Abstract
A corynebacterial clone, previously isolated by scoring repression of lacZYA fused to the aceB promoter of Corynebacterium glutamicum, was analyzed further. In the clone, an open reading frame designated glxR, consisting of 681 nucleotides and encoding a 24,957-Da protein, was found. The molecular mass of a native GlxR protein was estimated by gel filtration column chromatography to be 44,000 Da, suggesting that the protein formed dimers. The predicted amino acid sequence contained both cyclic AMP (cAMP)- and DNA-binding motifs and was homologous with the cAMP receptor protein family of proteins. The aceB-repressing activity of the glxR clone was markedly relieved in an Escherichia coli cya mutant, but the activity was restored in growth medium containing cAMP. In glucose medium, the intracellular cAMP concentration of C. glutamicum reached 22 nmol/mg of protein in the early exponential phase and then decreased further; but in acetate medium, the intracellular cAMP concentration was only 5 nmol/mg of protein and remained low throughout the growth phase. The expression of glxR was not affected by the carbon source. Binding of purified GlxR to the promoter region of aceB could be demonstrated only in the presence of cAMP. These data suggest that GlxR may form dimers which bind to the aceB promoter region in the presence of cAMP and repress the glyoxylate bypass genes.
Corynebacterium glutamicum, a gram-positive organism, is well known as a host organism for the industrial production of amino acids such as glutamate and lysine (19). Due to C. glutamicum's role in amino acid production, the catabolic and anabolic pathways leading to these industrially important amino acids have been studied in detail (for reviews, see references 24, 31, and 32). Although it is necessary to understand the metabolic pathways in detail, information on the regulatory mechanisms at the level of gene expression is very limited.
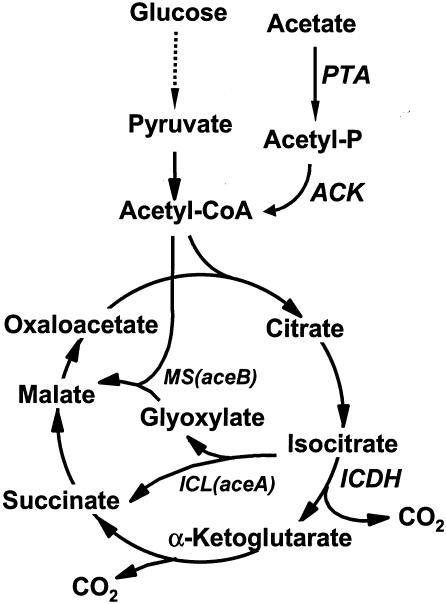
The glyoxylate bypass system of C. glutamicum is a good candidate for studying the regulation of gene expression because the expression of the isocitrate lyase and malate synthase enzymes that catalyze the bypass (Fig. 1) is quite responsive to the availability of carbon sources (43). Isocitrate lyase, encoded by aceA, catalyzes the conversion of isocitrate, a tricarboxylic acid cycle intermediate, into glyoxylate and succinate (29). Malate synthase, encoded by aceB, catalyzes the subsequent condensation of glyoxylate with acetyl-coenzyme A to produce malate, which is a tricarboxylic acid cycle intermediate (21, 30). The bypass conserves carbon to synthesize cell materials by bypassing the CO2-generating steps of the tricarboxylic acid cycle.
FIG. 1.
Glyoxylate bypass and associated pathways of C. glutamicum. The glyoxylate bypass is carried out by isocitrate lyase and malate synthase, which are encoded by the aceA and aceB genes, respectively. Abbreviations: ACK, acetate kinase; ICDH, isocitrate dehydrogenase; ICL, isocitrate lyase; MS, malate synthase; PTA, phosphotransacetylase; CoA, coenzyme A. Dashed arrows indicate multiple steps.
The aceA and aceB genes are derepressed when two-carbon compounds such as acetate are provided as a sole carbon source. Glucose, supplied as a carbon source, represses the aceA and aceB genes. Thus, expression of the glyoxylate bypass enzymes is regulated transcriptionally by the available carbon sources (43), but the mechanism by which this is achieved is not known, although Wendisch et al. suggested the involvement of acetyl-coenzyme A as a signaling molecule (43). In Escherichia coli, the IclR repressor controls the regulation of the aceBAK operon (6, 38) and hence bypasses expression. The expression of iclR is regulated by FadR, which is known to regulate the expression of genes involved in fatty acid metabolism. Although the structural organization of the aceA and aceB genes in C. glutamicum is different from that in Escherichia coli, they may have common features related to the regulation of gene expression by available carbon sources.
