Abstract
ETT2 is a second cryptic type III secretion system in Escherichia coli which was first discovered through the analysis of genome sequences of enterohemorrhagic E. coli O157:H7. Comparative analyses of Escherichia and Shigella genome sequences revealed that the ETT2 gene cluster is larger than was previously thought, encompassing homologues of genes from the Spi-1, Spi-2, and Spi-3 Salmonella pathogenicity islands. ETT2-associated genes, including regulators and chaperones, were found at the same chromosomal location in the majority of genome-sequenced strains, including the laboratory strain K-12. Using a PCR-based approach, we constructed a complete tiling path through the ETT2 gene cluster for 79 strains, including the well-characterized E. coli reference collection supplemented with additional pathotypes. The ETT2 gene cluster was found to be present in whole or in part in the majority of E. coli strains, whether pathogenic or commensal, with patterns of distribution and deletion mirroring the known phylogenetic structure of the species. In almost all strains, including enterohemorrhagic E. coli O157:H7, ETT2 has been subjected to varying degrees of mutational attrition that render it unable to encode a functioning secretion system. A second type III secretion system-associated locus that likely encodes the ETT2 translocation apparatus was found in some E. coli strains. Intact versions of both ETT2-related clusters are apparently present in enteroaggregative E. coli strain O42.
The species Escherichia coli contains a wide range of commensal strains and pathogenic varieties (pathotypes) in addition to the model laboratory organism, E. coli K-12 (16). At least six pathotypes are associated with human intestinal disease: they are enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enteroinvasive E. coli, enteroaggregative E. coli (EAEC), and diffusely adherent E. coli. Two pathotypes are associated with extraintestinal disease in humans, namely uropathogenic E. coli (UPEC) and neonatal meningitic E. coli (NMEC). In addition, it is now clear that on phylogenetic grounds, all members of the genus Shigella belong within the species of E. coli (50). Furthermore, this dazzling phenotypic variety is matched by remarkable variations in genome size, with the largest E. coli genomes possessing more than a megabase more DNA than the smallest ones (43).
Initial studies of UPEC, and later of other pathotypes, suggested that E. coli strains often acquire new complex pathogenic phenotypes in a single step by the acquisition of pathogenicity islands, which contain virulence genes clustered on the chromosome and which are acquired en bloc by horizontal gene transfer (21). Similar studies with the related bacterium Salmonella enterica have delineated several Salmonella pathogenicity islands (Spi-1, Spi-2, Spi-3, etc.) (2, 22). The horizontal transfer of DNA by mobile elements such as bacteriophages and plasmids is also known to play a role in the evolution of virulence in E. coli and Shigella (15). However, if bacterial genomes are subject to a continual ingress of novel DNA through horizontal gene transfer, the question is raised of how this DNA is removed from the genome should it cease to provide any selective advantage (33). Furthermore, some authors have questioned the utility of the pathogenicity island concept when in some cases the position, order, and clustering of virulence genes seem remarkably fluid (61).
Many strains of EHEC and EPEC, like S. enterica, utilize type III secretion to subvert eukaryotic signaling pathways by injecting bacterial effector proteins into the host cell cytoplasm (27, 30). Within these pathotypes of E. coli, a well-characterized type III secretion system (TTSS), similar to the Spi-2 system of S. enterica and encoded by a pathogenicity island termed the locus of enterocyte effacement (LEE), is responsible for the development of the attaching-effacing lesion and for other effects on enterocyte functions (21, 30, 37, 38, 48).
An analysis of the complete genome sequences of two strains of EHEC O157:H7 revealed genes that potentially encode a second cryptic TTSS, which has been termed ETT2 (for E. coli TTSS 2, with the term ETT1 reserved for the LEE-encoded TTSS) and which resembles the SPI-1 TTSS from S. enterica (24, 49). Three recent studies have shown that the ETT2 gene cluster is found in some pathogenic strains of E. coli in addition to O157:H7 (23, 35, 40). However, these studies failed to agree on the boundaries of the cluster, were limited in terms of the phylogenetic diversity of the strains they sampled (leading to the erroneous conclusion that ETT2 is largely absent from nonpathogenic strains), and did not describe any ETT2-associated chaperone or translocator genes.
Prompted by the discovery of ETT2, we wished to address several interrelated questions: how widespread is the ETT2 gene cluster among E. coli isolates, how has this pathogenicity island evolved, and where are the ETT2 chaperones and translocators? In pursuit of these goals, we performed in silico analyses of ETT2 gene clusters available from genome sequences and other sources and developed a PCR-based approach, called tiling-path PCR scanning (TP-PCR), that allowed us to construct a complete tiling path through a 40-kb fragment of the chromosome centered on the ETT2 gene cluster for 79 well-validated and phylogenetically diverse strains drawn from the ECOR collection and representatives of selected pathotypes. We were surprised to discover that the ETT2 gene cluster is present in whole or in part in the majority of E. coli strains, but that in almost all cases, including that of EHEC O157:H7, it has been subjected to varying degrees of mutational attrition. In addition, we found a second type III secretion-associated locus (eip) in some E. coli strains which we predict encodes the ETT2 translocation apparatus.
MATERIALS AND METHODS
Sequence analysis. Comparative analyses of the regions surrounding and containing ETT2 and eip gene clusters were performed and visualized by use of the coliBASE server (http://colibase.bham.ac.uk) (6) in September 2003 and covered the complete or near-complete genome sequences of the following 12 Escherichia or Shigella strains: UPEC strain CFT073; laboratory strains K-12 MG1655, K-12 W3110, and DH10B; EHEC strains O157:H7 EDL933 and O157:H7 RIMD 0509952 (“Sakai strain”); EAEC strain 042; EPEC1 strain E2348/69; Shigella flexneri strains 2a 2457T and 2a 301; Shigella dysenteriae strain M131649 (M131); and Shigella sonnei strain 53G (3, 6, 24, 28, 49, 60). The sequences were completed with the associated annotated protein-coding sequences for finished genomes or with our own predictions of coding sequences based on GLIMMER results for unfinished genomes (14). All genome-derived sequences described here can be retrieved from the coliBASE server (http://colibase.bham.ac.uk/ETT2). In addition, we included in our analyses the published sequence (35) of a fragment of the ETT2 gene cluster from EPEC2 strain B171-8 (O111:NM).
Each coding sequence annotated in the ETT2 cluster in the published E. coli O157:H7 Sakai genome sequence (24) was analyzed by BLAST searches on the coliBASE server, supplemented as needed by visualization of the genomic context of homologues, by G+C percentage plots using an Artemis applet (53) within coliBASE, by multiple alignments on the EBI Clustal server (http://www.ebi.ac.uk/clustal), by searches of the National Center for Biotechnology Information's conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), and by PSI-BLAST searches on the ViruloGenome server (http://www.vge.ac.uk). Any coding sequences in the Spi-1 TTSS that were not represented in the ETT2 cluster were subjected to similar analyses, as were those in the newly discovered eip gene cluster.
The length of each predicted coding sequence in the ETT2 gene cluster from each strain was compared with the lengths of homologous predicted coding sequences in every other E. coli strain and in the Spi-1 system. When two coding sequences in one strain were represented by a single longer sequence in another strain or when a single coding sequence in one strain was judged to be substantially longer than those in another (allowing for the fact that GLIMMER sometimes makes mispredictions of start sites or even of coding sequences), the longer sequence was assumed to represent the physiologically active ancestral state, while the shorter sequences were judged to be pseudogenes.
Phylogenetic analysis.
Homologues of EivC (from the Sakai strain) and EicA (from EAEC 042) were identified by a PSI-BLAST search of the NCBI nonredundant protein database supplemented with predicted protein products from unfinished genome sequences on the ViruloGenome web site (http://www.vge.ac.uk). These protein sequences were aligned with ClustalW, version 1.8 (58), with minor manual adjustments done with SeaView (20). All positions with gaps were removed from the alignment, and phylogenetic trees were generated by the neighbor-joining algorithm (54), as implemented in ClustalW. The topology of the EivC tree was assessed by using 1,000 bootstrap replicates.
Bacterial strains.
Details of the bacterial strains used for this study are provided in Table 1. The E. coli reference (ECOR) strain collection was kindly supplied by Thomas Whittam and has been described elsewhere (44; http://foodsafe.msu.edu/whittam/ECOR). Representatives of other pathotypes, including neonatal meningitic E. coli strain RS218, EAEC strain 042, enterotoxigenic E. coli (ETEC) strain H10407, EAEC strain EAEC25, UPEC strain CFT073, and E. coli strain K-12, were kindly provided by Ian Henderson (University of Birmingham), while an isogenic nontoxigenic derivative of the E. coli O157:H7 Sakai strain was a kind gift from Chihiro Sasakawa (University of Tokyo). Two sets of clinical isolates were obtained in early 2003 from the clinical microbiology laboratory of the Queen Elizabeth Hospital, Birmingham, United Kingdom: they were a series of 43 consecutive blood culture isolates of E. coli and a series of 36 putative commensal strains obtained by subculturing E. coli isolates present in less than significant numbers (<107/ml) from urine samples without pyuria or any other evidence of infection (i.e., presumed to be perineal contaminants).
