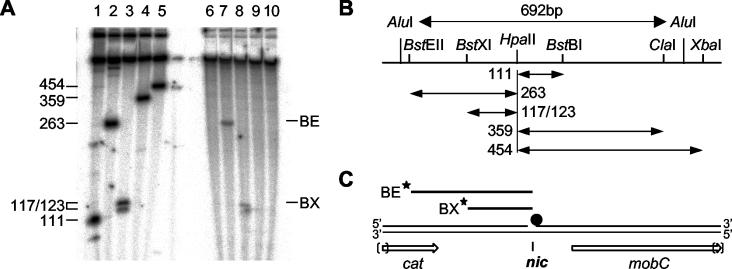
Initial studies with S1 nuclease suggested that the open circular form of pC221 obtained in whole-cell lysates was nicked at a consistent location, close to the HpaII site at nucleotide 3109 of pC221 and thus within the 692-bp oriT fragment (data not shown). This location was refined by restriction digestion, end labeling, denaturing gel electrophoresis, and autoradiography (Fig. 3). pC221cop903, isolated from whole-cell lysates and subjected to extensive pronase digestion, was digested variously with the single-cutting restriction endonucleases BstEII, BstXI, BstBI, ClaI, and XbaI. As a control, negatively supercoiled plasmid was first digested with HpaII and then by the same set of enzymes. All fragments were then labeled by using T4 polynucleotide kinase and [γ-32P]ATP prior to electrophoresis. The autoradiograph is presented in Fig. 3A. Control fragments larger than 500 nucleotides (owing to the presence of additional HpaII sites within the plasmid) are not resolved on the gel; the fragment between BstXI and HpaII sites is resolved as a doublet of 117 and 123 nucleotides due to the unequal 5′ and 3′ overhangs generated by these enzymes. This control restriction pattern is interpreted in Fig. 3B. In the case of the nicked material, no HpaII digestion was performed: thus the resolved fragments must arise from a conserved nick site. Fragments were only observed after digestion with BstEII and BstXI: this is consistent with a nick in the positive strand only and the 5′ side of the nick being blocked, presumably by a residual, covalently attached fragment of MobA. Moreover, the observed fragments map the nick site to a region of less than 50 nucleotides to the 3′ of the HpaII site (interpreted in Fig. 3C).
FIG. 3.
Location of the nic site within pC221. (A) Denaturing polyacrylamide gel electrophoresis of 5′-end-labeled restriction products. Negatively supercoiled pC221 previously digested with HpaII (lanes 1 to 5) or pC221 obtained from whole-cell lysates (lanes 6 to 10) was further digested with BstBI (lanes 1 and 6), BstEII (lanes 2 and 7), BstXI (lanes 3 and 8), ClaI (lanes 4 and 9), or XbaI (lanes 5 and 10). 5′-End-labeled fragments were detected by autoradiography. Fragment sizes are indicated in nucleotides; the doublet seen in lane 3 results from the uneven upper and lower strand lengths after digestion. (B) Location of restriction sites within the 692-bp AluI fragment previously shown to contain oriT (49). Larger fragments were not resolved on the gel shown. (C) Mapping of the labeled (★) fragments BE and BX locates the nic site 3′ to the HpaII site and in the sense strand with respect to cat and the mob genes; failure to generate similar fragments in lanes 6, 9, and 10 indicates the 5′ end at nic is blocked (•).