In our previous study, we constructed a reporter plasmid to isolate genes exerting regulatory effects on the aceB promoter of C. glutamicum (18). As reported here, a gene, glxR, was found in one of the isolated clones. In this report, we performed a functional analysis of the glxR gene and showed its involvement in regulating the expression of aceB.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains, plasmids, and primers used in this study are listed in Table 1. E. coli DH5αF′ was used for the construction and propagation of plasmids. E. coli JM105 was used to express GlxR from pKK-glxR. The operon fusion oPaceB-lacZYA constructed in plasmid pRS415 was transferred to the chromosome of E. coli strains with lambda phage λRS415 as described by Simons et al. (36). Unless otherwise stated, E. coli and C. glutamicum cells were cultured at 37°C in Luria broth (LB) (34) and at 30°C in MB (12), respectively. Minimal media for E. coli and C. glutamicum were M9 (34) and MCGC (42), respectively. Glucose and acetate as carbon sources were added to the minimal medium at 1% and 2%, respectively. Antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; tetracycline, 20 μg/ml; and kanamycin, 20 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) and cyclic AMP (cAMP) were added to the media at concentrations of 40 μg/ml and 8 mM, respectively. For protein expression, isopropylthiogalactopyranoside (IPTG) was added to a final concentration of 1 or 0.3 mM.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain plasmid or primer | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| DH5αF′ | F− φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169λ− | Bethesda Research Laboratories |
| JM105 | endA1 supE sbcB15 thi rpsL Δ(lac-proAB)/F′(traD36 proAB+lacIqlacZΔM15) | NIGb |
| TP2010 | xyl cya argH lacX74 recA ilvA srl::Tn10 | 7 |
| TP2139 | crp lacX74 xyl ilvA argH recA1 | 7 |
| DH5αF′-145 | DH5αF′ containing plasmid pSL145 | 18 |
| HL994 | DH5αF′ containing chromosomal PaceB-lacZYA fusion | This study |
| HL989 | TP2010 containing chromosomal PaceB-lacZYA fusion | This study |
| C. glutamicum | ||
| AS019 | Spontaneous rifampin-resistant mutant of C. glutamicum ATCC 13059 | 44 |
| AS019E12 | Restriction-deficient variant of AS019 | 12 |
| Plasmids | ||
| pBluescript SK(+) | Cloning vector; AprlacZ, 2,958 bp | Stratagene |
| pKK223-3 | Expression vector; Ptac Apr, 4,587 bp | Amersham Pharmacia Biotech |
| pMAL-c2 | Expression vector; AprPtac malE lac1qlacZα, 6,723 bp | New England Biolabs |
| pRS415 | Promoterless lac operon fusion vector; Apr, 10,752 bp | 36 |
| λRS45 | Phage vector for transferring genes into chromosome | 36 |
| pMT1 | Shuttle vector; Apr Kmr | 12 |
| pSL130 | pRS415 carrying 2.3-kb fragment including aceB promoter region, Apr | 18 |
| pSL145 | pACYC184 carrying 9.1-kb fragment including PaceB-lacZYA, Tcr | 18 |
| pSL329 | pMT1 carrying 7.8-kb fragment including glxR | 18 |
| pSL329-1 | pMT1 carrying 5.1-kb EcoRI-XbaI fragment of pSL329 | This study |
| pSL329-5 | pMT1 carrying 1.8-kb KpnI fragment of pSL329 | This study |
| pSL08 | pMT1 carrying 4.3-kb fragment including aceB | 21 |
| pSL08-glxR | pSL08 carrying 1.8-kb KpnI-BamHI fragment including glxR of pSL329-5 | This study |
| pKK-glxR | pKK223-3 carrying 0.83-kb fragment including glxR | This study |
| pMAL-glxR | pMAL-c2 carrying 0.89-kb fragment including glxR | This study |
| Primersc | ||
| C1 | 5′CGGAATTCGATGTGTGCAGATGAAGG3′ | |
| C2 | 5′CGGAATTCTATTCGTTACCTGCAGGC3′ | |
| D1 | 5′CGGAATTCGTGGAAGGTGTACAGG3′ | |
| D2 | 5′ACGCGTCGACTATTCGTTACCTGCAGG3′ | |
| E1 | 5′GGAAAATGCAGGCACCGC3′ | |
| E2 | 5′GACTACCTCTGGAATCTAGG3′ |
Ap, ampicillin; Km, kanamycin; Tc, tetracycline.
NIG, National Institute of Genetics, Japan.
Underlining indicates either an EcoRI or SalI site.
DNA technology.
Standard molecular cloning, transformation, and electrophoresis procedures were used (34). Plasmids were introduced into C. glutamicum cells by electroporation (12). Miniplasmid preparation for C. glutamicum cells was performed as described previously (44). Chromosomal DNA from C. glutamicum AS019E12 was prepared as described previously (40). Restriction enzymes and DNA-modifying enzymes were purchased from Takara Shuzo Co. (Shiga, Japan) and New England Biolabs (Beverly, Mass.) and used as recommended by the manufacturer.
Cloning and sequencing.
Plasmids pSL329-1 and pSL329-5 were constructed by ligating the 5.1-kb EcoRI-XbaI and 1.8-kb KpnI fragments of pSL329 into the pMT1 vector digested with SmaI plus XbaI and with KpnI, respectively. Plasmid pSL08-glxR was constructed by inserting the 1.8-kb KpnI-BamHI fragment of pSL329-5 into pSL08 which had been digested with XbaI and then blunt-ended with the Klenow fragment of DNA polymerase I. The direction of transcription of the cloned glxR gene was opposite that of the aceB gene. Plasmid pKK-glxR was constructed by amplifying the 828-bp fragment of pSL329-5 with primers C1 and C2 (Table 1), followed by insertion of the resulting fragment into the EcoRI site of pKK223-3. The 828-bp fragment carries glxR and its ribosome-binding site. Plasmid pMAL-glxR was constructed as follows. The glxR coding region was amplified with primers D1 and D2 (Table 1) with plasmid pSL329-5 as a template. The 0.89-kb PCR product was treated with EcoRI and SalI and ligated into EcoRI- and SalI-digested and dephosphorylated pMAL-c2.