TABLE 1.
Distribution of ETT2 island PCR fragments among E. coli strains
| Straina | Category | ETT2 LPCR pattern | Indel-specific PCR resultsb | Conclusions from deletion-spanning PCRsc |
|---|---|---|---|---|
| CFT073 | UPEC | ++−−−−−−−+ | CFT073-like | ETT2 absent |
| RS218 | NMEC | |||
| ECOR 23 | ECOR A | |||
| ECOR 51, 52, 54, 55, 56, 57, 59, 61, 62, 63, 64, 65, 66 | ECOR B2 | |||
| ECOR 4 | ECOR A | CFT073-like, with 2.4-kb insertion | ETT2 absent and something else inserted next to glyU or ∼25-kb deletion in ETT2 between 3F and 9R | |
| H10407 | ETEC | ++++++−−++ | B171-8-like | 8.7-kb deletion as in B171-8 sequence |
| EAEC25 | EAEC | |||
| ECOR 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, 12, 15, 18, 22, 24, 25 | ECOR A | |||
| ECOR 26, 27, 28, 29, 30, 32, 33, 34, 58, 67, 68 | ECOR B1 | |||
| ECOR 39 | ECOR D | |||
| ECOR 45 | ECOR B1 | |||
| ECOR 14 | ECOR A | B171-8-like, with 1-kb insertion | 8.7-kb deletion as in B171-8 sequence, with ∼1-kb insertion | |
| ECOR 13, 20, 21 | ECOR A | ++−−−−−−−+ | Negative | ∼23-kb deletion between 3F and 9R |
| ECOR 19 | ∼30-kb deletion between 2R and 10F | |||
| ECOR 53 | ECOR B2 | ∼30-kb deletion between 3F and 9R, i.e., ETT2 absent | ||
| ECOR 60 | ||||
| ECOR 69 | ECOR B1 | Unresolved deletion between 2R and 10F | ||
| ECOR 16 | ECOR A | ++−+−−−−−+ | Unresolved deletion between 2R and 4F, ∼18-kb deletion between 4R and 9R | |
| ECOR 17 | ECOR A | +++−−−−−−+ | 22-kb deletion between 4F and 9R | |
| ECOR 38 | ECOR D | ++−−−−−+++ | ∼21-kb deletion between 3F and 7R | |
| ECOR 35 | ECOR D | +++−++−−−− | No deletion between 3R and 5F, unresolved deletion after 6R | |
| ECOR 31 | ECOR E | ++−−−−++++ | ∼15-kb deletion between 2R and 7F | |
| K12 | Laboratory | ++++−−−−++ | 18-kb deletion as in K12 sequence | |
| ECOR 36 | ECOR D | +++−++−−+− | No deletion between 3R and 5F, 8-kb deletion between 7F and 8R, unresolved deletion after 9R | |
| ECOR 40 | ECOR D | +++−+−−+++ | No deletion between 3R and 5F, 6-kb deletion between 6F and 7R | |
| ECOR 41 | No deletion between 3R and 5F, 4-kb deletion between 6F and 7R | |||
| ECOR 70 | ECOR B1 | ++++++−−−+ | ∼5-kb deletion between 6R and 9R | |
| ECOR 72 | ECOR B1 | ++++++−−++ | ∼5-kb deletion between 7F and 8R | |
| ECOR 71 | ECOR B1 | ∼8-kb deletion between 6R and 9F | ||
| ECOR 42, 43 | ECOR E | |||
| ECOR 37 | ECOR E | +++++−++++ | ∼3-kb deletion between 6R and 7F | |
| ECOR 48 | ECOR D | +++−++++−+ | No deletion between 3R and 5F, no deletion between 8R and 10F, i.e., Sakai-like | |
| ECOR 44, 46, 47, 49, 50 | +++−++++++ | No deletion between 3R and 5F, i.e., Sakai-like | ||
| 0157 | EHEC | ++++++++++ | No deletions detected, i.e., Sakai-like | |
| 042 | EAEC | ++++++++++ | No deletions detected, i.e., Sakai-like |
Strains in bold possess the eip cluster.
An ∼200-bp PCR was used to detect a deletion point identical to that seen in B171-8, while an ∼600-bp PCR was used to detect a CFT078-like vacant ETT2 insertion point. Negative means that both indel-specific PCRs failed to give a product.
When the indel-specific PCRs failed to resolve a deletion, deletion-scanning long PCRs were performed with forward and reverse primers from the sets flanking the deletion. Deletion sizes were calculated by subtracting the deletion-scanning PCR product size from the expected product size from the Sakai strain. This approach cannot resolve deletions followed by insertions, e.g., of IS elements.
TP-PCR.
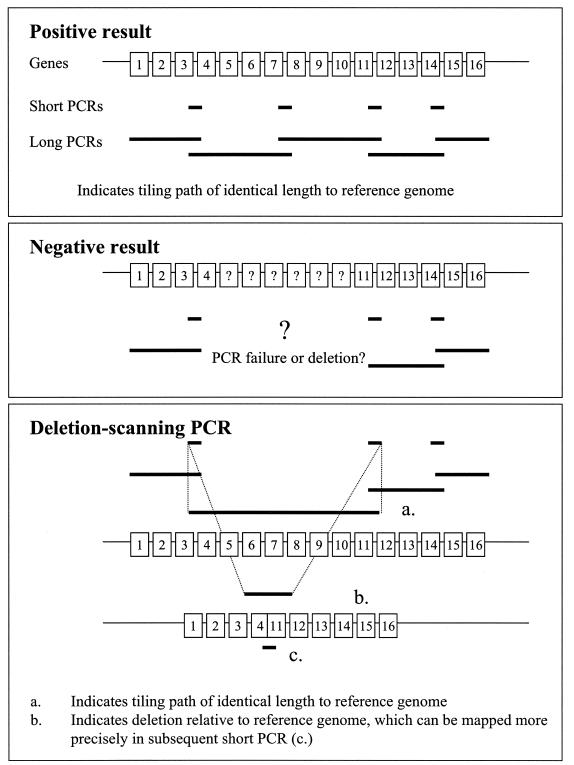
TP-PCR, which is essentially a scaled-down version of whole-genome PCR scanning (45), consists of several interlocking PCR-based approaches. A series of primers was designed to amplify ∼5-kb fragments that spanned the region of interest (Fig. 1 and 2) but that also had short overlaps of a few hundred base pairs. Long PCRs were performed to survey the region in question. Any negative results by long PCR for a given primer pair were followed up by amplification of the relevant short overlaps with the same primers and/or a deletion-scanning long PCR that employed primers flanking the lost segments (Fig. 1). For two strains for which there is published evidence of the precise site of an insertion or deletion (an 8.7-kb deletion in the EPEC2 B171-8 sequence or an unoccupied yqeG-glyU intergenic region in the CFT073 sequence), a short PCR centered on the insertion-deletion (in-del) was employed to screen other strains in our collection. In a few cases for which these approaches failed to produce coherent results, the original long PCRs and/or the deletion-scanning PCRs were repeated with a different enzyme preparation. In some supplementary experiments, for which we decided that a complete tiling path was not required (the surveys for ETT2 in clinical isolates and the surveys of the eip cluster), rapid short PCR scans using TP-PCR primer sets were performed.
FIG. 1.
TP-PCR.
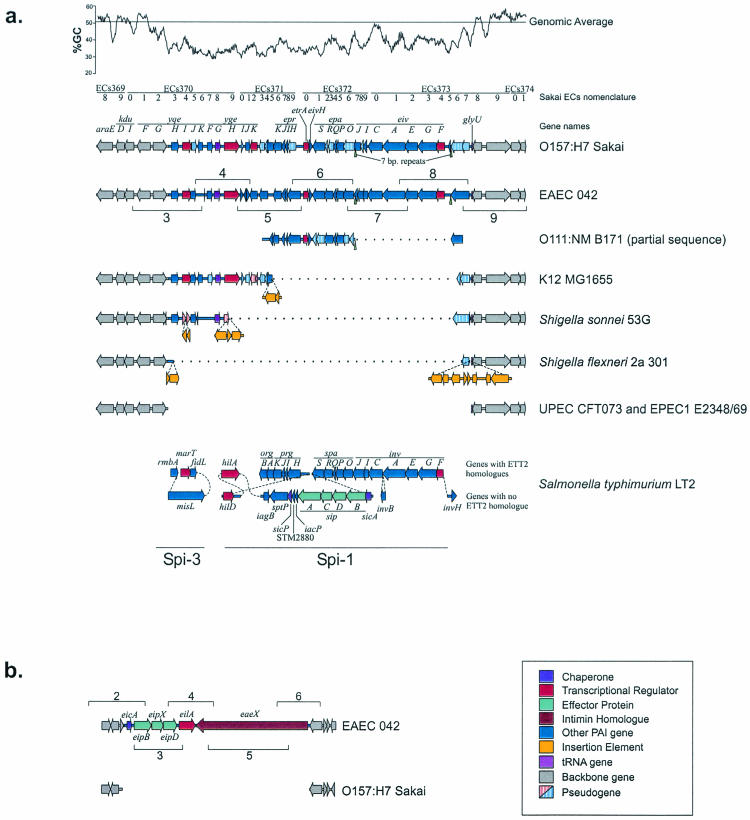
FIG. 2.