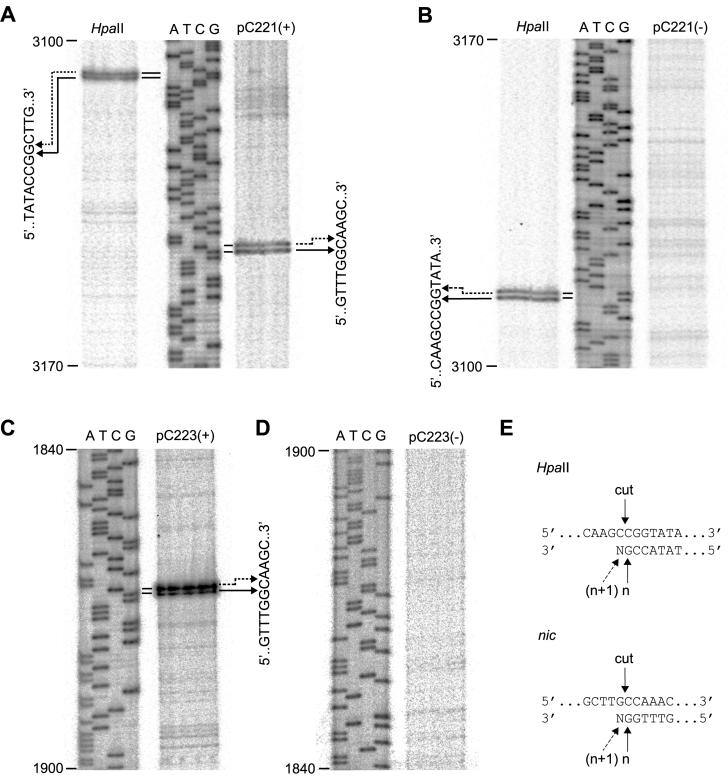
Having located the nick to this region, we identified the site in both pC221cop903 and pC223 by primer extension analysis (Fig. 4). Again, pC221cop903 previously digested with HpaII was used as a control; the characteristic banding pattern observed after extension against this template resulting from partial terminal transferase activity of the polymerase (T7 Sequenase) as observed elsewhere (33) (Fig. 4A and B, interpreted in Fig. 4E). Primer extension products were observed when a negative-strand primer was used for both pC221cop903 and pC223 (Fig. 4A and C); no such products were detected by using a positive-strand primer (Fig. 4B and D), indicating that the nick is in the positive strand of both plasmids. Taking into account the terminal transferase activity of the polymerase, the nick site maps to a unique location, hereafter referred to as nic (Fig. 4E). The surrounding sequence GCTTG′CCAAA is identical in both plasmids; nic is located between nucleotides 3150′3151 in pC221 and 1869′1870 in pC223.
FIG. 4.
Identification of the nic sites of pC221cop903 and pC223. (A) Primer extension analysis against the positive-strand of pC221cop903. The same negative-strand primer was used for all tracks. HpaII, plasmid template cut with HpaII; ATCG, sequencing ladder corresponding to coordinates 3100 to 3170 of the pC221 sequence; pC221(+), pC221cop903 obtained by whole-cell lysate procedure as a template. Doublets resulting from primer extension are correlated with nucleotide sequence by the arrows. (B) Same as for panel A, but with a positive-strand primer. (C and D) Same as for panels A and B, respectively, but with pC223 as a template; this plasmid does not possess a corresponding HpaII site. (E) Interpretation of the primer extension doublets: HpaII cuts the positive-strand as indicated; primer extension products of the expected length (n) and one nucleotide longer (n + 1) are generated by terminal transferase activity of the polymerase. The same interpretation is compatible with a unique nic site at the position indicated.
Plasmid specificity of the Mob proteins.
The origin of transfer, oriT, represents the functional target for the Mob proteins in vivo. In addition to the conserved sequence at nic and the retention of many restriction sites in common between the two plasmids, oriT includes many sequence differences between pC221 and pC223. The potential for plasmid specificity at oriT was investigated by exchange of the BstEII-BstBI fragment (Fig. 3) between the two plasmids: insertion of the 374-bp oriT fragment from pC221 into pC223 produced pC223oriT1, and the parallel insertion of the 360-bp fragment of pC223 into pC221 produced pC221oriT3. A similar exchange between pC221cop903 and pC223 resulted in pC223oriT2 and pC221oriT4, respectively. The latter allows contrast of the pC223 oriT in the context of a high copy number compared to pC221oriT3; since the oriT sequences of pC221 and pC221cop903 were found to be identical (see above), pC223oriT2 is identical in sequence to pC223oriT1.