For nucleotide sequence analysis of glxR, pSL329-5 was used as the template. The complete nucleotide sequence of glxR was determined at the Korea Research Center for Basic Sciences (Taejon, Korea) with universal and synthetic oligonucleotide primers. A search for similar nucleotide and amino acid sequences was performed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) with the Basic Local Alignment Search Tool (BLAST) (1, 2). Pairwise sequence alignments were performed at the ExPASy Proteomics Tools website (http://www.expasy.ch/tools/) with the ClustalW alignment method (39).
Purification of GlxR.
Plasmid pMAL-glxR was designed to express GlxR. The vector carries glxR fused to malE and expresses the maltose-binding protein (MBP)-GlxR fusion protein. The fusion protein was expressed and purified as suggested by the manufacturer (New England Biolabs). Cleavage of the fusion protein with factor Xa releases a free GlxR protein with a string of amino acids (Ile-Ser-Val-Phe) attached to the N terminus. GlxR was separated from MBP by Q-Sepharose Fast Flow column chromatography (Amersham Pharmacia Biotech; 2.8 by 11 cm) with a linear gradient of 25 to 500 mM NaCl. Fractions containing GlxR were pooled, and the protein was concentrated by dialysis against polyethylene glycol 8000.
Gel mobility shift assay.
The probe DNA was prepared as follows. The 200-bp DNA fragment which includes an upstream region of the aceB gene (from 120 to 320 bp upstream from the ATG start codon) was amplified with primers E1 and E2 (Table 1) and subsequently labeled with [γ-32P]dATP and T4 polynucleotide kinase; 10 μl of the binding reaction mixture contained the labeled DNA fragments, various amounts of purified GlxR, 10 mM phosphate buffer, 137 mM NaCl, 2.7 mM KCl, 3 μg of bovine serum albumin, and 1 μg of poly(dI-dC) · poly(dI-dC). GlxR binding to the probe DNA was performed at room temperature for 15 min, and the mixture was analyzed on 6% native polyacrylamide gels as described before (34). When necessary, 0.2 mM cAMP was included in the binding buffer, gels, and running buffer.
Biochemical analysis and preparation of antibody.
Corynebacterial cell extracts were prepared as described previously (16). The enzymatic activities of β-galactosidase (25), malate synthase (8, 15), isocitrate lyase (8, 15), isocitrate dehydrogenase (13), and acetate kinase (41) were determined as described previously. Protein concentration was measured by the method of Bradford, with bovine serum albumin as the standard (4). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis were performed as described previously (20). The anti-GlxR antiserum was prepared commercially at Koma Biotech (Seoul, Korea) with a rat as the host.
Measurements of cAMP and glucose.
For measurement of the intracellular cAMP concentration, cells were prepared as described previously (35). The cAMP concentration was measured with the Biotrak cAMP enzyme immunoassay system (Amersham Pharmacia Biotech; RPN 225). Glucose was measured with a glucose assay kit (Sigma Chemical Co. St. Louis, Mo.).
Nucleotide sequence accession number.
The nucleotide sequence of glxR was deposited in GenBank under accession number AF293334.
RESULTS
Isolation of the glxR gene.
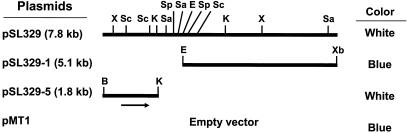
Plasmid pSL329 (Fig. 2), which carries a 7.8-kb insert, repressed expression of aceB-lacZYA (18). The DNA region responsible for modulating β-galactosidase activity was narrowed down to 1.8 kb by patching the cells harboring each subclone on LB plates containing X-Gal (Fig. 2). In accordance with the color test data, E. coli DH5αF′-145 cells carrying plasmid pSL329 or pSL329-5 showed only 10% of the β-galactosidase activity of the parental strain (data not shown). The data suggest that a gene (glxR, see below) in the cloned DNA expresses proteins which may bind to the aceB gene promoter region, interfering with the expression of lacZYA.
FIG. 2.
Schematic diagram of clones and subclones. The colony color (blue or white) of E. coli DH5αF′-145 cells carrying each clone was tested on LB plates containing X-Gal, tetracycline, and ampicillin. The arrow indicates the glxR gene. The cloning vector pMT1 is not shown. Abbreviations: B, BamHI; E, EcoRI; K, KpnI; Sa, SalI; Sc, ScaI; Sp, SphI; X, XhoI; Xb, XbaI.
Sequence analysis of the glxR gene.
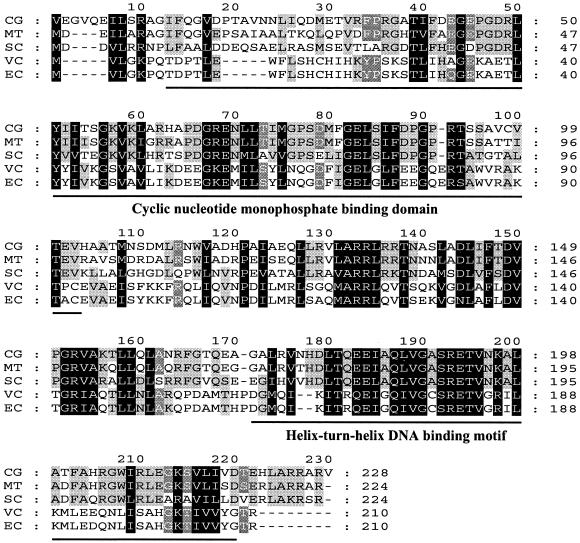
The complete nucleotide sequence of the clone was determined with pSL329-5 as the sequencing template. An open reading frame (ORF) composed of 684 bp was found at the central region of the clone (Fig. 2). Based on similarity to other proteins (see below), GTG was chosen as the start codon. A potential ribosome-binding site, AGGA, was found 9 bp upstream from the GTG. The GC content of the ORF was 59%, which is typical of C. glutamicum genes. Codon preference was very similar to that of previously reported corynebacterial genes and, interestingly, also indicated that the ORF could encode a protein expressed at a low level (10). The putative gene product of 227 amino acids had a predicted mass of 24,957 Da and a predicted isoelectric point of 7.0. In the protein database, the translated amino acid sequence of the ORF was used to search for similar and matching amino acid sequences. Among the known proteins in the database, the putative transcriptional regulatory proteins of Mycobacterium tuberculosis (E70790) and Streptomyces coelicolor (T36556) showed high identity scores of 78 and 53%, respectively. Among the proteins with known roles, cAMP receptor proteins (CRP) of Vibrio cholerae (NP232242), Salmonella enterica serovar Typhimurium (A26049), and E. coli (J01598) showed the highest identity score, approximately 27%.