ETT2 and eip structures. (a) Structure of the ETT2 pathogenicity island in a number of E. coli and Shigella strains and comparisons with regions of Spi-1 and Spi-3 from S. enterica serovar Typhimurium. Homologous genes are vertically aligned. Insertions relative to the complete ETT2 sequence (as seen in strains Sakai and EAEC 042) are indicated with dashed lines. Dotted lines indicate deletions. (b) Structure of the eip island in EAEC 042 and comparison with the backbone sequence seen in Sakai (and other sequenced E. coli strains). Numbered brackets in both sections indicate the positions of primer pairs used for long PCR (see the text for details).
TP-PCR primer sets to amplify regions of the ETT2 cluster, the eip cluster, and the LEE were designed with Primer 3 software on the coliBASE server (6, 51). The primer sequences are listed in Table 2. Long PCRs were performed with a KlenTaq mixture (DNA Polymerase Technology, Inc., St. Louis, Mo.) in the buffer supplied by the manufacturer. Each 20-μl long-PCR mixture contained 20 ng of genomic DNA as a template, 8 pmol of each primer, and a 250 μM concentration of each deoxynucleoside triphosphate (dNTP). Long-PCR conditions were 30 cycles of 10 s at 94°C, 30 s at 62°C, and 10 min at 68°C, followed by a 10-min extension at 68°C. For short PCRs, each 20-μl reaction mixture contained 1 U of Taq polymerase (Invitrogen, Renfrew, United Kingdom) in the buffer supplied by the manufacturer, 20 ng of genomic DNA, and a 250 μM concentration of each dNTP. Short-PCR conditions were 30 cycles of 30 s at 94°C, 30 s at 62°C, and 30 s at 72°C, followed by a 7-min extension at 72°C. Supplementary long PCRs were performed in a few cases, using TaKaRa LA Taq (Cambrex BioScience Wokingham, Ltd.) in the buffer supplied by the manufacturer. For these supplementary PCRs, each 20-μl reaction mixture contained 48 ng of template DNA, 4 pmol of each primer, a 200 μM concentration of each dNTP, and 1 U of TaKaRa LA Taq; the reaction conditions were 30 cycles of 20 s at 96°C and 10 min at 69°C, with a 10-min extension at 72°C. All PCRs were carried out in a PTC-225 DNA Engine Tetrad thermal cycler (Genetic Research Instrumentation, Braintree, Essex, United Kingdom). Long PCR fragments were analyzed by electrophoresis in a 0.7% agarose gel, while short products were analyzed in a 1.2% gel.
TABLE 2.
Primers used to detect ETT2 and eip gene clusters
| Primer | Annealing site | Sequence |
|---|---|---|
| ETT2-1F | ECs3693 | GACCCAGCGCACCTGAGTAAGT |
| ETT2-1R | ECs3697 | AAGAGCGCAGTGTTTTGCCTGT |
| ETT2-2F | ECs3696 | GTGTGTTACCTCCGGGTCATCC |
| ETT2-2R | ECs3701 | CGCCGGACGATTTAAAGATGAG |
| ETT2-3F | ECs3700 | CGCACTGTGGATGCTCTGTCTT |
| ETT2-3R | ECs3706 | CGACTCATGGATTTGCACCAGA |
| ETT2-4F | ECs3705 | AATGACCAGGGACGAGCAAATC |
| ETT2-4R | ECs3711 | TATCCATTGCAAAACCCGCATT |
| ETT2-5F | ECs3709 | ATGTGCCTAACCCGCTCAAAAA |
| ETT2-5R | Intergenic | ACCGACCCTGATCTGGTTGTAA |
| ETT2-6F | ECs3719 | GGGAAATTATCAGCAAGCCATGA |
| ETT2-6R | ECs3726 | GCAGAAGAGAGTGGCAGCTGGT |
| ETT2-7F | ECs3726 | AGCGCGCCATTTACACGTATCT |
| ETT2-7R | ECs3732 | TGCACTTGATGCGAGTTGTTCA |
| ETT2-8F | ECs3731 | GGTGGGCAATGGAATTATGAGC |
| ETT2-8R | ECs3737 | AAACAGCGGCAGAAACCCACTA |
| ETT2-9F | ECs3736 | TCGGTCACCTTTTTGCCAATCT |
| ETT2-9R | ECs3741 | TCCCGTTAATGGTGCATTCGAT |
| ETT2-10F | ECs3740 | AATTACGCCTGGCATTTGGTGT |
| ETT2-10R | ECs3744 | TCAGGCGAACGGTATCGTCATA |
| LEE-1F | ECs4602 | TTAAGGCATCGATGTGTCCTTC |
| LEE-1R | ECs4595 | GACATCTTGTTCTGCGCCATTA |
| LEE-2F | Intergenic | TTTTAAACTGCAGCGACCTTACC |
| LEE-2R | ECs4586 | GCCTGAGGATCTGTTTTTGCTT |
| LEE-3F | ECs4588 | CGGAACTCATCGAAAGGTGTTT |
| LEE-3R | ECs4579 | CAAAACAAACAAAAACGGAACG |
| LEE-4F | ECs4579 | GCATTATACGCACCAACTGCAT |
| LEE-4R | ECs4572 | CGACATCTTGCAACAATGAACA |
| LEE-5F | ECs4573 | CTGAATGACCGATGGTGCTAAG |
| LEE-5R | ECs4568 | CCCCATCGTGTACTCACCAATA |
| LEE-6F | ECs4568 | ACTTCCGCGATCAAGGTAAAAA |
| LEE-6R | ECs4559 | ACCAGGATTCGACTGCAGCTTA |
| LEE-7F | ECs4559 | CGACGATTTGGTCGTTGAATAA |
| LEE-7R | ECs4533 | CTCCCATGCCATAACCAATTTT |
| LEE-8F | ECs4533 | TTATCGGTCTCAGCACCCCTAT |
| LEE-8R | ECs4531 | CTTTCGCTCAATGATGTTCCTG |
| 8.7kb-F | ECs3726 | AGACCAGCTGCCACTCTCTTCT |
| 8.7kb-R | ECs3736 | GCTTGATTTAGGGGGAGAATCC |
| NoETT2-F | ygeG | CCTGATCGTGGGTATCCTGT |
| NoETT2-R | ygeR | GCTTGCATTTCCAGATTCGT |
| Eip-1F | Intergenic | CTGTGATCTTCCGCCAAAAT |
| Eip-1R | Intergenic | GCCTGAGAAATGGCTGAAAA |
| Eip-2F | yicL | CAATTCCTCTCACCGACGAT |
| Eip-2R | eipB | GGAACTGCAGCAAGAAGACC |
| Eip-3F | eipB | GAATTATGTCGGCACCGATT |
| Eip-3R | eilA | TTCGGTACGAAAGGGAAAAA |
| Eip-4F | eilA | AGGCGATGCGATAGTTTTTGAT |
| Eip-4R | eaeX | CTGATTGGTACTGGCAATCGTC |
| Eip-5F | eaeX | CGCCGACCATGTAGCAGTATAA |
| Eip-5R | eaeX | GGAGCTGTGTTGGCTTATTCCT |
| Eip-6F | eaeX | CGTAAAGGTGACGGTCTGCTC |
| Eip-6R | yicM | GGCAATGTTTGCCAGTTTGT |
| Eip-7F | yicM | CGCCGCATTAAAGACATTG |
| Eip-7R | yicP | ATTATGGCTGTCATGGCTGA |
RESULTS
The EHEC ETT2 gene cluster is larger than O-island 115 and includes homologues of genes from the Salmonella Spi-1, Spi-2, and Spi-3 pathogenicity islands, but it cannot encode a functional TTSS.
The ETT2 gene cluster was first identified as an apparent insertion in the EHEC O157:H7 genome relative to E. coli K-12 (called O-island 115 in Blattner's nomenclature) (24, 49), and close homologies have been noted between the proteins encoded by this O-island and those in the S. enterica Spi-1 TTSS (percent identities were recorded by Makino et al. [35]). However, previous authors have disagreed on the boundaries of the cluster: using the Sakai genome nomenclature, Makino et al. (35) placed the left boundary at ECs3714, delimiting what they considered to be a 17-kb insertion, while Hartleib et al. (23) place the boundary further upstream, at ECs3703 (rmbA/yqeH), making the cluster 29.9 kb in length. To resolve this issue, we used three independent methods to define the boundaries of the island, namely homologies to other TTSS genes, G+C content, and genomic comparisons (Fig. 2a).