Relaxation complex formation by these oriT recombinants is summarized in Table 1 and depicted in Fig. 5. In the context of the wild-type copy number replicon carrying the noncognate oriT relaxation complex formation is effectively abolished, indicating that the MobA or MobC proteins are specific for their cognate transfer origins. This is also reflected in their reduced relative mobilization frequencies. However, pC221oriT4 demonstrates partial nicking activity, suggesting that the higher gene dosage resulting from the higher copy number of this replicon allows the MobA/MobC proteins of pC221 to overcome the lower affinity for the noncognate oriT of pC223.
FIG. 5.
Specificity for oriT in relaxosome formation. Whole-cell lysates were analyzed by gel electrophoresis. Plasmids are shown as follows: lane 1, pC221; lane 2, pC221mobA4; lane 3, pC223; lane 4, pC221cop903; lane 5, pC223oriT1; lane 6, pC223oriT2; lane 7, pC221oriT3; and lane 8, pC221oriT4. CHR, chromosomal DNA; OC and SC, open circular and negatively supercoiled plasmid DNA, respectively. Loadings have been normalized to compensate for the higher copy number of the pC221cop903 replicon.
DISCUSSION
We have demonstrated the requirement for a MobC accessory protein in addition to the MobA relaxase for both relaxosome formation and efficient mobilization of pC221 and pC223. This requirement distinguishes these plasmids from another well-characterized mobilizable plasmid from a gram-positive host, pMV158 (48), which appears to encode a single polypeptide, MobM, required for mobilization. Moreover, determination of the nic site provides further justification for categorizing the mobility functions of pC221 and pC223 with the IncP plasmids of Mob family 1 (66).
The MobC proteins of pC221 and pC223 are closely related throughout most of their length; database searches reveal homologues in plasmids of S. aureus such as pS194 (52), pRJ6 (40), and pRJ9 (39), as well as many from S. epidermidis, including pSK639 (2) and pIP1629 and pIP1630 (4). These contain the conserved motif suggested by Apisiridej et al. (2) (L/FxxxG/SxNxNQxAxxxN), although we have not been able to assign a DNA-binding domain comparable to the functionally equivalent VirD1 (63) nor reveal significant sequence homology with the essential TraJ protein of RP4 (18). pSK639, pIP1629, and pIP1630 also encode an additional overlapping reading frame of ∼64 aa between MobC and MobA with unknown function: this may serve to provide translational coupling from MobC to MobA within a single transcript. Such coupling is likely in the overlap of MobC with MobA in our plasmids. In the case of pC221mobC3, we note that translation of the modified MobC reading frame terminates only 33 nt away from the start of MobA: the nicking activity observed for this mutant indicates that translation of MobA is still adequate.
Alignments of MobA sequences from the above plasmids show strong sequence conservation within the first 122 aa (corresponding to relaxase motifs I, II, and III) with significant differences in the remaining two-thirds of the proteins. This appears to be a common feature of the relaxases; in the context of the gram-positive plasmids, the pMV158-related MobM and pT181-related Pre (plasmid recombinase) functions show a similar pattern (19, 48). Conservation of a functional relaxase amino-terminal domain would account for the relaxase activity still evident in both our pC223mobAB1 fusion and the disruption at EcoRI created by Projan and Archer (49), represented here by pC221mobA4. The C-terminal domain shows no sequence motifs corresponding to helicase or primase functions; its role in (and requirement for) mobilization may involve interaction with the coupled MobB reading frame. Although the latter has proven to be the most divergent sequence across pC221, pC223, and related plasmids, we note the presence of extensive hydrophobic regions in each case, potentially locating the relaxosome to the cell wall prior to transfer.