Close analysis of the amino acid sequence revealed the presence of two conserved motifs that may be involved in the protein's regulatory activity (Fig. 3). Amino acid residues 13 to 102 showed a conserved motif which is supposed to be a cAMP-binding domain, having 31% identity with the consensus sequence for cyclic nucleotide monophosphate-binding domains. In addition, a helix-turn-helix DNA binding motif of the CRP/FNR (fumarate-nitrate reduction regulator) family was identified at the carboxy-terminal region (amino acid residues 170 to 218) of the encoded protein. The region showed 41% identity with the helix-turn-helix motif of CRP. Based on the characteristics of the cloned gene discussed below, we designated the corynebacterial gene glxR (glyoxylate bypass regulator).
FIG. 3.
Multiple sequence alignment of the GlxR protein of C. glutamicum with homologous sequences. Conserved and functionally similar amino acids are marked with black and shaded boxes, respectively. The cyclic nucleotide monophosphate-binding motif and helix-turn-helix DNA-binding motif are marked with solid bars. Abbreviations: CG, GlxR of C. glutamicum (accession no. AF220150); MT, putative transcriptional regulator Rv3676 of Mycobacterium tuberculosis H37RV (E70790); SC, putative transcriptional regulator of Streptomyces coelicolor (T36556); VC, CRP of Vibrio cholerae (NP232242); EC, CRP of Escherichia coli (J01598).
Involvement of cAMP.
Since a cAMP-binding motif was found in the nucleotide sequence of glxR, cAMP's involvement in modulating GlxR activity was examined. For the experiment, the PaceB-lacZYA fusion was introduced by phage-mediated transduction into the chromosome of an E. coli cya mutant strain. In the strain, the aceB-repressing activity decreased markedly with the introduction of a glxR clone, but the addition of 10 mM cAMP to the growth medium retrieved the repressible activity, indicating that GlxR needs cAMP for its repressible activity at the aceB promoter (Table 2). To investigate the functional similarities between the glxR and crp genes, a complementation test was carried out. E. coli crp mutants cannot utilize maltose as a carbon source because the maltose regulon is not expressed without a crp gene (7). E. coli crp mutant cells harboring plasmid pSL329-5 showed growth on plates containing maltose as a carbon source, indicating that GlxR can play a role equivalent to that of CRP in E. coli crp mutant cells (Table 3).
TABLE 2.
Effect of cAMP on repression of the glxR genea
| Host cells | Plasmid | Relevant genotype | cAMP | Relative β-galactosidase activity |
|---|---|---|---|---|
| E. coli HL994 | pMT1 | cya+ | − | 1 |
| pSL329-5 | cya+glxR | − | 0.52 ± 0.06 | |
| E. coli HL989 | pMT1 | cya | − | 1 |
| pSL329-5 | cya glxR | − | 0.91 ± 0.07 | |
| pSL329-5 | cya glxR | + | 0.37 ± 0.05 |
Cells were grown on LB medium, and cell extracts were prepared as described in Materials and Methods. PaceB-lacZYA was introduced into the chromosome as described in Materials and Methods. cAMP was added to a final concentration of 10 mM.
TABLE 3.
Complementation of the E. coli TP2139 crp mutant by glxRa
| Plasmid | Relevant genotype | Growthb
|
|
|---|---|---|---|
| Maltose | Glucose | ||
| pMT1 | crp | − | ++ |
| pSL329-5 | crp glxR+ | + | ++ |
Cells were grown on M9 minimal medium containing maltose or glucose as a carbon source. Growth was monitored on agar plates.
++, good growth; +, growth; −, no growth.
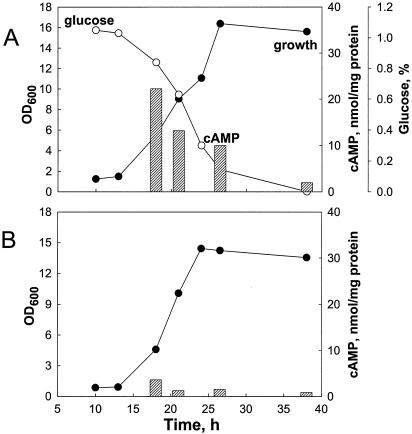
To examine the relationship between cAMP and GlxR, the intracellular cAMP concentration was measured in glucose- and acetate-grown C. glutamicum cells (Fig. 4). Unlike the case in E. coli, the intracellular concentration of cAMP was high (22 nmol/mg of protein) in the early exponential phase in glucose-grown C. glutamicum cells but decreased and was 11 nmol/mg of protein in the early stationary phase, indicating that the intracellular cAMP concentration in C. glutamicum cells appears to decrease as glucose is consumed. In acetate-grown cells, however, the initial cAMP concentration was considerably lower (5 nmol/mg of protein) in the early exponential phase and remained low throughout the growth phase.