Several genes that are homologous to components of TTSS gene clusters were found upstream of ECs3714 in the Sakai strain (Table 3): they are ECs3713 (a pseudogene, homologous to part of orgB from Spi-1 [19% identity at the protein level over a 101-amino-acid stretch]), ECs 3712/ygeK (encodes a homologue of the Spi-2 regulator SsrB [32% identity at the protein level over a 209-amino-acid stretch]), ECs3711/B2854 (a homologue of iagB from Spi-1 [43% identity at the protein level over a 122-amino-acid stretch]), ECs3709/ygeH (encodes a tetratricopeptide repeat TTSS regulator, homologous to hilA in Spi-1 [29% identity at the protein level over a 403-amino-acid stretch] [46]), and ECs3708/ygeG (encodes a tetratricopeptide repeat TTSS chaperone, homologous to sicA in Spi-1 [37% identity at the protein level over a 140-amino-acid stretch] [46]). These findings provide strong evidence that the ETT2 cluster extends upstream of Ec3714 and negate the claim (35) that the ETT2 gene cluster lacks chaperones (Table 3). Furthermore, they imply that the essential difference between EHEC and K-12 at this locus is a deletion in K-12 rather than an insertion into the EHEC genome, as claimed by Makino et al. (35), and that the originally commensal and now laboratory-adapted strain K-12 retains a remnant of an apparently virulence-associated type III secretion cluster.
TABLE 3.
Genes within the ETT2 gene cluster
| Gene in Sakai strain | Length (aa) | Gene in K-12 | Pseudogene(s) | Other homologies |
|---|---|---|---|---|
| ECs3703 | 230 | YqeH | None | RmbA regulator from Spi-3 |
| ECs3704 | 269 | YqeI | None | MarT from Spi-3 |
| ECs3705 | 159 | YqeJ | Sakai, EDL933, K-12, W3110 | FidL from Spi-3 |
| ECs3706 | 145 | YqeK | None | None |
| ECs3707 | 163 | YgeF | K-12 W3110, K-12 MG1655 | None |
| ECs3708 | 163 | YgeG | None | SicA-like TPR TTSS chaperone |
| ECs3709 | 458 | YgeH | None | HilA-like TPR TTSS regulator |
| ECs3710 | 72 | YgeI | Sakai, EDL933, K-12, W3110 | None |
| ECs3711 | 133 (167) | B2854 | K-12 W3110, K-12 MG1655 | IagB from Spi-1, PilT from S. enterica |
| ECs3712 | 210 | YgeK | K-12 W3110, K-12 MG1655, misidentification of start site in EDL933 | SsrB from Spi-2 |
| ECs3713 | 143 | B2857 | Sakai, EDL933, K-12 | OrgB, MxiN |
| ECs3174 | 86 | B2858 | W3110, K-12 MG1655 | |
| ECs3715 | 193 | B2859 | K-12 W3110, K-12 MG1655 | OrgA, MxiK |
| ECs3716/EprK | 244 | Absent | None | PrgK |
| ECs3717/EprJ | 110 | None | PrgJ, MxiI | |
| ECs3718/EprI | 79 | Numerous frame shifts in EDL933 | PrgI, MxiH | |
| ECs3719/EprH | 244 | Sakai, EDL933 | PrgH | |
| ECs3720/EtrA | 166 | None | Transcriptional regulator | |
| ECs3721/EpaS | 373 | Last three residues missing in EAEC, ? significant | SpaS | |
| ECs3722/EpaR2 | 78 | Sakai, EDL933 | SpaR | |
| ECs3723/EpaR3 | 155 | |||
| ECs3724/EpaQ | 86 | None | SpaQ | |
| ECs3725/EpaP | 221 | None | SpaP | |
| ECs3726/EpaO | 328 | Sakai, EDL933 | SpaO | |
| ECs3727/EivJ | 205 | EAEC or Sakai/EDL933 | SpaN/invJ | |
| ECs3728 | 77 | None | ||
| ECs3729/EivI | 111 | None | SpaM/InvI | |
| ECs3730/EivC | 439 | None | SpaI/InvC | |
| ECs3731/EivA | 686 | None | InvA | |
| ECs3732/EivE | 381 | None | InvE | |
| ECs3733/EivG | 567 | None | InvG | |
| ECs3734/EivF | 249 | None | InvF | |
| ECs3735 | 59 | None | Inner membrane proteins YjdO/YdcX | |
| ECs3736 | 352 | B2863 | Different frame shifts in K-12, O157, S. sonnei, S. flexneri 2a | Phosphorylase kinase and glucomamylases |
| ECs3737 | 201 | B2862 |
An examination of a G+C% plot revealed a region of lower-than-average G+C content that extended from ECs3703 to ECs3737 (Fig. 2a). Genomic comparisons identified two genome sequences, those of EPEC1 strain E2348/69 and UPEC strain CFT073, that lacked this region of G+C deviation in its entirety (and thus presumably showed the ancestral state for the species). Scrutiny of these sequences confirmed the assignments of ECs3702/yqeG and ECs3738/yqeR as the ancestral E. coli “backbone” protein-coding genes flanking the 27.5-kb ETT2 island (Fig. 2a) and indicated that the island had inserted into the intergenic region between yqeG and the tRNA glyU gene 142 bp upstream of the start of glyU.
Curiously, three genes from the leftmost extremity of the ETT2 pathogenicity island are homologues of uncharacterized genes from the Spi-3 pathogenicity island from S. enterica: yqeH (ECs3703) is homologous to rmbA (39% identity at the protein level over a 190-amino-acid stretch), yqeI (ECs3704) is homologous to marT (41% identity at the protein level over a 101-amino-acid stretch), and yqeJ (ECs3705) is homologous to fidL (43% identity at the protein level over a 139-amino-acid stretch). Furthermore, ygeK (ECs3712) encodes a homologue of the Spi-2 regulator SsrB (32% identity at the protein level over a 209-amino-acid stretch). Others have noticed the presence of Spi-1- and Spi-3-related genes in E. coli K-12 (2, 27), but their context as remnants of the ETT2 island only becomes clear through comparisons with E. coli O157 and other pathotypes (Fig. 2a).
A comparison of the ETT2 island with Spi-1 revealed a generally similar gene complement and arrangement, with a few notable differences (Fig. 2a; Table 3). For example, there were some additional putative transcriptional regulator genes, such as ECs3720, a lack of gene products similar to the Spi-1 secreted proteins AvrA, SptP, and SipABCD (despite the presence of the sicA-like chaperone gene ygeG), and an absence in the O157 annotation of an invH homologue, although a potential invH homologue or pseudogene can be discerned in the region between ECs3720 and EprS and has been annotated as orf7 by Makino et al. (35) for strain B171-8. However, the most striking finding was that three TTSS structural genes, the homologues of spaR, prgH, and orgB, were disrupted by frameshift mutations in both EHEC O157:H7 genome sequences (Fig. 2a; Table 3). Since any one of these mutations would, by analogy with Spi-1, abolish type III secretion (8, 31), we conclude that ETT2 is incomplete and inactive as a TTSS in EHEC O157:H7, despite claims to the contrary (35).
The alignment of EivC from ETT2 with its homologues from other TTSSs allowed the construction of a phylogenetic tree (Fig. 3a) which indicated that ETT2 belongs to the Spi-1/Mxi-Spa group of TTSSs, as do the recently discovered TTSSs from Chromobacterium violaceum, two insect endosymbionts, and Yersinia enterocolitica (11, 12, 19, 59).
FIG. 3.
Phylogenetic trees showing relationship of ETT2 to other TTSSs based on neighbor-joining analysis of EivC and its homologues (a) and of EicA and YgeG TPR chaperones to each other and to other chaperones (b). The numbers on the branches indicate the percentages of bootstrap support based on 1,000 replicates. Numbers in parentheses are GI numbers of published sequences.
The ETT2 gene cluster is present, at least in part, in the majority of genome-sequenced Escherichia and Shigella strains, but is usually disrupted or incomplete.