The pC221 nic site follows established precedents in that the 5′ end is blocked, presumably by covalent attachment of the relaxase as observed for TraI among others (47). It is also situated close enough to the mobC and mobA reading frames to make coordinated regulation of both nicking activity and mob expression a possibility. However, in the majority of gram-negative plasmids reported to date the nick is to be found in the antisense strand with respect to the adjacent relaxase; this is also true for nic in staphylococcal plasmid pGO1, found in the antisense strand upstream of the nes relaxase (13). Unusually, the nick site of pC221 and pC223 is in the sense strand with respect to the mobC and mobA reading frames. This may be a feature of small, mobilizable gram-positive plasmids, since we note that the nick sites of oriT and RSA in plasmids pMV158 and pT181, respectively, are also located in the sense strand with respect to the adjacent mobM or pre functions. Furthermore, the mob reading frames are typically followed by the single-strand (minus) origin of replication, ssoA (21), facing in the opposite orientation. This arrangement would permit synthesis of the complementary strand upon transfer of the sense strand into the recipient. Unlike the gram-negative examples indicated above, the genes encoding the relaxase and its adjacent accessory proteins are the first to enter the recipient cell and not the last: a mechanism enabling expression of these genes may be required to complete transfer. In this respect, ssoA is reminiscent of the ssi functions of ColIb-P9 (6).
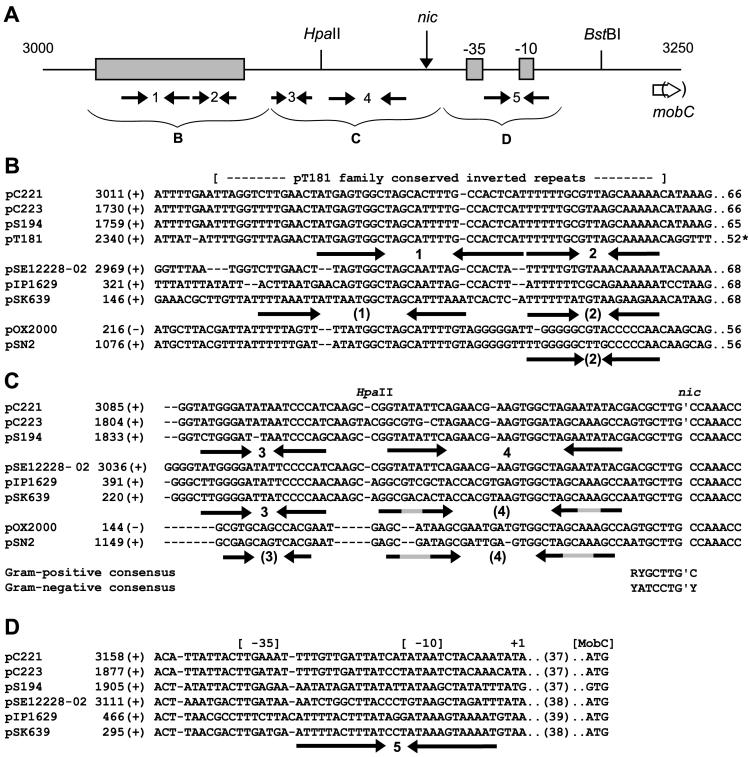
Exchange of oriT fragments between pC221 and pC223 revealed a Mob/oriT specificity reminiscent of the Rep/ori specificity of the pT181 family plasmids (53). In the case of pC221oriT4 this was overcome, the higher gene dosage of MobC and/or MobA of pC221 overcoming the sequence differences of the pC223 oriT. This resembles the effect of copy number on replication specificity: when provided at high intracellular concentrations, both RepC and RepD (of pT181 and pC221, respectively) are able to initiate replication from a range of noncognate origins (24). An analysis of the oriT region of pC221 is presented in Fig. 6. In common with the transfer origins of other plasmids (31) there are many inverted complementary repeat sequences (numbered 1 to 5 in the figure). These fall into three groups, discussed further below.
FIG. 6.