FIG. 4.
Measurement of intracellular cAMP in C. glutamicum. Cells were grown in MCGC containing glucose (A) or acetate (B) as a carbon source. Intracellular cAMP (shaded bars) was measured as described in Materials and Methods. The data represent three independent experiments. Symbols: •, growth; ○, glucose. OD, optical density at 600 nm.
Expression of glxR in C. glutamicum.
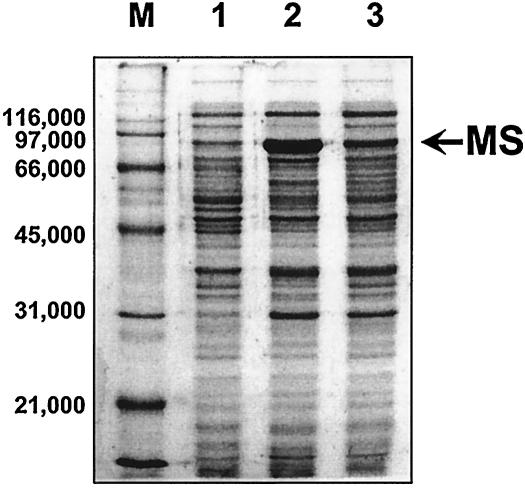
Next, after introducing plasmid pSL329-5, which is a glxR clone, into C. glutamicum AS019E12, its effects were monitored by measuring the activities of isocitrate lyase and malate synthase, which are glyoxylate bypass enzymes; acetate kinase, which converts acetylphosphate from acetate into acetyl-coenzyme A; and isocitrate dehydrogenase, which is a tricarboxylic acid cycle enzyme (Fig. 1). In glucose medium, only basal-level expression of the glyoxylate bypass enzymes was observed, and the activities were affected very little by the presence of the glxR clone (Table 4). The specific activity of isocitrate dehydrogenase increased significantly in C. glutamicum cells harboring plasmid pSL329-5 compared to the parental cells. In acetate medium, however, the specific activities of malate synthase and isocitrate lyase decreased significantly (92 to 93%) in C. glutamicum cells harboring plasmid pSL329-5 from the value in the parental cells (Table 4). Lowered malate synthase activity caused by the introduced glxR gene was due to decreased expression of malate synthase protein, as judged by SDS-PAGE (Fig. 5). For the experiment in Fig. 5, the glxR gene was inserted into the vector that carries and overexpresses the aceB gene (pSL08), and the resulting plasmid (pSL08-glxR) was introduced into C. glutamicum for PAGE analysis of the expressed proteins. Under growth with either glucose or acetate as the carbon source, the amount of GlxR protein expressed in C. glutamicum cells was unaffected, as evidenced by Western blot analysis (data not shown).
TABLE 4.
Activities of isocitrate lyase (ICL), malate synthase (MS), acetate kinase (ACK), and isocitrate dehydrogenase (ICDH) in cell extracts of C. glutamicum harboring a glxR clonea
| Carbon source | Plasmid | Description | Sp act (nmol min−1 mg−1)
|
|||
|---|---|---|---|---|---|---|
| ICL | MS | ACK | ICDH | |||
| Glucose | pMT1 | Empty vector | 9.0 ± 3.0 | 40 ± 5.0 | 267 ± 22 | 12 ± 2.0 |
| pSL329-5 | glxR | 10 ± 2.0 | 32 ± 7.5 | 155 ± 12 | 21 ± 3.0 | |
| Acetate | pMT1 | Empty vector | 971 ± 25 | 460 ± 20 | 478 ± 35 | 26 ± 3.0 |
| pSL329-5 | glxR | 68 ± 11 | 38 ± 10 | 337 ± 21 | 22 ± 2.0 | |
The enzymes were induced by growth of C. glutamicum AS019E12 cells to the stationary phase on MCGC minimal medium. Cells were harvested, disrupted, and assayed as described in Materials and Methods. Values are means ± standard deviations.
FIG. 5.
Effect of glxR on the expression of malate synthase (MS). Cells were grown on MB containing acetate. Crude extracts were prepared as described in Materials and Methods. For each lane, a total of 5 μg of protein was loaded. Lanes: M, molecular mass markers (sizes are shown in daltons); 1, C. glutamicum/pMT1; 2, C. glutamicum/pSL08; 3, C. glutamicum/pSL08-glxR.
Expression and purification of GlxR.
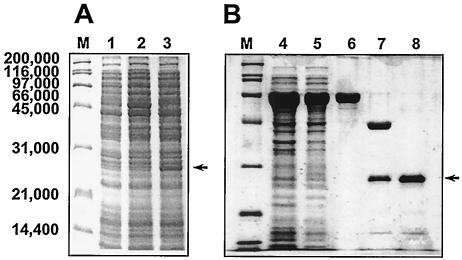
Introducing the glxR coding region including the ribosome-binding site into vector pKK223-3 and transforming the resulting vector into E. coli resulted in the expression of a 25,000-Da protein, as shown by SDS-PAGE (Fig. 6A). The observed molecular mass agreed with the predicted size of the protein. The GlxR protein was purified by MBP affinity column chromatography. As expected, the purified protein showed a mass of 25,000 Da on SDS-PAGE (Fig. 6B). The native molecular mass of the purified protein was 44,000 Da, as judged by gel filtration column chromatography (data not shown). The data suggest that the proteins are likely to form functional dimers, like most DNA-binding proteins.