A comparison of the ETT2 gene cluster from EHEC with the equivalent region of other Escherichia or Shigella genomes revealed several surprises. Nine of the 12 genome sequences showed evidence of ETT2 genes being inserted at the same chromosomal site, within the yqeG-glyU intergenic region, which is compatible with the idea that this island entered the species only once and has remained immobile ever since. However, only one other pathotype besides EHEC, namely EAEC strain O42, possessed the full complement of ETT2 genes. Tellingly, a comparison of the O42 ETT2 genes with their homologues in Spi-1 failed to identify any inactivating frameshift mutations (Table 3), suggesting that the EAEC O42 ETT2 gene cluster may encode a functional TTSS. Two genes, eivJ and eivH, were shorter than their equivalents in Spi-1. invH appears to be dispensable and highly variable in Spi-1, whereas invJ/spaN is known to be essential for Spi-1-mediated secretion (4, 52). However, there is considerable sequence diversity in SpaN, even within S. enterica, and a marked divergence between spaN and its homologue, spa32 from Shigella, even though they are functionally interchangeable, so this variation in length may not signal an inactivating mutation (4, 56). Furthermore, several of the O42 ETT2 genes were longer than, or spanned more than one of, their equivalents in EHEC, allowing the identification of additional pseudogenes in the EHEC ETT2 gene cluster (ECs3705, ECs3710, ECs3713/4, ECs3722/3, and ECs3736/7) (Fig. 2a; Table 3).
In contrast to EAEC O42, all other genome-sequenced Escherichia or Shigella pathotypes possessed either an incomplete ETT2 gene cluster or, as noted for the EPEC1 and UPEC strains, none at all. We noted an important distinction between the EPEC1 genome-sequenced strain E2348/69, which lacks the ETT2 island entirely, and EPEC2 strain B171-8, which contains an ETT2 cluster with an 8.7-kb deletion (35) centered on a 7-bp repeat (CC/ATCATT) (Fig. 2a). This finding conflicts with the report by Makino et al. (35) that E2348/69 and B171-8 both contain similar sets of ETT2 genes, but it is concordant with the known highly divergent origins of the EPEC1 and EPEC2 clades (17). Three additional patterns of deletion were noted for the ETT2 gene cluster. An identical 14.6-kb deletion, which removed almost all of the secretion apparatus genes but left some TTSS regulators and chaperones, was found in the two K-12 laboratory strains (Fig. 2a). A similar but slightly larger 18.3-kb deletion was seen for S. sonnei, while a 26.5-kb deletion resulting in an almost total loss of the gene cluster was seen for the two S. flexneri sequences, in which all that remained were fragments of the two genes at the extremities of the island. We could not find the yqeG and glyU genes in the most recent release of the S. dysenteriae M131649 genome sequence, which suggests that this strain has undergone such extensive deletions in this region of the chromosome that it is impossible to determine whether its lineage ever possessed ETT2. Additional frameshift mutations were present in some pathotypes (Fig. 2a; Table 3).
The observed spectrum of ETT2 genotypes could be explained either by a single insertion with subsequent gene loss, or far less plausibly, by assembly of the largest ETT2 gene clusters from the smaller clusters by gene acquisition. Several lines of evidence lead us to strongly favor the first scenario. (i) The indels are centered on structural genes, which form a functionally coherent unit, showing a large degree of conservation in gene order with Spi-1 and other TTSSs. It is highly unlikely that the same gene order would arise independently by gene acquisition within E. coli. (ii) In the smaller clusters, the indel boundaries are often marked by truncated genes, as judged by homology with Spi-1 (e.g., b2859 in K-12 is a truncation of ECs3715), suggesting that deletion occurred rather than insertion. (iii) Insertion sequences of several classes (IS1, IS2, and IS3) are found at the sites of indels in ETT2 gene clusters, suggesting that homologous recombination between such elements may account for multigene deletions. Similarly, homologous recombination between the two copies of the 7-bp repeat might account for the 8.7-kb deletion in B171-8 (Fig. 2a).
The ETT2 locus is present in whole or in part in the A, B1, D, and E groups of the ECOR commensal strains but was acquired after the divergence of the B2 group.
We wished to survey the phylogenetic distribution of the ETT2 pathogenicity island. However, unlike previous surveys, which sampled discontinuous fragments of the island from pathogenic E. coli strains allied to O157, we attempted (i) to sample the full range of phylogenetic diversity within the species E. coli, including commensal strains, and (ii) to determine the complete tiling path through this region of the chromosome for a large number of strains. We surveyed the well-characterized ECOR strain collection, which is richly diverse in terms of phylogeny and geographical, clinical, and zoological strain origins (Table 1) (44). We devised a method, TP-PCR (Fig. 1), that exploits short- and long-PCR protocols to construct a complete tiling path through the relevant chromosomal region. This method allowed us to construct a complete tiling path through the ETT2 gene cluster for 68 of 72 ECOR strains and for all of the representative pathotype strains (Table 1).
Using TP-PCR, we discovered that the ETT2 gene cluster was present in whole or in part in the majority of the E. coli strains sampled (50 of 72 [∼69% of the ECOR collection]), whether they were commensal or pathogenic (Table 1; Fig. 4). There was no evidence of large insertions or rearrangements within this cluster. Furthermore, the ETT2 gene cluster always occurred at the same chromosomal location, within the yqeG-glyU intergenic region, unlike the LEE, which can be inserted into selC, pheV, or pheU (17, 57), adding weight to our conclusion that the island entered an ancestral E. coli strain once and then was lost through mutational attrition in most strains.
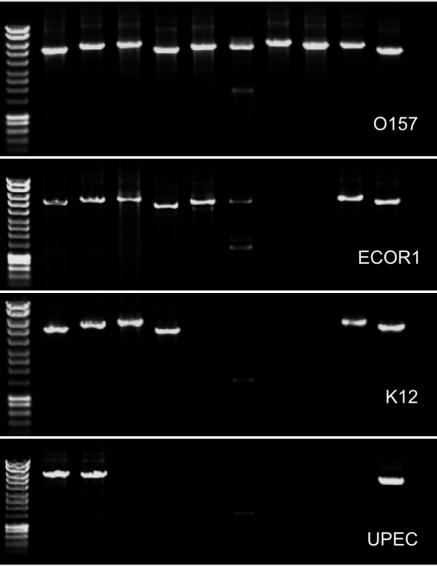
FIG. 4.
Illustrative PCR results for ETT2 gene cluster. Strains, from top to bottom: O157 Sakai strain (complete ETT2 gene set), ECOR1 (a B171-8-like strain with an 8.7-kb deletion), K-12 (14.6-kb deletion), and CFT073 (UPEC, with no ETT2). Lanes, from left to right: molecular weight markers, ∼5-kb amplicons obtained with ETT2 TP-PCR primer pairs 1 to 10 (see text for details), negative control (DNA, no primers), and molecular weight markers (HyperLadder I; Bioline).
Certain salient conclusions can be drawn from the TP-PCR amplicon patterns (Table 1; Fig. 5), particularly when they are interpreted in the light of the known phylogenetic structure of the ECOR collection and of the E. coli pathotypes, for example, the existence of two EPEC lineages, EPEC1 and EPEC2, and two EHEC lineages, EHEC1 and EHEC2 (EPEC1 and EHEC1 are highly divergent, whereas EPEC2 and EHEC2 are closely related to one another and fall into the B1 group of the ECOR collection) (17, 25, 29). The following three predominant patterns were detected: the unoccupied yqeG-glyU intergenic region (the ancestral state, as deduced from genome sequence analysis), the Sakai-like complete ETT2 genotype, and the EPEC2-like 8.7-kb deletion genotype (Fig. 5). The phylogenetic distribution of these genotypes was not random, but instead was largely congruent with the major phylogenetic divisions (A, B1, B2, D, and E), as deduced by multilocus enzyme electrophoresis (MLEE) and other methods (25, 29) (Fig. 5). Opinion is divided on whether the branching order of these major ECOR divisions has been or can ever be established (M. Achtman, personal communication). However, the distribution of ETT2 genotypes is consistent with the results of multilocus sequence typing (MLST) studies which imply that B2 is the earliest splitting branch of the tree, followed by the D group (34).
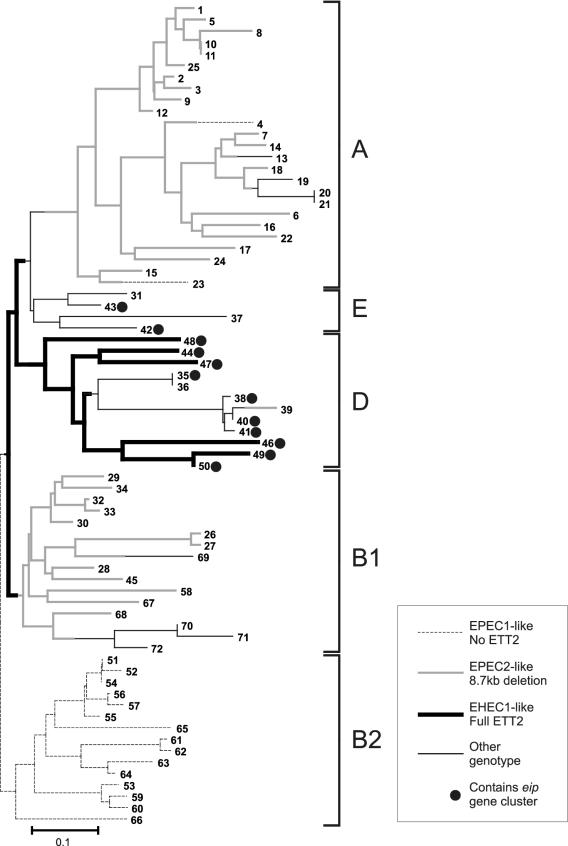
FIG. 5.