Alignment of oriT sequences. (A) Orientation of features between coordinates 3000 and 3250 of pC221. The locations of nic, restriction sites HpaII and BstBI, putative −35 and −10 signals, and the start of the mobC reading frames are shown, as are five prominent inverted repeats (labeled 1 to 5). The shaded region indicates the sequences present in pT181, shown in panel B. (B) Alignment of pC221-like, pSK639-like, and pSN2-like plasmids with pT181. Selected sequences identified by BLAST searches (1) were aligned by using CLUSTAL X (61). The coordinate (and strand) of the first nucleotide in each row are derived from the present study (pC221 and pC223) or GenBank accession numbers X06627 (pS194), J01764 (pT181), AE015931 (pSE12228-02), AF045240 (pIP1629), U40259 (pSK639), X55798 (pOX2000), and V01282 (pSN2). The numbers at the end of the line represent the distance to nic (or RSA in the case of pT181). (C) Alignment of sequences immediately upstream of nic (contiguous with panel B above). The consensus nic sequence obtained from these gram-positive plasmids is compared to the consensus for gram-negative plasmids of the IncP family (66). (D) Potential promoter region for mobC-encoding plasmids. The separation of the initiator codon from the start of the transcript is indicated.
Between 70 and 130 bp upstream of nic are two repeats (1 and 2) which lie within a region of ∼60 bp almost identical in pC221, pC223, pS194, and pT181 (Fig. 6B). Given its proximity to the end of the antibiotic resistance functions, this region has been named t1 (for termination of transcription) (53), but we note the similar juxtaposition with nic and RSA, even though the sequences of the latter are unrelated. However, the sequences are poorly conserved in the named plasmids of S. epidermidis. A third set of “cryptic” plasmids of S. aureus, pOX2000 (64) and pSN2 (26), as well as their relatives pSK3 and pSK6 (35), also possess the same nic site but are almost unrelated in this region, apart from a short sequence falling between the arms of repeat 1.
The conservation of complementary base substitutions in inverted repeat 3 (Fig. 6C) implies functional significance, but neither this region nor the adjacent arm of repeat 4 is strongly conserved in the cryptic plasmids. However, homology across the examples shown greatly improves closer to the nick site, which is identical in all examples shown and also found in the functional oriT sequences of other gram-positive plasmids, such as pNZ4000 of Lactococcus lactis (62). The consensus sequence for these gram-positive examples, RYGCTTG′C, compares favorably with that for gram-negative examples of Mob family 1, YATCCTG′Y. Immediately downstream of nic we note potential −35 and −10 sequences for transcription of the mob genes, which overlap with the location of repeat 5, representing a target for autoregulation (Fig. 6D); in the cryptic plasmids pOX2000 and pSN2 there are no such reading frames, and the ssoA single strand replication origin follows immediately.
Overall, the organization of oriT and adjacent mob genes by pC221 and relatives represents an efficient use of space both by overlapping of reading frames and in overall size of the proteins concerned and sets a precedent for the juxtaposition of nic in the sense strand with respect to the coding regions for these multicomponent mobilization functions. The ability to discriminate between closely related sequences is reminiscent of the organization of the replicons of these small, gram-positive plasmids; we suggest the term “mobilon” to describe these functional units for mobilization, which appear to be freely distributed on “cassettes” in a similar fashion to the replicons and resistance determinants of these plasmids. In MobC and MobA, our “mobilon” functionally resembles TraJ and TraI components of RP4, but we do not yet know if these two proteins alone are sufficient to nick at oriT, or whether additional, chromosomal factors are required. Nor has the present study yet characterized the individual activities of MobC and MobA in isolation. These topics are the subject of an accompanying study (12).
Acknowledgments
We thank Denise Ashworth and Emma Stanley for DNA sequence analysis and oligonucleotide synthesis and T. J. Foster (Dublin, Ireland) for strains used in mobilization studies.