FIG. 6.
Expression and purification of GlxR in E. coli. (A) The GlxR protein was expressed from the pKK-glxR vector as described in Materials and Methods. Lanes: 1, E. coli JM105/pKK223-3; 2, E. coli JM105/pKK-glxR (uninduced); 3, E. coli JM105/pKK-glxR (induced). (B) Purification of GlxR. The protein was expressed from pMAL-glxR. Lanes: 4, total protein; 5, soluble fraction; 6, purified MBP-GlxR fusion protein; 7, after factor X treatment; 8, purified GlxR protein. Lanes M, molecular mass standards (sizes are shown in daltons). Arrows indicate GlxR protein.
DNA-binding activity of GlxR.
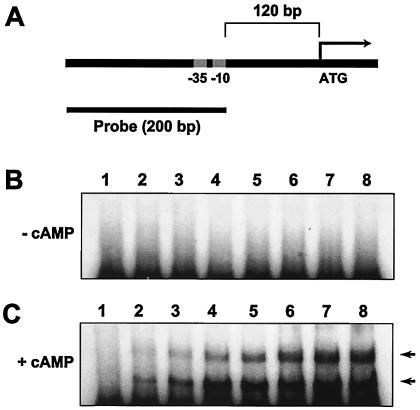
Identifying GlxR's involvement in controlling the expression of aceB and a helix-turn-helix DNA binding motif of the CRP/FNR family in the carboxy-terminal region (amino acid residues 170 to 218) of the encoded protein, we determined the ability of purified GlxR to bind to the promoter region of the aceB gene. A DNA fragment carrying the promoter region of aceB was used as a probe (Fig. 7A). In the absence (Fig. 7B) and presence (Fig. 7C) of cAMP, increasing amounts of purified GlxR protein were incubated with the probe DNA, and two shifted bands resulted when cAMP was present (Fig. 7C). It is uncertain whether the two bands are the result of GlxR protein oligomerization or if two binding sites with different affinities for the proteins exist in the probe DNA. Although an excess amount of purified GlxR protein was added to the assay mixture, a complete shift was not observed. The results indicate that some proteins might lose their activity or be degraded (Fig. 6B, lane 8). However, the finding that a band shift was observed only in the presence of cAMP may rule out the possibility that the band shift was caused by nonspecific interactions between the DNA and protein. Replacing cAMP with acetyl-coenzyme A, which has been suggested to be a modulator for the glyoxylate bypass enzymes (43), did not result in any DNA shifts with the probe (data not shown).
FIG. 7.
Gel shift assay with purified GlxR. Purified GlxR proteins were incubated with the probe, and the mixtures were analyzed by 6% PAGE. (A) Schematic diagram of the aceB promoter region. (B) Gel shift in the absence of cAMP. (C) Gel shift in the presence of cAMP. cAMP was added (to 0.2 mM) to the assay mixture, gels, and PAGE buffer. Lane 1 contained no protein; lanes 2 through 8 contained 30 ng, 60 ng, 130 ng, 250 ng, 500 ng, 1.0 μg, and 2.0 μg of purified GlxR protein, respectively. Arrows indicate shifted bands.
DISCUSSION
In this study, we found that GlxR proteins are involved in the regulation of the aceB gene, which in turn is concerned with the utilization of acetate as a carbon source. The evidence is as follows: the gene products of glxR clones modulate β-galactosidase activity in a reporter plasmid carrying lacZYA fused to the aceB promoter of C. glutamicum, multicopies of glxR lead to decreased activities in glyoxylate bypass enzymes, the GlxR protein contains a putative DNA-binding domain, and the protein has the ability to bind to the aceB promoter region. Like other DNA-binding proteins, the findings that the proteins are present as dimers and have a relatively basic isoelectric point also support a regulatory role for the protein. Although our data suggest that the glxR gene might be involved primarily in regulating gene expression in the glyoxylate bypass, such as aceA and aceB, we cannot rule out the possibility that the gene is also involved in the expression of other genes.
The growth retardation by glxR in multicopies in glucose or acetate minimal medium suggests that the GlxR protein may interact with other promoters (data not shown), although it could also be due to nonspecific binding of the GlxR protein to DNA. The fact that we were unable to construct a glxR mutant strain with the cloned DNA despite numerous attempts (data not shown) indicates that the gene is essential and that the glxR gene is involved in the regulation of other genes or functions as an activator of other genes, like CRP in E. coli. CRP plays a role in carbon catabolite repression, which is governed by cAMP in enteric bacteria such as E. coli (11). It is interesting that GlxR in C. glutamicum and CRP in E. coli have several common aspects: they have about 27% identity in amino acid sequence, they have a cAMP-binding motif, cAMP modulates their regulatory activities, and the glxR gene complements the E. coli crp mutation. Despite these similarities, however, GlxR appears to be distinct from CRP in E. coli. Our failure to construct a glxR mutant strain may also suggest different roles.