TP-PCR results superimposed on phylogenetic structure of E. coli. The tree was obtained by neighbor-joining analysis of the ECOR MLEE data (available at http://foodsafe.msu.edu/whittam/ecor) using the program Neighbor, part of the PHYLIP package (J. Felsenstein; available from http://evolution.genetics.washington.edu/phylip.html). Branches containing one of the three most common genotypes are indicated by bold, dashed, or gray lines. Filled circles indicate strains with eip clusters.
Consistent with the MLST-derived branching pattern, the ancestral ETT2-less long-PCR genotype was seen for all 15 ECOR strains belonging to the B2 group, for EPEC1 strain E2348/69, which is known to be extremely divergent from other groups (17), and for UPEC strain CFT073, which is closely affiliated with the B2 group (10). The long-PCR results for all but two of these strains were confirmed by a short PCR spanning the ETT2 insertion site (Table 1; Fig. 6). Furthermore, ETT2 was absent from four Escherichia spp. other than E. coli (E. blattae, E. fergunsonii, E. hermannii, and E. vulneris) (data not shown).
FIG. 6.
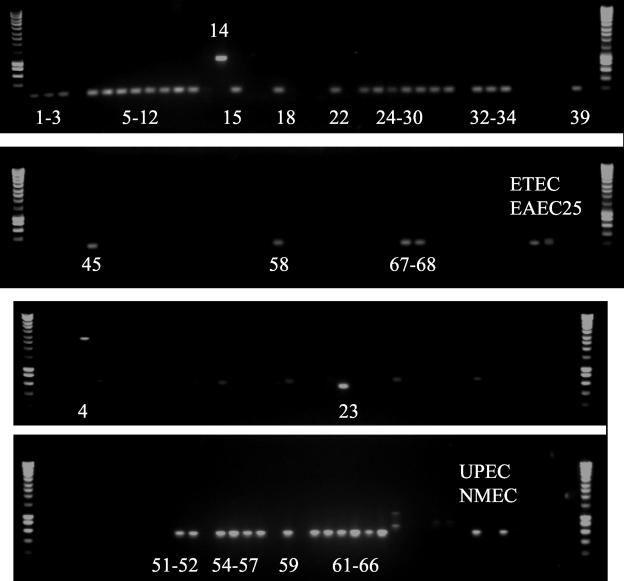
Indel-specific short PCRs. The first two rows show the results for ECOR strains from 200-bp PCRs across the deletion seen in strain B171-8. The second two rows show the results for ECOR strains from 600-bp PCRs across the ETT2 insertion site (see Table 1 for details). Positive results are labeled with ECOR strain numbers or pathotypes.
Interestingly, apparently intact ETT2 gene sets occurred in 6 of 10 strains, 4 of them commensals, within ECOR group D, to which EAEC O42 is most closely related (Table 1; Fig. 5). Relatively intact gene sets predominated among the rest of group D and group E (to which the genome-sequenced EHEC1 strains are closely related) (7, 17). This is in line with (i) predictions from MLST data that indicate that the D group was next to diverge from the ancestral state after B2 and (ii) MLEE data showing that the EHEC1 strains diverged from other E. coli strains after the divergence of the EPEC1 strains. Thus, we conclude that the ETT2 gene cluster entered an ancestral E. coli strain sometime after the divergence of the B2 group (and the EPEC1 strains) but before the divergence of the D group (and EHEC1 strains).
Overall, the most common TP-PCR pattern was that corresponding to the 8.7-kb deletion seen in the already sequenced ETT2 cluster from EPEC2 strain B171-8 (35). This TP-PCR pattern predominated in the A and B1 ECOR groups. We confirmed that the deletion was identical (to within a few base pairs) to that in B171-8 in 17 of 25 ECOR group A strains, 12 of 16 ECOR group B1 strains, and the pathogenic strains H10407 (ETEC) and EAEC25 (EAEC) by performing a short 200-bp PCR across the deletion site (Table 1; Fig. 6).
To confirm that the high prevalence of ETT2 gene fragments among the strains of the ECOR collection was a general phenomenon of human E. coli isolates, we performed short PCRs with the ETT2 TP-PCR primers and collections of 43 freshly collected local blood culture isolates of E. coli and 36 freshly collected local urine contaminants from patients with no laboratory evidence of urinary tract infections (presumptive commensal strains of E. coli). ETT2 gene fragments were detected in 16 of 43 (37%) bloodstream isolates and 10 of 36 (28%) commensal isolates (data not shown), confirming the high prevalence of ETT2 gene fragments in E. coli (albeit a lower one than in the ECOR collection) and the lack of any obvious link to virulence.
For comparison, we surveyed the ECOR collection for fragments of the LEE by using short PCR. Only two strains were positive for LEE fragments, namely ECOR25, an ECOR group A strain from a healthy dog (four of five fragments were positive), and ECOR37, an ECOR group E strain from a healthy marmoset (all five fragments were positive) (data not shown).
A second type III secretion locus, the eip locus, encodes homologues of Spi-1 translocators and additional TTSS-related proteins in a minority of E. coli strains.
Prompted by the lack of any sipABCD homologues in the ETT2 pathogenicity island, we searched the available Escherichia and Shigella genomes for similar genes. We discovered a novel 20.9-kb pathogenicity island—which we have termed the eip island—within the EAEC O42 genome. This island is inserted between the E. coli backbone genes yicM and nlpA and contains two homologues of sip genes that we have termed eipB (encodes a protein that shows 20% identity over a 527-amino-acid stretch with SipB) and eipD (encodes a product that shows 31% identity over a 266-amino-acid stretch with SipD) (Fig. 2b). Between these two eip genes lies a third gene (which we have termed eipX) that shows weak similarity to espD (the encoded product shows 20% identity over a 282-amino-acid stretch with EspD) and thus may encode an additional secreted translocator protein. In addition, the eip island contains genes for a novel SicA-like tetratricopeptide repeat chaperone (eicA), a novel HilA-like regulator (eilA) (46), and an invasin/intimin-like large outer membrane protein (eaeX) (Fig. 2b). Phylogenetic analysis of the tetratricopeptide repeat chaperones from O42 suggested that the chaperones from the eip and ETT2 gene clusters arose from a duplication after the ETT2 system diverged from Spi-1 (Fig. 3b).
A set of short PCRs, targeting fragments that were evenly spaced throughout the eip island, were applied to the ECOR collection and to our collections of clinical isolates (Fig. 2b). The distribution of the eip island mapped onto the phylogenetic structure of the ECOR collection and correlated well with the distribution of the most intact ETT2 clusters: the 13 eip-positive ECOR strains encompassed 11 of the 12 D strains and 2 of the 5 E strains (Fig. 5; Table 1). Significantly, all six ECOR strains that showed an intact Sakai-like ETT2 genotype possessed the eip cluster. This suggests that the Sakai and EDL933 strains are unusual in harboring an intact ETT2 cluster without an accompanying eip island. Among clinical isolates, the eip island was present in 7 of 15 ETT2-positive bloodstream isolates and 6 of 10 ETT2-positive commensal isolates but was never present in any isolates that lacked genes from the ETT2 cluster (data not shown). Furthermore, like the ETT2 gene cluster, it did not appear to be significantly more common in bloodstream isolates than in commensals (7 of 43 versus 6 of 36 isolates) (data not shown).
DISCUSSION
The sequence analysis and PCR-based studies reported here afford novel insights into the evolution of bacterial genomes and TTSSs, but they also raise new questions. Our most surprising conclusion was that the ETT2 gene cluster is present in whole or in part in the majority of E. coli strains. A previous study showed that ETT2 genes were found in 78 of 89 O-antigen serotypes selected from several different pathotypes (EPEC2, enteroinvasive E. coli, EAEC, and ETEC) and that ETT2 genes are more common in EHEC strains than the LEE-encoded ETT1 (23). However, among the ECOR strains we found the difference to be even more striking, with 50 of 72 strains containing ETT2 genes, but only 2 containing the LEE. Furthermore, we confirmed the presence of ETT2 genes in ETEC, EAEC, EPEC2, and EHEC strains and in two Shigella genomes. Thus, ETT2 is clearly the most prevalent nonflagellar type III secretion-associated gene cluster in E. coli. However, because ETT2-associated genes are distributed equally among pathogenic and commensal strains (they occur even in the originally commensal but now laboratory-adapted K-12 strain), it seems hard to accept claims (35) that ETT2-associated genes may be a marker of virulence, particularly as apparently complete Sakai-like ETT2 gene sets occur in several commensal strains from the ECOR collection. Instead, the ETT2 cluster appears to be a marker of phylogenetic origin, in that it is absent from all B2 strains but present in almost all others. We conclude that the genomes of the sequenced strains E2348/69 (EPEC1) and CFT073 (UPEC) and the ECOR B2 strains represent the ancestral state with no ETT2 cluster and that the ETT2 gene cluster entered an ancestral E. coli just once, sometime after the divergence of the B2 group (and the EPEC1 strains), but before the divergence of the D group (and EHEC1 strains).