M.C.A.S. was supported by a studentship from the BBSRC. C.D.T. was supported by the Wellcome Trust (041984/Z/94).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apisiridej, S., A. Leelaporn, C. D. Scaramuzzi, R. A. Skurray, and N. Firth. 1997. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 38:13-24. [DOI] [PubMed] [Google Scholar]
- 3.Archer, G. L., J. P. Coughter, and J. L. Johnston. 1986. Plasmid-encoded trimethoprim resistance in staphylococci. Antimicrob. Agents Chemother. 29:733-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubert, S., K. G. Dyke, and N. E. Solh. 1998. Analysis of two Staphylococcus epidermidis plasmids coding for resistance to streptogramin A. Plasmid 40:238-242. [DOI] [PubMed] [Google Scholar]
- 5.Augustin, J., and F. Gotz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 66:203-208. [DOI] [PubMed] [Google Scholar]
- 6.Bates, S., R. A. Roscoe, N. J. Althorpe, W. J. Brammar, and B. M. Wilkins. 1999. Expression of leading region genes on IncI1 plasmid ColIb-P9: genetic evidence for single-stranded DNA transcription. Microbiology 145:2655-2662. [DOI] [PubMed] [Google Scholar]
- 7.Becker, E. C., and R. J. Meyer. 2002. MobA, the DNA strand transferase of plasmid R1162: the minimal domain required for DNA processing at the origin of transfer. J. Biol. Chem. 277:14575-14580. [DOI] [PubMed] [Google Scholar]
- 8.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner, D. G., and W. V. Shaw. 1985. The use of synthetic oligonucleotides with universal templates for rapid DNA sequencing: results with staphylococcal replicon pC221. EMBO J. 4:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalyzed by a relaxase. Mol. Microbiol. 25:1011-1022. [DOI] [PubMed] [Google Scholar]
- 11.Byrd, D. R., J. K. Sampson, H. M. Ragonese, and S. W. Matson. 2002. Structure-function analysis of Escherichia coli DNA helicase I reveals non-overlapping transesterase and helicase domains. J. Biol. Chem. 277:42645-42653. [DOI] [PubMed] [Google Scholar]
- 12.Caryl, J. A., M. C. A. Smith, and C. D. Thomas. 2004. Reconstitution of a staphylococcal plasmid-protein relaxation complex in vitro. J. Bacteriol. 186:3374-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Climo, M. W., V. K. Sharma, and G. L. Archer. 1996. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J. Bacteriol. 178:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreher, J., and H. Matzura. 1988. The Staphylococcus aureus plasmid pC223 carries two open reading frames responsible for relaxation complex formation and conjugative mobilization. Hoppe-Seyler's Biol. Chem. 369:809.
- 15.Ehret, M., and H. Matzura. 1988. Replication control of the Staphylococcus aureus chloramphenicol resistance plasmids pC223 and pUB112 in Bacillus subtilis. Nucleic Acids Res. 16:2045-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filichkin, S. A., and S. B. Gelvin. 1993. Formation of a putative relaxation intermediate during T-DNA processing directed by the Agrobacterium tumefaciens VirD1,D2 endonuclease. Mol. Microbiol. 8:915-926. [DOI] [PubMed] [Google Scholar]
- 17.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furste, J. P., W. Pansegrau, G. Ziegelin, M. Kroger, and E. Lanka. 1989. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc. Natl. Acad. Sci. USA 86:1771-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gennaro, M. L., J. Kornblum, and R. P. Novick. 1987. A site-specific recombination function in Staphylococcus aureus plasmids. J. Bacteriol. 169:2601-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruss, A. D., H. F. Ross, and R. P. Novick. 1987. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc. Natl. Acad. Sci. USA 84:2165-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hickman, A. B., D. R. Ronning, R. M. Kotin, and F. Dyda. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol. Cell 10:327-337. [DOI] [PubMed] [Google Scholar]
- 23.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes, and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iordanescu, S. 1989. Specificity of the interactions between the Rep proteins and the origins of replication of Staphylococcus aureus plasmids pT181 and pC221. Mol. Gen. Genet. 217:481-487. [DOI] [PubMed] [Google Scholar]