cAMP is an important signaling molecule controlling the expression of many other genes. The involvement of cAMP in many bacteria has been reported, such as enteric bacteria, Bacillus spp., and Streptomyces spp. (3). In E. coli, the molecule plays important roles in regulating carbon catabolite genes, such as the lactose and arabinose operons (11). Under conditions of glucose starvation, the intracellular concentration of cAMP becomes elevated, a general phenomenon found in many bacteria with the exception of Brevibacterium species, which apparently keeps the concentration of cAMP high when cells are grown on glucose minimal medium (22, 27). Similarly, in C. glutamicum, the intracellular concentration of cAMP is kept low when cells are grown on acetate medium but high when they are grown on glucose medium, as evidenced in our growth experiment (Fig. 4). Addition of cAMP to the medium did not seem to affect the growth of C. glutamicum cells, whereas cAMP enhanced the effects of growth retardation found in C. glutamicum cells harboring multicopies of glxR clones in both glucose and acetate medium, but especially in acetate medium (data not shown). In enteric bacteria, the addition of cAMP induces the growth retardation of cells when the intracellular cAMP concentration is low, as in high-glucose medium (9). Although this contrasts with the established activities in E. coli, other researchers reported that cAMP is present at its highest levels during periods of rapid growth rather than in glucose-limited conditions in Streptomyces spp. and thus signals the availability rather than the lack of carbon sources (9, 14, 28). This also appears to be true for C. glutamicum, as shown in this study.
Although we isolated the glxR gene by its role in regulating the aceB gene, the activities of isocitrate lyase and malate synthase, which are involved in the glyoxylate bypass and active in acetate medium, decreased quite a lot in C. glutamicum cells harboring multiple copies of glxR. Since growth on acetate medium probably requires no repressors for the aceA and aceB genes, repression of the genes by the introduced glxR clone is as yet unexplained. As generally found with some transcriptional regulators, this effect appears to be caused by the glxR gene's multicopy effects. Under these conditions, the overexpressed GlxR proteins may scavenge intracellular cAMP and subsequently bind to the promoter region for repression of the aceA and aceB genes. However, we cannot rule out the possibility that growth retardation on acetate medium by the glxR gene may have been caused by other regulatory effects displayed by the gene.
Although the cAMP-independent mechanisms of catabolite repression have been studied in detail in low-GC gram-positive bacteria, such as Bacillus spp., there is a dearth of information on high-GC gram-positive bacteria, such as Corynebacterium species (for reviews, see references 3, 5, 26, 33, and 37). Therefore, this study is the first to present evidence describing cAMP's involvement in the expression of genes for carbon utilization, although the physiological apparatus determining the intracellular cAMP concentration has not been clarified yet.
Acknowledgments
This work was supported by grants from BASF Korea (to H.-S. Lee) and the Ministry of Science and Technology (via 21C Microbial Genomics and Applications Center to H.-S. Lee).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Bruckner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 6.Chung, T., D. J. Klumpp, and D. C. LaPorte. 1988. Glyoxylate bypass operon of Escherichia coli: cloning and determination of the functional map. J. Bacteriol. 170:386-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cossart, P., and B. Gicquel-Sansey. 1985. Regulation of expression of the crp gene of Escherichia coli K-12: in vivo study. J. Bacteriol. 161:454-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon, G. H., and H. L. Kornberg. 1959. Assay methods for key enzymes of the glyoxylate cycle. Biochem. J. 72:3P. [Google Scholar]
- 9.Dobravá, Z., Naprstek, J., Jiresová, M., and J. Janecek. 1984. cAMP and adenylate cyclase in Streptomyces granaticolor. FEMS Microbiol. Lett. 11:197-200. [Google Scholar]
- 10.Eikmanns, B. J. 1992. Identification, sequence analysis, and expression of a Corynebacterium glutamicum gene cluster encoding the three glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase, 3-phosphoglycerate kinase, and triosephosphate isomerase, J. Bacteriol. 174:6076-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epstein, W., L. B. Rothman-Denes, and J. Hesse. 1975. Adenosine 3′:5′-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc. Natl. Acad. Sci. USA 72:2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Follettie, M. T., O. Peoples, C. Agoropoulou, and A. J. Sinskey. 1993. Gene structure and expression of the Corynebacterium flavum N13 ask-asd operon. J. Bacteriol. 175:4096-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnak, M., and H. Reeves. 1979. Purification and properties of phosphorylated isocitrate dehydrogenase of Escherichia coli. J. Biol. Chem. 254:7915-7920. [PubMed] [Google Scholar]
- 14.Gersch, D., W. Romer, H. Bocker, and H. Thrum. 1978. Variations in cyclic adenosine 3′,5′-monophosphate and cyclic guanosine 3′,5′-monophosphate in antibiotic producing strains of Streptomyces hygroscopicus. FEMS Microbiol. Lett. 3:39-41. [Google Scholar]
- 15.Gubler, M., S. M. Park, M. Jetten, G. Stephanopoulos, and A. J. Sinskey. 1993. Effects of phosphoenolpyruvate carboxylase deficiency on metabolism and lysine production in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 40:857-863. [Google Scholar]
- 16.Hwang, B.-J., H.-J. Yeom, Y. Kim, and H.-S. Lee. 2002. Corynebacterium glutamicum utilizes both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis. J. Bacteriol. 184:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jetten, M. S. M., M. E. Gubler, S. H. Lee, and A. J. Sinskey. 1994. Structural and functional analysis of pyruvate kinase from Corynebacterium glutamicum. Appl. Environ. Microbiol. 60:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H.-J., Y. Kim, M.-S. Lee, and H.-S. Lee. 2001. Gene lmrB of Corynebacterium glutamicum confers efflux-mediated resistance to lincomycin. Mol. Cell 12:112-116. [PubMed] [Google Scholar]
- 19.Kinoshita, S. 1995. Glutamic acid bacteria, p. 115-142. In A. L. Demain and N. A. Solomon (ed.), Biology of industrial microorganisms. Benzamin/Cummings Publishing Company, London, England.