The second surprise was that in most strains, the ETT2 gene cluster has undergone mutational attrition, so that it can no longer encode a functioning TTSS. In most strains, genes have been deleted, with one particular deletion, that already described for EPEC2 strain B171-8 (35), being common to most strains in the ECOR A and B1 groups. However, even in the O157 genomes for which it was first described (24, 49), the ETT2 gene cluster contains numerous inactivating mutations which, by analogy with Spi-1, must abolish its functions. Thus, in contrast to a previous claim (35) that the ETT2 genes from the O157 Sakai strain encode a functioning TTSS that can mediate the secretion of EspB (now considered doubtful [T. Tobe, personal communication]), we conclude that ETT2 cannot function as a secretion system in either of the two genome-sequenced E. coli O157 strains. Instead, here, as in most strains, it represents a “rudimentary, atrophied or aborted organ” of the sort predicted to occur by Darwin as a consequence of the theory of evolution (13).
Several salient messages arise from the observation of ETT2 gene fragments in the laboratory model and nonpathogenic strain K-12. Firstly, despite the fact that K-12 has one of the smallest E. coli genomes (43), it is not safe to conclude that it represents the ancestral state for the species—in other words, apparent insertions relative to K-12 in other strains may in fact represent deletions in K-12 (47). Examples of this phenomenon other than ETT2 include O-island 82, of which K-12 has lost most of the gene for an iron-regulated outer membrane protein (1), and the mbhA and fhiA genes from K-12 which represent the residual boundaries of an ancient lateral flagellar gene cluster present in the ancestral E. coli (M. J. Pallen, unpublished data). Thus, as others have noted (47), the polarity of such intergenomic changes can only be inferred safely after comparisons with several diverse genomes.
Secondly, not every apparent coding sequence in K-12 should be expected to encode a functioning protein: as others have noted, many genes, particularly orphan open reading frames, may represent pseudogenes (26, 39), a fact that is likely to frustrate attempts for a global genome-wide functional characterization of all open reading frames of K-12 (41). Within the K-12 ETT2 cluster, we can confidently dismiss yqeJ, ygeF, ygeI, b2854, ygeK, b2858, b2859, b2863, and b2864 as pseudogenes. These observations on indel polarity and pseudogenes together imply that the accurate annotation of any E. coli genome, including that of K-12, depends on multiple strain comparisons. This is a compelling reason to support genome sequencing efforts with additional strains and is a point that is strongly made by the fact that only 1 of 12 completely or nearly completely sequenced Escherichia and Shigella genomes possesses an apparently intact ETT2. Similarly, this study emphasizes that it is necessary in laboratory-based strain comparisons to sample a wide range of phylogenetic diversity and to construct a complete tiling path through a region of interest in almost all strains before safe conclusions can be drawn about associations between the genotype of a gene cluster and virulence or other phenotypes.
The function of ETT2 remains a mystery, as do the niche and signals that might trigger its expression and the identity and nature of its effectors. Its presence in commensal strains suggests that it might now play or once played a role in symbiotic colonization rather than in the pathogenesis of human or mammalian disease, as has been suggested for some other TTSSs (11, 12, 18, 36), including Spi-1 (42). Alternatively, its target may not be the mammalian gut at all, but instead it may aid in survival in the struggle with microscopic eukaryotes in the external environment (9, 32). The discovery of an apparently intact ETT2 secretion system in EAEC O42 raises the hope that studies of this cryptic TTSS might mirror the successes of recent studies of a cryptic type II secretion system that was first discovered in the genome sequence of E. coli K-12 (19a).
We do not yet have any functional data to link the newly found eip cluster to the ETT2 cluster. However, the fact that this cluster is only ever found in ECOR strains that possess an apparently complete ETT2 gene complement, taken together with the homology to Spi-1 and the clear functional relationship between the type III secretion and translocation machinery in other TTSSs, makes this a compelling hypothesis. Furthermore, the configuration of the ETT2 and eip gene clusters and the identities of the genes carried within them shed light on the evolution of pathogenicity islands in general and TTSSs in particular and raise some interesting questions. The separation of translocon genes from genes encoding the needle complex is unusual and is shared only with chlamydial TTSSs (55). However, given the close similarities of ETT2 to the other TTSSs in the proteobacteria, it seems unlikely that this separation of genes represents the ancestral state for the Spi-1-like systems; instead, it appears to be a specific derived feature of this system. Intriguingly, the ETT2 gene cluster encodes a tetratricopeptide repeat chaperone, which would normally be expected to bind to a translocator (46); however, the ETT2 cluster lacks translocator genes, while the eip cluster encodes its own tetratricopeptide repeat chaperone, which presumably binds to the EipBXD proteins. Similarly, there are two hilA homologues: one in the ETT2 cluster and one in the eip cluster. This curious redundancy, together with phylogenetic data on the chaperones, suggests that the ETT2 and eip clusters were once part of a larger cluster that underwent fission, accompanied by the duplication of chaperones and regulators, after its divergence from its common ancestor with Spi-1.
The ETT2 cluster simultaneously provides a model of gene flux and mobility on the one hand and a model of genetic stasis and loss on the other. The shuffling of homologues of genes from three distinct Salmonella pathogenicity islands (Spi-1, Spi-2, and Spi-3) into one cluster in E. coli represents pathogenicity genes in motion. However, a single insertion followed by mutational attrition provides a model for how genes are lost from a genome once they no longer provide any selective advantage (33): frameshift mutations are followed by gene deletions and the arrival of insertion sequences that may then catalyze deletions through homologous recombination between nearby elements. With the ETT2 cluster, we see the whole spectrum of reductive evolution, from an apparently intact 27.5-kb cluster in O42 to just two residual gene fragments in S. flexneri (Fig. 2a). However, even though some TTSS genes within the cluster may be nonfunctional or missing in most strains, this does not mean that all genes within the cluster are without an effect. We have recently found evidence that some regulators encoded within the island exert a profound effect on other virulence-related loci in at least one strain (L. Zhang and M. J. Pallen, unpublished data). In this regard, we propose that a useful metaphor for the ETT2 cluster might be that of the grin of the Cheshire cat in Alice in Wonderland (5). We speculate that this metaphor—that powerful regulatory effects might outlive structural decay through mutational attrition—might also apply to other decaying prophages and pathogenicity islands.
Acknowledgments
We thank the BBSRC for funding the TP-PCR work through project grant D13414 and for funding the ViruloGenome and coliBASE sites through grants FGT11398 and EGA16107. We thank the Wellcome Trust and the Pathogen Sequencing Team at the Sanger Institute for funding and for making available unfinished E. coli and Shigella genome sequences.
We thank Gad Frankel, David O'Connor, and Mark Stevens for critical reading of the manuscript. We thank Arshad Khan for systems administration, and we are grateful to Michael Russell and Chengjie Liu for medium preparation. We thank Terry Alli for establishing PureGene chromosomal DNA preparations within our laboratory. We thank Debbie Mortiboy and other staff in the clinical microbiology laboratory of the Queen Elizabeth Hospital for help in obtaining local clinical isolates.
REFERENCES
- 1.Allen, N. L., A. C. Hilton, R. Betts, and C. W. Penn. 2001. Use of representational difference analysis to identify Escherichia coli O157-specific DNA sequences. FEMS Microbiol. Lett. 197:195-201. [DOI] [PubMed] [Google Scholar]
- 2.Blanc-Potard, A. B., F. Solomon, J. Kayser, and E. A. Groisman. 1999. The SPI-3 pathogenicity island of Salmonella enterica. J. Bacteriol. 181:998-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E. F., J. Li, H. Ochman, and R. K. Selander. 1997. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J. Bacteriol. 179:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll, L., and J. Tenniel. 2001. Alice in wonderland. Dover Publications, Mineola, N.Y.