- 25.Karl, W., M. Bamberger, and E. L. Zechner. 2001. Transfer protein TraY of plasmid R1 stimulates TraI-catalyzed oriT cleavage in vivo. J. Bacteriol. 183:909-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan, S. A., and R. P. Novick. 1982. Structural analysis of plasmid pSN2 in Staphylococcus aureus: no involvement in enterotoxin B production. J. Bacteriol. 149:642-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin, E. V., and T. V. Ilyina. 1993. Computer-assisted dissection of rolling circle DNA replication. Biosystems 30:241-268. [DOI] [PubMed] [Google Scholar]
- 28.Kornblum, J., B. J. Hartman, R. P. Novick, and A. Tomasz. 1986. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur. J. Clin. Microbiol. 5:714-718. [DOI] [PubMed] [Google Scholar]
- 29.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 30.Kupelwieser, G., M. Schwab, G. Hogenauer, G. Koraimann, and E. L. Zechner. 1998. Transfer protein TraM stimulates TraI-catalyzed cleavage of the transfer origin of plasmid R1 in vivo. J. Mol. Biol. 275:81-94. [DOI] [PubMed] [Google Scholar]
- 31.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 32.Llosa, M., S. Bolland, and F. de la Cruz. 1994. Genetic organization of the conjugal DNA processing region of the IncW plasmid R388. J. Mol. Biol. 235:448-464. [DOI] [PubMed] [Google Scholar]
- 33.Llosa, M., G. Grandoso, and F. de la Cruz. 1995. Nicking activity of TrwC directed against the origin of transfer of the IncW plasmid R388. J. Mol. Biol. 246:54-62. [DOI] [PubMed] [Google Scholar]
- 34.Llosa, M., G. Grandoso, M. A. Hernando, and F. de la Cruz. 1996. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J. Mol. Biol. 264:56-67. [DOI] [PubMed] [Google Scholar]
- 35.Lyon, B. R., J. W. May, and R. A. Skurray. 1983. Analysis of plasmids in nosocomial strains of multiple-antibiotic-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 23:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matson, S. W., and B. S. Morton. 1991. Escherichia coli DNA helicase I catalyzes a site- and strand-specific nicking reaction at the F plasmid oriT. J. Biol. Chem. 266:16232-16237. [PubMed] [Google Scholar]
- 37.Moncalian, G., G. Grandoso, M. Llosa, and F. de la Cruz. 1997. oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J. Mol. Biol. 270:188-200. [DOI] [PubMed] [Google Scholar]
- 38.Morton, T. M., D. M. Eaton, J. L. Johnston, and G. L. Archer. 1993. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J. Bacteriol. 175:4436-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Netz, D. J., R. Pohl, A. G. Beck-Sickinger, T. Selmer, A. J. Pierik, C. Bastos Mdo, and H. G. Sahl. 2002. Biochemical characterization and genetic analysis of aureocin A53, a new, atypical bacteriocin from Staphylococcus aureus. J. Mol. Biol. 319:745-756. [DOI] [PubMed] [Google Scholar]
- 40.Netz, D. J., H. G. Sahl, R. Marcelino, J. dos Santos Nascimento, S. S. de Oliveira, M. B. Soares, M. do Carmo de Freire Bastos, and R. Marcolino. 2001. Molecular characterization of aureocin A70, a multi-peptide bacteriocin isolated from Staphylococcus aureus. J. Mol. Biol. 311:939-949. [DOI] [PubMed] [Google Scholar]
- 41.Novick, R. 1976. Plasmid-protein relaxation complexes in Staphylococcus aureus. J. Bacteriol. 127:1177-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novick, R. P. 1989. Staphylococcal plasmids and their replication. Annu. Rev. Microbiol. 43:537-565. [DOI] [PubMed] [Google Scholar]
- 43.Novick, R. P., and D. Bouanchaud. 1971. The problems of drug-resistant pathogenic bacteria: extrachromosomal nature of drug resistance in Staphylococcus aureus. Ann. N. Y. Acad. Sci. 182:279-294. [DOI] [PubMed] [Google Scholar]
- 44.Pansegrau, W., and E. Lanka. 1991. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 19:3455. [DOI] [PMC free article] [PubMed]
- 45.Pansegrau, W., and E. Lanka. 1996. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acids Res. Mol. Biol. 54:197-251. [DOI] [PubMed] [Google Scholar]