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Lee, H.-S., and A. J. Sinskey. 1994. Molecular characterization of aceB, a gene encoding malate synthase in Corynebacterium glutamicum. J. Microbiol. Biotechnol. 4:256-263. [Google Scholar]
- 22.Lynch, T. J., E. A. Tallant, and W. Y. Cheung. 1975. Brevibacterium liquefaciens adenylate cyclase and its in vivo stimulation by pyruvate. J. Bacteriol. 124:1106-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malumbres, M., J. A. Gil, and J. F. Martín. 1993. Codon preference in corynebacteria. Gene 134:15-24. [DOI] [PubMed] [Google Scholar]
- 24.Malumbres, M., and J. F. Martin. 1996. Molecular control mechanisms of lysine and threonine biosynthesis in amino acid-producing corynebacteria: redirecting carbon flow. FEMS Microbiol. Lett. 143:103-114. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Paulsen, I. T. 1996. Carbon metabolism and its regulation in Streptomyces and other high GC Gram-positive bacteria. Res. Microbiol. 147:535-541. [DOI] [PubMed] [Google Scholar]
- 27.Peters, E. P., A. F. Wilderspin, S. P. Wood, M. J. J. M. Zvelebil, O. Sezer, and A. Danchin. 1991. A pyruvate-stimulated adenylate cyclase has a sequence related to the fes/fps oncogenes and to eukaryotic cyclases. Mol. Microbiol. 5:1175-1181. [DOI] [PubMed] [Google Scholar]
- 28.Ragan, C. M., and L. C. Vining. 1978. Intracellular cyclic adenosine 3′,5′-monophosphate levels and streptomycin production in cultures in Streptomyces griseus. Can. J. Microbiol. 24:1012-1015. [DOI] [PubMed] [Google Scholar]
- 29.Reinscheid, D. J., B. J. Eikmanns, and H. Sahm. 1994. Characterization of the isocitrate lyase gene from Corynebacterium glutamicum and biochemical analysis of the enzyme. J. Bacteriol. 176:3474-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinscheid, D. J., B. J. Eikmanns, and H. Sahm. 1994. Malate synthase from Corynebacterium glutamicum: sequence analysis of the gene and biochemical characterization of the enzyme. Microbiology 140:3099-3108. [DOI] [PubMed] [Google Scholar]
- 31.Sahm, H., L. Eggeling, B. Eikmanns, and R. Krämer. 1995. Metabolic design in amino acid producing bacterium Corynebacterium glutamicum. FEMS Microbiol. Rev. 16:243-252. [Google Scholar]
- 32.Sahm, H., L. Eggeling, and A. A. de Graaf. 2000. Pathway analysis and metabolic engineering in Corynebacterium glutamicum. Biol. Chem. 381:899-910. [DOI] [PubMed] [Google Scholar]
- 33.Saier, M. H., Jr. 1996. Cyclic AMP-independent catabolic repression in bacteria. FEMS Microbiol. Lett. 138:97-103. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Siegel, L. S., P. B. Hylemon, and P. V. Phibbs, Jr. 1977. Cyclic adenosine 3′,5′-monophosphate levels and activities of adenylate cyclase and cyclic adenosine 3′,5′-monophosphate phosphodiesterase in Pseudomonas and Bacteroides. J. Bacteriol. 129:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 37.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus subtilis. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 38.Sunnarborg, A., D. Klumpp, T. Chung, and D. C. LaPorte. 1990. Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J. Bacteriol. 172:2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomioka, N., K. Shinozaki, and M. Sugiura. 1981. Molecular cloning and characterization of ribosomal RNA genes from a blue-green alga, Anacystis nidulans. Mol. Gen. Genet. 184:359-363. [DOI] [PubMed] [Google Scholar]
- 41.van Dyk, T. K., and R. A. LaRossa. 1987. Involvement of ack-pta operon products in α-ketobutyrate metabolism by Salmonella typhimurium. Mol. Gen. Genet. 207:435-440. [DOI] [PubMed] [Google Scholar]
- 42.von der Osten, C. Gioannettie, and A. J. Sinskey. 1989. Design of a defined medium for growth of Corynebacterium glutamicum in which citrate facilitate iron uptake. Biotechnol. Lett. 11:11-16. [Google Scholar]
- 43.Wendisch, V. F., M. Spies, D. J. Reinschied, S. Schnicke, H. Sahm, and B. J. Eikmanns. 1997. Regulation of acetate metabolism in Corynebacterium glutamicum: transcriptional control of the isocitrate lyase and malate synthase genes. Arch. Microbiol. 168:262-269. [DOI] [PubMed] [Google Scholar]
- 44.Yoshihama, M., K. Higashiro, E. A. Rao, M. Akedo, W. G. Shanabruch, M. T. Follettie, G. C. Walker, and A. J. Sinskey. 1995. Cloning vector system for Corynebacterium glutamicum. J. Bacteriol. 162:591-597. [DOI] [PMC free article] [PubMed] [Google Scholar]