- 6.Chaudhuri, R. R., A. M. Khan, and M. J. Pallen. 2004. coliBASE: an online database for E. coli, Shigella and Salmonella comparative genomics. Nucleic Acids Res. 32:D296-D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clermont, O., C. Cordevant, S. Bonacorsi, A. Marecat, M. Lange, and E. Bingen. 2001. Automated ribotyping provides rapid phylogenetic subgroup affiliation of clinical extraintestinal pathogenic Escherichia coli strains. J. Clin. Microbiol. 39:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosson, P., L. Zulianello, O. Join-Lambert, F. Faurisson, L. Gebbie, M. Benghezal, C. Van Delden, L. K. Curty, and T. Kohler. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184:3027-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Culham, D. E., and J. M. Wood. 2000. An Escherichia coli reference collection group B2- and uropathogen-associated polymorphism in the rpoS-mutS region of the E. coli chromosome. J. Bacteriol. 182:6272-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dale, C., G. R. Plague, B. Wang, H. Ochman, and N. A. Moran. 2002. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc. Natl. Acad. Sci. USA 99:12397-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale, C., S. A. Young, D. T. Haydon, and S. C. Welburn. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. USA 98:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darwin, C. 1859. On the origin of species by means of natural selection, p. 454-459. J. Murray, London, United Kingdom.
- 14.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrindt, U., and J. Reidl. 2000. Pathogenicity islands and phage conversion: evolutionary aspects of bacterial pathogenesis. Int. J. Med. Microbiol. 290:519-527. [DOI] [PubMed] [Google Scholar]
- 16.Donnenberg, M. S. 2002. Escherichia coli: virulence mechanisms of a versatile pathogen. Academic Press, Amsterdam, The Netherlands.
- 17.Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Investig. 107:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ffrench-Constant, R. H., N. Waterfield, V. Burland, N. T. Perna, P. J. Daborn, D. Bowen, and F. R. Blattner. 2000. A genomic sample sequence of the entomopathogenic bacterium Photorhabdus luminescens W14: potential implications for virulence. Appl. Environ. Microbiol. 66:3310-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foultier, B., P. Troisfontaines, S. Muller, F. R. Opperdoes, and G. R. Cornelis. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J. Mol. Evol. 55:37-51. [DOI] [PubMed] [Google Scholar]
- 19a.Francetic, O., and A. P. Pugsley. 1996. The cryptic general secretion pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J. Bacteriol. 178:3544-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 21.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 22.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 23.Hartleib, S., R. Prager, I. Hedenstrom, S. Lofdahl, and H. Tschape. 2003. Prevalence of the new, SPI1-like, pathogenicity island ETT2 among Escherichia coli. Int. J. Med. Microbiol. 292:487-493. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain, K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 25.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homma, K., S. Fukuchi, T. Kawabata, M. Ota, and K. Nishikawa. 2002. A systematic investigation identifies a significant number of probable pseudogenes in the Escherichia coli genome. Gene 294:25-33. [DOI] [PubMed] [Google Scholar]
- 27.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, J. R. 2002. Evolution of pathogenic Escherichia coli, p. 55-77. In M. S. Donnenberg (ed.), Escherichia coli: virulence mechanisms of a versatile pathogen. Academic Press, Amsterdam, The Netherlands.
- 30.Kenny, B. 2002. Mechanism of action of EPEC type III effector molecules. Int. J. Med. Microbiol. 291:469-477. [DOI] [PubMed] [Google Scholar]
- 31.Klein, J. R., T. F. Fahlen, and B. D. Jones. 2000. Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect. Immun. 68:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labrousse, A., S. Chauvet, C. Couillault, C. L. Kurz, and J. J. Ewbank. 2000. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 10:1543-1545. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence, J. G., R. W. Hendrix, and S. Casjens. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9:535-540. [DOI] [PubMed] [Google Scholar]
- 34.Lecointre, G., L. Rachdi, P. Darlu, and E. Denamur. 1998. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 15:1685-1695. [DOI] [PubMed] [Google Scholar]
- 35.Makino, S., T. Tobe, H. Asakura, M. Watarai, T. Ikeda, K. Takeshi, and C. Sasakawa. 2003. Distribution of the secondary type III secretion system locus found in enterohemorrhagic Escherichia coli O157:H7 isolates among Shiga toxin-producing E. coli strains. J. Clin. Microbiol. 41:2341-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marie, C., W. J. Broughton, and W. J. Deakin. 2001. Rhizobium type III secretion systems: legume charmers or alarmers? Curr. Opin. Plant Biol. 4:336-342. [DOI] [PubMed] [Google Scholar]
- 37.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 39.Mira, A., L. Klasson, and S. G. Andersson. 2002. Microbial genome evolution: sources of variability. Curr. Opin. Microbiol. 5:506-512. [DOI] [PubMed] [Google Scholar]
- 40.Miyazaki, J., W. Ba-Thein, T. Kumao, H. Akaza, and H. Hayashi. 2002. Identification of a type III secretion system in uropathogenic Escherichia coli. FEMS Microbiol. Lett. 212:221-228. [DOI] [PubMed] [Google Scholar]
- 41.Mori, H., K. Isono, T. Horiuchi, and T. Miki. 2000. Functional genomics of Escherichia coli in Japan. Res. Microbiol. 151:121-128. [DOI] [PubMed] [Google Scholar]
- 42.Murray, R. A., and C. A. Lee. 2000. Invasion genes are not required for Salmonella enterica serovar Typhimurium to breach the intestinal epithelium: evidence that Salmonella pathogenicity island 1 has alternative functions during infection. Infect. Immun. 68:5050-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochman, H., and I. B. Jones. 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pallen, M. J., M. S. Francis, and K. Futterer. 2003. Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol. Lett. 223:53-60. [DOI] [PubMed] [Google Scholar]
- 47.Perna, N. T. 2002. The genomes of K12 and pathogenic Escherichia coli, p. 3-54. In M. S. Donnenberg (ed.), Escherichia coli: virulence mechanisms of a versatile pathogen. Academic Press, Amsterdam, The Netherlands.
- 48.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 50.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 52.Russmann, H., T. Kubori, J. Sauer, and J. E. Galan. 2002. Molecular and functional analysis of the type III secretion signal of the Salmonella enterica InvJ protein. Mol. Microbiol. 46:769-779. [DOI] [PubMed] [Google Scholar]
- 53.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 54.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 55.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 56.Tamano, K., E. Katayama, T. Toyotome, and C. Sasakawa. 2002. Shigella Spa32 is an essential secretory protein for functional type III secretion machinery and uniformity of its needle length. J. Bacteriol. 184:1244-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tauschek, M., R. A. Strugnell, and R. M. Robins-Browne. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44:1533-1550. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vasconcelos, A. T. R., D. F. de Almeida, F. C. Almeida, L. G. P. de Almeida, R. de Almeida, J. A. A. Goncalves, E. M. Andrade, R. V. Antonio, J. Araripe, M. F. F. de Araujo, S. A. Filho, V. Azevedo, A. J. Batista, L. A. M. Bataus, J. S. Batista, A. Belo, C. vander Berg, J. Blamey, M. Bogo, S. Bonato, J. Bordignon, C. A. Brito, M. Brocchi, H. A. Burity, A. A. Camargo, D. D. P. Cardoso, N. P. Carneiro, D. M. Carraro, C. M. B. Carvalho, J. C. M. Cascardo, B. S. Cavada, L. M. O. Chueire, T. B. C. Pasa, N. Duran, N. Fagundes, C. L. Falcao, F. Fantinatti, I. P. Farias, M. S. S. Felipe, L. P. Ferrari, J. A. Ferro, M. I. T. Ferro, G. R. Franco, N. S. A. Freitas, L. R. Furlan, R. T. Gazzinelli, E. A. Gomes, P. R. Goncalves, T. B. Grangeiro, D. Grattapaglia, E. C. Grisard, C. T. Guimaraes, E. S. Hanna, M. Hungria, S. N. Jardim, J. Laurino, L. C. T. Leoi, L. Fassarella, A. Lima, M. F. Loureiro, M. C. P. Lyra, M. Macedo, H. M. F. Madeira, G. P. Manfio, A. Q. Maranhao, W. S. Martins, S. M. Z. di Mauro, S. R. B. de Medeiros, R. D. V. Meissner, C. F. M. Menck, M. A. M. Moreira, F. F. Nascimento, M. F. Nicolas, J. G. Oliveira, S. C. Oliveira, R. F. C. Paixao, J. A. Parente, F. O. Pedrosa, S. J. D. Pena, J. O. Perreira, M. Perreira, L. S. R. C. Pinto, L. S. Pinto, J. I. R. Porto, D. P. Potrich, C. E. R. Neto, A. M. M. Reis, L. U. Rigo, E. Rondinelli, E. B. P. Dos Santos, F. R. Santos, M. P. C. Schneider, H. N. Seuanez, A. M. R. Silva, A. L. C. da Silva, D. W. Silva, R. Silva, I. C. Simoes, D. Simon, C. M. A. Soares, et al. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 100:11660-11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei, J., M. B. Goldberg, V. Burland, M. M. Venkatesan, W. Deng, G. Fournier, G. F. Mayhew, G. Plunkett III, D. J. Rose, A. Darling, B. Mau, N. T. Perna, S. M. Payne, L. J. Runyen-Janecky, S. Zhou, D. C. Schwartz, and F. R. Blattner. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]