- 46.Pansegrau, W., W. Schroder, and E. Lanka. 1994. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem. 269:2782-2789. [PubMed] [Google Scholar]
- 47.Pansegrau, W., G. Ziegelin, and E. Lanka. 1990. Covalent association of the traI gene product of plasmid RP4 with the 5′-terminal nucleotide at the relaxation nick site. J. Biol. Chem. 265:10637-10644. [PubMed] [Google Scholar]
- 48.Priebe, S. D., and S. A. Lacks. 1989. Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J. Bacteriol. 171:4778-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Projan, S. J., and G. L. Archer. 1989. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J. Bacteriol. 171:1841-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Projan, S. J., S. Carleton, and R. P. Novick. 1983. Determination of plasmid copy number by fluorescence densitometry. Plasmid 9:182-190. [DOI] [PubMed] [Google Scholar]
- 51.Projan, S. J., J. Kornblum, S. L. Moghazeh, I. Edelman, M. L. Gennaro, and R. P. Novick. 1985. Comparative sequence and functional analysis of pT181 and pC221, cognate plasmid replicons from Staphylococcus aureus. Mol. Gen. Genet. 199:452-464. [DOI] [PubMed] [Google Scholar]
- 52.Projan, S. J., S. Moghazeh, and R. P. Novick. 1988. Nucleotide sequence of pS194, a streptomycin resistance plasmid from Staphylococcus aureus. Nucleic Acids Res. 16:2179-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Projan, S. J., and R. Novick. 1988. Comparative analysis of five related staphylococcal plasmids. Plasmid 19:203-221. [DOI] [PubMed] [Google Scholar]
- 54.Rawlings, D. E., and E. Tietze. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65:481-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Schaberg, D. R., D. B. Clewell, and L. Glatzer. 1982. Conjugative transfer of R-plasmids from Streptococcus faecalis to Staphylococcus aureus. Antimicrob. Agents Chemother. 22:204-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94:133-138. [DOI] [PubMed] [Google Scholar]
- 58.Scherzinger, E., R. Lurz, S. Otto, and B. Dobrinski. 1992. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 20:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwarz, S., and M. Cardoso. 1991. Nucleotide sequence and phylogeny of a chloramphenicol acetyltransferase encoded by the plasmid pSCS7 from Staphylococcus aureus. Antimicrob. Agents Chemother. 35:1551-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Street, L. M., M. J. Harley, J. C. Stern, C. Larkin, S. L. Williams, D. L. Miller, J. A. Dohm, M. E. Rodgers, and J. F. Schildbach. 2003. Subdomain organization and catalytic residues of the F factor TraI relaxase domain. Biochim. Biophys. Acta 1646:86-99. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Kranenburg, R., and W. M. de Vos. 1998. Characterization of multiple regions involved in replication and mobilization of plasmid pNZ4000 coding for exopolysaccharide production in Lactococcus lactis. J. Bacteriol. 180:5285-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogel, A. M., and A. Das. 1994. Mutational analysis of Agrobacterium tumefaciens pTiA6 virD1: identification of functionally important residues. Mol. Microbiol. 12:811-817. [DOI] [PubMed] [Google Scholar]
- 64.Walters, J. A., and K. G. Dyke. 1990. Characterization of a small cryptic plasmid isolated from a methicillin-resistant strain of Staphylococcus aureus. FEMS Microbiol. Lett. 59:55-63. [DOI] [PubMed] [Google Scholar]
- 65.Willetts, N., and B. Wilkins. 1984. Processing of plasmid DNA during bacterial conjugation. Microbiol. Rev. 48:24-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, G. Lanka, W. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool: bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 67.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ziegelin, G., W. Pansegrau, B. Strack, D. Balzer, M. Kroger, V. Kruft, and E. Lanka. 1991. Nucleotide sequence and organization of genes flanking the transfer origin of promiscuous plasmid RP4. DNA Seq. 1:303-327. [DOI] [PubMed] [Google Scholar]